Abstract
It is known that siRNAs are capable of reducing expression of non-target genes due to the interaction of the siRNA guide strand with a partially complementary site on the ‘off-target’ mRNA. In the current study, we show that reduction of cellular Ago2 levels has no effect on off-target reduction of endogenous genes and that off-target degradation of mRNA can occur even in an Ago2 knockout cell line. Using antisense mediated reduction of Ago proteins and chemically modified cleavage- and binding-deficient siRNAs, we demonstrate that siRNA mediated off-target reduction is Ago2 cleavage independent, but does require siRNA interaction with either Ago1 or Ago2 and the RISC-loading complex. We also show that depletion of P-body associated proteins results in a reduction of off-target siRNA-mediated degradation of mRNA. Finally, we present data suggesting that a significant portion of on-target siRNA activity is also Ago2 cleavage independent, however, this activity does not appear to be P-body associated.
INTRODUCTION
RNA interference (RNAi) is a mechanism by which double-stranded RNA triggers the inhibition of target gene expression by inducing sequence-specific target mRNA degradation (1). Short interfering RNAs (siRNAs), exogenously administered RNA duplexes of 21–23 nucleotides, lead to degradation of specific mRNAs through the RNA-induced silencing complex (RISC) (2). After cellular delivery of synthetic siRNAs, the double stranded molecules are incorporated into the RISC-loading complex (RLC), which consists of Ago2, Dicer and the HIV trans-activating response RNA-binding protein (TRBP) (3). Prior to target mRNA recognition, an siRNA duplex goes through an ATP-dependent unwinding process and one of the two siRNA strands, referred to as the passenger strand, is released, while the other strand (guide strand) remains bound to the Argonaut protein, Ago2 (4,5). This strand then facilitates interaction of RISC with its complementary target RNA, which is finally cleaved at the site opposite the 10th and 11th positions of the siRNA guide strand by the RNase activity located in the Ago2 protein, triggering its destruction (6–11).
It has been reported that siRNAs can effectively reduce expression of mRNAs even when there are a few mismatches between the siRNA and the mRNA target sequence (12,13). Microarray analyses have shown that siRNAs with partial complementarity to an mRNA can cause a reduction in the levels of non-targeted messages (14–16). Additional studies demonstrated that six to seven consecutive matches between the 5′-end of siRNA guide strand and the ‘off-target’ RNA as well as the context of the matching sequence within the off-target RNA are important parameters that determine if an off-target mRNA is degraded (17,18).
Like siRNA mediated off-target RNA degradation, it has been reported that miRNAs, naturally produced small RNAs that work through RISC, can reduce the levels of partially complementary target genes (19–21). Consistent with the off-target activity observed with siRNAs, miRNA-mediated RNA degradation requires a homologous ‘seed region’ 2–7 nucleotides from the 5′-end of the miRNA (22–24). In addition, it has been reported that Drosophila embryos lacking Ago2, which are not capable of siRNA-directed cleavage, are still capable of miRNA-directed target RNA reduction (8) through deadenylation and subsequent degradation of target transcripts in a process which likely involves P-bodies (25–27). Recent studies employing reporter genes suggest that siRNA-mediated off-target mRNA reduction is also the result of Ago2-independent degradation processes (28,29); however, the mechanism of off-target mRNA degradation has not been fully elucidated.
The similarities between siRNA-mediated off-target and miRNA-mediated mRNA reduction suggest that they may share common mechanisms. However, since almost all siRNAs contain chemical modifications (30,31) it cannot simply be assumed that they have the same properties as endogenously produced miRNAs. In addition, the transfection process required for siRNA activity may also influence sub-cellular localization and activity of siRNA relative to miRNA (32). In the current study, we present data from experiments utilizing Ago2-reduced and Ago2 knockout cell lines, which confirm that siRNA mediated off-target degradation of endogenous mRNAs can occur independently of Ago2. In addition, our data also suggest that interaction of the siRNA with the RLC is required for off-target activity, as Dicer cleavage of long dsRNAs is required for activity. We also demonstrate that Ago binding, but not cleavage is necessary for siRNA-mediated off-target degradation of RNA and show that Ago1 is capable of mediating off-target mRNA downregulation. Reductions in the levels of siRNA and miRNA related proteins suggest that siRNA off-target degradation and miRNA-mediated target degradation share common pathways; namely degradation of the mRNA is associated with cellular P-bodies. Finally, our data strongly suggest that at least some siRNA on-target activity is also Ago2 independent, however, this activity appears to occur independently of the P-body proteins that we have evaluated.
MATERIALS AND METHODS
Preparation of antisense oligonucleotides and siRNAs
Synthesis and purification of phosphorothioate/2′-MOE oligonucleotides was performed using an Applied Biosystems 380B automated DNA synthesizer as described previously (33). All RNaseH-dependent ASOs used for target mRNA reduction were 18–20 bases in length, full phosphorothioate with 2′-O-methoxyethyl (2′MOE) substitutions at the positions indicated by bold type. The sequences of the ASOs are as follows: PIK3CB (NM_006219, RefSeq ID: PIK3CB): TAGGATTCATATTAGGAG (ISIS 405249); Ago2 (NM_012154, RefSeq ID: EIF2C2): CTGCTGGAATGTTTCCACTT (ISIS 136764); Dicer (NM_030621, RefSeq ID: DICER1): GCTGACCTTTTTGCTTCTCA (ISIS 138648); FEN1 (NM_004111.4, RefSeq ID:FEN1): TGGCTGGTGGTCTCACCCTC (ISIS 366372); mouse Ago1 (NM_153403, RefSeq ID: Eif2c1): CTGGACTATTTGCATAGCAA (ISIS 395392); mouse Ago2 (NM_153178, RefSeq ID: EIF2C2): CGTCTCATGTTCGATGCTGG (ISIS 395507); mouse DDX6 (NM_007841, RefSeq ID: Ddx6): CTCGGGTAAACAGATCAGTG (ISIS 339800); mouse DCP1A (NM_133761, RefSeq ID: Dcp1a): CACCATGTTGCTCAGGGAGG (ISIS 368473); mouse DCP2 (NM_027490, RefSeq ID: Dcp2): CTGCATGTAGAAATCCAAGT (ISIS 365128); mouse GW182 (NM_144925, RefSeq ID: Tnrc6a): TCCTCAGTAGATTTCCATCC (ISIS 443335). Silencer Pre-designed siRNAs for knockdown of RISC and P-Body proteins were obtained from Ambion (Austin, TX). Ago2: siRNA ID# 133832; GW182: siRNA ID# 135276; Lsm-1 siRNA ID# s84653. PIK3CB (15), YY1 and mPIK3CB synthetic unmodified siRNAs were purchased from Dharmacon Research, Inc (Boulder, CO). Sequences of the siRNA sense strands are as follows:
PIK3CB: 5′-UCUCCUAAUAUGAAUCCUAU-3′
YY1: 5′-AUUUUAAAAAUGAAUCCUAC-3′
mPIK3CB: 5′-CUCCCAAUUUGAAUCCUAU-3′
MOE modified PIK3CB siRNAs were synthesized as previously described (34). The positions of the MOE substituted nucleotides on the antisense strand are underlined: AUAGGAUUCAUAUUAGGAGU (siPIK3CB 12–14) and AUAGGAUUCAUAUUAGGAGU (siPIK3CB 1–3). The siRNA sense strand is unmodified RNA. The sequence of the PIK3CB 40-mer ‘dicer-substrate’ siRNA is CUCCUAAUAUGAAUCCUAUCAAAGUAAAUGAAUUGGCAAU (sense) and AUUGCCAAUUCAUUUACUUUGAUAGGAUUCAUAUUAGGAGAC (antisense). The sequence of the 19-mer is highlighted within the 40-mer. A two base overhang was added to the antisense strand to ensure directionality of Dicer cleavage.
siRNA/ASO treatment
Tissue culture medium, trypsin, Lipofectamine RNAiMAX and Lipofectamine 2000 were purchased from Invitrogen (Carlsbad, CA). HeLa cells were obtained from the American Type Tissue Collection (Manassas, VA). Wild-type and Ago2 knockout mouse embryonic fibroblasts (MEF) were a kind gift of Dr Gregory J. Hannon. Cells were cultured in DMEM supplemented with 10% fetal calf serum, streptomycin (0.1 µg/ml), and penicillin (100 U/ml). For the knockdown of RISC or P-Body associated genes, cells were treated at 30–100 nM with the indicated ASO/siRNA in Opti-MEM media (Invitrogen) containing 4 µg/ml Lipofectamine 2000 for ASOs or 6 µg/ml Lipofectamine RNAiMAX for 4 h at 37°C, as described previously (35). Cells were split 24–72 h after the start of ASO treatment and seeded in 96 -ell plates at 4500 cells/well. Cells were allowed to adhere to the plates for 4 h then treated with the siRNA or ASO targeting PIK3CB. For the generation of IC50 curves, cells treated as above at concentrations ranging from 0.1 to 300 nM in half-log serial dilutions (N = 3–4/dose). The next day, total RNA was purified from 96-well plates using an RNeasy 3000 BioRobot (Qiagen, Valencia, CA) and PIK3CB mRNA levels assessed by quantitative reverse transcriptase polymerase chain reaction (qRT–PCR).
Reduction of target mRNA expression was determined by qRT–PCR performed essentially as described elsewhere (36). Briefly, 200 ng of total RNA was analyzed in a final volume of 50 µl containing 200 nM gene-specific PCR primers, 0.2 mM of each dNTP, 75 nM fluorescently labeled oligonucleotide probe, 5 µl RT–PCR buffer, 5 mM MgCl2, 2 U of Platinum Taq DNA Polymerase (Invitrogen) and 8 U of RNase inhibitor. Reverse transcription was performed for 30 min at 48°C followed by PCR: 40 thermal cycles of 30 s at 94°C and 1 min at 60°C using an ABI Prism 7700 Sequence Detector (Applied Biosystems). To avoid artifacts based upon well-to-well variation in cell number, mRNA levels were normalized to the total amount of RNA present in each reaction as determined by Ribogreen assay (37) (Invitrogen). IC50 curves and P-values were generated using Prism 4 software (GraphPad). Sigmoidal dose response was calculated according to Y = Bottom + (Top–Bottom)/{1 + 10 [(logEC50 − X)]}; where X is the logarithm of concentration and Y is the response. The sequence for the human PIK3CB primer/probe set used in the RT–PCR reaction is GGGAAGGAGGCCATGCAT for the forward primer, CAACATAGAGTTCTGAGAGGATAACACA for the reverse primer and TGACCTGCAGTCACCCCTGAACCC for the probe. The sequence for the human YY1 primer/probe set GCGAATCCATACCGGAGACA for the forward primer, AGGTTAGTTGACTGAGCAAACTTCTTATT for the reverse primer and CCCTATGTGTGCCCCTTCGATGGTT for the probe. The sequence for the human FADD primer/probe set GTCATGGAACTCAGACGCATCT for the forward primer, TCCACCAGCGCAAAGCA for the reverse primer and CCTCCGAAGCGTCCTGATGGGC for the probe. The sequence for the mouse YY1 primer/probe set is ACAACCAGTGAAAAGAAGAGAGAAGAC for the forward primer, AGAGGCATATTTATTCCCAATCACA for the reverse primer and TTCTCGACCCGGGAAGCCTCTTCA for the probe.
Primer probe sets used to analyze the levels of targeted RISC/P-body genes are as follows. Human Ago2: CCAGCTACACTCAGACCAACAGA (forward primer), GAAAACGGAGAATCTAATAAAATCAATGAC (reverse primer) and CGTGACAGCCAGCATCGAACATGAGA (probe). Human Ago1: GAGCCTATGTTCCGGCATCTC (forward primer), AGAGTGTATCTCCGACACGTTTCAC (reverse primer) and AGCATACACCGGCGTCTTCCCTGG (probe). Human Dicer: ATTAACCTTTTGGTGTTTGATGAGTGT (forward primer), GCGAGGACATGATGGACAAATT (reverse primer) and ATCTTGCAATCCTAGACCACCCCTATCGAGAA (probe). Human Lsm1: TGTGCATTGCAGCATTATTTCA (forward primer), CTTTTTGTCAATGTCCTCGATGAG (reverse primer) and CAAAATGAACTATATGCCTGGCACCGCC (probe). Human GW182: CAAACAACGGTGGCCTGAAT (forward primer), AATCGCTGTAACTGGGAGATCTG (reverse primer) and CCTCAACAGGTAGCCATGCTGAACCAG (probe). Mouse Ago1: AGTGATGCCAACACTCCATCTG (forward primer), AGGGTGATGGAACACGAAGTAGA (reverse primer) and CCTGTCCATCGCCTTCATAGACA (probe). Mouse Ago2: CCATCTAGCTGTGAAGGCTCTGA (forward primer), TTCTTAGGGCCAGGCTTTAAAA (reverse primer) and CATGAAAGCCACACCATTTCCATTGGG (probe). Mouse DDX6: GCAAAAGCGGTGCCTACCT (forward primer), CACCATTGCTTGAATATTGTCCTT (reverse primer) and TCCCCTTACTTGAAAGGCTGGACCTGA (probe). Mouse GW182: GACAATGTCATGCCCCACACT (forward primer), CAAGCCTATAGGAAAGTTGCTGAAA (reverse primer) and CACCCGAGCTGCAGAAAGGGCC (probe).
Recombinant human Ago2 activity assay
Cloning and purification of human recombinant Ago2 is detailed in Supplementary Data. For human GST-Ago2 cleavage activity, 1 ng recombinant Ago2 was incubated at 37°C with 10 nM of either PIK3CB siRNA guide strand in cleavage buffer (10 mM Tris, pH 7.5, 100 mM KCl, 2 mM MgCl2, Protease Inhibitor and 0.5 mM DTT). After 2 h, 0.1 nM 32P labeled 40-mer target RNA was added. The sequence of the PIK3CB target was CUUUCAAGUGUCUCCUAAUAUGAAUCCUAUCAAAGUAAAU (siRNA compliment in bold) and the sequence of the YY1 target was UCUAGGAAGAAUUUUAAAAAUGAAUCCUACACACCUAAGG (seed region underlined). After 30 min cleavage reactions were quenched in gel loading buffer (Ambion, TX). Cleavage products were resolved by denaturing PAGE and quantitated with Storm 850 Phosphorimager (Molecular Dynamics).
Human Ago2-binding assay
The binding affinity of siRNA antisense strands to human GST-Ago2 was determined by homologous saturation using recombinant Ago2. 32P-labeled oligoribonucleotide was incubated with 1 ng GST-Ago2 enzyme in the presence of increasing concentrations of unlabeled PIK3CB, PIK3CB-M1-3 or PIK3CB-M12-14 siRNA guide strand ranging in concentration from 100 pM to 10 µM. The enzyme was added to 50 µl of Miltenyi Biotec µMACS Anti-GST Microbeads (Auburn, CA) and allowed to bind for 2 h. The beads were then loaded onto paramagnetic columns supplied by Miltenyi and unbound 32P-labeled oligoribonucleotide was washed away with 1 ml of cleavage buffer. The remaining, i.e. bound, radio-activity was counted in a scintillation counter and the bound counts were plotted as a function of the concentration of the unlabeled oligoribonucleotide. The dissociation constants (Kd) were calculated from non-linear least-squares fit of the data to the equation for the one-site binding hyperbola (Y = Bmax* X/Kd + X; where Bmax is maximum binding and Kd is the concentration of unlabeled oligoribonucleotide at half-maximal binding). The errors reported for the binding affinities are based on three trials.
RLM-RACE mapping of Ago2 cleavage sites
HeLa cells were transfected with 100 nM PIK3CB or YY1 siRNA in Opti-MEM media containing 6 µg/ml Lipofectamine RNAiMAX for 16 h at 37°C, as described above. Total RNA was harvested 16 h after transfection using an RNAeasy kit (Qiagen). One microgram of total RNA was ligated to FirstChoice 5′ RACE Adapter RNA oligo (Applied Biosystems, Foster City, CA). The ligated products were reverse transcribed using random decamers and MMLV-RT according to the manufacturer’s guidelines. The resulting cDNAs were amplified by successive rounds of PCR with nested pairs of primers. The first round of PCR was performed for 30 cycles using the 5′ RACE adaptor outer FP and either the PIK3CB outer RP 5′-CCAGCCCTGACATGAACTTT-3′ or YY1 outer RP 5′-CAACCAAAATTAAGATAGCAAACAAA-3′. The second round of PCR was performed for 40 cycles using the 5′ RACE adaptor inner FP and either the PIK3CB inner RP 5′-TTTGGTGGTAATGGAAGAGGA-3′ or YY1 inner RP 5′-AACCTCCAGAAAAATCACACG-3′. RT–PCR production were run on a 2% agaraose gel in 1× TBE buffer and visualized by ethidium bromide staining. PCR products were excised from the gel, purified, then cloned into the pCR2.1-TOPO vector (Invitrogen) according to the manufacturer’s protocol. Clones were sequenced to confirm putative siRNA cleavage sites.
Gene expression array analysis
Wild-type and Ago2−/− knockout MEF cells in 6-well plates were treated PIK3CB siRNA at a concentration of 10nM using ml Lipofectamine RNAiMAX as detailed above. The following day total RNA was purified from mock-transfected and siRNA treated cells using RNeasy mini columns according to the manufacturer’s protocol (Qiagen). Microarray analysis using Affymetrix Mouse Genome 430 2.0 arrays was performed by Expression Analysis (Durham, NC). Microarray data were analyzed using Array Studio (Omicsoft, Durham, NC). Log2 expression ratios were derived by subtracting log2 hybridization intensities to each probe in samples from mock-transfected cells from log2 hybridization intensities in samples from transfected cells. Log2 expression ratios were sorted and sets of associated 3′UTR regions were analyzed for over or under-represented hexamer word strings using Sylamer (38) (http://www.sanger.ac.uk/Teams/Team101/sylamer/). Log2 expression ratios were sorted by degree of down-regulation and binned into sets of 400–500 probes. The frequency of transcripts containing each hexamer motif in 3′UTRs from each bin was compared with the frequency of that motif in the background set of 3′UTRs representing the entire microarray. The significance of enrichment of each hexamer motif was calculated using a hypergeometric P-value.
RESULTS
Ago2 is not required for PIK3CB off-target and some on-target siRNA activity
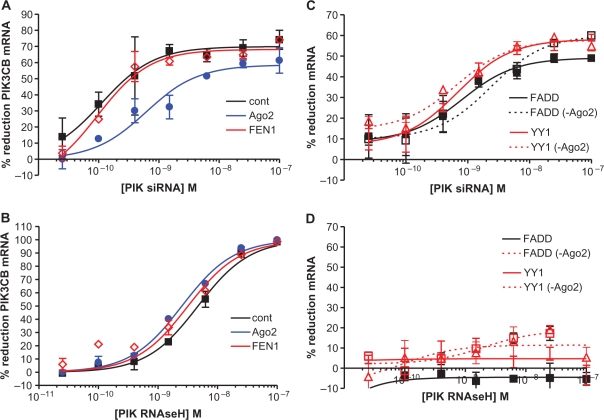
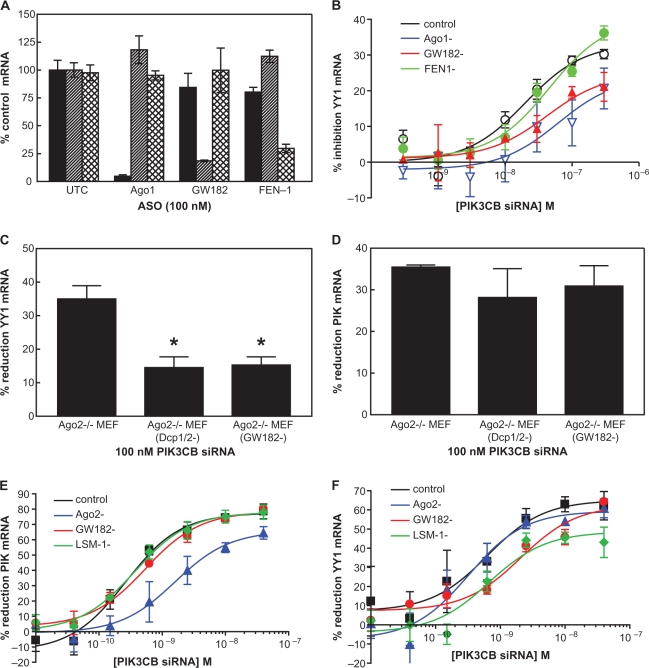
It has recently been demonstrated that siRNA-dependent off-target mRNA reduction of stably and transiently expressed reporter genes is attributable to Ago2-independent degradation processes (28,29). To better understand the role of Ago2 in siRNA-mediated off-target degradation of endogenous mRNAs, we used a previously described siRNA targeting human PIK3CB which was demonstrated to reduce mRNA for two non-targeted transcripts, FADD and YY1, putatively via sequence complementarity to the seed region of the PIK3CB siRNA (15). PIK3CB siRNA or an RNaseH-dependent chimeric phosphorothioate/2′-MOE antisense oligonucleotide (ASO) of the same sequence (ISIS 405249) were administered to control HeLa cells and cells that had been treated for 24 h with ASOs targeting either Ago2 or FEN-1. Depletion of Ago2 resulted in an ∼6-fold reduction in the potency of the siRNA for PIK3CB and had no effect on the potency of the RNaseH ASO (Figure 1A and B, blue lines). As expected reduction of the nuclease FEN-1, had no effect on the target-specific activity of either the siRNA or ASO (red lines). Although reduction of Ago2 was ∼80% under our transfection conditions (Supplementary Figure S1), siRNA target-specific reduction of PIK3CB was still quite potent (IC50: 60 nM), suggesting that even for siRNA on-target effects, the ‘slicer’ activity of Ago2 may not be the only mechanism involved in reducing levels of the targeted RNA. Off-target reduction of FADD and YY1 mRNA were also evaluated. While neither FADD (black lines) nor YY1 (red lines) mRNA was reduced by the RNaseH ASO (Figure 1D), both FADD and YY1 were reduced in a concentration-dependent manner by PIK3CB siRNA (Figure 1C). In contrast to the on-target reduction of PIK3CB mRNA, no change in potency was observed with Ago2 reduction for the off-target transcripts (dotted lines).
Figure 1.
Ago2 reduction attenuates target-specific, but not off-target PIK3CB activity. ASO-mediated reduction of Ago2 or FEN1 was performed for 24 h prior to PIK3CB siRNA/ASO treatment. Cells were then split and seeded in 96-well plates. Cells were allowed to adhere to the plates for 4 h then treated with siRNA targeting PIK3CB or an RNAseH-dependent chimeric phosphorothioate/2′-MOE oligonucleotide of the same sequence (ISIS 405249). Cells were harvested the following day, total RNA purified and PIK3CB mRNA levels assessed by qRT–PCR (A) Percent reduction of PIK3CB mRNA by PIK3CB siRNA in control (black), Ago2-reduced (blue) and FEN1-reduced (red) HeLa cells. (B) Percent reduction of PIK3CB mRNA by PIK3CB RNAseH ASO. (C) Percent reduction of FADD (black) or YY1 (red) off-target mRNA by PIK3CB siRNA in control (solid lines) and Ago2-reduced (dashed lines) cells. (D) Percent reduction of off-target mRNA by PIK3CB RNAseH ASO in control and Ago2-reduced cells.
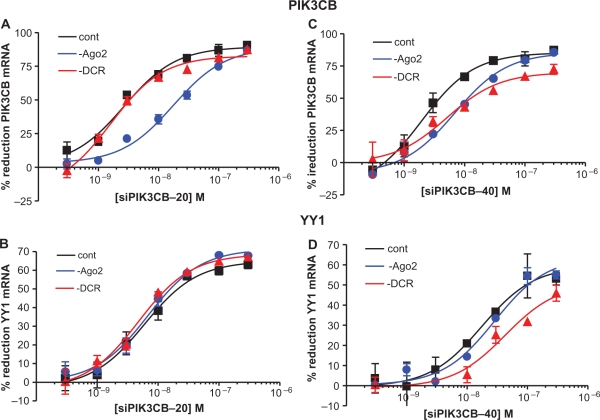
On- and Off-target activities require ‘dicing’ of a 40-mer RNA duplex
Since our results indicated that siRNA off-target activity may occur independently of Ago2, we wondered whether off-target activity may also occur independently of other RISC associated proteins. It has previously been shown that RNA duplexes 27 base pairs and greater in length require the presence of Dicer for cleavage into 21–22 base duplexes capable of activating RISC (39). To investigate the effect of Dicer reduction on both on- and off-target activity, the potency of the 19-mer PIK3CB siRNA was compared with a 40-mer RNA duplex targeting the same site. HeLa cells were first treated with 50 nM ASO targeting either Ago2 or Dicer. Twenty-four hours later, the cells were seeded in 96-well plates then transfected with 19- or 40-mer PIK3CB siRNAs. On- and off-target IC50 values for the PIK3CB siRNA and Dicer substrate were assessed the following day. The reductions of Ago2 and Dicer were also evaluated by qRT–PCR and found to be >80% for both mRNAs. As previously observed, Ago2 reduction (Figure 2A, blue lines) resulted in decreased on-target potency of the 19-mer siRNA, however, reduction of Dicer (red lines) had no effect. Off-target potency of the 19-mer siRNA, as measured by reduction of the YY1 transcript, was not affected by either Dicer or Ago2 reduction (Figure 2B). For the 40-mer siRNA, both Ago2 and Dicer appear to be required for on-target activity, as reduction of either RISC component resulted in a similar attenuation of activity (Figure 2C). In contrast, off-target siRNA activity of the 40-mer was diminished only by Dicer reduction (Figure 2D), while reduction of Ago2 had no effect on potency. Again, these data suggest that off-target, as well as a portion of on-target activity; of either a 19- or 40-mer siRNA may occur independently of Ago2 cleavage. However, the requirement of ‘dicing’ of the 40-mer siRNA suggests that RISC loading is necessary for both on- and off-target activities.
Figure 2.
Off-target activity requires ‘dicing’, but not Ago2 cleavage. The effect of Dicer reduction on the potency of the 20-mer PIK3CB siRNA was compared with a 40-mer RNA duplex targeting the same site. HeLa cells were treated with 50 nM ASO targeting either Ago2 or Dicer. Twenty-four hours later, the cells were seeded in 96-well plates and transfected with PIK3CB siRNA at concentration ranging from 330 pM to 300 nM. The following day on-target and off-target IC50’s for PIK3CB siRNA were assessed for the 19- or 40-mer PIK3CB siRNAs by qRT–PCR. (A) IC50 curves for the reduction of PIK3CB with 20-mer siRNA in control (black), Ago2-reduced (blue) and Dicer-reduced (red) HeLa cells. (B) IC50 curves for the reduction of YY1 off-target mRNA in cells treated with 20-mer PIK3CB siRNA. (C) 40-mer siRNA on-target PIK3CB reduction. (D) Off-target reduction of YY1 mRNA by PIK3CB 40-mer siRNA.
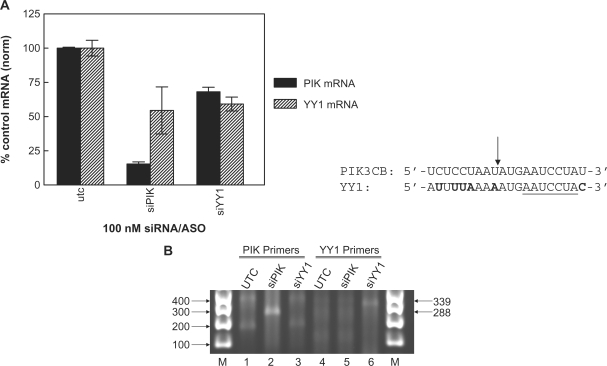
RACE product is detected only for target-specific, Ago2-dependent cleavage
We used a modified RLM-RACE protocol to further confirm that off-target mRNA degradation is not associated with cognate Ago2 cleavage. HeLa cells were transfected with 30 nM of either PIK3CB siRNA or a siRNA designed with perfect complementarity to the YY1 off-target site. Eighteen hours later total RNA was isolated. Analysis of on- and off-target mRNA reduction by qRT–PCR confirmed that both PIK3CB and YY1 were reduced by treatment with the PIK3CB siRNA, while only YY1 was reduced by the YY1 specific siRNA (Figure 3A). Mapping of degradation products was performed using rapid amplification of cDNA ends (5′ RACE) on the mRNA (28). Canonical siRNA-mediated cleavage by Ago2 at position 10 results in a 3′-cleavage product that leaves a 5′ phosphate which is ligation competent and can therefore act as a substrate for 5′ RACE. The RACE adapter ligated products were reverse transcribed, then amplified using nested PCR using primers complementary to the RACE RNA adapter sequence and the PIK3CB or YY1 sequences 3′ of the purported Ago2 cleavage site. A PCR product of ∼288 bp, consistent with the size of the expected amplified PIK3CB cleavage product and the location of the primers used during nested PCR, was observed with RNA from cells transfected with the PIK3CB siRNA (Figure 3B, lane 2). Sequencing of the cloned PCR product established cleavage at the expected base, confirming that at least some of the initial cleavage of the targeted mRNA by the siRNA was a direct consequence of Ago2/RISC activity. Likewise, PCR amplification with YY1 specific primers resulted in a detectable PCR product of the expected size of ∼339 bp for cells treated YY1 siRNA (lane 6). The expected YY1 cleavage site was also confirmed by sequencing of the PCR product. However, despite approximately equal reduction of the YY1 mRNA with the PIK3CB siRNA than with the YY1 siRNA, no cleavage product was observed for the YY1 off-target mRNA (lane 5), again suggesting that off-target degradation of an endogenous gene can occur independently of Ago2 cleavage, and that these observations cannot be explained by the inability to amplify low abundance degradation products.
Figure 3.
Ago2-dependent cleavage product is not detected for off-target siRNA. A modified RLM-RACE protocol was used to determine if off-target mRNA degradation was Ago2 slicer/cleavage independent. HeLa cells were transfected with 30 nM of either PIK3CB siRNA or an siRNA designed with perfect complementarity to the YY1 off-target site. Eighteen hours later total RNA was isolated. (A) Percent reduction of PIK3CB or YY1 mRNA was evaluated by qRT–PCR with gene specific primers. Solid bars, percent untreated control PIK3CB mRNA; Striped bars, percent untreated control YY1 mRNA. The sequences of the PIK3CB on-target and the YY1 off-target mRNA transcripts are shown. Bold text indicates mismatched nucleotides. Nucleotides complementary to the PIK3CB siRNA seed region are underlined and the siRNA cleavage site is indicated by an arrow. (B) One microgram of total RNA was ligated to an RNA oligo. The ligated products were reverse transcribed, then subject to nested PCR using primers specific for either PIK3CB or YY1. PCR products were visualized by electrophoresis on a 2% agarose gel and ethidium bromide staining. The expected size of the PIK3CB (288) and YY1 (339) PCR products are shown.
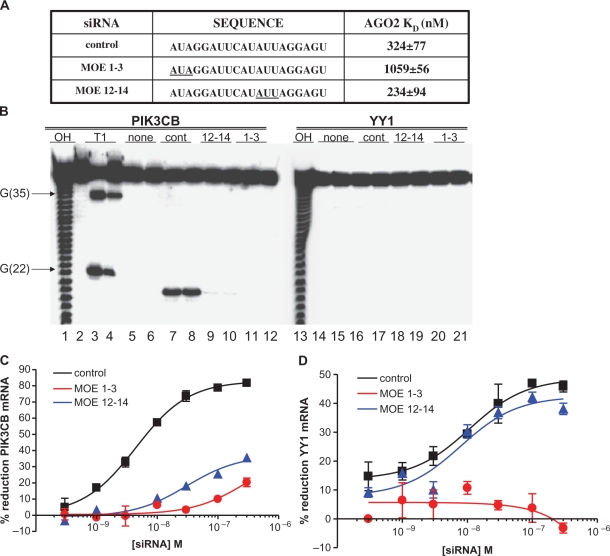
Cleavage-deficient siRNAs are able to promote off-target and a fraction of target-specific siRNA degradation
It has previously been demonstrated that substitution of unmodified ribonucleotides with 2′MOE modifications at positions 1–3 or 12–14 of the siRNA guide strand results in a reduction in target specific siRNA activity (34). We synthesized PIK3CB siRNAs with three 2′MOE modified nucleotides at either the 5′-end (PIK3CB-M1-3) or near the center (PIK3CB-M12-14) of the guide strand in an attempt to understand the role of Ago2 binding verses cleavage on both target-specific and off-target siRNA activity. As it has been shown that the Ago2 PIWI domain is involved in 5′-end recognition of the siRNA guide strand (10,40), we expected that the increased size of the 2′MOE modification relative to unmodified RNA would affect binding when placed at positions 1–3, but not 12–14. Binding affinities of each guide strand for recombinant Ago2 were determined by homologous saturation using recombinant Ago2 as detailed in ‘Materials and Methods’ section. While the unmodified guide strand and the guide strand with MOE substitutions at positions 12–14 were found to have similar affinity for Ago2, the binding affinity of the guide strand with MOE substitutions at positions 1–3 was ∼4-fold lower (Figure 4A).
Figure 4.
MOE modification at positions 12–14 of the siRNA attenuates some target-specific, but not off-target activity. PIK3CB siRNAs were synthesized with 2′MOE modified nucleotides at positions 1–3 (MOE 1–3) or 12–14 (MOE 12–14) of the guide strand. (A) In vitro binding affinity for recombinant Ago2 was determined as detailed in ‘Materials and Methods’ section. (B) An Ago2 based in vitro RNA cleavage assay was utilized to evaluate cleavage of the targeted mRNA by the parent and MOE-substituted PIK3CB siRNAs. RNA cleavage was performed using a 40 nucleotide 32P-labeled RNA substrate corresponding to the region containing the PIK3CB on-target or YY1 off-target site for the PIK3CB siRNA. (C) HeLa cells were treated with unmodified or MOE-substituted PIK3CB siRNAs at concentrations ranging from 300 pM to 300 nM. The following day target-specific PIK3CB (C) and off-target YY1 (D) mRNA reduction was evaluated by qRT–PCR. Control PIK3CB siRNA (black), PIK3CB-M1-3 siRNA (red), PIK3CB-M12-14 siRNA (blue).
In vitro RNA cleavage was performed in the presence of recombinant Ago2 using 40 nucleotide, P32-labeled RNA substrates corresponding to the region containing the target site for the PIK3CB or YY1 siRNAs. No cleavage of the YY1 off-target RNA was observed with either the unmodified or the MOE substituted siRNAs (Figure 4B, lanes 13–21). The unmodified siRNA generated the expected PIK3CB cleavage product at position 20 of the 40-mer target RNA, 10 nt from the 5′-end of the antisense-strand (lanes 7–8). In total, ∼15% substrate cleavage was observed. For PIK3CB-M12-14 only 0.25% cleavage of the targeted RNA was observed (lanes 9–10), while incubation with PIK3CB-M1-3 siRNA resulted in no detectable cleavage of the targeted RNA.
Next, HeLa cells were treated with PIK3CB, PIK3CB-M1-3 or PIK3CB-M12-14 siRNAs at concentrations ranging from 300 pM to 300 nM. The following day levels of PIK3CB target-specific and YY1 off-target mRNA reduction were evaluated by qRT–PCR. While the unmodified siRNA (black lines) efficiently reduced PIK3CB messenger RNA, the siRNA with 2′MOE substitutions at position 12–14 (blue lines) was less potent with a maximum target reduction of only 30% at the highest concentration tested, as compared to ∼80% for the unmodified siRNA (Figure 4C). The siRNA with 2′MOE substitutions at position 1–3 (red lines) was even less active, but did reduce PIK3CB mRNA by ∼20% at the highest concentration. As previously observed, the unmodified siRNA also reduced YY1 mRNA (Figure 4D), although the potency and activity at the highest concentration tested were reduced compared to PIK3CB on-target mRNA reduction. Reduction of the YY1 off-target message by the PIK3CB-M12-14 siRNA was equivalent to the unmodified siRNA, while no off-target activity was observed for the PIK3CB-M1-3 siRNA. Similar results were obtained by analyzing off-target reduction of FADD mRNA (data not shown). As the PIK3CB-M12-14 siRNA supports only very limited Ago2 mediated cleavage, these data again suggest that Ago2 cleavage is not required for siRNA off-target activity and that a fraction of on-target activity is Ago2 independent. However, Ago2 binding or binding to a similar protein does appear to be required for both target-specific and off-target activity, as no meaningful activity was observed with the PIK3CB-M1-3 siRNA, which does not promote target cleavage and binds to Ago2 only very weakly. It should also be noted that in our system, commercially available, chemically modified siRNAs of the same sequence did not reduce off-target effects in a specific manner (Supplementary Figure S2).
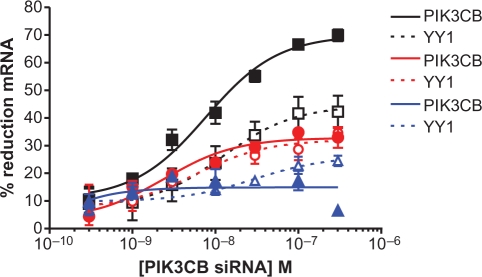
Off-target and some on-target siRNA activity are maintained in Ago2–/– knockout cells
To address the possibility that the observed activities could be accounted for by the fraction of Ago2 protein remaining in HeLa cells following siRNA or RNaseH ASO mediated Ago2 reduction, we evaluated both on- and off-target siRNA activity in mouse embryonic fibroblasts (MEFs) derived from Ago2 knockout (Ago2–/–) mice (41). While the mouse and human YY1 off-target sites are perfectly complementary, the mouse PIK3CB on-target site contains two base mismatches relative to the human sequence. We therefore synthesized a PIK3CB siRNA fully complementary to the homologous mouse PIK3CB target site. Both wild-type and Ago2–/– MEF cells were treated with mPIK3CB siRNA at concentrations ranging from 300 pM to 300 nM. The following day PIK3CB and YY1 mRNA reduction were evaluated by qRT–PCR (Figure 5). In wild-type cells (black lines) the efficacies of target-specific reduction of PIK3CB (solid lines) and off-target reduction of YY1 (dotted lines) were similar to that previously observed in HeLa cells. In the Ago2–/– knockout cells (red lines), the level of off-target reduction was nearly identical to that observed in the wild-type cells and very similar to that observed in the human cell line (Figure 4D), confirming that off-target mRNA degradation can occur even in the absence of Ago2. In contrast, PIK3CB was reduced less efficiently in the knockout as compared to the wild-type cells. The extent of on-target reduction in the knockout cells was very similar to that observed for off-target reduction, suggesting that fully complementary on-target siRNAs are able to promote mRNA reduction even in the absence of Ago2, albeit less efficiently than in wild-type cells.
Figure 5.
PIK3CB off-target and some on-target activity is maintained in mouse Ago2–/– cells. MEF cells were treated with mPIK3CB siRNA at concentrations ranging from 300 pM to 300 nM as detailed in ‘Materials and Methods’ section. The following day total RNA was isolated. Percent reduction of mPIK3CB (solid lines) or YY1 (dashed lines) mRNA was evaluated by qRT–PCR with gene specific primers in wild-type (black), Ago2–/– (red), or Ago2–/– MEF cells treated 24 h prior to the siRNA transfections with an RNAseH ASO to reduce Ago1 (blue). The level of Ago1 reduction was evaluated by qRT–PCR and western blot and found to be <75% control following treatment with the RNAseH ASO (Supplementary Figure S4).
One possible explanation for the off-target activity in Ago2–/– MEFs is that another Ago may be able to be used mechanistically to duplicate the function of Ago2 in off-target activity. All four Ago proteins have been shown to be associated with small RNAs in mammalian cells (42). Although only Ago2 has been determined to have slicer activity (41), previous studies have shown Dcr1 and Ago1 to be essential for the miRNA pathway (8,43). Alternatively, Supplementary Figure S3 suggests that in the presence of adequate levels of Ago2, reduction of any one of the other Agos has little effect. Therefore, we evaluated the effect of Ago1 reduction on siRNA on- and off-target activity in MEF Ago2–/– cells. Ago2–/– MEF cells were treated for 48 h with an RNaseH-dependent ASO targeting Ago1, followed by treatment with mPIK3CB siRNA as detailed above. Ago1 reduction was confirmed by both qRT–PCR and western blot (Supplementary Figure S4) and resulted in a decrease in both on-target reduction of PIK3CB and off-target reduction of YY1 mRNA treated with the mPIK3CB siRNA (Figure 5, blue lines). These data suggest that either Ago1 or Ago2 can bind an siRNA and mediate off-target mRNA reduction. In addition, a significant proportion of on-target mRNA reduction appears to use an Ago2 slicer-independent mechanism and is apparently independent of Ago2. We also evaluated the effect of reducing Ago3 on activity in Ago2–/– MEF cells as well (Supplementary Figure S5). However, while reduction of Ago3 did attenuate off-target mRNA reduction, the effect was not as significant as that observed with Ago1 reduction. We were unable to evaluate the effect of Ago4 reduction, however, as the levels of Ago4 mRNA were more than 20-fold less than those of Ago1 and Ago3.
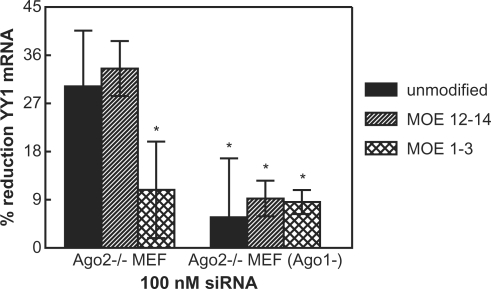
The off-target activity of PIK3CB-M1-3 or PIK3CB-M12-14 siRNAs was also compared in MEF Ago2–/– cells (Figure 6). While these siRNAs were designed to the human PIK3CB sequence, they are capable of promoting off-target reduction of mouse YY1 which is perfectly complementary to the human sequence at the YY1 off-target site. MEF Ago2–/– cells treated at 100 nM with siRNA PIK3CB-M12-14 (striped bars) or the unmodified PIK3CB siRNA (solid bars) showed similar levels of activity in reducing the YY1 off-target message, while siRNA PIK3CB-M1-3 (hatched bars) was significantly less active. Pre-treatment with an RNAseH ASO targeting Ago1 reduced YY1 off-target activity of PIK3CB and PIK3CB-M12-14 siRNAs to approximately equal levels, as observed for siRNA PIK3CB-M1-3 in the untreated cells, confirming that the MOE substituted siRNA is also capable of mediating off-target reduction via either Ago1 or Ago2.
Figure 6.
Activity of MOE modified siRNAs in mouse Ago2–/– cells. Ago2–/– MEF or Ago2–/– MEF pre-treated for 24 h an RNAseH ASO to reduce Ago1 were transfected with PIK3CB (solid bars), PIK3CB-M1-3 (crosshatched) or PIK3CB-M12-14 (striped) siRNAs at 100 nM. Percent reduction of mouse YY1 mRNA as evaluated by qRT–PCR is shown (asterisk, confidence level ≥95%).
Affymetrix expression analysis of off-target effects in wild-type and Ago2–/– MEF cells
To further confirm that siRNA mediated off-target mRNA reduction can occur in the absence of Ago2 and to expand our observations beyond a single off-target transcript, gene expression analysis was performed in siRNA treated MEF cells. Wild-type and Ago2–/– knockout cells were treated with PIK3CB siRNA at 10 nM. The following day RNA was purified and Affymetrix Mouse Genome 430 2.0 arrays used to determine mRNA profiles and identify genes whose expression levels were changed in siRNA-treated, compared to mock transfected cells as detailed in ‘Materials and Methods’ section.
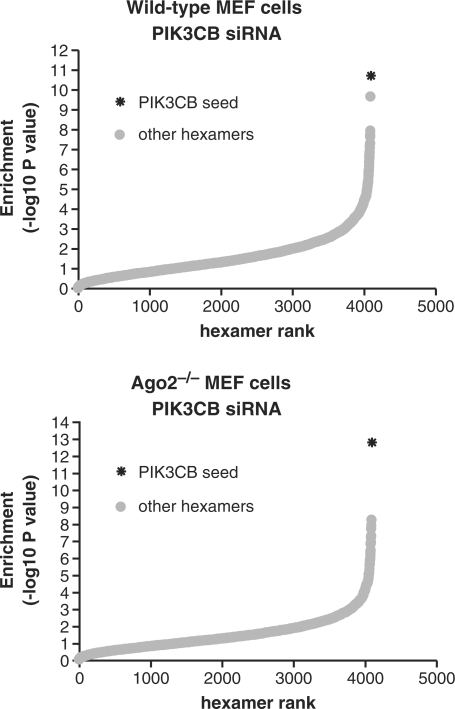
In the wild-type MEF cells treated with PIK3CB siRNA, a hexamer (ATCCTA), corresponding to nucleotides 2–7 of the siRNA guide strand was strongly over-represented in the population of down-regulated transcripts (Figure 7A). The results were similar in MEF Ago2–/– cells, where again transcripts containing the ATCCTA hexamer were significantly over-represented in the population of down-regulated transcripts (Figure 7B). In addition, transcripts containing the hexamer AATCCT, complementary to position 3–8 of the siRNA, were also reduced (not shown). Further, PIK3CB was down-regulated ∼44% (average of five probes). Since on- and off-target silencing follow similar dose-response curves, it is likely that off-target silencing was also suboptimal in this experiment.
Figure 7.
Off-target transcripts matching PIK3CB siRNA seed hexamer are down-regulated in MEF wild type and Ago2−/− knockout cells. Wild-type and Ago2−/− MEF cells were treated with 10 nM PIK3CB siRNA. The following day, RNA was purified and subjected to microarray analysis. 3′UTR regions associated with probes from each bin were analyzed for increased or reduced expression of all 4096 possible hexamer words. Shown are the maximum rank and enrichment in downregulation (−log10 hypergeometric P-value) for each possible hexamer motif in expression signatures from wild type (top) and Ago2–/– knockout (bottom) MEFs. PIK3CB seed: the hexamer matching residues 2–7 of the PIK3CA siRNA seed region (ATCCTA); other hexamers: hexamers other than ATCCTA.
P-body associated proteins are involved in off-target degradation of messenger RNA
Our data suggested that off-target siRNA activity and a fraction of on-target activity require the presence of Ago1 and/or Ago2; yet do not require Ago2 cleavage. Given the importance of 5′ seed region homology for both off-target mRNA down-regulation and miRNA mediated RNA degradation, it seems likely that both of these effects share common mechanisms. It has been demonstrated that miRNAs promote mRNA degradation by directing deadenylation then subsequent decapping and exonucleolytic degradation of messages to which they are partially complementary (21). Several groups have shown that both Ago1 and Ago2 proteins localize to mammalian P-bodies where they associate with other P-body components known to be involved in mRNA turnover (44–46). One suggested model for the mechanism by which miRNAs promote mRNA degradation is that miRNAs may bind to either Ago1 or Ago2 in a common complex. Ago1 and Ago2, along with associated proteins, then release the message from a translation pathway and allow P-bodies to form where the mRNA is subsequently subject to decapping and exonucleolytic degradation (47). By this mechanism, degradation of the message may occur even in the absence of Ago2-mediated cleavage. We reasoned that the binding and subsequent P-body sequestration functions may in fact be redundant between Ago1 and Ago2. We therefore performed experiments to evaluate the effect of reducing P-body associated proteins on off-target activity.
Ago2–/– MEF cells were treated with an RNaseH-dependent ASOs targeting Ago1 or GW182, a P-body component whose depletion has previously been demonstrated to impair gene silencing by miRNAs (27). After 72 h, the cells were treated with PIK3CB siRNA at concentrations ranging from 300 pM to 300 nM. Reduction of Ago1 and GW182 mRNA was evaluated by qRT–PCR and found to be >85% for both proteins (Figure 8A). YY1 off-target mRNA expression was assessed the following day. Significant YY1 off-target mRNA reduction was observed in the mock treated Ago2–/– MEF cells with ∼60% off-target reduction at the 100 nM concentration (Figure 8B, blue line). In contrast, in the Ago1 or GW182 depleted Ago2–/– MEF cells, levels of YY1 mRNA reduction were significantly less; in both cases less than ∼15% reduction in YY1 message was observed at a concentration of 100 nM.
Figure 8.
P-body associated protein reduction attenuates siRNA off-target activity. (A) Reduction of total RNA after 72 h ASO treatment. Relative expression levels of Ago1 (solid bars) and GW182 (striped bars) were measured by qRT–PCR. (B) Ago2−/− MEF cells were treated with RNAseH ASOs targeting Ago1 or GW182. Seventy-two hours later cells were treated with PIK3CB siRNA at concentrations ranging from 300 pM to 300 nM levels of YY1 mRNA were assessed by qRT–PCR the following day. (C) Ago2−/− MEF cells were treated with RNAseH ASOs targeting GW182 or DCP1:2. Seventy-two hours later cells were treated with PIK3CB siRNA at 100 nM. Levels of YY1 mRNA reduction were assessed as above. (D) As in (B), but levels of PIK3CB reduction were measured. (E) IC50 curves for PIK3CB on-target mRNA reduction. HeLa cells were treated with siRNAs targeted to P-body components LSM-1 or GW182 as above. An siRNA targeting Ago2 was also included as a control. Twenty-two hours later cells were seeded in 96-well plates, then treated with PIK3CB siRNA at concentrations ranging from 10 pM to 40 nM. Levels of PIK3CB mRNA were assessed by qRT–PCR the following day for control (black), Ago2 reduced (blue), GW182 reduced (red), or LSM1 reduced (green) cells. (F) IC50 curves for YY1 off-target mRNA reduction. Levels of YY1 mRNA were assessed by qRT–PCR using RNA isolated from HeLa cells above.
DCP1 and DCP2 are P-body associated proteins which have previously been reported to play a role in miRNA-mediated gene silencing (25). Ago2–/– MEF cells were treated with ASOs targeting GW182 at 100 nM or DCP1 and DCP2 at 50 nM each. Three days later the cells were treated with 100 nM PIK3CB siRNA and levels of YY1 off-target mRNA subsequently assessed by qRT–PCR. Reduction of DCP1:2 and GW182 mRNA was assayed by qRT–PCR and found to be >70% and the ability of the ASOs to disrupt P-body formation was also confirmed by immunofluorescence (Supplementary Figure S6). Depletion of both DCP1:2 and GW182 resulted in a >2-fold reduction in off-target activity (Figure 8C). In contrast, DCP1:2 and GW182 depletion had no significant effect on target-specific, Ago2-independent mRNA reduction, as there was no significant difference in the amount of PIK3CB siRNA reduction in the same cells (Figure 8D). Depletion of DDX6, which has been demonstrated to be contained within P-bodies and has previously been reported to play a role in miRNA-mediated gene silencing (46), was also found to attenuate off-target activity in Ago2–/– MEF cells (Supplementary Figure S7).
The effect of reduction of P-body associated proteins on target-specific as well as off-target activity was evaluated in HeLa cells as well. Cells were treated with siRNAs targeted to P-body components LSM-1 or GW182. An siRNA targeting Ago2 was also included as a control. Twenty-four hours later cells were seeded in 96-well plates, then treated with PIK3CB siRNA at concentrations ranging from 10 pM to 40 nM. ASO-mediated mRNA reduction of the targeted P-body proteins and subsequent disruption of P-body formation was confirmed by immunofluorescence (Supplementary Figure S8). Levels of PIK3CB and YY1 mRNA were assessed by qRT–PCR the following day. As previously observed, reduction of Ago2 (blue lines) resulted in a 6-fold reduction in on-target siRNA activity (Figure 8E), but had no effect on off-target potency as measured by the reduction of YY1 mRNA (Figure 8F). Conversely, reduction of the P-body proteins (red and green lines) had no effect on PIK3CB on-target mRNA reduction, but did reduce off-target potency by ∼3-fold suggesting that off-target RNA degradation is dependent on some of the same P-body associated protein previously demonstrated to be involved in miRNA mediated RNA degradation.
DISCUSSION
In this article, we have demonstrated that siRNA-mediated reduction of an off-target messenger RNA is Ago2 cleavage independent, but does require siRNA interaction with either Ago1 or Ago2 and the RLC. We also show that off-target siRNA-mediated degradation of mRNA is associated with proteins that have previously been shown to localize to P-bodies. Finally, our data suggest that a portion of on-target siRNA mediated mRNA degradation is also Ago2 cleavage independent, but may use mechanism different than that used for off-target mRNA degradation.
Experiments in both human and mouse cells demonstrated that siRNAs can reduce target-specific and off-target mRNAs via a mechanism that is independent of Ago2 slicer activity. It has previously been reported that siRNA mediated off-target mRNA reduction of a stably expressed reporter gene is likely the result of Ago-2 independent degradation processes (28). Another recently published article reported that translational repression mediated by siRNAs with full complementarity to a reporter mRNA can also occur independently of Ago2 (29). We extended these observations, demonstrating in HeLa cells that RNAseH ASO mediated reduction of Ago2 has no effect on siRNA off-target degradation of endogenous mRNAs (Figure 1C). Moreover, reduction of Ago2 decreased the maximum reduction of the target, PIK3CB, from 70 to 50% and resulted in a 6-fold shift in the IC50 but did not completely ablate the activity (Figure 1A). We confirmed and extended this observation by demonstrating that Dicer activity is required for both on- and off-target activities if Dicer substrates are used (Figure 2). This suggests that although Ago2 is not responsible for all of the on-target activities and very little if any of the off-target activities of the siRNAs used in this study, the RLC is likely involved in all the activities. Less of an effect of Ago2 reduction on PIK3CB silencing was observed at higher siRNA concentrations. This may also suggest that at higher siRNA concentrations activity may be mediated by an Ago2-independent mechanism.
It is known that cleavage of a target by Ago2 leaves a 5′ phosphate on the cleaved mRNA. To further confirm that Ago2 cleavage is not required for off target siRNA activities, we attempted to detect the cleavage fragments expected for Ago2 cleavage utilizing a modified RLM-RACE technique. As expected, for on-target activity the Ago2 cleavage product was detected. However, we were unable to detect any Ago2 specific fragments corresponding to the off-target mRNA reductions (Figure 3B). It could be argued that we simply did not detect a RACE product due to the lower efficacy of off-target as compared to target-specific activity; however, we were able to detect a RACE product for the YY1 siRNA which had much lower activity than either the target-specific or off-target activity of the PIK3CB siRNA (Figure 3A). A second site with homology to the PIK3CB siRNA seed region was found in the YY1 mRNA; however, we were unable to design PCR primers for this site since it ends a single nucleotide from the 3′ end of the message. If cleavage occurs via siRNA interactions with this site, it would not be detected with the primers used. However, our results suggest that if degradation of off-target transcripts occurs via siRNA interactions at the putative cleavage site for which the RACE primers were designed, it occurs via an Ago2 slicer-independent mechanism.
Studies with siRNAs modified in key positions with 2′-MOE nucleotides provided additional confirmation and suggested that binding to a member of the Ago family may be required for off-target activity. It has previously been shown that 2′-O-methyl ribosyl substitution of the siRNA guide strand at position 2 reduces off-target activity but has little to no effect on target-specific mRNA reduction (48), indicating that on- and off-target mechanisms may be differentially sensitive to chemical modifications of the siRNA. Modified siRNAs substituted with 2′-MOE nucleotides at various positions have been shown to result in reduction of siRNA activity (34), however, the effect of these modifications on off-target activity has not been evaluated. PIK3CB siRNAs with three 2′MOE modified bases at either the 5′-end or near the center of the guide strand were synthesized, then evaluated for Ago2 binding and cleavage as well as on- and off-target activities. Based upon the crystal structure of an siRNA guide strand with the Ago2 PIWI domain (10,40), we predicted that the increased size of the 2′MOE modification at positions 1–3 of the siRNA guide strand relative to unmodified or 2′-O-methyl RNA would interfere with binding to Ago2. As expected, inserting 2′-MOEs in positions 1–3 of the guide strand reduced affinity for Ago2 by 3-fold and consequently, ablated cleavage activity (Figure 4A and B). Even so, the siRNA modified at positions 1–3 displayed some siRNA activity, albeit very substantially less than the parent. However, off-target activity was essentially ablated in cells treated with PIK3CB-M1-3 siRNA. In contrast to PIK3CB-M1-3 siRNA, the siRNA with 2′-MOE substitutions at positions 12–14 had essentially the same binding affinity for recombinant Ago2 as the unmodified siRNA (Figure 4A), but a nearly 60-fold reduction in cleavage activity (Figure 4B). This was associated with a 2 log drop in potency for on-target activity and a significant reduction in the maximum effect (Figure 4C). In contrast to PIK3CB-M1-3 siRNA, PIK3CB-M12-14 siRNA resulted in the same amount of off-target activity as the unmodified siRNA, confirming that off-target activity is independent of Ago2 cleavage activity, but suggesting a requirement for binding to Ago2 or a similar protein.
Because siRNAs/ASOs reduced Ago2 levels in Hela cells by 80–90%, but did not eliminate all Ago2, it is possible that the activities observed could be mediated by the residual Ago2 activity that remained in ASO or siRNA treated cells. To evaluate this possibility and to begin to try to understand the mechanisms by which these effects are mediated, we studied on- and off-target mRNA degradation in Ago2–/– knockout MEF cells (41). Again, on-target activity was less potent in the knockout as compared to the wild-type cells, but still substantial for the siRNA studied and off-target activity was virtually identical in the wild type and knockout cells (Figures 5). These observations were confirmed by the behaviors of the 2′-MOE modified siRNAs in the knockout cells (Figure 6). Furthermore, to exclude the possibility that off-target activities for mRNAs other than YY1 are mediated by a mechanism different from the process suggested by our data, we performed a gene expression array analysis for all potential off-target effects. In both wild-type and Ago2–/– MEF cells treatment with PIK3CB siRNA resulted in the reduction of a set of transcripts containing the ATCCTA hexamer which is complementary to the PIK3CB siRNA seed region hexamer (Figure 7).
While slicer activity, which contributes to a substantial proportion of target-specific siRNA mediated cleavage, is associated with Ago2 only (9), the high degree of similarity of the other Ago proteins suggests a substantial degree of redundancy in the Ago protein family (49). Analysis of Ago1 and Ago2 containing protein complexes suggests that both Ago1 and Ago2 proteins may be components of miRNA-containing mRNA targeting complexes (50). Given the similarity between off-target silencing by siRNAs and transcript regulation by miRNAs, we explored the possibility that Ago1 might be capable of mediating off-target activity. Reduction of Ago1 with an RNaseH dependent ASO resulted in a further reduction of off-target as well as target-specific activity in Ago2–/– MEF cells with both unmodified and 2′-MOE modified siRNAs (Figures 5, 6 and 8). These results suggest that some on-target mRNA degradation may be, in part, mediated by Ago1 and that Ago1 can play a significant role in off-target degradation activities, and are consistent with a recently published paper describing overlapping functions for all four mammalian Ago proteins in miRNA silencing (51). We also observed attenuation of off-target activity resulting from reduction of Ago3 (Supplementary Figure S5), however, levels of Ago4 expression were too low in the Ago2–/– MEF cells to conclusively observe an effect on activity.
Several studies that have demonstrated that miRNA can induce degradation of mRNA targets despite imperfect miRNA-mRNA base-pairing (19,20,25,52). Like our observations for off-target activity, this miRNA-mediated mRNA degradation is Ago-dependent, however, it does not require the direct Ago2-mediated endonucleolytic cleavage of the target mRNA. Target degradation is instead a consequence of decapping and exonucleolytic processing of repressed mRNAs (53). To further understand the mechanism supporting Ago2 independent effects of siRNAs, we reduced the levels of several P-body associated proteins (54) in Ago2–/– MEF cells and in Hela cells. In Ago2–/– MEF cells, reduction of DDX6, GW182 and DCP1:2 caused a loss of off-target activity comparable to that achieved with Ago1 reduction (Figures 8B and C, Supplementary Figures S6 and S7). In Hela cells, we showed that reduction of P-body associated proteins, LSM1 and GW182 also reduced off-target activity (Figure 8F). These data suggest that off-target mRNA degradation is P-body associated. However, this correlation does not prove that P-bodies are the site of off-target degradation, as P-body formation and off-target degradation may simply require some of the same proteins. In addition, one would expect that P-body reduction would also reduce the fraction of Ago2-independent on-target activity. It is interesting to note, however, that P-body reduction in wild-type MEF and HeLa cells had no effect on target-specific activity (Figure 8D and E). While Ago2 independent on-target RNA degradation clearly involves the RLC and an Ago protein, it may not be affected via P-body associated proteins. Alternatively, any effects of P-body reduction on Ago2 independent on-target activity may be masked by the more robust slicer-dependent activity in these cells or degradation may be occurring in some other cellular compartment such as the exosome (55).
In conclusion, we have demonstrated that siRNA induced off-target activities do not require Ago2 slicer activity, but do require interaction with Ago1 and P-body associated proteins. Moreover, a fraction of siRNA mediated on-target activity is independent of Ago2, dependent on Ago1, and unaffected by reduction of the P-body proteins that we studied. Nevertheless, both on- and off-target activities are mediated by the RLC. These results are consistent with the previously published observation that RISC assembly can occur in the absence of passenger strand cleavage (5). Our results are also consistent with observations that the purified enzyme can only engage in a single round of cleavage (11). Given the inability of Ago2 to release product and the absence of cleavage by the other Ago proteins, one is forced to wonder if the slicer activity of Ago2 accounts for only a portion of observed activities, and, like the other Ago proteins, Ago2 activity may also be the result of binding to and transport of targeted RNAs to sites of degradation employing other enzymatic processes.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Funding for open access charge: ISIS Pharmaceuticals, Inc.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors are grateful to Dr Gregory J. Hannon (Cold Spring Harbor Laboratory) for providing the wild-type and Ago2 knockout MEF cells. They also thank Dr C. Frank Bennett and Dr Eric Swayze for critical reading of this manuscript and Tracy Reigle for help in preparation of the figures.
REFERENCES
- 1.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 2.Zamore PD, Tuschl T, Sharp PA, Bartel DP. RNAi: double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell. 2000;101:25–33. doi: 10.1016/S0092-8674(00)80620-0. [DOI] [PubMed] [Google Scholar]
- 3.Gregory RI, Chendrimada TP, Cooch N, Shiekhattar R. Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell. 2005;123:631–640. doi: 10.1016/j.cell.2005.10.022. [DOI] [PubMed] [Google Scholar]
- 4.Tomari Y, Zamore PD. Perspective: machines for RNAi. Genes Dev. 2005;19:517–529. doi: 10.1101/gad.1284105. [DOI] [PubMed] [Google Scholar]
- 5.Matranga C, Tomari Y, Shin C, Bartel D, Zamore P. Passenger-strand cleavage facilitates assembly of siRNA into Ago2-Containing RNAi enzyme complexes. Cell. 2005;123:607–620. doi: 10.1016/j.cell.2005.08.044. [DOI] [PubMed] [Google Scholar]
- 6.Hammond SM, Caudy AA, Hannon GJ. Post-transcriptional gene silencing by double-stranded RNA. Nat. Rev. 2001;2:110–119. doi: 10.1038/35052556. [DOI] [PubMed] [Google Scholar]
- 7.Martinez J, Patkaniowska A, Urlaub H, Lührmann R, Tuschl T. Single-stranded antisense siRNAs guide target RNA cleavage in RNAi. Cell. 2002;110:563–574. doi: 10.1016/s0092-8674(02)00908-x. [DOI] [PubMed] [Google Scholar]
- 8.Okamura K, Ishizuka A, Siomi H, Siomi M. Distinct roles for Argonaute proteins in small RNA-directed RNA cleavage pathways. Genes Dev. 2004;18:1655–1666. doi: 10.1101/gad.1210204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meister G, Landthaler M, Patkaniowska A, Dorsett Y, Teng G, Tuschl T. Human Argonaute2 Mediates RNA Cleavage Targeted by miRNAs and siRNAs. Mol. Cell. 2004;15:185–197. doi: 10.1016/j.molcel.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 10.Song JJ, Smith SK, Hannon GJ, Joshua-Tor L. Crystal structure of Argonaute and its implications for RISC slicer activity. Science. 2004;305:1434–1437. doi: 10.1126/science.1102514. [DOI] [PubMed] [Google Scholar]
- 11.Rivas FV, Tolia NH, Song J-J, Aragon JP, Liu J, Hannon GJ, Joshua-Tor L. Purified Argonaute2 and an siRNA form recombinant human RISC. Nat. Struct. Mol. Biol. 2005;12:340–349. doi: 10.1038/nsmb918. [DOI] [PubMed] [Google Scholar]
- 12.Persengiev SP, Zhu X, Green MR. Nonspecific, concentration-dependent stimulation and repression of mammalian gene expression by small interfering RNAs (siRNAs) RNA. 2004;10:12–18. doi: 10.1261/rna5160904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haley B, Zamore PD. Kinetic analysis of the RNAi enzyme complex. Nat. Struct. Mol. Biol. 2004;11:599–606. doi: 10.1038/nsmb780. [DOI] [PubMed] [Google Scholar]
- 14.Jackson AL, Bartz SR, Schelter J, Kobayashi SV, Burchard J, Mao M, Lin B, Cavet1 G, Linsley PS. Expression profiling reveals off-target gene regulation by RNAi. Nat. Biotechnol. 2003;21:635–637. doi: 10.1038/nbt831. [DOI] [PubMed] [Google Scholar]
- 15.Jackson AL, Burchard J, Schelter J, Chau BN, Cleary M, Lim L, Linsley PS. Widespread siRNA “off-target” transcript silencing mediated by seed region sequence complementarity. RNA. 2006;12:1179–1187. doi: 10.1261/rna.25706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tschuch C, Schulz A, Pscherer A, Werft W, Benner A, Hotz-Wagen A, Barrionuevo LS, Lichter P, Mertens D. Off-target effects of siRNA specific for GFP. BMC Mol. Biol. 2008;9:60. doi: 10.1186/1471-2199-9-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin X, Ruan X, Anderson MG, McDowell JA, Kroeger PE, Fesik SW, Shen Y. siRNA-mediated off-target gene silencing triggered by a 7 nt complementation. Nucleic Acids Res. 2005;33:4527–4535. doi: 10.1093/nar/gki762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Birmingham A, Anderson EM, Reynolds A, Ilsley-Tyree D, Leake D, Fedorov Y, Baskerville S, Maksimova E, Robinson K, Karpilow J. 3′ UTR seed matches, but not overall identity, are associated with RNAi off-targets. Nat. Methods. 2006;3:199–204. doi: 10.1038/nmeth854. [DOI] [PubMed] [Google Scholar]
- 19.Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 20.Bagga S, Bracht J, Hunter S, Massirer K, Holtz J, Eachus R, Pasquinelli A. Regulation by and miRNAs results in target mRNA degradation. Cell. 2005;122:553–563. doi: 10.1016/j.cell.2005.07.031. [DOI] [PubMed] [Google Scholar]
- 21.Wu L, Belasco JG. Let me count the ways: mechanisms of gene regulation by miRNAs and siRNAs. Mol. Cell. 2008;29:1–7. doi: 10.1016/j.molcel.2007.12.010. [DOI] [PubMed] [Google Scholar]
- 22.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are MicroRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 23.Brennecke J, Stark A, Russell RB, Cohen SM. Principles of microRNA-target recognition. PLoS Biol. 2005;3:e85. doi: 10.1371/journal.pbio.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Doench JG, Sharp PA. Specificity of microRNA target selection in translational repression. Genes Dev. 2004;18:504–511. doi: 10.1101/gad.1184404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Behm-Ansmant I, Rehwinkel J, Doerks T, Stark A, Bork P, Izaurralde E. mRNA degradation by miRNAs and GW182 requires both CCR4:NOT deadenylase and DCP1:DCP2 decapping complexes. Genes Dev. 2006;20:1885–1898. doi: 10.1101/gad.1424106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu L, Fan J, Belasco JG. MicroRNAs direct rapid deadenylation of mRNA. Proc. Natl Acad. Sci. USA. 2006;103:4034–4039. doi: 10.1073/pnas.0510928103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu J, Rivas FV, Wohlschlegel J, Yates JR, Parker R, Hannon GJ. A role for the P-body component GW182 in microRNA function. Nat. Cell Biol. 2005;7:1261–1266. doi: 10.1038/ncb1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alemán LM, Doench J, Sharp PA. Comparison of siRNA-induced off-target RNA and protein effects. RNA. 2007;13:385–395. doi: 10.1261/rna.352507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu L, Fan J, Belasco JG. Importance of translation and nonnucleolytic Ago proteins for on-target RNA interference. Curr. Biol. 2008;18:1327–1332. doi: 10.1016/j.cub.2008.07.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chiu YL, Rana TM. siRNA function in RNAi: a chemical modification analysis. RNA. 2003;9:1034–1048. doi: 10.1261/rna.5103703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chounga S, Kimb YJ, Kima S, Parka HO, Choi YC. Chemical modification of siRNAs to improve serum stability without loss of efficacy. Biochem. Biophys. Res. Commun. 2006;342:919–927. doi: 10.1016/j.bbrc.2006.02.049. [DOI] [PubMed] [Google Scholar]
- 32.Lavigne C, Thierry AR. Specific subcellular localization of siRNAs delivered by lipoplex in MCF-7 breast cancer cells. Biochimie. 2007;89:1245–1251. doi: 10.1016/j.biochi.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 33.Baker BF, Lot SS, Condon TP, Cheng-Flournoy S, Lesnik EA, Sasmor HM, Bennett CF. 2′-O-(2-methoxy)ethyl-modified anti-intercellular adhesion molecule 1 (ICAM-1) oligonucleotides selectively increase the ICAM-1 mRNA level and inhibit formation of the ICAM-1 translation initiation complex in human umbilical vein endothelial cells. J. Biol. Chem. 1997;272:11994–12000. doi: 10.1074/jbc.272.18.11994. [DOI] [PubMed] [Google Scholar]
- 34.Prakash TP, Allerson CR, Dande P, Vickers TA, Sioufi N, Jarres R, Baker BF, Swayze EE, Griffey RH, Bhat B. Positional effect of chemical modifications on short interference RNA activity in mammalian cells. J. Med. Chem. 2005;48:4247–4253. doi: 10.1021/jm050044o. [DOI] [PubMed] [Google Scholar]
- 35.Vickers TA, Zhang H, Graham MJ, Lemonidis KM, Zhao C, Dean NM. Modification of MyD88 mRNA splicing and inhibition of IL-1beta signaling in cell culture and in mice with a 2′-O-methoxyethyl-modified oligonucleotide. J. Immunol. 2006;176:3652–3661. doi: 10.4049/jimmunol.176.6.3652. [DOI] [PubMed] [Google Scholar]
- 36.Winer J, Kwang C, Jung S, Shackel I, Williams PM. Development and validation of real-time quantitative reverse transcriptase±polymerase chain reaction for monitoring gene expression in cardiac myocytes in vitro. Anal. Biochem. 1999;270:41–49. doi: 10.1006/abio.1999.4085. [DOI] [PubMed] [Google Scholar]
- 37.Hashimoto JG, Beadles-Bohling AS, Wiren KM. Comparison of RiboGreen and 18S rRNA quantitation for normalizing real-time RT-PCR expression analysis. BioTechniques. 2004;36:58–60. doi: 10.2144/04361BM06. [DOI] [PubMed] [Google Scholar]
- 38.Dongen Sv, Abreu-Goodger C, Enright AJ. Fast assessment of microRNA binding and siRNA off-target effects from expression data. Nat. Methods. 2008;5:1023–1025. doi: 10.1038/nmeth.1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim DH, Behlke MA, Rose SD, Chang MS, Choi S, Rossi JJ. Synthetic dsRNA Dicer substrates enhance RNAi potency and efficacy. Nat. Biotechnol. 2004;23:222–226. doi: 10.1038/nbt1051. [DOI] [PubMed] [Google Scholar]
- 40.Ma JB, Yuan YR, Meister G, Pei Y, Tuschl T, Patel DJ. Structural basis for 5[prime]-end-specific recognition of guide RNA by the A. fulgidus Piwi protein. Nature. 2005;434:666–670. doi: 10.1038/nature03514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song J-J, Hammond SM, Joshua-Tor L, Hannon GJ. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- 42.Landthaler M, Gaidatzis D, Rothballer A, Chen PY, Soll SJ, Dinic L, Ojo T, Hafner M, Zavolan M, Tuschl T. Molecular characterization of human Argonaute-containing ribonucleoprotein complexes and their bound target mRNAs. RNA. 2008;14:2580–2596. doi: 10.1261/rna.1351608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee Y, Nakahara K, Pham J, Kim K, He Z, Sontheimer E, Carthew R. Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. Cell. 2004;117:69–81. doi: 10.1016/s0092-8674(04)00261-2. [DOI] [PubMed] [Google Scholar]
- 44.Liu J, Valencia-Sanchez MA, Hannon GJ, Parker R. MicroRNA-dependent localization of targeted mRNAs to mammalian P-bodies. Nat. Cell Biol. 2005;7:719–723. doi: 10.1038/ncb1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pillai RS, Bhattacharyya SN, Artus CG, Zoller T, Cougot N, Basyuk E, Bertrand E, Filipowicz W. Inhibition of translational initiation by Let-7 microRNA in human cells. Science. 2005;309:1573–1576. doi: 10.1126/science.1115079. [DOI] [PubMed] [Google Scholar]
- 46.Chu CY, Rana TM. Translation repression in human cells by microRNA-induced gene silencing requires RCK/p54. PLoS Biol. 2006;4:e210. doi: 10.1371/journal.pbio.0040210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rossi JJ. RNAi and the P-body connection. Nat. Cell Biol. 2005;7:643–644. doi: 10.1038/ncb0705-643. [DOI] [PubMed] [Google Scholar]
- 48.Jackson AL, Burchard J, Leake D, Reynolds A, Schelter J, Guo J, Johnson JM, Lim L, Karpilow J, Nichols K, et al. Position-specific chemical modification of siRNAs reduces “off-target” transcript silencing. RNA. 2006;12:1197–1205. doi: 10.1261/rna.30706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Peters L, Meister G. Argonaute proteins: mediators of RNA silencing. Mol. Cell. 2007;26:611–623. doi: 10.1016/j.molcel.2007.05.001. [DOI] [PubMed] [Google Scholar]
- 50.Höck J, Weinmann L, Ender C, Rüdel S, Kremmer E, Raabe M, Urlaub H, Meister G. Proteomic and functional analysis of Argonaute-containing mRNA–protein complexes in human cells. EMBO Rep. 2007;8:1052–1060. doi: 10.1038/sj.embor.7401088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Su H, Trombly MI, Chen J, Wang X. Essential and overlapping functions for mammalian Argonautes in microRNA silencing. Genes Dev. 2009;23:304–317. doi: 10.1101/gad.1749809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Linsley PS, Schelter J, Burchard J, Kibukawa M, Martin MM, Bartz SR, Johnson JM, Cummins JM, Raymond CK, Dai H, et al. Transcripts targeted by the microRNA-16 family cooperatively regulate cell cycle progression. Mol. Cell Biol. 2007;27:2240–2252. doi: 10.1128/MCB.02005-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schmitter D, Filkowski J, Sewer A, Pillai RS, Oakeley EJ, Zavolan M, Svoboda P, Filipowicz W. Effects of Dicer and Argonaute down-regulation on mRNA levels in human HEK293 cells. Nucleic Acids Res. 2006;17:4801–4815. doi: 10.1093/nar/gkl646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Meister G, Landthaler M, Peters L, Chen PY, Urlaub H, Lührmann R, Tuschl T. Identification of novel Argonaute-associated proteins. Current Biol. 2005;15:2149–2155. doi: 10.1016/j.cub.2005.10.048. [DOI] [PubMed] [Google Scholar]
- 55.Schmid M, Jensen TH. The exosome: a multipurpose RNA-decay machine. Trends Biochem. Sci. 2008;33:501–510. doi: 10.1016/j.tibs.2008.07.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.