Abstract
Human Adenovirus type 5 encodes two short RNA polymerase III transcripts, the virus-associated (VA) RNAI and VA RNAII, which can adopt stable hairpin structures that resemble micro-RNA precursors. The terminal stems of the VA RNAs are processed into small RNAs (mivaRNAs) that are incorporated into RISC. It has been reported that VA RNAI has two transcription initiation sites, which produce two VA RNAI species; a major species, VA RNAI(G), which accounts for 75% of the VA RNAI pool, and a minor species, VA RNAI(A), which initiates transcription three nucleotides upstream compared to VA RNAI(G). We show that this 5′-heterogeneity results in a dramatic difference in RISC assembly. Thus, both VA RNAI(G) and VA RNAI(A) are processed by Dicer at the same position in the terminal stem generating the same 3′-strand mivaRNA. This mivaRNA is incorporated into RISC with 200-fold higher efficiency compared to the 5′-strand of mivaRNAI. Of the small number of 5′-strands used in RISC assembly only VA RNAI(A) generated active RISC complexes. We also show that the 3′-strand of mivaRNAI, although being the preferred substrate for RISC assembly, generates unstable RISC complexes with a low in vitro cleavage activity, only around 2% compared to RISC assembled on the VA RNAI(A) 5′-strand.
INTRODUCTION
RNA interference (RNAi) is a diverse, conserved regulatory mechanism in eukaryotic cells, which silences the target gene expression in a homology-dependent manner (1). It is well documented that RNAi is an antiviral mechanism in plants and insects, although it is still unclear whether RNAi naturally limits viral infections in vertebrates (2,3). Viruses are masters of using different strategies to subvert cellular defense mechanisms. They can not only suppress the effects of defensive RNA silencing, but also evade or exploit RNAi for their own benefits.
Several recent studies have shown that during a human adenovirus infection the activity of the two key enzyme systems involved in RNAi, Dicer and the RNA-induced silencing complex (RISC) are suppressed (4–7). The virus-associated RNAs (VA RNAs) function as suppressors of RNAi by binding Dicer through their terminal stems and squelching Dicer as competitive substrates (4,6). They are cleaved by Dicer into functional small RNAs (mivaRNAs) that are incorporated into RISC (4,5,7,8). The effect of an adenovirus infection on cellular miRNA function could be dramatic since the virus produces large amounts of the mivaRNAs. Thus, at late times of infection ∼80% of Ago2-containing RISC is hijacked by mivaRNAs (8).
Group C adenovirus encodes two VA RNAs, VA RNAI (major species, 108 molecules/cell) and VA RNAII (minor species, 5 × 106 molecules per cell). Both VA RNAs are about 160 nucleotides long, GC-rich and can adopt stable secondary structures that are important for their function (9). Besides their roles as RNAi suppressors, VA RNAI plays a crucial role as a suppressor of the interferon response. Thus, VA RNAI rescues the translational capacity of adenovirus-infected cells by blocking the activity of the dsRNA-dependent protein kinase (PKR), which inhibits translation in virus-infected cells by phosphorylating the eukaryotic initiation factor 2 (eIF-2) (10).
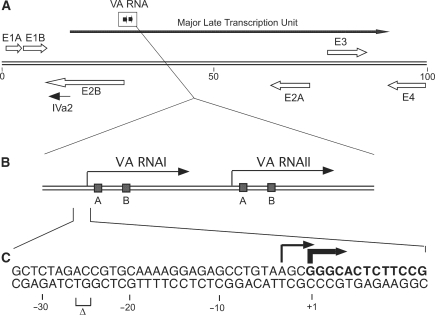
Both VA RNAs are transcribed by RNA polymerase III (11,12) from tRNA like promoters, which are characterized by having internal promoter elements (13,14). The location of the A and the B boxes (Figure 1B) have been mapped by genetic analysis in Ad2 VA RNAI. VA RNAI is heterogeneous at both its 5′- and 3′-ends, because of the variation in both transcription initiation and transcription termination (15,16). Adenovirus-infected cells contain two VA RNAI species, VA RNAI(A) and VA RNAI(G) (17). VA RNAI(A) initiates transcription 3 nucleotides upstream of the start site for VA RNAI(G) (Figure 1C). VA RNAI(G) is the major species and accounts for 75% of the total pool of VA RNAI. It has been reported that a two base pair deletion [nucleotide −25 and −26 (Figure 1C)] results in a complete loss of the VA RNAI(A) species (16).
Figure 1.
Schematic drawing showing the organization of the VA RNA genes on the adenovirus chromosome. (A) A simplified transcription map of the Ad5 genome. Arrows denotes the position of viral early (open) and late (thin) transcription units. (B) Expansion of the genome position encoding the VA RNA genes. (C) Nucleotide sequence at the promoter site of VA RNAI showing the position of the start site for VA RNAI(G) (bold arrow) and VA RNAI(A) (thin arrow). The sequence of the 5′-end of the major VA RNAI(G) start is indicated in bold letters. The nucleotides deleted in viruses lacking VA RNAI(A) expression are indicated by a delta sign (Δ).
In our previous work, we observed that the strand bias of mivaRNA incorporation into RISC varied when we used different viable Ad5 mutant viruses (4,8). A closer inspection of the DNA sequence surrounding the VA RNAI gene demonstrated that in the virus expected to produce both VA RNAI(G) and VA RNAI(A) both strands of mivaRNAI could guide RISC to cleave a synthetic target RNA, whereas only the 3′-strand of mivaRNAI generated specific RISC activity in the virus expected to produce VA RNAI(G) only (i.e. the two base pair deletion). Based on this finding, we suspected that the heterogeneity at 5′-end of VA RNAI could contribute to the variation of VA RNAI derived small RNA-associated RISC activity. Here, we show that in Ad5 infected cells the 3′-strand of VA RNAI is the preferred strand to be incorporated into RISC and exceeds the 5′-strand in RISC by >200-fold. However, these RISC complexes are inefficient in guiding in vitro RISC cleavage of a target RNA, showing around 2% of the activity of the 5′-strand of VA RNAI(A). Further, we show that the 5′-strand of VA RNAI(A) is incorporated into RISC and generates active RISC complexes, whereas the 5′-strand of VA RNAI(G), which is inefficiently incorporated into RISC generates inactive RISC complexes. Dicer cleaves VA RNAI(A) and VA RNAI(G) at the same position in the terminal stem resulting in the production of a 5′-strand from VA RNAI(A) that is 3 nucleotides longer than the corresponding strand processed from VA RNAI(G).
MATERIALS AND METHODS
Cell culture and DNA transfection
A 293 and 293-Ago2 cells were grown in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% newborn calf serum (NCS), 100 U/ml PEST at 37°C in 5% CO2. All cell culture reagents were purchased from Gibco/BRL. DNA was transfected using the calcium phosphate co-precipitation technique (18). Plasmid DNA was prepared using the Maxiprep kit (Qiagen).
Plasmid construction
Plasmid pVA RNAI G63A contains a point mutation at position 63 in the VA RNAI gene. The plasmid was constructed by PCR amplification using primer G63A Forw (5′-ATTCGAGCCCCGTATCCGGCC-3′) and primer G63A Rev (5′-CCCGGTCGTCCGCCATGATAC-3′). The first nucleotide in the forward primer introduces the mutation. Both primers were phosphorylated and then used to PCR amplify the VA RNAI gene and the entire pdl309HindB plasmid. The full-length PCR product was gel purified and circularized by religation. Plasmids pVA RNAI(G) and pVA RNAI(A) were created by PCR amplification using different forward primers: primer VAI-G Start (5′-CTTGTGGAAAGGACGAAACACCGGGCACTCTTCCGTGGTCTGG-3′) and VAI-A Start (5′-CTTGTGGAAAGGACGAAACACCAGCGGGCACTCTTCCGTGGTCTGG-3′) in combination with primer VAI Rev (5′-GGTGTTTCGTCCTTTCCACAAG-3′). Both forward primers contain 22 nucleotides from the U6 promoter sequence. The U6 promoter was PCR amplified with primers U6 Forw (5′-ACGGTACCAAGGTCGGGCAGGAAGAGG-3′) and U6 Rev (5′-CGGCAAGCTTAAAAGGAGCACTCCCCCGTTGTCTG-3′). The promoter sequence was fused to the VA RNAI A and G start through the flanking sequence in a final PCR amplification step. In all PCR amplification steps, PfuUltra™ High-Fidelity DNA Polymerase (Stratagene) was used. Plasmid dl309HindB was constructed by cloning the HindIII-B restriction fragment from virus dl309 into the HindIII site in pBR322. The relevant portion of all plasmids were verified by DNA sequencing.
Virus infection
All viruses used are derivatives of Ad5 and have previously been described. Both wt900 (19) and dl703 (20) are variants of Ad5. Mutant dl328 is a derivative of dl327, which does not express VA RNAII (21). Cells were infected with adenovirus at a multiplicity of 10 fluorescence forming units (FFU) per cell in DMEM containing 2% NCS. After 45 min, the medium was replaced with fresh medium containing 10% NCS. The uninfected control cells were treated identically except that virus was omitted. Twenty four hours later, the cells were harvested as described below.
Cytoplasmic S15 extract preparation
Cytoplasmic extracts were prepared as previously described (4). Briefly, cells were disrupted by 20–30 strokes in a 23-gauge syringe needle. The nuclei were pelleted, the supernatant centrifugated at 15 000 g for 60 min, supplemented with 5% glycerol, frozen in liquid nitrogen and stored at −80°C. The protein concentration was typically 6–8 μg/μl.
RISC assay
To generate target RNAs for mivaRNAI, the VA RNAI gene was separated near the apical loop into two halves and cloned into a pGL4-Luciferase reporter mRNA in the reverse orientation, generating target regions complementary to the 5′-(nucleotide 10614–10690, 22) and 3′-halves (nucleotide 10684–10779, 22) of VA RNAI. Target RNAs complementary to miR-18a, miR-93 and miR-106 b were constructed by cloning 28 bp annealed complementary oligonucleotides into the unique XbaI and FseI restriction cleavage sites in pGL3-control (Promega). The 32P-labeled target RNAs were incubated with S15 cytoplasmic extracts or the anti-FLAG M2 agarose beads (Sigma) containing purified RISC under the assay conditions previously described (4,8). Synthetic short RNA strands corresponding to the predicted mivaRNAI(A) and mivaRNAI(G) were purchased from Ambion and annealed to form duplexes. Reaction mixtures were incubated for 2 h at 30°C and RNA isolated and separated on a denaturing 8% polyacrylamide gel.
Immunopurification of RISC
Cytoplasmic cell extracts were prepared by treatment of cells on ice for 20 min with IsoB-NP-40 (10 mM Tris–HCl pH 7.9, 150 mM NaCl, 1.5 mM MgCl2, 1% NP-40) followed by a centrifugation at 16 000g for 10 min at 4°C. The supernatant from one 10 cm culture plate was incubated with 10 μl of anti-FLAG M2 agarose beads (Sigma) with constant rotation for 3 h at 4°C. The beads were washed three times in NET-1 buffer (50 mM Tris–HCl pH 7.5, 150 mM NaCl, 2.5% Tween 20). Co-precipitated RNA was extracted using phenol–chloroform and subsequently precipitated from the aqueous phase using ethanol.
Primer extension
Five micrograms of total RNA or small RNA were incubated with 0.5 pmol 5′-end labeled primer (5′-TCCACCAGACCACGGAAGAG-3′) in 1.3 × Superscript buffer (65 mM Tris–HCl, pH 8.3, 97.5 mM KCl, 3.9 mM MgCl2) at 70°C for 10 min. Then, the sample was allowed to cool down slowly to 45°C and 200 U Superscript II reverse transcriptase (Invitrogen) were added. The primer extension reaction was incubated at 45°C for 1 h in a buffer containing 1 × Superscript buffer (50 mM Tris–HCl, pH 8.3, 75 mM KCl, 3 mM MgCl2), 10 mM DTT and 500 µM dNTP. The reaction was terminated by addition of 1 µl 5M NaOH followed by an incubation at 65°C for 20 min. The cDNA was precipitated and analyzed on a denaturing 15% polyacrylamide gel.
Enzymatic assays of small RNAs
To analyze the 5′-end modification on mivaRNAI, small RNAs extracted from immuno-purified RISC complexes were treated with different enzyme combinations in a total reaction volume of 23 µl under the reaction conditions recommended by the manufacturer. RNAs were treated with 20 U of calf intestine alkaline phosphatase (CIAP, Fermentas) for 30 min at 37°C, 10 U of T4 polynucleotide kinase (PNK, Fermentas) for 20 min at 37°C and 1 U of Terminator 5′-exonuclease (TE, Epicentre) for 1 h at 30°C. The reactions were stopped by addition of 1 µl 0.5 M EDTA, an equal volume of loading buffer was added and the RNAs separated on a polyacrylamide gel as described below.
Northern blot analysis
Small RNAs were separated on a denaturing 15% polyacrylamide gel, and either transferred to a Hybond NX membrane (Amersham), chemically crosslinked and hybridized as described (23) (Figure 5), or transferred to a Hybond XL membrane (Amersham), UV-crosslinked and hybridized as described (4) (Figure 6). Hybridization probes were generated by 5′-end labeling of DNA oligonucleotides complementary to the 5′- or 3′-strand of mivaRNAI. (mivaRNAI 5′-probe: 5′-GAATTTATCCACCAGACCACGGAAGAGTGCC-3′; mivaRNAI 3′-probe: 5′-AAAAGGAGCACTCCCCCGTTGTCTGACGTCGCA-3′)
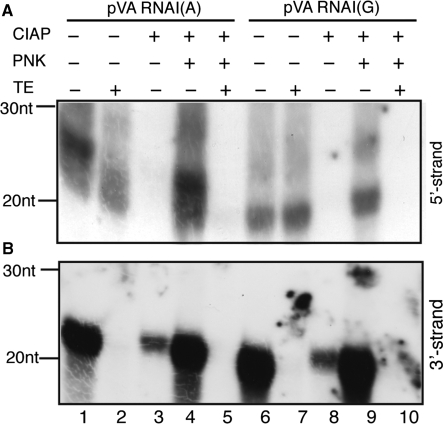
Figure 5.
The phosphorylation status of mivaRNAIs derived from VA RNAI(A) and VA RNAI(G). The RISC associated RNAs from pVA RNAI(A) or pVA RNAI(G) transfected cells were treated with combinations of calf intestinal alkaline phosphatase (CIAP), T4 polynucleotide kinase (PNK) and Terminator 5′-exonuclease (TE). After enzyme treatment, the 5′- (A) or 3′-strands (B) of mivaRNAI were analyzed by northern blotting.
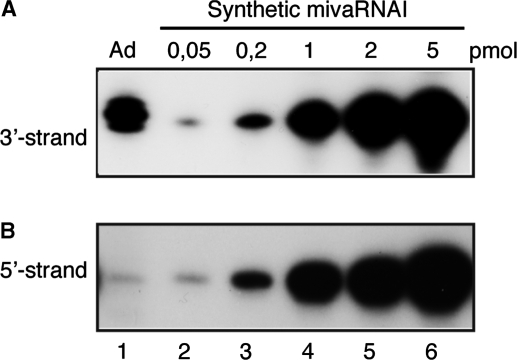
Figure 6.
The 3′-strand of mivaRNAI is efficiently incorporated into RISC. The RISC associated RNAs from wild-type Ad5 infected cells were analyzed for the abundance of 3′-strand (A) and 5′-strand (B) of mivaRNA I by northern blotting. Different amounts (from 0.05 to 5 pmol) of chemically synthetic mivaRNAI were phosphorylated and used as standards for quantification.
RESULTS
The strand bias of mivaRNAI association with functional RISC differs in Ad5 mutant virus infections
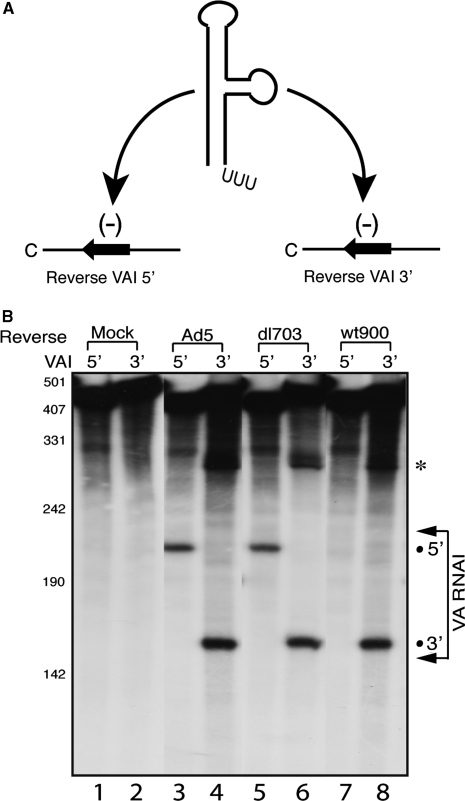
By analyzing the RISC activity against substrate RNAs harboring sequences complementary to the 5′ or 3′ half of VA RNAI (Figure 2A), it is possible to determine which strand of mivaRNAI is incorporated into functional RISC (4). Comparing the results from our previous work showed an unexpected asymmetry in strand incorporation when two different viable Ad5 mutant viruses were tested in our standard RISC assay system (4,8). In dl703 mutant infected cells, both strands of the terminal stem of VA RNAI were associated with functional RISC (Figure 2B, lanes 5 and 6). Also, both strands of VA RNAI generated active RISC in wild type Ad5-infected cells (Figure 2B, lanes 3 and 4). In contrast, in wt900 mutant infected cells only the 3′-strand generated active RISC (Figure 2B, lanes 7 and 8). DNA sequence analysis of the VA RNAI gene demonstrated that they were of the expected wild-type sequence in all three viruses. However, we noted that the wt900 virus lacks the 2 base pairs upstream of the transcription initiation start of the VA RNAI(G) gene (Figure 1C) that previously have been shown to be required for transcription initiation from the A start site (16). In Ad5 and the dl703 virus, the flanking sequences were of wild-type origin and therefore both VA RNAI(G) and VA RNA(A) would be expected to be produced (16).
Figure 2.
The strand bias of mivaRNAI incorporating into RISC differs in wild-type Ad5, dl703 and wt900 infections. (A) The schematic diagram of RISC substrate RNAs. The VA RNAI gene was separated within the apical loop into two halves and cloned in anti sense orientation into a reporter mRNA. The 32P-labeled target RNAs containing the sequence complementary to the 5′ or 3′ halves of VA RNAI were generated by in vitro transcription. (B) S15 cytoplasmic extracts prepared from uninfected 293-Ago2 cells (Mock) or cells infected with wild-type Ad5, dl703 or wt900 were assayed for RISC activity against the reverse VAI 5′ or VAI 3′ target RNAs. Arrows indicate the span of the VA RNAI target regions in respective transcripts. The positions of cleavage products are indicated by dots. The bands labeled with ‘asterisks’ indicate the 3′-end cleavage products of the substrate RNAs, which is sometimes seen but usually degraded in S15 extracts.
To determine whether these two nucleotides are responsible for the asymmetry observed in RISC activity, we identified the status of mivaRNAI strand incorporation in Ad5 and different Ad5 mutant infected cells. The results are summarized in Table 1 and show that in viruses expected to produce both VA RNAI(G) and VA RNAI(A) (i.e. dl703, dl327, dl328 and wild type Ad5) both the 5′ and the 3′-strands of the processed VA RNAI were incorporated into active RISC, whereas only the 3′-strand was incorporated in wt900 infected cells (two nucleotide deletion). Also, transfection of a plasmid derived from Ad5 mutant dl309 (2 nucleotide deletion) that contained a DNA fragment spanning the VA RNAI gene generated only RISC complexes derived from the 3′-strand of VA RNAI.
Table 1.
VA RNAI(A) expression status and the strand bias of mivaRNAI incorporation into RISC
| VA RNAI(A) | VA RNAI specific RISC |
||
|---|---|---|---|
| 5′ | 3′ | ||
| dl703 | Yes | + | + |
| wild-type Ad5 | Yes | + | + |
| dl327 | Yes | + | + |
| dl328 | Yes | + | + |
| wt900 | No | − | + |
| pdl309HindB | No | − | + |
5′ mivaRNAI associated RISC activity derives from VA RNAI(A), but not VA RNAI(G)
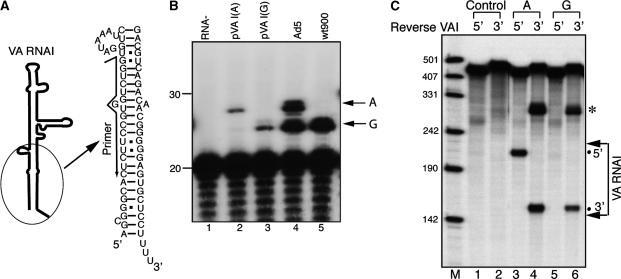
Based on the results presented in Table 1, we suspected that the heterogeneity at the 5′-end of VA RNAI was responsible for the difference in the assembly of active RISC in Ad5 mutant infected cells. To test this hypothesis, we constructed plasmids expressing VA RNAI starting either from the ‘A’ position [pVA RNAI(A)] or the ‘G’ position [pVA RNAI(G)]. In order to ensure that only one species of VA RNAI was transcribed in each case, we placed the expression of VA RNAI under the control of the constitutively active U6 promoter, which like the native VA RNAI promoter is transcribed by RNA polymerase III, but from a promoter that has the regulatory elements positioned upstream of the transcription initiation site (24). To further ensure that the native VA RNAI promoter was not used, we introduced a point mutation (G63A) in the critical B-box. This mutation has previously been shown to reduce transcription activity of VA RNAI to 1% in vitro (25). The transcription initiation start sites were verified for both pVA RNAI(A) and pVA RNA(G) by primer extension (Figure 3B, lanes 2 and 3).
Figure 3.
VA RNAI (A), but not VA RNAI (G), generates active RISC containing the 5′-strand of mivaRNAI. (A) Schematic representation of the terminal sequence of VA RNAI and the oligonucleotide used for primer extension. (B) Primer extension on total RNAs prepared from control 293-Ago2 cells (lane 1) or cells transfected with plasmid pVA RNAI(A) (lane 2) or pVA RNAI(G) (lane 3) or infected with virus Ad5 (lane 4) or wt900 infected cells (lane 5), were analyzed by primer extension for the transcription initiation start sites of VA RNAI. The products generated from ‘A’ start and ‘G’ start VA RNAI are indicated in the Figure. (C) The 293-Ago2 cells were transfected with plasmids pVA RNAI(A), pVA RNAI(G) or the empty vector as a control. The immunopurified RISC complexes from S15 cytoplasmic extracts were assayed for activity against the reverse VAI 5′ or VAI 3′ target RNAs.
The 293 cells stably expressing a FLAG tagged Ago2 protein [293-Ago2; (8)] were transfected with either pVA RNAI(A) or pVA RNAI(G). S15 cytoplasmic extracts were prepared 48 h posttransfection and Ago2-containing complexes immuno-purified by binding to an anti-FLAG M2 agarose resin, and the RISC activity assayed using our standard RISC assay protocol. The results show that both strands of VA RNAI(A) derived small RNAs [mivaRNAI(A)] generated active RISC (Figure 3C, lanes 3 and 4). In contrast, only the 3′-strand of VA RNAI(G) derived small RNAs [mivaRNAI(G)] generated active RISC complexes (Figure 3, lanes 5 and 6). These results are in perfect agreement with the data obtained from infections using viruses expected to produce both VA RNAI species or VA RNAI(G) only (Table 1). Taken together our data demonstrate that mivaRNAI(A) generates small RNAs from both strands of the terminal stem that are incorporated into RISC whereas mivaRNAI(G) generates active RISC complexes only from the 3′-strand of the terminal stem.
Mapping the Dicer processing site in VA RNAI(A) and VA RNAI(G)
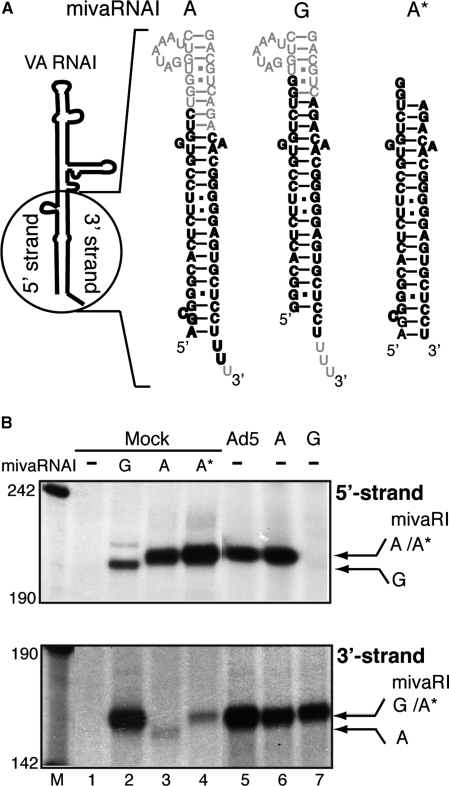
Since the RNAi effector complex will cleave a target RNA between nucleotide 10 and 11 relative to the 5′-end of the guiding siRNA, the exact cleavage position in a target RNA will help to fine map the 5′-end of both strands of the mivaRNAs produced in transfected or adenovirus-infected cells. As control RNAs, we used annealed synthetic siRNAs designed to mimic the predicted ‘G’ start and ‘A’ start mivaRNAI (Figure 4A mivaRNAI(A) and mivaRNAI(G)). Although these siRNAs were unphosphorylated they are expected to become rapidly phosphorylated at the 5′-end by the human RNA kinase hClp1 (26). Using this strategy, we could fine map the start nucleotide of both the 5′- and 3′-strands of mivaRNAI(A) and mivaRNAI(G). As shown in Figure 4B, strand loading was highly asymmetric, and opposite, for the two synthetic short RNA duplexes with the 5′-strand of mivaRNAI(A) and the 3′-strand of mivaRNAI(G) being preferentially assembled into RISC. In part, this variation in strand loading efficiency between the two siRNAs may result from differences in the thermodynamic stability of the short RNA ends. As expected, the synthetic mivaRNAI(A) generated a 5′-strand cleavage product that was detectable longer (three nucleotides) than the cleavage product produced from the synthetic mivaRNAI(G) (compare Figure 4B, lanes 2 and 3). Importantly, in Ad5 infected cells and in cells transfected with plasmid pVA RNAI(A) the 5′-strand cleavage product was identical in size to the synthetic mivaRNAI(A). This result suggests that the 5′-end of the 5′-strand of mivaRNAI(A) was not processed.
Figure 4.
Mapping the Dicer processing site in VA RNAI(A) and VA RNAI(G). (A) Schematic drawing showing the difference in nucleotide sequence at the terminal stems of VA RNAI(A) and VA RNAI(G). The synthetic short double-stranded RNAs (shown in bold letters) were designed to mimic the predicted ‘A’ start and ‘G’ start mivaRNAI. Note that the structure of the extra base pairs at the beginning of the stem in the VA RNAI(A) structure were predicted. (B) S15 cytoplasmic extracts prepared from uninfected 293-Ago2 cells (Mock, lanes 1–4) or cells infected with wild-type Ad5 (lane 5) or cells transfected with plasmids pVA RNAI(A) (lane 6) or pVA RNAI(G) (lane 7) were assayed for RISC activity against the reverse VAI 5′ (upper panel) or reverse VAI 3′ (lower panel) target RNAs. The uninfected cytoplasmic extracts were mixed with water (lane 1), 2 pmol synthetic ‘G’ start mivaRNAI (lane 2), 2 pmol short version (lane 3) or the experimentally verified version of the ‘A’ start mivaRNAI (lane 4). The cleavage products from different mivaRNAIs are indicated by arrows.
Since mivaRNAI(A) initiates transcription 3 nucleotides upstream of VA RNAI(G) the predicted start position of the 3′-strand would be expected to be shifted down three nucleotides on the terminal stem compared to mivaRNAI(G) (Figure 4A). As shown in Figure 4B, the synthetic siRNAs reproduced the expected result with mivaRNAI(A) generating a 3 nucleotide shorter 3′-strand cleavage product compared to mivaRNAI(G) (lanes 2 and 3). Importantly the 3′-strand cleavage site was identical in pVA RNAI(A), pVA RNAI(G) transfected cells and Ad5-infected cells suggesting that Dicer processing of VA RNAI(A) and VA RNAI(G) occurs at the same position in the terminal stem (Figure 4A). As a consequence the mivaRNAI(A) 5′-strand is expected to be three nucleotides longer than the corresponding 5′-strand from mivaRNAI(G). Also, the 3′-strand in both mivaRNAs produced in Ad5-infected cells will be identical (Figure 4A). To confirm this, another synthetic short RNA duplex mivaRNAI(A*) (Figure 4A), which was designed to mimic the actual mivaRNAI(A) produced in infected cells, was tested for its ability to guide RISC to cleave the target RNA. As shown in Figure 4B, the mivaRNAI(A*) 3′- and 5′-strand cleavage sites were the same as in those produced in pVA RNAI(A), pVA RNAI(G) transfected cells and Ad5-infected cells, confirming our hypothesis.
The 5′-end phosphorylation status differs between mivaRNAs generated from VA RNAI(A) and VA RNAI(G)
Since the VA RNAs are transcribed by RNA polymerase III, a large proportion of both VA RNAI(A) and VA RNAI(G) would be expected to have a tri-phosphorylated 5′-end. A direct measurement of the phosphorylated status of the 5′-end of VA RNAI showed that only about 50% of VA RNAI carries a tri-phosphate end. The rest of the population has di-phosphate (40%) or mono-phosphate (10%) ends (17). Functional siRNAs and miRNAs have a 5′-mono-phosphate that is generated by Dicer cleavage (27,28). Commercial siRNAs are usually unphosphorylated but become rapidly phosphorylated after transfection (26).
To test whether the 5′-strand of mivaRNAI produced in plasmid pVA RNAI(G) and pVA RNAI(A) transfected cells had a 5′-mono-phosphate or a 5′ multiple phosphate group, we used a combination of enzymes to treat the small RNA pool prepared from immuno-purified RISC complexes. The RNA samples were treated with calf intestinal alkaline phosphatase (CIAP) that removes all types of 5′-phosphates, T4 polynucleotide kinase (PNK), which adds a monophosphate to a free 5′-end and Terminator 5′-exonuclease (TE), which only degrades RNAs containing a 5′-mono-phosphate. After enzyme treatment, the 5′- or 3′-strands of mivaRNAI were analyzed by northern blotting.
As shown in Figure 5A, the 5′-strand northern blot analysis showed one predominant band in pVA RNAI(A) transfected cells (lane 1). This band was sensitive to TE treatment (lane 2), suggesting that it had a mono-phosphate at its 5′-end. A small fraction of the RNA was TE resistant and could represent mivaRNAI(A) species that had tri-phosphorylated 5′-ends. Much to our surprise, we could also detect the 5′-strand of mivaRNAI(G) in immuno-purified RISC complexes (Figure 5A, lane 6). This was unexpected since the 5′-strand of VA RNAI(G) does not generate active RISC complexes (Figure 3C, lane 5; Figure 4B, lane 7). This species was resistant to TE treatment showing that it did not have a 5′-mono-phosphate. CIAP treatment of the small RNA samples results in a complete loss of the signal for both VA RNAI(A) and VA RNAI(G) (Figure 5A, lanes 3 and 8). This result was expected since the method we use to cross-link the RNA to the membrane requires a phosphate group on the RNA (23). As expected, PNK treatment restores a 5′-mono-phosphate to the CIAP treated small RNAs from pVA RNAI(A) and pVA RNAI(G) transfected cells (Figure 5A, lanes 4 and 9), a phosphate that makes the RNAs susceptible to degradation by TE treatment (Figure 5A, lanes 5 and 10). Collectively, these results show that mivaRNAI(A) generates active 5′-strand RISC complexes that contain small RNAs with a 5′-end mono-phosphate, whereas mivaRNAI(G) generates a low frequency of inactive 5′-strand RISC complexes that contain small RNAs that do not have a 5′-mono-phosphate. Potentially, they may have multiple phosphate groups at their 5′-end.
The same analysis of 3′-strand incorporation in pVA RNAI(A) and pVA RNAI(G) transfected cells demonstrated that only one type of small RNA was incorporated into RISC (Figure 5B). Compared to the 5′-strand these small RNAs were very efficiently incorporated into RISC and sensitive to TE treatment indicating that they have a mono-phosphate at their 5′-end and therefore, most likely, are the products of Dicer cleavage. Note that the alkaline phosphatase treatment was not 100% effective in this experiment (Figure 5, lanes 3 and 8).
The 3′-strand of mivaRNAI is efficiently incorporated into RISC
To measure the relative amounts of the 5′- and 3′-strands of mivaRNAI in RISC purified from Ad5 infected 293-Ago2 cells we quantitated the abundance of respective strand by northern blot analysis. As a standard we used fixed amounts of the two strands of a chemically synthesized mivaRNAI (Figure 6, lanes 2–6). After transfer, the filters were hybridized with 5′-end labeled DNA oligonucleotides complementary to the 5′- or 3′-strands of mivaRNAI. As shown in Figure 6, the physical amount of the 3′-strand of mivaRNAI is dramatically higher compared to the 5′-strand. Quantification of this type of gels showed that the 3′-strand is in >200-fold excess compared to the 5′-strand. This result was surprising since functional RISC with the 3′-strand in Ad5 infected cells was only around 4-fold higher compared to the 5′-strand (Figure 2, lanes 3 and 4). Taken together, our results show that the 3′-strand of mivaRNAI is the preferred substrate for RISC assembly. However, these RISC complexes appear to have a very low specific activity, only around 2% compared to the RISC complexes assembled on the 5′-strand.
The 5′-strand of mivaRNAI(A) generates more stable RISC complexes compared to the 3′-strand
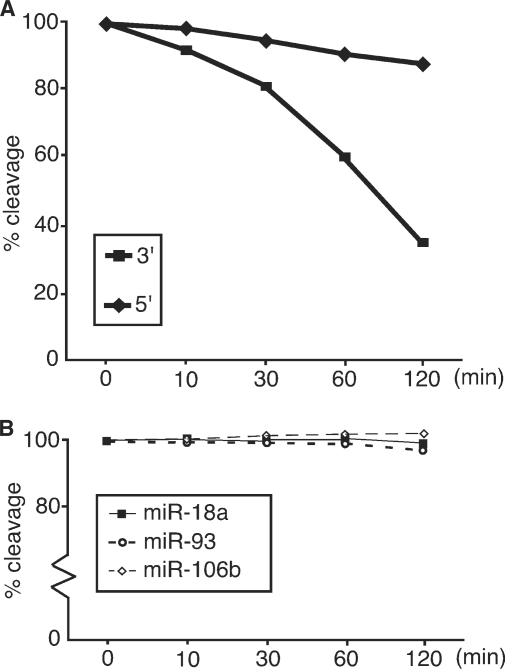
To determine the stability of the RISC complexes formed with the 5′- and 3′-strands of mivaRNAI we pre-incubated S15 cytoplasmic extracts from Ad5 infected 293 cells with a constant amount of a non-specific competitor siRNA for 0, 10, 30, 60 or 120 min. The pre-incubation was terminated by quick-freezing the extract in liquid nitrogen. As shown in Figure 7A, the activity of the 3′-strand RISC decreased more rapidly than that of the 5′-strand. Thus, whereas the 5′-strand RISC complexes remained essentially unaffected during the 2 h pre-incubation period the 3′-strand RISC complexes were reduced to <40% of the control. Taken together, these results suggest that the active RISC complexes formed with the 5′-strand of mivaRNAI are significantly more stable than the corresponding complexes formed with the 3′-strand.
Figure 7.
RISC complexes formed on the 5′-strand of mivaRNAI are more stable than RISC assembled on the 3′-strand. (A) S15 cytoplasmic extracts prepared from Ad5 infected 293 cells were pre-incubated with 10 µM of a non-specific siRNA for 0, 10, 30, 60 or 120 min before RISC activity directed against the reverse VAI 5′ or reverse VAI 3′ target RNAs was measured. The cleavage ratio of target RNAs was quantitated by PhosphorImager scanning. The cleavage activity of the zero minute sample was arbitrarily set as 100%. (B) Target RNAs complementary to miR-18a, miR-93 and miR-106b were incubated in S15 extracts prepared from 293 cells as described in (A). RISC activity was quantitated by PhosphorImager scanning and the cleavage activity of the zero minute sample set as 100%.
Cellular miRNAs are believed to form stable RISC complexes (32, 33). To determine whether the 3′-strand RISC complexes are unusually unstable, we assessed the stability of RISC complexes assembled on three cellular miRNAs: miR-18a, miR-93 and miR-106b. We selected these miRNAs because they belong to different miRNA clusters and have been annotated in different databases as expressed in 293 cells. As shown in Figure 7B, all three miRNAs were essentially unaffected by the 2 h pre-incubation confirming that cellular miRNAs form stable RISC complexes. This result also demonstrates that the 5′-strand mivaRNAI(A) generates RISC complexes with a stability similar to cellular miRNAs, whereas the 3′-strand form highly unstable complexes.
DISCUSSION
In our previous work, we showed that small RNAs generated from both strands of the terminal stem of VA RNAI were incorporated into functional RISC during a dl703 infection (4). This result is not controversial since it has been observed by other groups as well (5,7). However, in a follow-up study we noted that in cells infected with the wt900 virus, only the 3′-strands of VA RNAI were loaded into active RISC (8). This result was at first sight confusing.
Here, we have analyzed the basis for this difference in RISC activity. Our results show that the decisive parameter determining strand incorporation results from which VA RNAI species is expressed during the infection. Thus, VA RNAI comes in two variants, VA RNAI(G) and VA RNAI(A). These two species differ from each other in that VA RNAI(A) initiates transcription 3 nucleotides upstream of VA RNAI(G) (Figure 1C). In Ad5-infected cells, both strands of the terminal stem of VA RNAI(G) and VA RNAI(A) were assembled into RISC, albeit with different efficiencies and consequences for RISC activity. The 3′-strand processed from both VA RNAI(A) and VA RNAI(G) are identical in sequence (Figure 4A), and also the preferred strand used in RISC assembly (Figure 5), exceeding the 5′-strand by >200-fold in a lytic Ad5-infected cell (Figure 6). It should be noted that although the VA RNAI(A) 5′-strand is three nucleotides longer than the corresponding strand from VA RNAI(G) Dicer is expected to cleave the terminal stem of both VA RNAs at the same position since Dicer measures the distance to cleavage site based on the 3′-end of the dsRNA (29,30). Also, it is noteworthy that the 5′-strand of both mivaRNAI(A) and mivaRNAI(G) were assembled into RISC although our analysis showed that the 5′-strand of mivaRNAI(A) was assembled more effectively (Figure 5A, lanes 1 and 6) and generated RISC complexes that were functional in cleaving a target RNA in vitro (Figure 3C, lanes 3 and 4). The 5′-strand of mivaRNAI(G) is functionally inactive and contains a 5′-end that does not have a mono-phosphate, whereas the active 5′-strand of mivaRNAI(A) has a mono-phosphate (Figure 5).
Although the 3′-strand of mivaRNAI was assembled into RISC with a >200-fold preference compared to the 5′-strand (Figure 6) it generated unstable RISC complexes (Figure 7) with a surprisingly low cleavage activity, only around 2% of that of the 5′-strand. This result could mean that VA RNAI, with the exception of the 5′-strand of mivaRNAI(A), generates small RNAs that functionally inactivate RISC, thereby potentially contributing to the suppressive effect of the VA RNAs on RNAi (4–7). Such a mechanism would be attractive since it resembles the way VA RNAI binds and sequesters PKR. Thus, VA RNAI may have dual function during virus replication. It may antagonize the cellular defense pathways against both long dsRNA by binding and sequestrating PKR and short dsRNA by saturating RISC with mivaRNAs that do not generate enzymatically active complexes. Although this model is attractive, we can not exclude the possibility that the 3′-strand of mivaRNAI is a highly efficient viral miRNA that in the extract we prepare already is occupied by binding to cellular mRNAs. Since our assay system relies on the cleavage of an added reporter transcript such engaged RISC complexes will be scored as inactive in our RISC experiments. In addition, it is possible that structural constrains limit target accessibility for the 5′- and 3′-strand mivaRNAs. Also, the lower stability of the RISC complexes formed with the 3′-strand (Figure 7) contributes to the lower activity in the RISC assays. A high priority in our future work will be to clarify this point.
Curiously VA RNAI(A) appears to produce a 5′ mivaRNAI carrying a mono-phosphate instead of the tri-phosphate incorporated during transcription initiation. The RISC cleavage mapping experiment (Figure 4B) suggests that the 5′-end of this small RNA is not generated by RNA processing. More likely, the 5′-strand of mivaRNAI(A) is susceptible to dephosphorylation. Note that, 50% of the total pool of VA RNAI has mono- or di-phosphorylated 5′-termini in vivo (17). In fact, a closer inspection of this article indicates that a larger fraction of the VA RNAI(A) species has lost 5′ phosphates compared to VA RNAI(G) (see Figure 2 in ref. 17).
There are numerous studies that emphasize that the sequence and the structural features of the siRNA duplex are important and critical determinants for strand selection and RISC loading (27,31). The thermodynamic stability at the ends of the siRNA duplex appears to be important for RISC loading. Thus, experiments in Drosophila embryonic extracts have shown that the strand that is more loosely paired at its 5′-end is selectively assembled into RISC (32,33). However, in mammals there is no definite correlation between siRNA thermodynamic stability and strand selection (34). It seems more probable that the selection may be mediated by a combination of thermodynamic stability and a, yet to be described, structural feature component of the dsRNA. The predicted thermodynamic stability of the 5′-end of the 3′-strand is significantly lower compared to the 5′-ends of the 5′-strands in both mivaRNAI(G) and mivaRNAI(A) (Figure 4A). Thus, it seems reasonable that this difference in strand stability explains the highly asymmetric overrepresentation of the 3′-strand in RISC. Furthermore, this work suggests that RISC stability could be an important parameter to control RISC functionality. Thus, the 3′-strand of mivaRNAI assembled efficiently into RISC but generated unstable complexes (Figure 7). We do not know whether this represents a general mechanism regulating small RNA function in mammalian cells or if it is a virus-specific mechanism. However, it does not seem unreasonable to predict that future work will show that the functionality of both cellular and viral small RNA complexes will be regulated at the level of stability.
An important aspect of our work that we have not yet addressed is that the virus mutant defective in VA RNAI(A) production grows equally well as wild-type Ad5 in Hela cells (data not shown). This suggests that the production of the 5′ mivaRNA from VA RNAI(A) is not essential for virus growth at least not in tissue culture cells (16). Both VA RNAI(A) and VA RNAI(G) are made during the early and late phase of viral life cycle, but the proportion of ‘A’ start VA RNAI appears to increase late (17). It is possible that VA RNAI(A) and VA RNAI(G) have distinct roles in the RNAi/miRNA pathways, a function that may be important for adenovirus growth in nature. Previous work has suggested that mivaRNAs can function as miRNAs and regulate translation of cellular mRNAs (7,8). Therefore, RISC loading of the 5′-strand of mivaRNAI(A), in addition to the 3-strand, will yield additional potential target genes that might be regulated by the mivaRNAs (see Supplementary Material for potential target mRNAs). It is possible that such targets may contribute to the establishment of a benign long-term infection, or be related to tissue-specific acute infections.
Recombinant adenovirus vector systems have become widely used in gene therapy and gene transfer applications (35). However, adenovirus vectors designed to express short-hairpin RNAs (shRNAs) may suffer from negative effects caused by expression of large amounts of competing mivaRNAs. The basic function of VA RNAI during an adenovirus infection is to inhibit PKR (10) and this function is not dispensable for high-titer virus growth (21). Uncovering the exact mechanism involved in VA RNAI terminal stem processing and strand selection together with the fine-mapping of the mivaRNAIs expressed in Ad5-infected cells might be helpful in the design of a viral vector that retains the wild type PKR inhibitory activity but lacks competing mivaRNA expression. For example, based on the fact that so far it seems that strand selection is based on thermodynamic stability of the produced 5′-ends, it should be possible to increase the strength of the 3′-strand hybridization by changing the two A–T base pairs (5′-AGAC) to G–C base pairs (5′-GGGC) in the terminal stem. In combination with the 2 bp deletion restricting VA RNAI(A) synthesis such a VA RNAI gene would retain its PKR inhibitory activity and produce a Dicer cleavage product were both strands would have difficulties to enter RISC efficiently.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Swedish Cancer Society (07 0448 to G.A.); the Swedish Research Council through a grant to the Uppsala RNA Research Centre (URRC) (2006-5038-36531-16 to G.A.). Funding for open access charge: The Swedish Cancer Society.
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Mello CC, Conte D., Jr Revealing the world of RNA interference. Nature. 2004;431:338–342. doi: 10.1038/nature02872. [DOI] [PubMed] [Google Scholar]
- 2.Berkhout B, Jeang KT. RISCy business: microRNAs, pathogenesis, and viruses. J. Biol. Chem. 2007;282:26641–26645. doi: 10.1074/jbc.R700023200. [DOI] [PubMed] [Google Scholar]
- 3.van Rij RP, Andino R. The silent treatment: RNAi as a defense against virus infection in mammals. Trends Biotechnol. 2006;24:186–193. doi: 10.1016/j.tibtech.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 4.Andersson MG, Haasnoot PC, Xu N, Berenjian S, Berkhout B, Akusjärvi G. Suppression of RNA interference by adenovirus virus-associated RNA. J. Virol. 2005;79:9556–9565. doi: 10.1128/JVI.79.15.9556-9565.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aparicio O, Razquin N, Zaratiegui M, Narvaiza I, Fortes P. Adenovirus virus-associated RNA is processed to functional interfering RNAs involved in virus production. J. Virol. 2006;80:1376–1384. doi: 10.1128/JVI.80.3.1376-1384.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lu S, Cullen BR. Adenovirus VA1 noncoding RNA can inhibit small interfering RNA and MicroRNA biogenesis. J. Virol. 2004;78:12868–12876. doi: 10.1128/JVI.78.23.12868-12876.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sano M, Kato Y, Taira K. Sequence-specific interference by small RNAs derived from adenovirus VAI RNA. FEBS Lett. 2006;580:1553–1564. doi: 10.1016/j.febslet.2006.01.085. [DOI] [PubMed] [Google Scholar]
- 8.Xu N, Segerman B, Zhou X, Akusjärvi G. Adenovirus virus-associated RNAII-derived small RNAs are efficiently incorporated into the rna-induced silencing complex and associate with polyribosomes. J. Virol. 2007;81:10540–10549. doi: 10.1128/JVI.00885-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Akusjärvi G, Mathews MB, Andersson P, Vennström B, Pettersson U. Structure of genes for virus-associated RNAI and RNAII of adenovirus type 2. Proc. Natl Acad. Sci. USA. 1980;77:2424–2428. doi: 10.1073/pnas.77.5.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mathews MB, Shenk T. Adenovirus virus-associated RNA and translation control. J. Virol. 1991;65:5657–5662. doi: 10.1128/jvi.65.11.5657-5662.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Price R, Penman S. Transcription of the adenovirus genome by an -amanitine-sensitive ribonucleic acid polymerase in HeLa cells. J. Virol. 1972;9:621–626. doi: 10.1128/jvi.9.4.621-626.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weinmann R, Roeder RG. Role of DNA-dependent RNA polymerase 3 in the transcription of the tRNA and 5S RNA genes. Proc. Natl Acad. Sci. USA. 1974;71:1790–1794. doi: 10.1073/pnas.71.5.1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fowlkes DM, Shenk T. Transcriptional control regions of the adenovirus VAI RNA gene. Cell. 1980;22:405–413. doi: 10.1016/0092-8674(80)90351-7. [DOI] [PubMed] [Google Scholar]
- 14.Guilfoyle R, Weinmann R. Control region for adenovirus VA RNA transcription. Proc. Natl Acad. Sci. USA. 1981;78:3378–3382. doi: 10.1073/pnas.78.6.3378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harris B, Roeder RG. Structural relationships of low molecular weight viral RNAs synthesized by RNA polymerase III in nuclei from adenovirus 2-infected cells. J. Biol. Chem. 1978;253:4120–4127. [PubMed] [Google Scholar]
- 16.Thimmappaya B, Jones N, Shenk T. A mutation which alters initiation of transcription by RNA polymerase III on the Ad5 chromosome. Cell. 1979;18:947–954. doi: 10.1016/0092-8674(79)90207-1. [DOI] [PubMed] [Google Scholar]
- 17.Vennström B, Pettersson U, Philipson L. Two initiation sites for adenovirus 5.5S RNA. Nucleic Acids Res. 1978;5:195–204. doi: 10.1093/nar/5.1.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Current Protocols in Molecular Biology. Boston, MA: JohnWiley & Sons, Inc.; 2006. [Google Scholar]
- 19.Ulfendahl PJ, Linder S, Kreivi JP, Nordqvist K, Sevensson C, Hultberg H, Akusjärvi G. A novel adenovirus-2 E1A mRNA encoding a protein with transcription activation properties. EMBO J. 1987;6:2037–2044. doi: 10.1002/j.1460-2075.1987.tb02468.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bhat RA, Thimmappaya B. Adenovirus mutants with DNA sequence perturbations in the intragenic promoter of VAI RNA gene allow the enhanced transcription of VAII RNA gene in HeLa cells. Nucleic Acids Res. 1984;12:7377–7388. doi: 10.1093/nar/12.19.7377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thimmappaya B, Weinberger C, Schneider RJ, Shenk T. Adenovirus VAI RNA is required for efficient translation of viral mRNAs at late times after infection. Cell. 1982;31:543–551. doi: 10.1016/0092-8674(82)90310-5. [DOI] [PubMed] [Google Scholar]
- 22.Davison AJ, Benko M, Harrach B. Genetic content and evolution of adenoviruses. J. Gen. Virol. 2003;84:2895–2908. doi: 10.1099/vir.0.19497-0. [DOI] [PubMed] [Google Scholar]
- 23.Pall GS, Codony-Servat C, Byrne J, Ritchie L, Hamilton A. Carbodiimide-mediated cross-linking of RNA to nylon membranes improves the detection of siRNA, miRNA and piRNA by northern blot. Nucleic Acids Res. 2007;35:e60. doi: 10.1093/nar/gkm112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schramm L, Hernandez N. Recruitment of RNA polymerase III to its target promoters. Genes Dev. 2002;16:2593–2620. doi: 10.1101/gad.1018902. [DOI] [PubMed] [Google Scholar]
- 25.Rohan RM, Ketner G. A comprehensive collection of point mutations in the internal promoter of the adenoviral VAI gene. J. Biol. Chem. 1987;262:8500–8507. [PubMed] [Google Scholar]
- 26.Weitzer S, Martinez J. The human RNA kinase hClp1 is active on 3′ transfer RNA exons and short interfering RNAs. Nature. 2007;447:222–226. doi: 10.1038/nature05777. [DOI] [PubMed] [Google Scholar]
- 27.Elbashir SM, Lendeckel W, Tuschl T. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev. 2001;15:188–200. doi: 10.1101/gad.862301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- 29.MacRae IJ, Zhou K, Doudna JA. Structural determinants of RNA recognition and cleavage by Dicer. Nat. Struct. Mol. Biol. 2007;14:934–940. doi: 10.1038/nsmb1293. [DOI] [PubMed] [Google Scholar]
- 30.Zhang H, Kolb FA, Jaskiewicz L, Westhof E, Filipowicz W. Single processing center models for human Dicer and bacterial RNase III. Cell. 2004;118:57–68. doi: 10.1016/j.cell.2004.06.017. [DOI] [PubMed] [Google Scholar]
- 31.Vickers TA, Koo S, Bennett CF, Crooke ST, Dean NM, Baker BF. Efficient reduction of target RNAs by small interfering RNA and RNase H-dependent antisense agents. A comparative analysis. J. Biol. Chem. 2003;278:7108–7118. doi: 10.1074/jbc.M210326200. [DOI] [PubMed] [Google Scholar]
- 32.Schwarz DS, Hutvagner G, Du T, Xu Z, Aronin N, Zamore PD. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- 33.Tomari Y, Matranga C, Haley B, Martinez N, Zamore PD. A protein sensor for siRNA asymmetry. Science. 2004;306:1377–1380. doi: 10.1126/science.1102755. [DOI] [PubMed] [Google Scholar]
- 34.Hong J, Wei N, Chalk A, Wang J, Song Y, Yi F, Qiao RP, Sonnhammer EL, Wahlestedt C, Liang Z, et al. Focusing on RISC assembly in mammalian cells. Biochem. Biophys. Res. Commun. 2008;368:703–708. doi: 10.1016/j.bbrc.2008.01.116. [DOI] [PubMed] [Google Scholar]
- 35.Imperiale MJ, Kochanek S. Adenovirus vectors: biology, design, and production. Curr. Top. Microbiol. Immunol. 2004;273:335–357. doi: 10.1007/978-3-662-05599-1_10. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.