Abstract
Adenosine-to-inosine (A-to-I) RNA editing alters the original genomic content of the human transcriptome and is essential for maintenance of normal life in mammals. A-to-I editing in Alu repeats is abundant in the human genome, with many thousands of expressed Alu sequences undergoing editing. Little is known so far about the contribution of Alu editing to transcriptome complexity. Transcripts derived from a single edited Alu sequence can be edited in multiple sites, and thus could theoretically generate a large number of different transcripts. Here we explored whether the combinatorial potential nature of edited Alu sequences is actually fulfilled in the human transcriptome. We analyzed datasets of editing sites and performed an analysis of a detailed transcript set of one edited Alu sequence. We found that editing appears at many more sites than detected by earlier genomic screens. To a large extent, editing of different sites within the same transcript is only weakly correlated. Thus, rather than finding a few versions of each transcript, a large number of edited variants arise, resulting in immense transcript diversity that eclipses alternative splicing as mechanism of transcriptome diversity, although with less impact on the proteome.
INTRODUCTION
Diversity at the posttranscriptional stages has been suggested to explain much of the discrepancy between gene number and organismal complexity. For example, Caenorhabditis elegans and humans share a similar number of genes, while the human is considered to be a much more complex organism. The most highly studied mechanism capable of achieving this diversity by increasing the number of different transcripts derived from a single gene is alternative splicing, wherein different sets of the pre-RNA molecular regions (introns) are removed to form the mature RNA molecule. However, an additional, less-explored mechanism for transcriptome diversity exists, known as RNA editing.
Adenosine-to-inosine (A-to-I) RNA editing, resulting in nucleoside modification, is performed by the adenosine deaminases (ADAR) family of proteins, which act on RNA. The splicing and translational machineries recognize inosine (I) as guanosine (G). Therefore the result of ADAR editing consists of genomically encoded adenosines that are read as guanosines in the RNA sequence (1). Until recently, this modification was considered to be rare in the human genome, with only a handful of sites known. In recent years, however, it was revealed that thousands of human genes are subjected to A-to-I RNA editing, mainly within their untranslated regions and introns (2–5). The editing events occur primarily within Alu repeats—primate-specific repetitive elements that comprise about 10% of the human genome (6)—due to their tendency to form dsRNA structures that are considered necessary for editing to take place.
Virtually all editing locations found so far [currently more than 15 000 sites (2,4,5)] were detected while aligning cDNA/EST sequences to their corresponding genomic loci, where clusters of A-to-G mismatches indicate editing events. This approach, while powerful for detection of edited Alu, suffers from two shortcomings. Editing sites discovered by this method are typically supported by only a single RNA sequence. The low coverage gives little information about the editing patterns in any particular Alu sequence (2–5,7–9). Specifically, it is not known how Alu editing contributes to transcriptome diversity; a cluster of editing events can potentially generate a large number of different transcripts, up to 2N different transcripts (N representing the number of adenosine sites in the Alu), if the individual editing events within a given Alu are not correlated. On the other hand, if editing sites are highly correlated, similar to the pattern of genomic SNPs which are grouped together in haplotype clusters, they may contribute much less to transcriptome complexity; in the extreme case of maximally correlated editing events, only two versions of the transcript will be found—a fully edited transcript and a non-edited one. No systematic efforts have been done so far to determine which of the two scenarios is closer to the biological reality.
Moreover, due to the low coverage, it is not clear how representative the detected editing sites are, and what is the actual number of editing sites per Alu. Clearly, this RNA-based approach is biased toward sites edited with high efficiency. However, the efficacy of many known editing events is much lower than 50%, leaving most of the editing sites invisible for detection by this method. In contrast, direct sequencing approaches used to detect sites subject to low-level editing can reveal the average editing level per site, but do not provide the ‘digital’ picture, i.e. the editing status for individual transcripts; thus, information about the transcript diversity generated by Alu editing is still largely missing and is needed in order to promote our understanding of the biological roles of RNA editing. Therefore, higher-coverage sequencing is desired in order to better characterize RNA editing.
Here we provide a first comprehensive description of diversity generated by Alu editing combinations, based on analysis of available genome-wide editing data and sequencing information of one particular Alu sequence. We found that the number of editing sites per Alu was much higher than reported previously, with many weakly edited sites and with each transcript having a different set of editing sites. This leads to a large number of different transcripts from each edited gene. Since thousands of genes contain edited Alu sequences, we suggest that the transcriptome diversity derived from editing is highly underestimated and eclipses alternative splicing as mechanism of transcriptome diversity, although probably with less impact on the proteome. Moreover, the availability of large sets of edited clones allowed us to reconstruct an ‘editing tree’, in which we can describe a possible model of the editing paths that generated the different edited transcripts from a single locus.
METHODS
Analysis of genome-wide editing data
In order to investigate the diversity of transcripts in the human transcriptome, we used a publicly available (5) dataset (accessible at http://www.cgen.com/research/Publications/AtoIEditing/RNAEDITING.html).
This set was derived from alignment of more than 5 million ESTs to the human genome, when clusters of A to G mismatches were detected as a signature of editing. Using such a large dataset allowed, in many cases, the identification of more than one edited transcript per locus of edited Alu. However, due to the high rate of errors in dbEST [∼3% sequencing errors (10)] substantial cleaning procedures were employed to the EST/genome alignment in order to achieve reliable prediction of editing sites, and many ESTs were discarded [full details of the procedure are available in (5)]. For the analysis of transcript diversity, only clusters of edited Alu with more than four identified editing sites that are supported by at least four different ESTs were used. ESTs that did not overlap with all sites in a cluster were not considered. For each cluster, the subcluster with the largest number of ESTs was selected. RefSeq sequences were removed from the analysis since they represent exact copies of one of the RNA sequences.
Multiple alignment
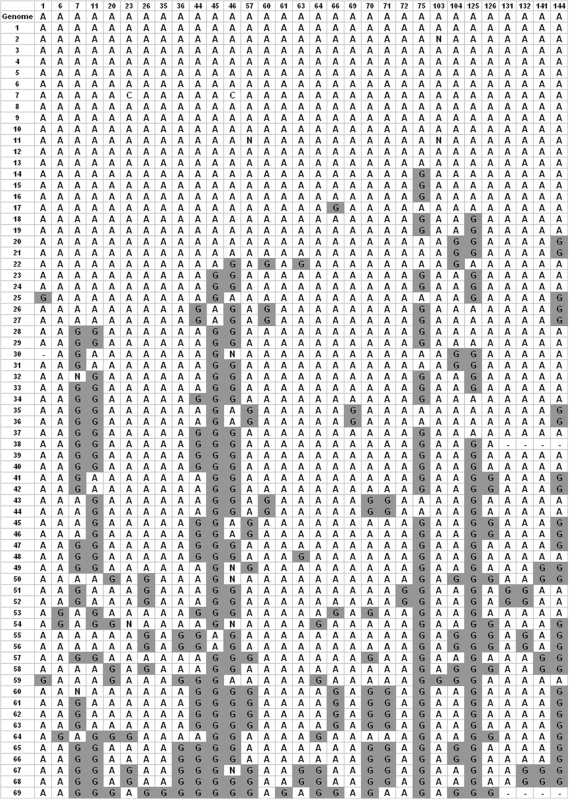
Multiple alignment of the clones from the direct sequencing of an Alu sequence located in intron 9 of the MED13 (THRAP1) gene was performed using the ClustalW program (11) between the RNA segments and the corresponding DNA with the A-to-I editing locations. We used data from normal control and cancer patients (the cancer analysis is given in the Supplementary Data). Details of the preparation of the clones are available in (12), which describes the source of the raw data. For the normal control 69 RNA clones were used and for the cancer patient 39 clones were studied. Although the cDNA were generated from two different sources, the DNA sequences for the short segment under investigation were identical. Editing locations were found within a span of 144 genomic bases. All clones were found, even after editing, to be specific for the same locus, without any other potential genomic locus that could have provided the source sequence. Editing locations are defined as locations that have A in the genome and G in at least one RNA segment, since I is read as G by the sequencing reaction after transformation to cDNA. By grouping all the editing locations, ignoring all the locations that are not edited in any RNA clone, we generated a dense matrix with all the editing data (Figure 1).
Figure 1.
Multiple alignment of editing locations. Graphical representation of the alignment with the edited clones. The first row represents the DNA root sequence and the following rows represent clones of RNA. Only locations that have at least one editing event are shown. The filled rectangles with ‘G’ represent editing events. Column numbering gives the location in the complete alignment.
We counted the number of editing events per location and their standard deviation with and without the non-edited sequences. For every editing location, we summed the total number of editing events. The percentage of editing events in every location was also calculated.
Motif analysis
Motif analysis was done by analyzing the nucleotides located directly preceding and following each editing and potential editing location. Due to the small number of editing locations, there were not enough data to do motif analysis for wider nucleotide ranges. For two preceding and following nucleotides, only 74 nucleotide combinations out of the possible 256 were found in the sequences.
Editing tree
We define an ‘Editing Tree’ as a tree representing the different editing paths. An editing path includes all the editing events that separate the sequence of the edited Alu from that of the parental DNA. In the editing tree, the root of the tree is the DNA data and every node in the tree represents an editing event. The trees were created using a version of the IgTree© program (13) that was originally developed for creating lineage trees of immunoglobulin genes (14,15). The new version was tailored for creating editing trees. The length of the RNA sequences checked for A-to-I editing was on the order of a few hundred bases. The data used to create the tree were derived from a single patient, so the root (genomic DNA sequence) is the same for all RNA sequences and the number of unique RNA sequences is 31. All the sampled sequences were incorporated into the tree, some as leaves and some as internal nodes of the tree. This corresponds to processes in which parent and descendent sequences can coexist. For A-to-I editing, those nodes are the different RNA sequences extracted from the cells. The parental relationship in the editing tree is such that a node’s sequence contains all the editing events that are present in the parent node sequence plus an additional editing event. This relationship does not necessarily suggest that RNA editing has a sequential nature.
For creating the editing tree, the aligned sequences were used. The requirements from the editing tree were: (i) every node in the tree should be separated from its immediate ancestor by only one editing event; (ii) all editing events in the parent appear in all of its immediate and non-immediate descendents and (iii) the tree will be the minimal tree in terms of number of nodes. The first demand ensures that every editing event is represented in the tree, the second that editing cannot revert to the original sequence in the editing path and the last condition ensures finding a minimal number of editing paths. To create the tree, the distance (in terms of editing events) is calculated for every pair of sequences and the possibility of their having an ancestor–progeny relationship is found. A preliminary tree is created using the derived parent–child relationships that had the minimum parent–child distance. The next stage is to fill the tree with nodes that represent individual editing events until the condition that only one editing event separates every two connected nodes in the tree is filled.
Editing tree analysis can show the relative popularity of different editing paths and the missing stages between the different editing stages. The editing tree can indicate if there is a sequential order to the editing as reflected in tree shape—sequential order would generate relatively few long branches, with more than one identical sequences found in most nodes. A random order of editing will generate a highly branched tree with a small number of copies of every sequence.
RESULTS
Identification of the variety of different transcripts per Alu in a genome-wide editing database
To explore the diversity of transcripts derived from Alu editing, we used a dataset of editing sites that were found using alignment of EST sequences to the genome. Although the data of full-length RNA comparisons are much more reliable and therefore were used as the default for most editing discovery studies (2,4,8,16), ESTs (10) have the advantage of representing a vast dataset with more than 5 million human EST sequences. Thus, even after extensive cleaning procedures (5), designed to remove less reliable sequences that have discrepancies with the reference genome due to technical artifacts or incorrect genome alignment, we found edited Alus which are supported by more than one sequence, allowing us to test the variety of edited transcripts.
We searched the RNA editing database (see ‘Materials and Methods’ section) for editing clusters with at least four editing sites that are all supported by at least four different ESTs. We found 173 such clusters (average 7.4 ESTs per cluster, median of five ESTs) containing 1106 editing sites in 1287 ESTs. In total, we detected 843 unique transcripts in this set, much more (almost 5-fold) than the 173 genomic sequences that exist without editing. When looking at the number of different transcripts per cluster, we found that the number of different transcripts increases with the number of ESTs (Figure 2). This observation supports the scenario of large diversity due to editing. Moreover, it seems that the diversity is much larger than the number of sequences available in the databases, since in most clusters with relatively few ESTs, the number of the different transcripts was close to the maximum possible number of potential transcripts (up to seven ESTs per cluster, total of 124 clusters). In many of the cases where we did find sequences with identical editing patterns, they were derived from the same EST library, raising the possibility that they represent a technical duplication of the same clone rather than true independent events with the same editing pattern.
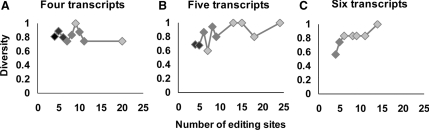
Figure 2.
Transcriptome complexity is increased by A-to-I editing. Using a genome-wide dataset, we found groups with equal numbers of transcripts. The number of different transcripts due to editing, for an Alu with four (A), five (B), or six (C) available transcripts in total, shows that editing increases the diversity in the transcriptome. The figures show the diversity—the ratio between the number of different transcripts and maximum possible number of different transcripts—as a function of the number of editing sites. The colors represent the number of sequences found: full black represents more than 15 sequences and light gray represents one sequence—the other shades are values between 1 and 15. Error bars appear in the figure, but are usually too small relative to the data points to be visible.
When we looked at the way in which the ratio between the number of different transcripts and number of total transcripts changes as a function of the number of editing sites, we found, as expected, that the increase in the number of editing sites increases the diversity in the transcript population (Figure 2). Given that the data are biased toward a small number of transcripts, even a small number of editing sites created large diversity. Typically the number of different transcripts is lower than the total number of transcripts (even when the latter is much smaller than 2N, the total number of possible combinations), reflecting the fact that some combinations are far more likely than others. However, the number of different transcripts does increase with the total number of transcripts, consistent with the possibility that all 2N combinations would be realized given a sufficiently large theoretical number of transcripts. Moreover, as different transcripts do not usually display partial editing patterns of other transcripts in their cluster, they are probably different transcripts and not incompletely edited and processed transcripts captured before the editing process was over. We expect the diversity to increase up to a threshold that will reflect the level of complexity induced by RNA editing. The threshold value will probably depend on the number of editing sites in the Alu. Although the set under investigation represents only a fraction of the known edited Alus, which are probably only a small fraction of the actual edited Alus (2–5), our analysis of a large set of edited Alu sequences suggests that diversity derived from Alu editing is widespread in the human genome and is not limited to a small number of loci.
Editing patterns and diversity detected in the analysis of edited Alu
After observing the diversity derived from editing on a genomic scale, we chose to focus on a single edited Alu repeat for which a large sequence dataset of clones is available, and the library of clones represents transcripts that actually coexist in the same tissue. We used data of an Alu sequence located in intron 9 of the MED13 (THRAP1) gene (see ‘Materials and Methods’ section and (12) for more information including genomic location). The signature of editing is an A residue in the genome and G in some of the cDNA molecules at the same location. Thus, in order to determine the location of editing sites, multiple alignments of all the available 69 cDNA clones from a normal brain were performed against the corresponding genomic sequence.
The genomic sequence used in this study has 41 genomic encoded As and most of them, 31, were found to be edited, while only 10 As have no evidence of editing in any of the clones. In total, counting all editing events in all 69 clones, 407 editing events were found. The complete sequence of this Alu length is 296 nucleotides long, while here we analyzed only the 145 nucleotides for which sequence information was available for all clones (Figure 1). Thus the total number of editing sites in this Alu sequence is probably higher. For comparison, only two editing sites were detected in the same sequence during a previous large-scale genomic screening for edited Alu (5). Those sites were predicted based on the availability of only one RNA molecule that aligns to this locus (AK123839—five editing events in total for this RNA; the other three are located in adjacent Alus). This demonstrates how the genomic screening methods used in previous studies are far from sufficient to describe the full editing picture of Alu sequences and supports the finding of high editing diversity (described in the previous section) based on the whole database.
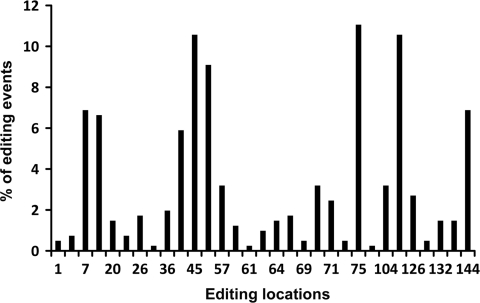
Editing rates per location vary widely (Figure 3) with the average number of editing events (that is, the editing level) per editing location being 13.1 with a range of one to 45 editing events (out of 69 clones; hence the average editing rate was 19%). About 2/3 of all editing events are found in eight ‘hot spot’ locations. For every hot spot, at least 34% of the clones are edited. Indeed, in former studies on global editing in Alu sequences (2–5) it was found that the Alu consensus has preferred editing positions. To achieve maximum diversity the editing probability for a site should be 50%, hence, hot spots, which has the editing probability closest to this value, are the main contributors to the diversity. In addition, editing events outside the hot spots are more common in clones with a large number of editing events, while clones with a low number of hot spot editing events show lower editing activity in the less-preferred spots as well. In total, 77% of the editing events in the less-edited clones are at hot spots. Still, non-hot spot also contributes to the overall diversity. It is not likely that the editing at the hot spots occurs at a predetermined order, as there are clones with different editing combinations within the hot spots.
Figure 3.
Editing locations and level. Thirty-one editing sites were found in the clones derived from normal brain. The distribution of the editing is not even and eight locations are ‘hot spots’ for editing, with more than 20 editing events in each. About two-thirds of all editing events are found within these hot spots.
Editing locations
The distribution of editing sites is not random in the Alu sequence that we studied. Editing sites tend to come in clusters, where neighboring editing sites within a cluster are separated by five or fewer nucleotides. In addition, there is a gap of 19 nucleotides without any observed editing locations, although four A residues are located in this gap. A possible explanation for the reduced editing in this region is that when aligned to all the nearby Alu repeats (in order to predict the structure of the dsRNA formed when the Alu is hybridized with the reverse-oriented Alus), the edited Alus contain a deletion of one nucleotide at this gap (see Supplementary Data). This deletion interferes with the formation of the double-stranded structure which is needed for ADAR enzyme activity. Interestingly, the other deletion within the sequence, only 9 bp downstream, seems to have little or no effect on the editing sites nearby, probably due to its larger size (four nucleotides), which can form a bulge that is less rigid than that of the single nucleotide, enabling the surrounding region to maintain binding to their counterpart Alus.
Mismatches do not seem to reduce the level of editing and an A–C mismatch is actually preferred at the most edited sites (see Supplementary Data), in agreement with previous reports (3,5,17). In addition, we observed a higher number of editing locations in the left arm of the Alu repeat.
Editing creates a diverse clone population
Not all the editing sites are actually edited in all clones and distinct editing patterns generate different transcripts. In theory, a locus with N independent editing sites can generate up to 2N different transcripts. Having the set of edited Alu clones allows us to probe the actual number of distinct transcripts and the dependencies, if any, of the editing sites on one another.
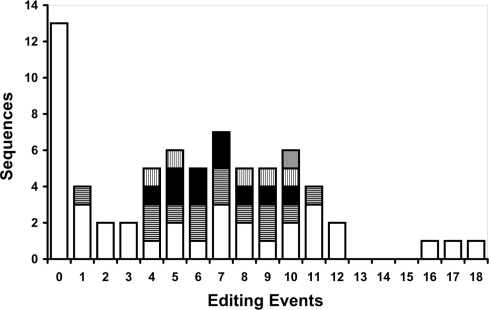
In order to achieve diversity, only a subset of the editing sites should be edited in each transcript, and indeed, we found that the average number of editing events per clone is about one-fourth of the possible editing sites, 7.27 ± 3.76 editing events per clone on average, out of 31 editing sites (a site is defined as an A residue with evidence for editing in at least one clone) in the tested locus. The most highly edited clone has 18 editing sites (Figure 4).
Figure 4.
Different populations of clones have the same number of editing sites. The columns in the graph represent the number of RNA sequences, shown for each number of editing events. There is a large number of unedited sequences—13 clones—that are identical to the DNA root (zero editing events). The different color patterns in some of the other columns represent different clone groups (each clone is identical to the others in the group). It can be seen that, for a given number of editing events, there is usually more than one edited transcript.
Counting the transcripts with defined numbers of editing events, we found 56 out of 69 transcripts that underwent editing, resulting in 30 unique edited transcripts. Thus, a large number of different transcripts indeed arise, but there is also some redundancy. Most of the edited patterns are repeated more than once; the same exact editing pattern can be found in up to four clones in the database. Of the 56 edited clones, 9 are found only once, 18 twice, 1 thrice and only 2 clones appear four times. The large fraction of clones that appear only once suggests that the complete repertoire of edited clones is not fully represented in the current set. However, the unbalanced distribution strongly suggests that most transcripts derived from the same Alu belong to a relatively small group of transcripts and not every possible transcript is created in the transcriptome.
Reconstructing the editing path
RNA editing takes place in double-stranded RNA (dsRNA). In humans, most of the target RNA structures are formed between two inverted Alu repeats. It was observed that nearly all A-to-I substitutions result in altered stability of the dsRNA structure (3,5). Therefore, each editing event alters the potential for editing of the surrounding As. Edited transcripts can be edited again at additional sites due to this dynamic process. Those sites may not have been accessible targets for the ADAR in the initial dsRNA structure. Thus, some of the different clones may represent RNA molecules sampled at different phases of the editing process. Another aspect of this unique dynamic process is that the editing reaction occurs only in the ‘forward’ direction, with no known mechanism to reverse the process and restore the editing site back to A.
Based on this mechanistic knowledge, we can reconstruct parts of the different editing paths. Two clones that have editing events at exactly the same locations, with only one of them having additional editing sites, were considered to be members of the same editing path, with the one that has additional sites belonging to a later stage in the path’s development. We constructed the editing paths (see the ‘Materials and Methods’ section for more detail). This algorithm is optimized toward creating the minimal path, in terms of number of editing steps, and therefore it probably cannot reconstruct the actual editing history; however, it allows us to visualize the editing stages in a clear and intuitive manner.
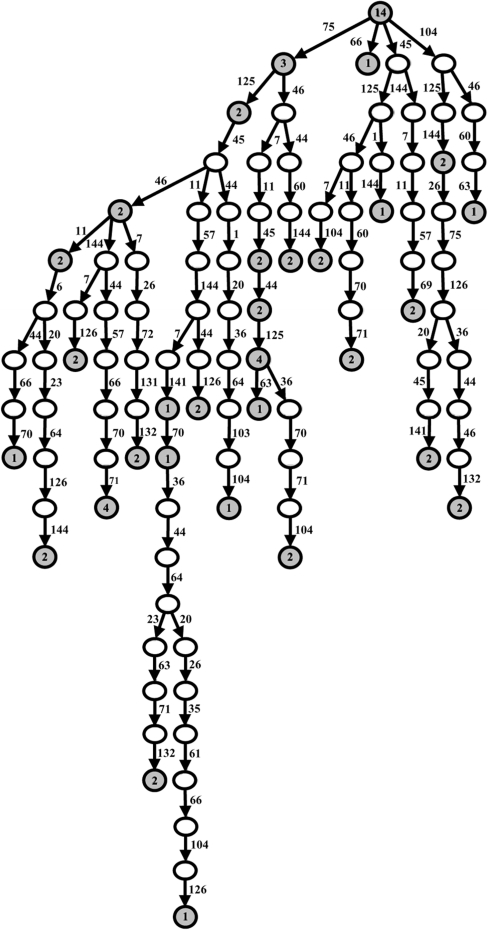
Based on the information in Figure 4, it is clear that there is more than one path for editing in the Alu locus. We can also see that some of the editing patterns occur more frequently than others, and indeed, when constructing the editing tree (Figure 5), some paths have more sequence copies in them. This may indicate that there are not only hot spots of editing and more common editing patterns, but also some sort of ‘hot paths’.
Figure 5.
A-to-I editing tree of a normal control sequence. The editing tree was created from the alignment of the cDNA sequences with the DNA sequence. The root of the tree is the DNA sequence and the gray nodes are the edited RNA sequences. The empty nodes are intermediate stages between the DNA and RNA, that were not detected in the library, but were deduced by the program. The numbers in the grey nodes are the numbers of copies found in the alignment where the root of the tree also includes the number of exact copies of the DNA sequence. The numbers beside the edges represent the location of the editing event separating each node from its parents. Every editing event represented by a node is found in all its direct and indirect descendents.
It is important to note that the editing path does not represent the actual order of the editing reactions, but the different potential combinations of editing events. The more highly utilized an editing location, the nearer it will be to the root of the tree. The accuracy of the tree is reduced for sequences that include many editing events, since they have a greater number of paths available to them.
The clones were derived from normal brain tissue and this repertoire probably resembles the repertoire of edited RNA molecules in a wide range of brain cells and in different ranges of time since their transcription. Thus, it is reasonable to assume that end nodes indeed represent, in most cases, final edited RNA, and not intermediate RNA sequences before the editing reaction was terminated. Moreover, A-to-I RNA editing is believed to occur in the nucleus immediately after transcription, prior to intron removal (18). The clones represent the total population of RNA in the cells; thus, the sample clones are probably enriched in final editing product.
Indeed, based on the tree, we found that the 30 clone groups belong to 20 different subpaths, and most of the clones, 35 out of 69, are in end nodes, while only 21 are in internal nodes. The end nodes most likely represent final editing products as most end nodes are supported by more than one clone. Although the tree model yields only the approximate structure of the actual editing path tree, it is very useful for gaining insight into the editing process. Empty circles in Figure 5 represent editing stages in the path that have no intermediate representative; such empty circles represent sites where the previous editing events dramatically increase the subsequent editing potential, while filled circles represent editing events that appear to negatively influence subsequent editing events. Clustering of such events between node positions occupied by clones suggests that editing occurs in bursts, in which several sites are edited in proximate times. However, not all such groups of sites are physically adjacent in the sequence, which may suggest that several ADARs proteins may simultaneously edit those sites, in agreement with the observation that ADAR acts as a dimer (19). Thus, each ADAR may bind to slightly different regions of the target dsRNA.
Three hot paths were discovered, containing 9, 11 and 13 events. If we interpret the paths as an approximate description of editing stages, then the subsequent path is highly dependent on where the first editing event took place. For example, if the initial editing event takes place at position 75, it opens a cascade of subsequent editing events, but if the first editing occurs at position 66, a complete cessation of the process generally results.
There are sequences that share the same path (the number of copies in a node is more than one), and others that are intermediate stages of sequences that are subsequently subjected to additional editing, but in most cases, there is only a small overlap in the editing paths (Figure 5). Overall it seems that editing in one location does not predict editing at another location and from examining the highly branched A-to-I tree, it is clear that there is no fixed editing path for all the sequences.
The starting point of editing seems to be mainly in the hot spots. Sequences with a small number of editing events (1–3) have, in most cases, at least one editing event in a hot spot. This makes them a probable location for the editing stating point. Sequences with more editing events are more likely to have some events in locations that are not hot spots.
Flanking sequence motifs
A-to-I RNA editing occurs in the context of a duplex structure and ADAR enzymes bind to their targets through their dsRNA-binding domains. However, the mechanism of selection of specific adenosine residues to be edited has not been defined to date. By analyzing several ADAR targets (17,20,21), and recently by exploring the large set of Alu editing (2–5) in human and mouse repeats (8), some sequence preferences were determined.
Elucidation of the editing motif in Alu was based on the detection of edited Alu in a large-scale alignment of cDNA sequences to the genome. However, although the consensus was calculated based on a large number of different RNA molecules, this approach has two main drawbacks. The first is that only the most highly edited sites in Alu are generally represented in the large set of RNA molecules. Moreover, since the RNA was derived from diverse genomic locations of Alu repeats, motif detection could vary based on the background sequence. In the case of the MED13 Alu, we have a comprehensive coverage of the editing sites with detailed information of their editing level coupled with a fixed background. These properties provide a realistic simulation of the actual editing process, as the editing enzyme faces multiple copies of the same RNA derived from the same Alu locus. This allowed us to search for a set of motifs that are related to editing events in this member of the AluSx family.
We found a correlation only between the nucleotides adjacent to editing sites and the editing level (Table 1). An A or T preceding the edited A increases the probability of an editing event versus a C or G at this location. Correspondingly, A and G are more common following the editing site. We could not detect any specific motif that characterizes hot spots. On the other hand, no editing occurs at potential editing locations having G both preceding and following the editing site. If G precedes the edit site and C or T follows it, then the frequency of editing is low. The editing frequency is also low when the preceding nucleotide is C and the following one is not G, in general agreement with the motifs previously detected for large-scale editing in Alu.
Table 1.
Editing locations and potential editing location motifs
| Before | After | Frequency | Events | Percentage of edited |
|---|---|---|---|---|
| A | A | 7 | 73 | 15.11 |
| A | C | 3 | 51 | 24.64 |
| A | G | 3 | 67 | 32.37 |
| A | T | 4 | 27 | 9.78 |
| C | A | 3 | 1 | 0.48 |
| C | C | 1 | 2 | 2.9 |
| C | G | 4 | 40 | 14.49 |
| C | T | 1 | 0 | 0 |
| G | A | 3 | 26 | 12.56 |
| G | C | 1 | 3 | 4.35 |
| G | G | 3 | 0 | 0 |
| G | T | 3 | 3 | 1.45 |
| T | A | 3 | 49 | 23.67 |
| T | C | 3 | 20 | 9.66 |
| T | G | 1 | 45 | 65.22 |
| T | T | 0 | 0 | 0 |
For every appearance of A in the DNA, the preceding and following nucleotides were characterized. We counted frequency of every nucleotide arrangement in the DNA (third column) and the number of editing events, out of the 407 total editing events, that occurred in locations with each arrangement in the entire database (fourth column). The last column shows the percentage of edited motifs out of the total number of appearances of the motif in the alignment. The distribution of the editing events is not even for all nucleotide arrangements.
DISCUSSION
The understanding of the biological implication of Alu editing is in its initial stage, thus detailed characterization is needed. In this study, we provide evidence that the actual number of A-to-I RNA editing sites in the human genome is probably much larger than previously found. Moreover, based on analysis of the dataset of edited Alu loci and a clone set of a single Alu locus we demonstrate that editing generates a large number of different isoforms. Finally, based on data from available clones, we constructed a consistent model that describes the different paths by which editing patterns are generated in a single locus.
The numbers of editing sites in the human genome, reported so far using alignments of cDNA (2,4,16) and ESTs (5) to the genome, probably reflects an underestimate due to limited sampling. Indeed, there is evidence for an elevated level of editing sites in human (3) and rat brain (22). Our results suggest, assuming that the Alu sequence chosen for this study is a typical one, that the number of editing sites in any edited Alu is much higher than previously presupposed. In particular, the numbers of sites in this Alu locus are 20-fold higher, when examined in a set of 69 clones, relative to the number of sites detected earlier. It is possible that higher coverage will detect an even larger set of editing sites where the upper limit is, of course, the number of ‘A’s in the Alu locus. With such a large amount of editing sites, the possible number of different transcripts that can be derived from one edited Alu exponentially increases.
In many ways, RNA editing is reminiscent of single nucleotide polymorphisms (SNPs). In both cases, a nucleotide in the consensus genome is replaced by another in some of the sequences generated from various specimens. Indeed, many of the known RNA editing sites were mistakenly deposited into SNP databases (dbSNP) (23–25). Based on the similarity between those two sources of genomic diversity, one would expect to find that, as in SNPs, clusters of nearby events will form a few highly correlated haplotype blocks (26), thereby dramatically reducing the potential diversity from an exponential number of possible combinations to a much lower number of combinations. Surprisingly, the picture that emerges for Alu editing seems to be very different from that of SNPs. Although some of the transcripts do appear multiple times, we observed 30 distinct sequences derived from only 56 edited transcripts. Many additional low-abundance transcripts will probably be detected with an increased sample size. This result is unexpected, since editing events have a significant impact on dsRNA structure. Editing of one site should have impact on editing in nearby sites; thus one may expect to see a reduced number of different editing events based on the dependencies between editing locations. However, the above analysis suggests that such modifications of editing probabilities are weak. Accordingly, no single ‘chain reaction’ path for the editing events is observed in the editing tree. Possibly, higher-coverage data, employing next-generation sequencing methods (27) could reveal site–site correlations. Further research is needed in order to determine the exact mechanism(s) governing the use of editing to generate multiple transcripts. Such a model should take into consideration that two ADAR isozymes may play a role in Alu editing, each with a somewhat different substrate specificity, as was observed for the mouse Alu counterpart—the B1 and B2 SINEs repeats (28).
The most common mechanism for transcript diversification is alternative splicing, with big majority of the genes in the human genome estimated to have more than one variant (29,30). In an extreme example, up to 38 016 (=12 * 48 * 33 * 2) different isoforms are theoretically feasible in the fly DSCAM gene (31). However, these numbers seem modest compared to the diversity that can be achieved in even one Alu locus through editing. For example, the Alu studied here has at least 31 editing sites, enabling the generation of up to 2 147 483 648 (=231) different isoforms. The real number of transcripts produced is certainly much lower, but when taking into account that thousands of genes have edited Alu loci, and in many cases several edited sites, one may conclude that diversity in the human transcriptome due to editing is much higher than that caused by alternative splicing. It is not known yet if this diversity has biological implications, and indeed, most of the editing events do not lead to production of different proteins, since they are usually located in noncoding parts of a gene. However, some of the Alu repeats do change the proteomic outcome and, in those cases, such as the NARF gene (32), many different proteins could potentially emerge. Additionally, some cases of editing clusters were recently discovered that are located in non-Alu repeats in the coding sequences of proteins (24,33). For example, in the BLCAP protein, editing was shown to change at least three amino acids and can generate proteomic diversity. However, the only RNA with cluster of editing sites in the coding region studied so far is the 5-HT2CR gene, where five edited sites were discovered, leading to up to 24 receptor isotypes, some of them with defined functional diversity (34,35). Diversity of Alu can influence not only the mature protein coding sequences, but can also play a role in the regulation of a gene’s expression by alteration of the signature sequences of miRNA targets. Indeed it was recently demonstrated that the Alu consensus contains such sequence motifs (36). Multiple editing sites within miRNA were detected as well (37,38).
Several studies link RNA editing to brain functionality; editing is much more prevalent in primates (16), takes place to the greatest degree in the brain, mediates diversification of neuronal proteins and is linked to neuronal/behavioral phenotypes (22,24,34,39–44). Together, these findings suggest that Alu editing may be involved, through an as yet an unknown mechanism, with human brain activity (16,45–47).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
EYL was supported by Rothschild fellowship from Yad Hanadiv foundation and by the Machiah Foundation. EE’s research was supported by the Israel Science Foundation, grant number 365/06. GR holds the Djerassi Chair in Oncology at the Sackler Faculty of Medicine, Tel Aviv University and his research was supported by the Kahn Family Foundation and the Flight Attendants Medical Research Institute (FAMRI). RM’s research was supported in parts by the following grants: Israel Science Foundation grants numbers 759/01-1 and 546, an Israel Cancer Research Fund project grant, a Systems Biology prize grant from Teva Pharmaceuticals, Human Frontiers Science Program—a Young Investigator Grant and a Research Grant, and a Swedish Foundation for Strategic Research grant funding the Strategic Research Center for studies on Integrative Recognition in the Immune System (IRIS), Karolinska Institute, Stockholm, Sweden. Funding for open access charge: The Israel Science Foundation.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors are grateful to Ms Hanna Edelman for help in manuscript preparation and graphics. The work was part of MB’s studies toward the PhD degree in The Mina and Everard Goodman Faculty of Life Sciences, Bar Ilan University. NP’s work was done in partial fulfillment of the requirements for a Ph.D. degree from the Sackler Faculty of Medicine, Tel Aviv University.
REFERENCES
- 1.Bass BL. RNA editing by adenosine deaminases that act on RNA. Annu. Rev. Biochem. 2002;71:817–846. doi: 10.1146/annurev.biochem.71.110601.135501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Athanasiadis A, Rich A, Maas S. Widespread A-to-I RNA editing of Alu-containing mRNAs in the human transcriptome. PLoS Biol. 2004;2:e391. doi: 10.1371/journal.pbio.0020391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blow M, Futreal PA, Wooster R, Stratton MR. A survey of RNA editing in human brain. Genome Res. 2004;14:2379–2387. doi: 10.1101/gr.2951204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim DD, Kim TT, Walsh T, Kobayashi Y, Matise TC, Buyske S, Gabriel A. Widespread RNA editing of embedded alu elements in the human transcriptome. Genome Res. 2004;14:1719–1725. doi: 10.1101/gr.2855504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Levanon EY, Eisenberg E, Yelin R, Nemzer S, Hallegger M, Shemesh R, Fligelman ZY, Shoshan A, Pollock SR, Sztybel D, et al. Systematic identification of abundant A-to-I editing sites in the human transcriptome. Nat. Biotechnol. 2004;22:1001–1005. doi: 10.1038/nbt996. [DOI] [PubMed] [Google Scholar]
- 6.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 7.Kawahara Y, Nishikura K. Extensive adenosine-to-inosine editing detected in Alu repeats of antisense RNAs reveals scarcity of sense-antisense duplex formation. FEBS Lett. 2006;580:2301–2305. doi: 10.1016/j.febslet.2006.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Neeman Y, Levanon EY, Jantsch MF, Eisenberg E. RNA editing level in the mouse is determined by the genomic repeat repertoire. RNA. 2006;12:1802–1809. doi: 10.1261/rna.165106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen LL, DeCerbo JN, Carmichael GG. Alu element-mediated gene silencing. EMBO J. 2008;27:1694–1705. doi: 10.1038/emboj.2008.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hillier LD, Lennon G, Becker M, Bonaldo MF, Chiapelli B, Chissoe S, Dietrich N, DuBuque T, Favello A, Gish W, et al. Generation and analysis of 280,000 human expressed sequence tags. Genome Res. 1996;6:807–828. doi: 10.1101/gr.6.9.807. [DOI] [PubMed] [Google Scholar]
- 11.Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, Thompson JD. Multiple sequence alignment with the Clustal series of programs. Nucleic acids Res. 2003;31:3497–3500. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Paz N, Levanon EY, Amariglio N, Heimberger AB, Ram Z, Constantini S, Barbash ZS, Adamsky K, Safran M, Hirschberg A, et al. Altered adenosine-to-inosine RNA editing in human cancer. Genome Res. 2007;17:1586–1595. doi: 10.1101/gr.6493107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barak M, Zuckerman NS, Edelman H, Unger R, Mehr R. IgTree: creating Immunoglobulin variable region gene lineage trees. J. Immunol. Methods. 2008;338:67–74. doi: 10.1016/j.jim.2008.06.006. [DOI] [PubMed] [Google Scholar]
- 14.Manske MK, Zuckerman NS, Timm MM, Maiden S, Edelman H, Shahaf G, Barak M, Dispenzieri A, Gertz MA, Mehr R, et al. Quantitative analysis of clonal bone marrow CD19+ B cells: use of B cell lineage trees to delineate their role in the pathogenesis of light chain amyloidosis. Clin. Immunol. 2006;120:106–120. doi: 10.1016/j.clim.2006.01.008. [DOI] [PubMed] [Google Scholar]
- 15.Steiman-Shimony A, Edelman H, Hutzler A, Barak M, Zuckerman NS, Shahaf G, Dunn-Walters D, Stott DI, Abraham RS, Mehr R. Lineage tree analysis of immunoglobulin variable-region gene mutations in autoimmune diseases: chronic activation, normal selection. Cell Immunol. 2006;244:130–136. doi: 10.1016/j.cellimm.2007.01.009. [DOI] [PubMed] [Google Scholar]
- 16.Eisenberg E, Nemzer S, Kinar Y, Sorek R, Rechavi G, Levanon EY. Is abundant A-to-I RNA editing primate-specific? Trends Genet. 2005;21:77–81. doi: 10.1016/j.tig.2004.12.005. [DOI] [PubMed] [Google Scholar]
- 17.Wong SK, Sato S, Lazinski DW. Substrate recognition by ADAR1 and ADAR2. RNA. 2001;7:846–858. doi: 10.1017/s135583820101007x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Higuchi M, Single FN, Kohler M, Sommer B, Sprengel R, Seeburg PH. RNA editing of AMPA receptor subunit GluR-B: a base-paired intron-exon structure determines position and efficiency. Cell. 1993;75:1361–1370. doi: 10.1016/0092-8674(93)90622-w. [DOI] [PubMed] [Google Scholar]
- 19.Cho DS, Yang W, Lee JT, Shiekhattar R, Murray JM, Nishikura K. Requirement of dimerization for RNA editing activity of adenosine deaminases acting on RNA. J. Biol. Chem. 2003;278:17093–17102. doi: 10.1074/jbc.M213127200. [DOI] [PubMed] [Google Scholar]
- 20.Lehmann KA, Bass BL. Double-stranded RNA adenosine deaminases ADAR1 and ADAR2 have overlapping specificities. Biochemistry. 2000;39:12875–12884. doi: 10.1021/bi001383g. [DOI] [PubMed] [Google Scholar]
- 21.Dawson TR, Sansam CL, Emeson RB. Structure and sequence determinants required for the RNA editing of ADAR2 substrates. J. Biol. Chem. 2004;279:4941–4951. doi: 10.1074/jbc.M310068200. [DOI] [PubMed] [Google Scholar]
- 22.Paul MS, Bass BL. Inosine exists in mRNA at tissue-specific levels and is most abundant in brain mRNA. EMBO J. 1998;17:1120–1127. doi: 10.1093/emboj/17.4.1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eisenberg E, Adamsky K, Cohen L, Amariglio N, Hirshberg A, Rechavi G, Levanon EY. Identification of RNA editing sites in the SNP database. Nucleic Acids Res. 2005;33:4612–4617. doi: 10.1093/nar/gki771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Levanon EY, Hallegger M, Kinar Y, Shemesh R, Djinovic-Carugo K, Rechavi G, Jantsch MF, Eisenberg E. Evolutionarily conserved human targets of adenosine to inosine RNA editing. Nucleic Acids Res. 2005;33:1162–1168. doi: 10.1093/nar/gki239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gommans WM, Tatalias NE, Sie CP, Dupuis D, Vendetti N, Smith L, Kaushal R, Maas S. Screening of human SNP database identifies recoding sites of A-to-I RNA editing. RNA. 2008;14:2074–2085. doi: 10.1261/rna.816908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, Higgins J, DeFelice M, Lochner A, Faggart M, et al. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- 27.Wahlstedt H, Daniel C, Enstero M, Ohman M. Large-scale mRNA sequencing determines global regulation of RNA editing during brain development. Genome Res. 2009;19:978–986. doi: 10.1101/gr.089409.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Riedmann EM, Schopoff S, Hartner JC, Jantsch MF. Specificity of ADAR-mediated RNA editing in newly identified targets. RNA. 2008;14:1110–1118. doi: 10.1261/rna.923308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Johnson JM, Castle J, Garrett-Engele P, Kan Z, Loerch PM, Armour CD, Santos R, Schadt EE, Stoughton R, Shoemaker DD. Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science. 2003;302:2141–2144. doi: 10.1126/science.1090100. [DOI] [PubMed] [Google Scholar]
- 30.Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schmucker D, Clemens JC, Shu H, Worby CA, Xiao J, Muda M, Dixon JE, Zipursky SL. Drosophila Dscam is an axon guidance receptor exhibiting extraordinary molecular diversity. Cell. 2000;101:671–684. doi: 10.1016/s0092-8674(00)80878-8. [DOI] [PubMed] [Google Scholar]
- 32.Lev-Maor G, Sorek R, Levanon EY, Paz N, Eizenberg E, Ast G. RNA-editing-mediated exon evolution. Genome Biol. 2007;8:R29. doi: 10.1186/gb-2007-8-2-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Clutterbuck DR, Leroy A, O'C;onnell MA, Semple CA. A bioinformatic screen for novel A-I RNA editing sites reveals recoding editing in BC10. Bioinformatics. 2005;21:2590–2595. doi: 10.1093/bioinformatics/bti411. [DOI] [PubMed] [Google Scholar]
- 34.Burns CM, Chu H, Rueter SM, Hutchinson LK, Canton H, Sanders-Bush E, Emeson RB. Regulation of serotonin-2C receptor G-protein coupling by RNA editing. Nature. 1997;387:303–308. doi: 10.1038/387303a0. [DOI] [PubMed] [Google Scholar]
- 35.Kawahara Y, Grimberg A, Teegarden S, Mombereau C, Liu S, Bale TL, Blendy JA, Nishikura K. Dysregulated editing of serotonin 2C receptor mRNAs results in energy dissipation and loss of fat mass. J. Neurosci. 2008;28:12834–12844. doi: 10.1523/JNEUROSCI.3896-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smalheiser NR, Torvik VI. Alu elements within human mRNAs are probable microRNA targets. Trends Genet. 2006;22:532–536. doi: 10.1016/j.tig.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 37.Blow MJ, Grocock RJ, van Dongen S, Enright AJ, Dicks E, Futreal PA, Wooster R, Stratton MR. RNA editing of human microRNAs. Genome Biol. 2006;7:R27. doi: 10.1186/gb-2006-7-4-r27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kawahara Y, Zinshteyn B, Sethupathy P, Iizasa H, Hatzigeorgiou AG, Nishikura K. Redirection of silencing targets by adenosine-to-inosine editing of miRNAs. Science. 2007;315:1137–1140. doi: 10.1126/science.1138050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sommer B, Kohler M, Sprengel R, Seeburg PH. RNA editing in brain controls a determinant of ion flow in glutamate-gated channels. Cell. 1991;67:11–19. doi: 10.1016/0092-8674(91)90568-j. [DOI] [PubMed] [Google Scholar]
- 40.Hoopengardner B, Bhalla T, Staber C, Reenan R. Nervous system targets of RNA editing identified by comparative genomics. Science. 2003;301:832–836. doi: 10.1126/science.1086763. [DOI] [PubMed] [Google Scholar]
- 41.Ohlson J, Pedersen JS, Haussler D, Ohman M. Editing modifies the GABA(A) receptor subunit alpha3. RNA. 2007;13:698–703. doi: 10.1261/rna.349107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Palladino MJ, Keegan LP, O'C;onnell MA, Reenan RA. A-to-I pre-mRNA editing in Drosophila is primarily involved in adult nervous system function and integrity. Cell. 2000;102:437–449. doi: 10.1016/s0092-8674(00)00049-0. [DOI] [PubMed] [Google Scholar]
- 43.Singh M, Kesterson RA, Jacobs MM, Joers JM, Gore JC, Emeson RB. Hyperphagia-mediated obesity in transgenic mice misexpressing the RNA-editing enzyme ADAR2. TJ. Biol. Chem. 2007;282:22448–22459. doi: 10.1074/jbc.M700265200. [DOI] [PubMed] [Google Scholar]
- 44.Jepson JE, Reenan RA. RNA editing in regulating gene expression in the brain. Biochim. Biophys. Acta. 2008;1779:459–470. doi: 10.1016/j.bbagrm.2007.11.009. [DOI] [PubMed] [Google Scholar]
- 45.Mehler MF, Mattick JS. Non-coding RNAs in the nervous system. J. Physiol. 2006;575:333–341. doi: 10.1113/jphysiol.2006.113191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Levanon K, Eisenberg E, Rechavi G, Levanon EY. Letter from the editor: Adenosine-to-inosine RNA editing in Alu repeats in the human genome. EMBO Rep. 2005;6:831–835. doi: 10.1038/sj.embor.7400507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mattick JS, Mehler MF. RNA editing, DNA recoding and the evolution of human cognition. Trends Neurosci. 2008;31:227–233. doi: 10.1016/j.tins.2008.02.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.