Abstract
Small non-coding RNAs (sRNAs) are an emerging class of post-transcriptional regulators of bacterial gene expression. To study sRNAs and their potential protein interaction partners, it is desirable to purify sRNAs from cells in their native form. Here, we used RNA-based affinity chromatography to purify sRNAs following their expression as aptamer-tagged variants in vivo. To this end, we developed a family of plasmids to express sRNAs with any of three widely used aptamer sequences (MS2, boxB, eIF4A), and systematically tested how the aptamer tagging impacted on intracellular accumulation and target regulation of the Salmonella GcvB, InvR or RybB sRNAs. In addition, we successfully tagged the chromosomal rybB gene with MS2 to observe that RybB-MS2 is fully functional as an envelope stress-induced repressor of ompN mRNA following induction of sigmaE. We further demonstrate that the common sRNA-binding protein, Hfq, co-purifies with MS2-tagged sRNAs of Salmonella. The presented affinity purification strategy may facilitate the isolation of in vivo assembled sRNA–protein complexes in a wide range of bacteria.
INTRODUCTION
Throughout their life cycle, small non-coding RNAs (sRNAs) that function as regulators of gene expression may interact with a variety of proteins. These include proteins that govern the maturation, turnover or intracellular transport of sRNAs; target proteins whose in vivo functions are modulated by an sRNA; and the auxiliary factors that help sRNAs recognize non-protein targets such as RNA, DNA or small metabolites. Over the years, various strategies have been used to isolate proteins along with RNA by means of RNA-based affinity chromatographic methods. The most direct approach is the immobilization of the RNA of interest on a column to which then cellular protein fractions are applied. For immobilization, in vitro-synthesized unmodified bait RNA is covalently coupled to cyanogen bromide (CNBr)-activated sepharose (1). Alternatively, modification of an in vitro-synthesized RNA can be used to facilitate its covalent or non-covalent attachment to the solid phase (2–5). Collectively, these methods rely upon RNA–protein interaction to take place after cellular lysate preparation, and therefore do not allow the isolation of in vivo assembled RNA–protein complexes.
To identify RNA-associated proteins, it remains desirable to purify a given RNA from its native source. Therefore, other strategies were developed in which a highly specific and non-covalent interaction with the RNA moiety is used for selection of, for example, eukaryotic ribonucleoprotein particles (RNPs) under non-denaturating conditions. Initially, biotinylated antisense oligonucleotides complementary to accessible, single-stranded regions of the RNP-contained RNA were used as affinity baits (6,7). More recently, the addition of diverse natural or in vitro selected RNA aptamer sequences to the RNA of interest has provided a convenient tool for RNP separation and purification; these aptamers bind their ligands with an affinity similar to that observed for natural antibodies. Aptamer tags specific for small molecules, e.g. streptavidin (8), streptomycin (9,10), or tobramycin (11), or for proteins and peptides, e.g. MS2 coat protein (12–14) and λN22 peptide (15,16), were successfully used for the isolation of a variety of RNPs. Recently, two RNA aptamers, one specific for the Pseudomonas phage 7 coat protein and the other for tobramycin, were used in combination for RNP purification (17).
Bacteria are known to express many regulatory sRNAs with diverse physiological functions (18–23). These sRNAs are typically 50–250 nucleotides in length, differ in structure from each other, and are generally untranslated. Depending on their mode of action, some bacterial sRNAs directly target proteins, e.g. 6S RNA which binds to and modifies the sigma factor affinity of RNA polymerase (24), or the family of CsrB-like RNAs which antagonizes CsrA/RsmA-like regulatory proteins (25,26).
The major class of regulatory sRNAs in bacteria targets trans-encoded mRNAs by antisense pairing (27). Target pairing is generally short and imperfect, and most often requires the bacterial Sm-like protein, Hfq, for successful target interaction (28,29). Recent co-immunoprecipitation experiments showed that up to half of all known Escherichia coli or Salmonella sRNAs may bind Hfq in vivo (30–32). However, we generally know little about the nature and number of proteins bacterial sRNAs associate with throughout their life time as regulators (33).
In this work, we have studied whether aptamer-tagging can be used to recover in vivo expressed regulatory sRNAs of Salmonella along with the commonly associated Hfq protein. We selected for study three structurally unrelated Salmonella sRNAs, i.e. GcvB, InvR and RybB (Figure 1A). Collectively, these sRNAs repress trans-encoded target mRNAs by Hfq-dependent base-pairing mechanisms. GcvB is a widely conserved 200 nt RNA which was originally identified in E. coli (34) and recently shown to directly target many Salmonella ABC transporter mRNAs (35); one of its targets is the oppA mRNA encoding a function in the major oligopeptide transport system. InvR (80 nt) RNA is expressed from the Salmonella-specific invasion gene island, and targets ompD mRNA encoding the major outer membrane protein of Salmonella under conditions of eukaryotic host cell invasion (36). The 80 nt RybB RNA also down-regulates outer membrane protein (OMP) synthesis yet in a rather global fashion; when overproduction of OMPs causes envelope instability, RybB expression is activated by the specialized sigma factor, σE, and the sRNA then promotes the rapid decay of all major and many minor omp mRNAs of Salmonella to restore envelope homeostasis (37).
Figure 1.
Regulatory sRNAs. Sequences of (A) the Salmonella RybB, InvR and GcvB sRNAs, and (B) the various aptamer tags specific for phage MS2 coat protein (MS2 and MS2*), for λN22 peptide (1xboxB and 4xboxB), or the eukaryotic initiation factor 4A (eIF4A) used in the present study. Shadowed nucleotides denote residues of the three Salmonella sRNAs that are involved in base-pairing to target mRNAs.
We have tested how the addition of any of three well-investigated aptamer sequences (MS2, boxB, eIF4A; Figure 1B) impacts on the expression levels and regulatory properties of sRNAs, and have successfully used MS2 aptamer-tagged GcvB, InvR or RybB RNAs for Hfq co-purification from Salmonella extracts. We provide an optimized protocol for affinity purification of MS2 aptamer-tagged sRNAs that may facilitate the isolation of in vivo assembled sRNA–protein complexes in a wide range of bacteria. Our protocol further proves useful for the subsequent proteome analysis of the purified complexes by mass spectrometry.
MATERIALS AND METHODS
Plasmid construction
DNA oligonucleotides and plasmids used in this study are listed in Supplementary Tables S1 and S2. In the following, a detailed description of the construction of GcvB aptamer expression plasmids and the control plasmids is given. The details of the RybB/InvR expression plasmids are found in the Supplementary Text and Table S2.
For GcvB plasmids pNS-7 and pNS-8, plasmid pTP005 was amplified with primer pairs JVO-2660/-2661 (introduce MS2 tag), and JVO-2658/-2659 (introduce eIF4A tag) and the PCR product was re-ligated.
To construct the general vectors, two complementary oligodesoxynucleotides, JVO-2750 and JVO-2751 containing the 1xboxB aptamer flanked by NheI and XbaI cleavage sites and the vrrA sequence flanked by the AvrII restriction site, were annealed and digested with AvrII. The DNA fragment was inserted by blunt-end/AvrII-XbaI cloning into plasmid vector pJV752. The resulting plasmid, pNS-18, which now harbored the 1xboxB aptamer cassette, subsequently served as the parental plasmid for construction of additional general aptamer plasmids that carry 4xboxB (pNS-20), eIF4A (pNS-19) or MS2* (pNS-21) cassettes. To this end, the 1xboxB cassette was excised from pNS-18 by NheI/XbaI digest, and replaced with NheI/XbaI treated DNA fragment with the new aptamer sequence. Inserts were obtained by PCR with primer JVO-2754/2755 for eIF4A, or JVO-2756/-2757 for MS2*, and using plasmids pNS-3 or pMS2* respectively, as PCR templates. The 4xboxB aptamer fragment was obtained by annealing complementary oligodesoxynucleotides JVO-2752 and JVO-2753.
To construct plasmids in which GcvB sRNA is 3′-end-tagged with an aptamer, the gcvB gene of plasmid pTP005 was amplified with primer pair pZE-A/JVO-2956, which yielded the sRNA gene but without the native gcvB terminator, and with a NheI restriction site at the 3′ of the gene fragment. These fragments were digested with XhoI and NheI, and inserted into the aforementioned general aptamer vectors (pNS-18, pNS-19, pNS-20 or pNS-21) (note that XhoI cloning remove the plasmid-borne PLlacO-1 promoter, thus the sRNA fragment must contain the native sRNA promoter). This gave yield to plasmids pNS-22, pNS-27, pNS-31 and pNS-32.
Control vector, pNS-17, was generated by NheI/XbaI digest of plasmid pNS-18, followed by re-ligation of the vector backbone. This control plasmid expresses a ∼80 nt nonsense RNA that corresponds to the rrnB terminator of the plasmid and vector sequences upstream of the terminator.
To construct the MS2 control plasmid pRR-05, which expresses a short MS2 tag RNA, plasmid pNS-10 was amplified with primer pair JVO-1745/-3446, and religated.
All plasmids used throughout this study were introduced into Salmonella by electroporation as described (38).
Bacterial strains and growth conditions
Salmonella strain JVS-2999, which is a non-infectious aroA mutant of Salmonella enterica serovar Typhimurium, is referred to as the Salmonella wild-type strain throughout this study. It corresponds to S. typhimurium SL7207 strain (2337–65 (WRAY) hisG46ΔaroA407 (TcsaroA544::Tn10) as published (39), and was kindly provided by the Department of Immunology at the MPI for Infection Biology, Berlin, Germany. For experiments in Figure 7B, the virulent Salmonella strain SL1344 (JVS-0007) was used.
Figure 7.
Analysis of chromosomally tagged RybB sRNA. (A) A PCR-product containing the MS2-tagged sRNA flanked by homologous regions (H1 and H2) to the sRNA locus is integrated into the bacterial chromosome via λRed-mediated recombination. The integrated sRNA is selected by the attached antibiotic resistance gene. (B) Northern blot analysis of chromosomally expressed RybB RNAs (under control of σE) and ompN target mRNA. Wild-type Salmonella (JVS-0007) and Salmonella with chromosomally MS2-tagged RybB (rybB-MS2, JVS-4892) carrying a control plasmid (pBAD) or an arabinose-inducible σE expression plasmid (pBAD–RpoE) were induced in early stationary phase. RNA was prepared prior to (0 min) and at 2, 5 and 10 min following induction, and probed as indicated. (C) Affinity purification. Prior to collection and lysis, Salmonella strains JVS-4279 and JVS-3617, both carrying the pBAD–RpoE expression plasmid, were grown to late exponential phase (OD600 of 1) and σE expression was induced for 5 min. Affinity purification and northern (RybB probe) and western (via the FLAG tag of Hfq) blot analyses were performed as in Figure 5 but loading eluate samples equivalent to 20 OD600 of bacterial culture.
The derivatives of JVS-2999 carrying sRNA deletions (JVS-3008, ΔinvR::KmR; JVS-3142, ΔrybB::KmR; JVS-3143, ΔgcvB::KmR) and/or an epitope-tagged hfq gene (JVS-3618, hfq::3xFLAG ΔinvR::KmR; JVS-3789, hfq::3xFLAG ΔrybB::KmR; JVS-3790, hfq::3xFLAG ΔgcvB::KmR) were constructed by phage P22 transduction of previously published strains as outlined in Supplementary Table S3.
To express endogenous RybB-MS2, we adapted the λRed recombinase method for one-step inactivation of chromosomal genes (40). The DNA fragment to be integrated (contains the aptamer-tagged sRNA of pNS-14 fused to the kanamycin resistance cassette of pKD4) was generated via overlapping PCR: the two DNA fragments obtained by PCR with primer pair JVO-906/3516 on pNS-14 and primer pair JVO-0203/3517 on pKD4 were mixed in equimolar amounts and used as template for a PCR with primer pairs JVO-3515/3521. The chromosomal RybB-MS2 mutation was subsequently transferred to strains JVS-0007 and JVS-3617, which results in JVS-4892 (used for functional studies in Figure 7B) and JVS-4279 (used for affinity purification in Figure 7C).
Bacterial strains were grown in LB (Lennox) broth at 37°C, 220 r.p.m., throughout this study. Antibiotics (where appropriate) were used at the following concentrations: 100 µg/ml ampicillin, 50 µg/ml kanamycin.
Sample preparation
Whole cell protein samples were prepared as follows: bacteria were cultured to an OD600 of 1 (for GcvB samples) or an OD600 of 2 (InvR, RybB) and samples equivalent to 0.5 OD600 (e.g. 0.5 ml of an OD600 of one culture) were removed and centrifuged for 2 min at 15 800g at 4°C. The pellet was suspended in 100 µl 1× protein loading buffer (Fermentas; #R0891). Proteins were denatured for 5 min at 96°C, and separated by sodium dodecyl sulfate (SDS)–polyacrylamide gelelectrophoresis (PAGE) using 15% polyacrylamide gels.
For isolation of total RNA, samples equivalent to 2 OD600 were centrifuged as described earlier.
The pellet was dissolved in 1 ml TRIzol Reagent (Invitrogen), and RNA was isolated and precipitated as described (38). The precipitated RNA was directly dissolved in 20 µl loading buffer II (Ambion, #AM8548G) and denatured for 3 min at 95°C. Ten microliters of each RNA sample were separated on 6% polyacrylamide/8.3 M urea gels.
To analyze chromosmal MS2-tagged RybB (Figure 7B), bacteria were cultered to an OD600 of 1, at which sRNA expression was induced with l-arabinose for 10 min. Culture aliquots equivalent to 4 OD600 were removed, mixed with 0.2 volumes of stop solution (5% water-saturated phenol, 95% ethanol) and snap-frozen in liquid nitrogen. After thawing on ice, bacteria were pelleted and the RNA was isolated as described earlier. The purified RNA was quantified on a Nanodrop Machine (NanoDrop Technologies). RNA samples (∼6 µg) were denatured for 3 min at 95°C in loading buffer II and separated on 6% polyacrylamide/8.3 M urea gels.
Purification of MS2-MBP fusion protein
For use as an affinity tag, MS2 coat protein fused to maltose-binding protein (MS2-MBP) was expressed in E. coli. This fusion places the maltose binding protein N-terminal to MS2 coat protein which carries a double mutation that prevents oligomerization (41). The fusion protein was purified first over an amylose (NEB) column in 20 mM HEPES pH 7.9, 200 mM KCl, 1 mM EDTA, and then over a heparin column using a KCl gradient in the same buffer (42). Purity of the protein was evaluated by SDS–PAGE and Commassie brilliant blue staining.
Affinity purification
The Salmonella strains indicated in figure legends were grown under standard laboratory conditions to an OD600 of 1 (GcvB) or 2 (InvR, RybB). Samples for the whole cell protein and the total RNA (culture samples) were prepared as described earlier. For affinity purification, cells equivalent to 50 OD600 were chilled on ice for 20 min and harvested (20 min, 2900g, 4°C). Cells were resuspended in 1 ml Buffer A (20 mM Tris–HCl pH 8.0, 150 mM KCl, 1 mM MgCl2, 1 mM DTT) and then cells were subsequently centrifuged (5 min, 11 200g, 4°C) and the pellets were shock-frozen in liquid nitrogen and stored at −80°C.
Frozen pellets were thawed on ice, resuspended in 2 ml Buffer A, followed by cell breakage using a French press (French Pressure Cell Press, SLM instruments). A pressure of 800 psi was applied to the cells for three times; the lysate was chilled on ice between every step. The lysate was cleared by centrifugation (30 min, 30 000g, 4°C) and the soluble fraction subjected to affinity chromatography.
All steps for affinity purification were performed at 4°C. For preparation of the affinity column, 50µl amylose resin (New England Biolabs, #E8021S) was applied to Bio-Spin disposable chromatography columns (BioRad, #732-6008). The prepared column was washed three times with 2 ml Buffer A. Next, 100 pmol MS2-MBP coat protein (diluted in 1 ml Buffer A) was immobilized on the amylose resin, and the column was washed with 2 ml of Buffer A (for the experiments in Figure 6, 100 µl amylose resin and 1200 pmol MS2-MBP were used). Subsequently, the cleared bacterial lysate was loaded onto the column, followed by three washes with 2 ml Buffer A. For RNA analysis, aliquots equivalent to 2 OD600 of the lysate, the flow-through and the wash fractions were treated with 1 ml TRIzol reagent as described earlier. For protein analysis, aliquots equivalent to 0.5 OD600 were mixed with protein loading buffer to a final volume of 100 µl.
Figure 6.
Several proteins co-purify with MS2-tagged InvR sRNA. Proteins of the lysate, flow-through, wash (0.1 OD600) and eluate (50 OD600) fractions were separated on an SDS-gel and stained with silver. Positions of the eluted MS2-MBP, the endogenous MBP of Salmonella (marked by an asterisk) and the co-purified FLAG-tagged Hfq are indicated right to the gel. Lysis with French press and affinity chromatography were performed with buffer A containing either the standard concentration of 150 mM KCl, or 1 M KCl. The eluate fractions of the affinity chromatography using the increased KCl concentration were analysed by mass spectrometry. Positions of co-migrating size marker proteins (in kDa) are shown to the left.
Finally, RNA and proteins were eluted from the column with 900 µl of Buffer A containing 12 mM maltose. Eluted RNA was extracted with phenol–chloroform–isoamylalcohol [25 : 24 : 1 (v/v), Roth], followed by ethanol (3 vol) precipitation of the aqueous phase in the presence of 20 µg yeast tRNA. Once re-dissolved in 25 µl water, the RNA was stored at −20°C. For protein isolation, the organic phase was subjected to acetone precipitation at −20°C over night. The pellet was washed twice with acetone and air-dried. The pellet was solved in 50 µl 1× protein loading buffer. For analysis of influence of different parameters on affinity purification changing conditions were used as indicated in the legend of Figure 6 and Supplementary Figure S4.
For the isolation of endogenous expressed RybB-MS2 (Figure 7C), some parameters were modified. JVS-2999 and JVS-4279, both harboring pBAD-RpoE, were grown to an OD600 of 1 nm, at which sRNA expression was induced with l-arabinose for 5 min. Cells equivalent to 200 OD600 were chilled at −10°C for 5 min and harvested (20 min, 2900g, 4°C). The lysate was prepared as described above, cleared by centrifugation (15 min, 2900g, 4°C) and the soluble fraction was subjected to affinity chromatography in which 100 µl amylose resin and 300 pmol MS2-MBP coat protein were used.
Isolation of tagged sRNAs and co-purification of Hfq was performed in triplicates, and results were consistent. The results of representative experiments are shown.
Mass spectrometry
For mass spectrometric analysis of InvR associated proteins, precipitated proteins were dissolved in 1× protein loading buffer and loaded onto a 4–12% NuPAGE-Gel (Invitrogen). After Coomassie stain gel lanes of affinity purified InvR-MS2 and InvR were cut into 23 slices and proteins therein are digested with Trypsin according to (43). Extracted peptides were analyzed by LC-MSMS on an Orbitrap XL instrument (ThermoFisherScientific) under standard conditions. Peptide fragment spectra were searched against a target-decoy database for Salmonella typhimurium strain LT2 (M. Selbach, personal communication) using MASCOT as search engine.
Northern blot analysis
After polyacrylamide gel electrophoresis, the RNA was transferred to Hybond-XL membranes (GE Healthcare) by electro-blotting (1 h, 50 V, 4°C) in a tank electro blotter (Peqlab, #52-WEB-20), and cross-linked to membrane by exposure to UV light (302 nm) for 4 min. The GcvB, InvR, RybB sRNA and the RNA with the MS2 tag alone were detected using γ-[32P] ATP 5′-end-labeled oligodeoxyribonucleotides JVO-0749 (antisense to nucleotides 4–20 of GcvB RNA), JVO-0222 (antisense to nucleotides 19–44 of InvR RNA), JVO-2644 (antisense to nucleotides 5–24 of RybB RNA), and JVO 3562 (antisense to nucleotides 8–31 of the MS2 aptamer), respectively. 5S rRNA (loading control) was detected with oligo JVO-0322; ompN mRNA was detected as described previously (37).
Prehybridization and hybridization of membranes with oligonucleotides was carried out each for 1 h in Roti-Hybri-Quick buffer (Roth) at 42°C. Following hybridization, membranes were rinsed in 5× SSC, followed by three washing steps at 42°C for 15 min in SSC (5×, 1× and 0.5×)/0.1% SDS solutions. Signals were visualized on a phosphorimager (Phosphorimager, FLA-3000 Series, Fuji), and band intensities quantified with AIDA software (Raytest, Germany).
Western blot analysis and silver staining
For western blot analysis, polyvinylidene difluoride membrane (PVDF, PerkinElmer) was activated by incubation in methanol (90 s), H2O (5 min) and transfer buffer (5 min), respectively. After SDS–PAGE, the gel was blotted for 2 h at 2 mA/cm2 and 4°C in a semi-dry electro blotter (Peqlab, #52–2020) onto the PVDF membrane in transfer buffer. After rinsing in TBST20, the membrane was incubated over night in 10% dry milk in TBTS20. OppA or Hfq3×FLAG proteins, and the major OMPs, were detected using a polyclonal serum against OppA (1 : 3000 in TBST20-BSA), porin antiserum, which detects major porin such as OmpA/C/D/F (1 : 20000 in TBST20-BSA, provided by R. Misra) or a monoclonal anti-FLAG antibody (Sigma-Aldrich, #F2555, 1 : 1000 in TBST20-BSA), for 1 h at room temperature under agitation. Membranes were washed 5×6 min in TBST20, incubated with either anti-mouse–horseradish peroxidase or anti-rabbit–horseradish peroxidase (GE Healthcare, 1:5000 in TBST20-BSA) for 1 h at room temperature and washed again 6×10 min in TBST20. Blots were analysed using Western Lightning Reagent (Perkin Elmer), and signals detected with a Fuji LAS-3000 CCD camera. Silver staining of the 15% polyacrylamide gel was performed according to ref. (44).
RESULTS
Construction of plasmid-based aptamer-tagged sRNA genes
To isolate in vivo expressed GcvB, InvR and RybB sRNAs from Salmonella by affinity chromatography, the sRNA genes were cloned on plasmids (Figure 2 and Supplementary Figure S1) and modified with any of three RNA affinity tags (Figure 1B). We used the following RNA affinity tags: (i) a tandem repeat of the 43 (45) or 50 nt (46,47) variant of phage MS2 coat protein binding motif, to which we will refer as MS2 or MS2*, respectively; (ii) monomeric (15 nt) or tetrameric (72 nt) versions of the λN22 peptide binding sequence (16,48), here referred to as 1xboxB or 4xboxB, respectively; (iii) a 60 nt SELEX-derived aptamer sequence specific for the eukaryotic initiation factor 4A (49) which will be referred to as the eIF4A aptamer.
Figure 2.
Cloning strategies for plasmids to express aptamer-tagged sRNAs. (A) Strategy I: an aptamer tag is introduced by addition of the sequence to PCR primers and amplification of a plasmid that carries the sRNA gene of interest. This strategy permits the insertion of the aptamer at any position in the sRNA sequence. (B) Strategy II: alternatively, sRNA fragments are amplified by PCR and introduced into the general aptamer cloning vectors, pNS-18, pNS-19, pNS-20 and pNS-21, using unique NheI and XbaI sites. This strategy permits to fuse the aptamer to either the 5′-end (insertion at XbaI site) or 3′-end (NheI) of the sRNA. Cloned fragment will be expressed from a PLlacO-1 promoter located immediately upstream of the NheI site. Alternatively, cloning in the XhoI–NheI region permits expression of an sRNA from its own promoter.
Based on prior knowledge of which region in the three sRNAs were required for target regulation, the aptamer sequences were introduced at various positions (Figure 3 and Supplementary Figure S1). In GcvB RNA, the binding site for oppA mRNA is located between stem loops 1 and 2 [Figure 1A and (35)]; consequently, aptamer sequences were added to the 5′-end of GcvB (MS2 and eIF4A-a constructs), or replaced the gcvB transcription terminator (MS2*, eIF4A-b, 1xboxB, 4xboxB). If an aptamer replaced the native transcription terminator, transcription was expected to terminate at a plasmid-borne terminator which derives from the vrrA sRNA gene of Vibrio species (50) and is located downstream of the sRNA cloning site. We used the heterologous vrrA terminator rather than a Salmonella or E. coli terminator to minimize possible recognition of the RNA 3′-end by Salmonella proteins. Concerning InvR, residues 33–66 of this RNA constitute the core interaction site for pairing with the ompD target mRNA [(36), and V. Pfeiffer and J. Vogel, unpublished data]. Therefore, aptamers were either fused to the 5′-end of InvR (MS2, eIF4A-a), inserted upstream of the invR terminator (1xboxB-a), or cloned such that the aptamer replaced the invR terminator (MS2*, eIF4A-b, 1xboxB-b, 4xboxB). The first 16 residues of RybB are stringently required for pairing to all omp target mRNAs (51); thus, aptamers were only cloned at positions downstream of RybB residue 31, by either insertion (MS2, 1xboxB) or terminator replacement (MS2*, eIF4A, 4xboxB).
Figure 3.
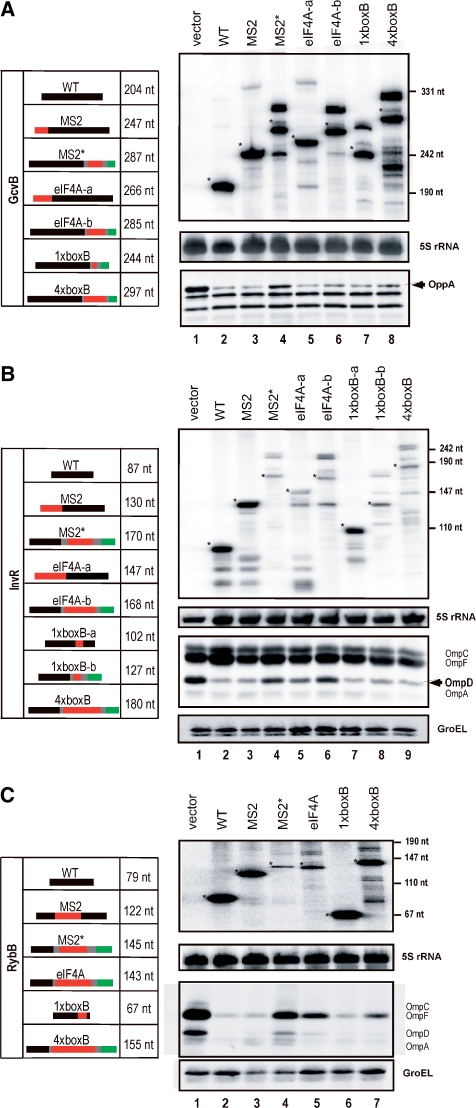
Expression and target regulation of aptamer-tagged sRNAs. (A) Northern and western blot analyses of Salmonella ΔgcvB (strain JVS-3143) carrying an ‘empty’ control vector (lane 1, pNS-17), or plasmids expressing wild-type (lane 2, pTP005) or aptamer-tagged GcvB sRNAs (lanes 3–8). The schematic drawing to the left shows the sRNA sequences in black, aptamer sequence in red, and (where applicable) the vrrA transcription terminator sequence in green. Co-transcribed linker sequences unrelated to aptamer, sRNA or vrrA terminator sequences are shown in grey. The expected length (in nucleotides) of the expressed RNA is given. Supplementary Figure S1 presents the sequence of the RNAs and connects the insert names to the expression plasmids. The top panel shows the relevant section of a northern blot probed with GcvB-specific oligonucleotide, JVO-0749. Asterisks indicate bands of expected full-length sRNAs. Positions of a co-migrating size marker are shown to the right of the blot. The middle panel shows probing for 5S rRNA to confirm equal loading. The bottom panel shows the relevant section of a western blot probed with a polyclonal serum against OppA protein, whose synthesis is repressed by GcvB sRNA. The OppA-specific band is indicated by an arrowhead. RNA and protein samples were prepared as described in ‘Materials and Methods’ section. (B) Analyses as above but of Salmonella ΔinvR (JVS-3008) and InvR expression plasmids. Wild-type InvR sRNA was expressed from plasmid pNS-9, and the sRNAs were detected with oligonucleotide probe, JVO-0222 (top panel). The third panel from top shows the section of a western blot probed with a general anti-porin serum in order to detect down-regulation of OmpD synthesis by InvR sRNAs. Bottom panel: the same blot probed for GroEL protein as loading control. (C) Analyses of Salmonella ΔrybB (JVS-3142) and RybB plasmids (pNS-13 for expression of wild-type RybB). Detection of the sRNA (top panel) was done with oligonucleotide probe, JVO-2644. Regulatory activity of the sRNAs was evaluated by western blot probing of major porins (third panel from top) whose synthesis is negatively controlled by RybB.
The pZ plasmid series (52) served as building blocks for construction of our aptamer-tagged sRNA expression plasmids; the p15A origin used here ensures 20–30 plasmid copies/cell. InvR and RybB sRNAs were expressed from a constitutive PLlacO-1 promotor (52) In contrast, GcvB was expressed from its native promotor since high expression of this sRNA is lethal in Salmonella (35).
We used two different cloning strategies (Figure 2). The first strategy provides plasmids carrying the wild-type sRNA genes which were amplified using PCR primers that introduced the aptamer tag at the desired sRNA position (Figure 2A). Second, we constructed general vectors harboring the 1xboxB (pNS-18), 4xboxB (pNS-20), eIF4A (pNS-19) or MS2* (pNS-21) aptamer sequences flanked by unique NheI and XbaI cleavage sites, and followed by the vrrA transcription terminator (Figure 2B). These vectors facilitate the parallel cloning of the sRNA of interest and aptamer-tagging at either the 5′- or 3′-end. The cloned sRNA will then be transcribed from the constitutive PLlacO-1 promotor; alternatively, an XhoI site further upstream in the plasmid facilitates the insertion of the sRNA gene with its native promoter. Cloning strategies for the individual constructs are summarized in the ‘Materials and Methods’ and Supplementary Methods sections.
Aptamer-tagged sRNAs are stably expressed and functional in vivo
To test if and how the aptamers impacted on sRNA stability and function in vivo, Salmonella carrying sRNA expression plasmids were grown in standard media to early stationary phase, and steady-state levels of the aptamer-tagged versus wild-type sRNAs, both expressed from plasmid in the corresponding sRNA deletion strain, were compared on northern blots. In parallel, target regulation was monitored by western blot detection of OmpD (InvR), OppA (GcvB) or the major porins (RybB). The results of these analyses are shown in Figure 3.
Aptamer-tagged GcvB sRNAs (Figure 3A, lanes 3–8) were found to be expressed at levels comparable to wild-type GcvB (lane 2); in all cases the most prominent GcvB-derived RNA species corresponded with the expected increase in RNA length upon aptamer-tagging. Additional prominent higher molecular weight RNA species were observed with four of the GcvB constructs that are supposed to terminate with the vrrA terminator (lanes 4, 6–8); these are likely to result from transcripts terminating at another terminator (rrnB) located ∼50 bp downstream on the plasmid. The smaller bands observed for the MS2* and 4xboxB variants (lanes 4 and 8) may indicate aberrant RNA processing events or premature termination. Except for MS2* (lane 4) all RNAs reduced OppA protein levels to the same degree as wild-type GcvB, which suggests that aptamer tagging did not impair the regulatory properties of these sRNAs. However, even the MS2* variant facilitated partial OppA repression.
In contrast to GcvB, aptamer-tagging strongly impacted on the steady-state level of InvR RNA (Figure 3B); in fact, only two (lanes 3 and 7) out of seven tagged InvR RNAs accumulated as a single species and at levels comparable to wild-type InvR expressed from plasmid. Interestingly, in both constructs (MS2 and 1xboxB-a) transcription terminates at the native invR terminator whereas all but one (eIF4A-a; lane 5) of the less abundant RNAs are supposed to terminate with the vrrA sequence. Strikingly, in spite of varying RNA levels and prominent RNA processing all but two variants (MS2* RNA; lane 4 and eIF4A-b RNA; lane 6) down-regulated OmpD protein synthesis comparably to wild-type InvR.
We constructed five aptamer versions of RybB RNA (Figure 3C). Whilst three (MS2, 1xboxB and 4xboxB; lanes 3, 6 and 7, respectively) accumulated at or close to wild-type levels, the MS2* (lane 4) and eIF4A (lane 5) RNAs were unstable. In contrast to GcvB and InvR, however, almost all RybB variants accumulated as a single RNA species of the predicted size. Regarding target (major porin) repression, there was good but not strictly linear correlation between steady-state levels of the regulatory RNA and the degree of repression. That is both the highly expressed MS2 and 1xboxB alleles decreased OmpA/C/D/F protein levels to the same degree as wild-type RybB (compare lanes 2, 3 and 6), whereas the poorly expressed MS2* RNA (lane 4) largely failed to deplete the major porins. The eIF4A and 4xboxB RNA (lanes 5 and 7) both facilitated the same degree of intermediate porin expression, although 4xboxB RNA was considerably more abundant than eIF4A RNA.
When the same type of aptamer was used, the most drastic difference in terms of RNA abundance and regulation was observed with the MS2 and MS2* RNAs (lanes 3 and 4). The aptamer position is similar in these two RNAs (fusion to residue +31 or +37, respectively, of RybB; Figure 3 and Supplementary Figure S1). However, whereas the MS2 aptamer was merely inserted, i.e. without deleting any RybB nucleotides, the MS2* aptamer was cloned such that it replaced the 3′-end of RybB. The similarly composed MS2* versions of GcvB and InvR were also strongly impaired in target regulation. Thus, the plasmid-borne cassette of MS2* and vrrA terminator may not consistently yield stable regulators, at least in Salmonella.
Taken together, these in vivo experiments suggest that the successful tagging of bacterial sRNA regulators with aptamer sequences requires empirical testing of both the tagging strategy and the insertion site in the sRNA. For the experiments to follow, we selected MS2-tagged sRNAs constructed according to cloning strategy I (Figure 2A) because these yielded the most homogenous transcripts, and efficient target regulation (Figure 3).
Affinity purification of in vivo expressed sRNAs
In order to test whether inserted aptamer sequences enable the purification of in vivo expressed sRNAs, we adapted a previously established protocol for the isolation of in vitro assembled spliceosomal RNPs (53) which employs MS2-MBP. Figure 4 gives an overview of the experimental steps. Salmonella expressing MS2-tagged sRNAs from plasmids are collected by centrifugation, followed by rapid lysis. A cleared lysate is then directly applied to an amylose column that is non-covalently coupled with MS2-MBP protein (via amylose binding of the MBP moiety). Upon binding of the MS2-tagged RNA species to MS2-MBP, the column is extensively washed to remove non-specifically bound RNA. Finally, the tagged sRNA and with it the associated protein(s) are eluted with a maltose-containing buffer that disrupts the MBP–amylose interaction.
Figure 4.
Experimental strategy to purify MS2 aptamer-tagged sRNAs and possibly in vivo formed RNPs under non-denaturing conditions from bacterial lysates. Red lines denote aptamer tags, yellow and green circles the MS2 coat protein and the maltose-binding protein (MPB) moieties of the MS2–MBP fusion protein that binds to the subunits of the amylose column (black circles).
All experiments described below were carried out in Salmonella strains in which the chromosomal copy of the sRNA of interest was deleted. We used plasmids expressing the 247 nt GcvB-MS2, the 130 nt InvR-MS2, or the 122 nt RybB-MS2 sRNAs (Figure 3 and Supplementary Figure S1). Parallel experiments with plasmids expressing the cognate wild-type sRNAs were used to determine the degree of unspecific column binding.
Comparative northern blot analysis of the eluted fractions (Figure 5A, lane 5) showed that the MS2 tag permitted the specific isolation of a tagged sRNA as compared to the cognate wild-type sRNA. Quantification of the northern blot signals in the lysate (input) and eluted fractions suggested that 9% of InvR-MS2 RNA was recovered. In contrast, only 0.14% of wild-type InvR RNA (which is expressed from the plasmid at the same level; Figure 3B) was recovered, arguing that InvR-MS2 RNA was enriched 65-fold as compared to wild-type InvR. Of the other two MS2-tagged sRNAs, GcvB and RybB, 3.5 and 22%, respectively, were recovered from the original lysate. Based on comparison to the eluted fractions of the wild-type sRNA samples, this suggested 55-fold (GcvB) or 135-fold (RybB) enrichment. Regardless of the enrichment, we note a considerable loss of RNA already in the lysate samples; this may be caused by rapid RNA degradation in the course of cell disruption. In addition, although the bait protein was in molar excess over expressed RNAs (see quantification below), comparison of the flow-through fractions of the tagged versus untagged RNAs showed that generally <20% of the MS2-tagged sRNAs bound to the column.
Figure 5.
(A) Affinity purification of in vivo expressed MS2-tagged GcvB, InvR and RybB sRNAs. Shown are northern blot analyses of RNA prepared before, during or after the affinity purification described in the ‘Materials and Methods’ section. We analyzed Salmonella strains hfq3×FLAGΔsRNA or hfq3×FLAG harboring the RNA expression plasmids (from top to bottom: pNS-7 and pTP005 in strain JVS-3790, pNS-10 and pNS-9 in strain JVS-3618, pNS-14 and pNS-13 in strain JVS-3789 and pRR-05 in strain JVS-3616). RNA samples were prepared from intact cells before lysate preparation (lane 1), from the cleared lysate shortly before loading onto the column (lane 2), from the flow-through fraction collected after loading on a column pre-incubated with MS2-MBP protein (lane 3), from the combined column wash fractions (lane 4) and of the eluate (lane 5). Note that the amount of RNA loaded in lanes 1–4 each corresponds to 1 OD600 of bacterial culture, but in lane 5 the eluate corresponds to 10 OD600 of culture. Oligonucleotide probes to detect sRNAs were as in Figure 3; the MS2 control RNA was detected using JVO-3562. Comparison of the sRNA signal in the eluates of a wild-type sRNA versus its MS2 variant proves the specific isolation of the aptamer-tagged sRNAs. (B) Co-purification of Hfq with MS2-tagged sRNAs. Protein fractions of the samples shown in (A) were analyzed on western blot using a monoclonal antibody directed against the FLAG-epitope tag of the Hfq3×FLAG protein. Loaded protein samples correspond to 0.05 OD600 of bacterial culture for lanes 1–4 and to 10 OD600 for the eluate (lane 5). Enrichment of Hfq3×FLAG in the eluate samples of MS2-tagged GcvB, InvR and RybB, but not of MS2 control RNA argues that the protein was bound to the sRNA rather than the MS2 sequence.
To address in a more quantitative manner which amounts of the MS2-tagged RNAs were recovered by the above affinity purifications, sRNA-specific northern blot signals of eluted RNA fractions were compared to the signal obtained with known quantities of in vitro transcribed sRNA (Supplementary Figure S3). From a starting material of 5 × 1010 (A600 of 50) bacteria, 0.35 pmol (∼25 ng) of GcvB-MS2, 2.5 pmol (∼100 ng) of InvR-MS2, and 2.1 pmol (∼80 ng) of RybB-MS2 were purified. Thus, the MS2-tagged sRNAs were recovered in quantities that should permit downstream biochemical analysis.
Co-purification of Hfq protein
InvR, GcvB and RybB belong to a class of bacterial sRNAs that strictly depend on the Sm-like protein, Hfq, for both intracellular stability and target regulation (35,36,54). All three sRNAs are enriched by co-immunoprecipitation (coIP) of Hfq in Salmonella lysates (30,32), suggesting that they form stable complexes with Hfq in vivo. Therefore, if the MS2-MBP affinity approach indeed recovered sRNAs in their native form, Hfq protein should not only be detectable in the eluted fraction, but also considerably enriched with the MS2-tagged sRNAs as compared to untagged wild-type sRNAs.
We expressed the sRNAs in a Salmonella hfq3×FLAG strain which provided a means to detect Hfq protein on western blots by way of the added FLAG epitope (30). Figure 5B shows that Hfq was recovered with all three MS2-tagged sRNAs, whereas little (InvR) or no (GcvB, RybB) Hfq was detected in the elute fraction of the untagged counterparts of these sRNAs. This suggested that Hfq was purified along with the RNA, rather than by unspecific interaction with the column-bound MS2-MBP protein.
Next, Hfq3×FLAG protein was over-expressed in E. coli and purified to homogeneity. Dilution series of the purified protein were then probed along with the protein samples from the elute fractions of MS2-tagged sRNAs, in order to determine the number of Hfq molecules that were co-purified with these sRNAs (Supplementary Figure S3). We determined 1.0, 2.6 or 1.1 pmol Hfq (monomer) in the elute fractions of GcvB-MS2, InvR-MS2, or RybB-MS2, respectively. Our successful affinity purification of Hfq protein along with MS2-tagged RNAs (Figure 5) was in agreement with earlier results for the GcvB, InvR and RybB wild-type sRNAs showing (i) their enrichment by Hfq coIP (30,31,55) and (ii) their lower nanomolar binding affinities for purified Hfq in vitro (35–37). However, none of this excluded the possibility that Hfq primarily associated with the MS2 moiety rather than the sRNA sequence. To address this, we expressed in Salmonella a short RNA comprised of the MS2 tag followed by the RNA sequence of the rrnB transcription terminator (plasmid pRR-05). This ∼80 nt MS2 RNA was stable in vivo, and could be recovered by MS2-MBP affinity purification in amounts comparable with the MS2-tagged sRNAs (Figure 5). Western blot analysis of the elute fraction determined 0.1 pmol Hfq. Given this weak association of Hfq with MS2 alone, the significantly higher recovery of Hfq with MS2-tagged sRNAs argues that the latter were purified as an Hfq-containing RNP that specifically assembled on the tagged sRNAs.
Optimized affinity purification
The successful co-purification of Hfq with MS2-tagged InvR sRNA was taken as a departure point to improve the initial protocol. Specifically, we asked whether the incubation time with MS2-MBP fusion protein influences the recovery of tagged sRNA from the cell lysate. Cell extracts containing MS2-tagged InvR or native InvR were subjected to affinity chromatography either without any preincubation with the protein or after 20, 50 or 70 min, respectively (Supplementary Figure S4A). The results indicate that specific and efficient binding occurs immediately and preincubation is not necessary. Therefore, the cell lysate can be loaded directly on the prepared MS2-MBP/amylose resin to decrease the possibility of RNA degradation and RNP complex dissociation.
We further asked which parameters can be changed to increase the isolation yield of sRNA associated proteins. To test the influence of the quantity of bait protein, columns were loaded with 10, 50, 100 and 200 pmol MS2-MBP protein. Supplementary Figure S4B shows that increasing MS2-MBP amounts do increase the yield of Hfq3×FLAG co-purified with InvR-MS2 RNA, yet that 10 pmol of MS2-MBP suffice for efficient co-purification, at least for this sRNA. Note that high amounts of MS2-MBP protein seem to increase unspecific binding, i.e. Hfq3×FLAG protein becomes detectable in the elute fraction of wild-type InvR RNA loaded onto a column with 200 pmol MS2-MBP protein (Supplementary Figure S4B).
Salt concentration can have a profound positive effect on RNA folding and the stability of non-covalent RNA–protein interactions during the course of purification (56–58). We tested the effects of KCl by increasing its concentration in all buffers from 150 mM to 1 M. Figure 6 shows that 1 M KCl permitted better co-purification of Hfq3×FLAG with InvR-MS2 RNA and revealed that the increase in salt concentration did not necessarily increase background protein levels. Interestingly, the silver-stained gels also revealed that the elute fraction equivalent to 5×1010 Salmonella cells (or 25 ml of a stationary phase bacterial culture) were sufficient to visualize the Hfq3×FLAG protein that co-purified with InvR-MS2 RNA. The amount of associated proteins was sufficient to perform a first proteome analysis by liquid chromatography tandem mass spectrometry (LC-MS/MS) of the tagged (InvR-MS2) RNP complexes. Table S4 summarizes the proteins that were identified. Hfq could exclusively be detected in the MS2 tagged sample but not in the non-tagged control.
MS2-tagging of a chromosomal sRNA gene
Additionally to the aptamer-tagging of sRNAs on plasmids, we have developed a strategy for MS2-tagging of chromosomal loci in order to purify sRNAs without any over-expression. To integrate the aptamer tag into the sRNA gene, we adapted a gene replacement strategy based on the one-step inactivation of chromosomal genes (40). Using a two-step PCR approach, a DNA fragment is generated that contains the aptamer-tagged sRNA and an antibiotic resistance gene, and which is then used to replace the wild-type sRNA gene by λRed-mediated recombination (Figure 7A). The rybB gene is directly activated by the envelope stress sigma factor, σE (37). We determined that within 10 min following σE induction, MS2-tagged RybB accumulated to comparable RNA levels as RybB of a wild-type strain (Figure 7B), and that the tagged RNA down-regulated the well-established ompN mRNA target (37,51) with the same kinetics as wild-type RybB (Figure 7B), arguing that the MS2 tag did not compromise RybB’s activity as an ompN repressor.
For RNA affinity purification (Figure 7C), samples were withdrawn 5 min after σE induction. As before, we could recover the MS2-tagged RybB sRNA along with Hfq protein. Specifically, we obtained ∼1.2 pmol (∼50 ng) MS2-tagged RybB and ∼7 pmol (∼100 ng) Hfq from a lysate of 200 OD600 of bacterial culture. This indicates that, on average, each purified RybB-MS2 molecule was associated with one Hfq hexamer, which is in perfect agreement with the stochiometry of a RybB/Hfq complex formed in vitro (37).
DISCUSSION
There have been several studies to identify sRNA-binding proteins by way of affinity purification from lysates of E. coli cells. Wassarman and Storz (59) used a DNA antisense oligonucleotide directed against an extended loop region of the otherwise highly structured 6S RNA, which led to the discovery of the tight association of 6S RNA with RNA polymerase (RNAP). The Schroeder laboratory have extensively used in vitro-transcribed variants of E. coli sRNAs which carried a streptomycin aptamer tag in order to purify proteins that would bind to these sRNA upon incubation with E. coli lysates (9,60). Mass spectrometry analysis of the purified protein fractions confirmed many predicted sRNA–protein interactions, e.g. 6S RNA/RNAP, CsrB/CsrA or OxyS/Hfq, and also revealed unexpected interactions of several sRNAs with RNAP or ribosomal S1 protein (60). In a further study, in vitro-transcribed ∼70 nt Rcd RNA of plasmid ColE1 was cross-linked to CNBr-activated sepharose, followed by incubation with E. coli lysates; step-wise elution of the retained proteins identified the enzyme, tryptophanases, as a novel target of this small RNA regulator (61).
The present study aimed at establishing whether bacterial sRNAs can be successfully modified by the addition of aptamer sequences such that they would (i) retain regulatory properties in vivo, and (ii) be recovered as an in vivo assembled RNP with their natural binding partner, Hfq, via RNA-based affinity purification. The results obtained with three model sRNAs of Salmonella suggest that successful tagging in terms of maintaining a homogenous RNA population will require empirical testing of several aptamers and insertion points, and that of the aptamer sequences tested here, the MS2 tag is likely to work best.
The in vivo expression of the aptamer-tagged sRNAs may offer advantages in comparison to solely in vitro-based methods. First, the functionality of the tagged sRNA can be tested by first scoring its native regulatory function in vivo. That is the integration of relatively large aptamers (40–80 nt) into bacterial sRNAs that are mostly ≤150 nt in length (62) inevitably comes with the risk of intramolecular structural interference, impairing the folding of the sRNA or shielding its target interaction regions. Second, in vivo transcription of sRNA and its target mRNA(s) might be crucial for correct assembly of active RNPs, whereas in vitro transcribed RNA could be trapped in an inactive fold due to absence of auxiliary factors that act co-transcriptionally in vivo. Third, reversible formaldehyde cross-linking in vivo which is commonly used to identify RNA–protein interactions may capture very transient RNPs that promote key kinetic steps in sRNA-mediated target recognition/regulation; the cross-linking may limit the loss of weakly associated factors. Last, expression of aptamer-tagged sRNAs may not only be useful for RNP purification but may also permit visualization of spatial expression pattern of an sRNA in vivo. That is GFP fusion proteins to MS2 or eIF4A have been successfully used to visualize the in vivo distribution of mRNA or 5S rRNA, respectively, in E. coli (49,63).
To isolate in vivo assembled sRNA–protein complexes, antisense oligonucleotides binding to regions that remain accessible in the wild-type RNA after complex formation has been amply used. This method has the advantage that tagging of the RNA is not required, and it has been successfully applied to, e.g. the isolation of well-established spliceosomal and telomerase complexes (6,7). However, purification via affinity oligonucleotides are usually more dependent on the proper salt concentrations than an aptamer approach. In general, little is known about the structure and thus the accessible regions of bacterial sRNAs in vivo. Therefore, the aptamer and the oligonucleotide approach equally bear the risk of missing crucial protein interactions if the targeted region is only accessible in a subcomplex.
Purification of in vivo formed RNPs using aptamer-tags was not previously described in prokaryotes. Whilst our study focused on three aptamer tags for the MS2-coat protein, λN22 peptide, and the eukaroytic initiation factor 4A, other aptamers remain to be tested. For example, active RNase P was successfully isolated from yeast following tagging of the RNA component of RNase P with a streptavidin aptamer (8). Recently, RNA affinity in tandem (RAT) was described (17). Here, the combinational use of the aptamer sequence binding to the Pseudomonas phage 7 coat protein and the tobramycin aptamer allowed the purification of human 7SK RNPs.
However, there are several important issues to be considered for any sRNAs to be studied with our tagging approach in the future. In the present study, we have often relied upon overexpression of the regulator (of InvR and RybB; Supplementary Figure S2) to ensure sufficient and comparable expression of the various sRNAs under the growth conditions used here. It is important to stress, though, that over-expression of the tagged sRNAs may allow them to function even if they are not as active as the endogenous sRNAs, or if they comprise a mixed pool of active and non-active sRNAs. In addition, the sRNA may form complexes with mRNAs and/or proteins that would not be targeted by native expression levels; may act indirectly by titrating Hfq, thereby affecting the stability of other molecules; and might dilute a relevant protein in the pool of isolated RNPs. In this respect, our demonstration that a chromosomally encoded RybB-MS2 sRNA is fully functional as an envelope stress-induced ompN repressor should encourage integration of tagged sRNAs in the chromosome after initial testing on plasmids.
Homogeneity of the expressed sRNAs is another issue. In this regard, if the insertion of the aptamer yield in an sRNA that retained its natural terminator sequences (Figures 2A and 3), the RNAs are mostly expressed as homogenous species and in levels comparable the wild-type sRNAs. However, for constructs using the plasmid-borne vrrA terminator we often observe aberrant processing and transcriptional read-through into the downstream located rrnB terminator (Figures 2B and 3). We do not currently understand the reason for the observed heterogeneity of transcripts in those latter cases. That is, the terminator of the Vibrio VrrA sRNA gene was chosen because it represents a heterologous sRNA terminator expected to have little influence on sRNA function in Salmonella, and which had been known to produce a single-sized VrrA RNA in both Vibrio and Salmonella (50). Although most of our tagged sRNAs to terminate at vrrA seem to be still functional in vivo, at least when over-expressed (Figure 3), it seems advisable to integrate the aptamer such that the original sRNA terminator is retained.
Whilst the general yield of purified RNA and protein is in the practical range for biochemical studies (see below), we note that generally <20% of MS2-tagged sRNA present in the lysate were captured on the MS2-MBP column (compare lanes 2 and 3 in Figure 5). In comparison, others have reported recovery rates between 12 and 90% when using in vitro transcribed streptotag-modified E. coli sRNAs and the respective matrix (60). However, differences in the tested sRNAs as well as the fact that sRNAs were bound to the column prior to lysate application preclude a direct comparison. Regardless, if a given sRNA is only partially retained on the column (because of shielding of the aptamer by a protein or by improper folding), we must at least consider the possibility that the purified sRNA may not fully represent the active regulatory complex.
Protein visualization and comparative quantification by standard laboratory techniques, e.g. SDS–PAGE, requires minimal concentrations of co-purified proteins. The lower detection limit for proteins using standard gel staining methods is ∼2–10 ng (silver staining) or ∼10–100 ng (Coomassie) per protein band (64). It is worthwhile to mention that we could easily detect an Hfq band upon co-purification with plasmid-expressed InvR-MS2 RNA (Figure 6). Therefore, our approach should be successful to recover at least the major binding partner of a sRNA of interest. Indeed a first MS-based proteome analysis revealed proteins associated with InvR-MS2 in addition to the expected Hfq protein (Table S4). Further studies of the detected proteins are ongoing.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
YIP grant of the EURANET within the 6th EU framework to H.U. Work in the Vogel lab is supported by the Deutsche Forschungsgemeinschaft (DFG) Priority Program SPP1258 Sensory and Regulatory RNAs in Prokaryotes. Funding for open access charge: Deutsche Forschungsgemeinschaft.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Monika Raabe for excellent technical assistance in MS; Matthias Selbach for providing the Salmonella LT database; Rajev Misra for porin antiserum; K. Igarashi for anti-Oppa serum.
REFERENCES
- 1.Gerber CA, Relich A, Driscoll DM. Isolation of an mRNA-binding protein involved in C-to-U editing. Methods Mol. Biol. 2004;265:239–249. doi: 10.1385/1-59259-775-0:239. [DOI] [PubMed] [Google Scholar]
- 2.Allerson CR, Martinez A, Yikilmaz E, Rouault TA. A high-capacity RNA affinity column for the purification of human IRP1 and IRP2 overexpressed in Pichia pastoris. RNA. 2003;9:364–374. doi: 10.1261/rna.2143303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Caputi M, Mayeda A, Krainer AR, Zahler AM. hnRNP A/B proteins are required for inhibition of HIV-1 pre-mRNA splicing. EMBO J. 1999;18:4060–4067. doi: 10.1093/emboj/18.14.4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bindereif A, Green MR. An ordered pathway of snRNP binding during mammalian pre-mRNA splicing complex assembly. EMBO J. 1987;6:2415–2424. doi: 10.1002/j.1460-2075.1987.tb02520.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grabowski PJ, Sharp PA. Affinity chromatography of splicing complexes: U2, U5, and U4 + U6 small nuclear ribonucleoprotein particles in the spliceosome. Science. 1986;233:1294–1299. doi: 10.1126/science.3638792. [DOI] [PubMed] [Google Scholar]
- 6.Blencowe BJ, Sproat BS, Ryder U, Barabino S, Lamond AI. Antisense probing of the human U4/U6 snRNP with biotinylated 2′-OMe RNA oligonucleotides. Cell. 1989;59:531–539. doi: 10.1016/0092-8674(89)90036-6. [DOI] [PubMed] [Google Scholar]
- 7.Lingner J, Cech TR. Purification of telomerase from Euplotes aediculatus: requirement of a primer 3′ overhang. Proc. Natl Acad. Sci. USA. 1996;93:10712–10717. doi: 10.1073/pnas.93.20.10712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Srisawat C, Engelke DR. Streptavidin aptamers: affinity tags for the study of RNAs and ribonucleoproteins. RNA. 2001;7:632–641. doi: 10.1017/s135583820100245x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Windbichler N, Schroeder R. Isolation of specific RNA-binding proteins using the streptomycin-binding RNA aptamer. Nature Protoc. 2006;1:637–640. doi: 10.1038/nprot.2006.95. [DOI] [PubMed] [Google Scholar]
- 10.Locker N, Easton LE, Lukavsky PJ. Affinity purification of eukaryotic 48S initiation complexes. RNA. 2006;12:683–690. doi: 10.1261/rna.2227906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hartmuth K, Urlaub H, Vornlocher HP, Will CL, Gentzel M, Wilm M, Luhrmann R. Protein composition of human prespliceosomes isolated by a tobramycin affinity-selection method. Proc. Natl Acad. Sci. USA. 2002;99:16719–16724. doi: 10.1073/pnas.262483899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bardwell VJ, Wickens M. Purification of RNA and RNA-protein complexes by an R17 coat protein affinity method. Nucleic Acids Res. 1990;18:6587–6594. doi: 10.1093/nar/18.22.6587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou Z, Sim J, Griffith J, Reed R. Purification and electron microscopic visualization of functional human spliceosomes. Proc. Natl Acad. Sci. USA. 2002;99:12203–12207. doi: 10.1073/pnas.182427099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bessonov S, Anokhina M, Will CL, Urlaub H, Luhrmann R. Isolation of an active step I spliceosome and composition of its RNP core. Nature. 2008;452:846–850. doi: 10.1038/nature06842. [DOI] [PubMed] [Google Scholar]
- 15.Czaplinski K, Kocher T, Schelder M, Segref A, Wilm M, Mattaj IW. Identification of 40LoVe, a Xenopus hnRNP D family protein involved in localizing a TGF-beta-related mRNA during oogenesis. Dev. Cell. 2005;8:505–515. doi: 10.1016/j.devcel.2005.01.012. [DOI] [PubMed] [Google Scholar]
- 16.Duncan K, Grskovic M, Strein C, Beckmann K, Niggeweg R, Abaza I, Gebauer F, Wilm M, Hentze MW. Sex-lethal imparts a sex-specific function to UNR by recruiting it to the msl-2 mRNA 3′ UTR: translational repression for dosage compensation. Genes Dev. 2006;20:368–379. doi: 10.1101/gad.371406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hogg JR, Collins K. RNA-based affinity purification reveals 7SK RNPs with distinct composition and regulation. RNA. 2007;13:868–880. doi: 10.1261/rna.565207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Majdalani N, Vanderpool CK, Gottesman S. Bacterial small RNA regulators. Crit. Rev. Biochem. Mol. Biol. 2005;40:93–113. doi: 10.1080/10409230590918702. [DOI] [PubMed] [Google Scholar]
- 19.Romby P, Vandenesch F, Wagner EG. The role of RNAs in the regulation of virulence-gene expression. Curr. Opin. Microbiol. 2006;9:229–236. doi: 10.1016/j.mib.2006.02.005. [DOI] [PubMed] [Google Scholar]
- 20.Görke B, Vogel J. Noncoding RNA control of the making and breaking of sugars. Genes Dev. 2008;22:2914–2925. doi: 10.1101/gad.1717808. [DOI] [PubMed] [Google Scholar]
- 21.Waters LS, Storz G. Regulatory RNAs in bacteria. Cell. 2009;136:615–628. doi: 10.1016/j.cell.2009.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Massé E, Salvail H, Desnoyers G, Arguin M. Small RNAs controlling iron metabolism. Curr. Opin. Microbiol. 2007;10:140–145. doi: 10.1016/j.mib.2007.03.013. [DOI] [PubMed] [Google Scholar]
- 23.Livny J, Waldor MK. Identification of small RNAs in diverse bacterial species. Curr. Opin. Microbiol. 2007;10:96–101. doi: 10.1016/j.mib.2007.03.005. [DOI] [PubMed] [Google Scholar]
- 24.Wassarman KM. 6S RNA: a regulator of transcription. Mol. Microbiol. 2007;65:1425–1431. doi: 10.1111/j.1365-2958.2007.05894.x. [DOI] [PubMed] [Google Scholar]
- 25.Lapouge K, Schubert M, Allain FH, Haas D. Gac/Rsm signal transduction pathway of gamma-proteobacteria: from RNA recognition to regulation of social behaviour. Mol. Microbiol. 2008;67:241–253. doi: 10.1111/j.1365-2958.2007.06042.x. [DOI] [PubMed] [Google Scholar]
- 26.Babitzke P, Romeo T. CsrB sRNA family: sequestration of RNA-binding regulatory proteins. Curr. Opin. Microbiol. 2007;10:156–163. doi: 10.1016/j.mib.2007.03.007. [DOI] [PubMed] [Google Scholar]
- 27.Vogel J, Wagner EG. Target identification of regulatory sRNAs in bacteria. Curr. Opin. Microbiol. 2007;10:262–270. doi: 10.1016/j.mib.2007.06.001. [DOI] [PubMed] [Google Scholar]
- 28.Aiba H. Mechanism of RNA silencing by Hfq-binding small RNAs. Curr. Opin. Microbiol. 2007;10:134–139. doi: 10.1016/j.mib.2007.03.010. [DOI] [PubMed] [Google Scholar]
- 29.Valentin-Hansen P, Eriksen M, Udesen C. The bacterial Sm-like protein Hfq: a key player in RNA transactions. Mol. Microbiol. 2004;51:1525–1533. doi: 10.1111/j.1365-2958.2003.03935.x. [DOI] [PubMed] [Google Scholar]
- 30.Sittka A, Lucchini S, Papenfort K, Sharma CM, Rolle K, Binnewies TT, Hinton JC, Vogel J. Deep sequencing analysis of small noncoding RNA and mRNA targets of the global post-transcriptional regulator, Hfq. PLoS Genet. 2008;4:e1000163. doi: 10.1371/journal.pgen.1000163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang A, Wassarman KM, Rosenow C, Tjaden BC, Storz G, Gottesman S. Global analysis of small RNA and mRNA targets of Hfq. Mol. Microbiol. 2003;50:1111–1124. doi: 10.1046/j.1365-2958.2003.03734.x. [DOI] [PubMed] [Google Scholar]
- 32.Sittka A, Sharma CM, Rolle K, Vogel J. Deep sequencing of Salmonella RNA associated with heterologous Hfq proteins in vivo reveals small RNAs as a major target class and identifies RNA processing phenotypes. RNA Biol. 2009;6:e1000163. doi: 10.4161/rna.6.3.8332. [DOI] [PubMed] [Google Scholar]
- 33.Pichon C, Felden B. Proteins that interact with bacterial small RNA regulators. FEMS Microbiol. Rev. 2007;31:614–625. doi: 10.1111/j.1574-6976.2007.00079.x. [DOI] [PubMed] [Google Scholar]
- 34.Urbanowski ML, Stauffer LT, Stauffer GV. The gcvB gene encodes a small untranslated RNA involved in expression of the dipeptide and oligopeptide transport systems in Escherichia coli. Mol. Microbiol. 2000;37:856–868. doi: 10.1046/j.1365-2958.2000.02051.x. [DOI] [PubMed] [Google Scholar]
- 35.Sharma CM, Darfeuille F, Plantinga TH, Vogel J. A small RNA regulates multiple ABC transporter mRNAs by targeting C/A-rich elements inside and upstream of ribosome-binding sites. Genes Dev. 2007;21:2804–2817. doi: 10.1101/gad.447207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pfeiffer V, Sittka A, Tomer R, Tedin K, Brinkmann V, Vogel J. A small non-coding RNA of the invasion gene island (SPI-1) represses outer membrane protein synthesis from the Salmonella core genome. Mol. Microbiol. 2007;66:1174–1191. doi: 10.1111/j.1365-2958.2007.05991.x. [DOI] [PubMed] [Google Scholar]
- 37.Papenfort K, Pfeiffer V, Mika F, Lucchini S, Hinton JC, Vogel J. sigma(E)-dependent small RNAs of Salmonella respond to membrane stress by accelerating global omp mRNA decay. Mol. Microbiol. 2006;62:1674–1688. doi: 10.1111/j.1365-2958.2006.05524.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sittka A, Pfeiffer V, Tedin K, Vogel J. The RNA chaperone Hfq is essential for the virulence of Salmonella typhimurium. Mol. Microbiol. 2007;63:193–217. doi: 10.1111/j.1365-2958.2006.05489.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hoiseth SK, Stocker BA. Aromatic-dependent Salmonella typhimurium are non-virulent and effective as live vaccines. Nature. 1981;291:238–239. doi: 10.1038/291238a0. [DOI] [PubMed] [Google Scholar]
- 40.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl Acad. Sci. USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.LeCuyer KA, Behlen LS, Uhlenbeck OC. Mutants of the bacteriophage MS2 coat protein that alter its cooperative binding to RNA. Biochemistry. 1995;34:10600–10606. doi: 10.1021/bi00033a035. [DOI] [PubMed] [Google Scholar]
- 42.Jurica MS, Licklider LJ, Gygi SR, Grigorieff N, Moore MJ. Purification and characterization of native spliceosomes suitable for three-dimensional structural analysis. RNA. 2002;8:426–439. doi: 10.1017/s1355838202021088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- 44.Jungblut PR, Seifert R. Analysis by high-resolution two-dimensional electrophoresis of differentiation-dependent alterations in cytosolic protein pattern of HL-60 leukemic cells. J. Biochem. Biophys. Methods. 1990;21:47–58. doi: 10.1016/0165-022x(90)90044-d. [DOI] [PubMed] [Google Scholar]
- 45.Batey RT, Kieft JS. Improved native affinity purification of RNA. RNA. 2007;13:1384–1389. doi: 10.1261/rna.528007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Das R, Zhou Z, Reed R. Functional association of U2 snRNP with the ATP-independent spliceosomal complex E. Mol. Cell. 2000;5:779–787. doi: 10.1016/s1097-2765(00)80318-4. [DOI] [PubMed] [Google Scholar]
- 47.Deckert J, Hartmuth K, Boehringer D, Behzadnia N, Will CL, Kastner B, Stark H, Urlaub H, Luhrmann R. Protein composition and electron microscopy structure of affinity-purified human spliceosomal B complexes isolated under physiological conditions. Mol. Cell Biol. 2006;26:5528–5543. doi: 10.1128/MCB.00582-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Daigle N, Ellenberg J. LambdaN-GFP: an RNA reporter system for live-cell imaging. Nat. Methods. 2007;4:633–636. doi: 10.1038/nmeth1065. [DOI] [PubMed] [Google Scholar]
- 49.Valencia-Burton M, McCullough RM, Cantor CR, Broude NE. RNA visualization in live bacterial cells using fluorescent protein complementation. Nat. Methods. 2007;4:421–427. doi: 10.1038/nmeth1023. [DOI] [PubMed] [Google Scholar]
- 50.Song T, Mika F, Lindmark B, Liu Z, Schild S, Bishop A, Zhu J, Camilli A, Johansson J, Vogel J, et al. A new Vibrio cholerae sRNA modulates colonization and affects release of outer membrane vesicles. Mol. Microbiol. 2008;70:100–111. doi: 10.1111/j.1365-2958.2008.06392.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bouvier M, Sharma CM, Mika F, Nierhaus KH, Vogel J. Small RNA binding to 5′ mRNA coding region inhibits translational initiation. Mol. Cell. 2008;32:827–837. doi: 10.1016/j.molcel.2008.10.027. [DOI] [PubMed] [Google Scholar]
- 52.Lutz R, Bujard H. Independent and tight regulation of transcriptional units in Escherichia coli via the LacR/O, the TetR/O and AraC/I1-I2 regulatory elements. Nucleic Acids Res. 1997;25:1203–1210. doi: 10.1093/nar/25.6.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhou Z, Reed R. Purification of functional RNA-protein complexes using MS2-MBP. In: Ausubel FM, editor. Current Protocols in Molecular Biology. Inc, New York: John Wiley & Sons; 2003. Chapter 27, Unit 27. [DOI] [PubMed] [Google Scholar]
- 54.Johansen J, Rasmussen AA, Overgaard M, Valentin-Hansen P. Conserved small non-coding RNAs that belong to the sigma(E) regulon: role in down-regulation of outer membrane proteins. J. Mol. Biol. 2006;364:1–8. doi: 10.1016/j.jmb.2006.09.004. [DOI] [PubMed] [Google Scholar]
- 55.Wassarman KM, Repoila F, Rosenow C, Storz G, Gottesman S. Identification of novel small RNAs using comparative genomics and microarrays. Genes Dev. 2001;15:1637–1651. doi: 10.1101/gad.901001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Talbot SJ, Altman S. Kinetic and thermodynamic analysis of RNA-protein interactions in the RNase P holoenzyme from Escherichia coli. Biochemistry. 1994;33:1406–1411. doi: 10.1021/bi00172a017. [DOI] [PubMed] [Google Scholar]
- 57.Lohman TM, Overman LB, Ferrari ME, Kozlov AG. A highly salt-dependent enthalpy change for Escherichia coli SSB protein-nucleic acid binding due to ion-protein interactions. Biochemistry. 1996;35:5272–5279. doi: 10.1021/bi9527606. [DOI] [PubMed] [Google Scholar]
- 58.Baumann C, Otridge J, Gollnick P. Kinetic and thermodynamic analysis of the interaction between TRAP (trp RNA-binding attenuation protein) of Bacillus subtilis and trp leader RNA. J. Biol. Chem. 1996;271:12269–12274. doi: 10.1074/jbc.271.21.12269. [DOI] [PubMed] [Google Scholar]
- 59.Wassarman KM, Storz G. 6S RNA regulates E. coli RNA polymerase activity. Cell. 2000;101:613–623. doi: 10.1016/s0092-8674(00)80873-9. [DOI] [PubMed] [Google Scholar]
- 60.Windbichler N, von Pelchrzim F, Mayer O, Csaszar E, Schroeder R. Isolation of small RNA-binding proteins from E. coli: evidence for frequent interaction of RNAs with RNA polymerase. RNA Biol. 2008;5:30–40. doi: 10.4161/rna.5.1.5694. [DOI] [PubMed] [Google Scholar]
- 61.Chant EL, Summers DK. Indole signalling contributes to the stable maintenance of Escherichia coli multicopy plasmids. Mol. Microbiol. 2007;63:35–43. doi: 10.1111/j.1365-2958.2006.05481.x. [DOI] [PubMed] [Google Scholar]
- 62.Hershberg R, Altuvia S, Margalit H. A survey of small RNA-encoding genes in Escherichia coli. Nucleic Acids Res. 2003;31:1813–1820. doi: 10.1093/nar/gkg297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Golding I, Cox EC. RNA dynamics in live Escherichia coli cells. Proc. Natl Acad. Sci. USA. 2004;101:11310–11315. doi: 10.1073/pnas.0404443101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Winkler C, Denker K, Wortelkamp S, Sickmann A. Silver- and Coomassie-staining protocols: detection limits and compatibility with ESI MS. Electrophoresis. 2007;28:2095–2099. doi: 10.1002/elps.200600670. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.