Abstract
Glycogen synthase kinase 3β (GSK3β) is involved in several cellular signaling systems through regulation of the activity of diverse transcription factors such as Notch, p53 and β-catenin. Mastermind-like 1 (MAML1) was originally identified as a Notch coactivator, but has also been reported to function as a transcriptional coregulator of p53, β-catenin and MEF2C. In this report, we show that active GSK3β directly interacts with the MAML1 N-terminus and decreases MAML1 transcriptional activity, suggesting that GSK3β might target a coactivator in its regulation of gene expression. We have previously shown that MAML1 increases global acetylation of histones, and here we show that the GSK3 inhibitor SB41, further enhances MAML1-dependent histone acetylation in cells. Finally, MAML1 translocates GSK3β to nuclear bodies; this function requires full-length MAML1 protein.
INTRODUCTION
The protein Mastermind-like 1 (MAML1) (1–3), belongs to the MAML family that also contains MAML2 and MAML3 (4,5). The MAML proteins are transcriptional coactivators for Notch signaling (1–5), an evolutionarily conserved pathway that plays several key roles in development and in human disease (6–9). The Notch ICD (intracellular domain) activates expression of genes controlled by Notch signaling by interacting with the conserved DNA-binding protein CSL (10) and recruiting coactivators such as PCAF, GCN5 (11), p300 (12) and MAML (1–5). More recently, MAML1 has been shown to be involved in other cell signaling pathways, and to function as a coactivator for the tumor suppressor p53 (13), the MADS box transcription enhancer factor (MEF) 2C (14), and β-catenin (15). The function of MAML1 as a coactivator for diverse activators also suggests that MAML1 might be a key molecule that connects various signaling pathways to regulate cellular processes in normal cells and in human disease.
MAML1 has been shown to be important for recruitment of coregulators, such as the histone acetyltransferase (HAT) p300 (16,17) and the cyclin-dependent kinase (CDK) 8 (18). Recruitment of CDK8 by MAML1 leads to phosphorylation of Notch1 and subsequent degradation via the Fbw7/Sel10 ubiquitin ligase (18). Previous studies reported that MAML1 recruitment of p300 to a DNA-CSL-Notch complex potentiates Notch ICD transcription from chromatin templates in vitro (16,17), and the p300-MAML1 complex specifically acetylates histone H3 and H4 tails in chromatin in vitro (19). In addition, MAML1 enhances p300 autoacetylation and HAT activity and this coincides with the translocation of MAML1, p300 and acetylated histones to nuclear bodies (20).
Glycogen synthase kinase 3β (GSK3β) is a multifunctional kinase found in all eukaryotes, and its activity is regulated by serine (inhibitory) and tyrosine (stimulatory) phosphorylation (21,22). GSK3β regulates many diverse cellular processes including proliferation, differentiation and apoptosis (23). Aberrant regulation of GSK3β has been suggested to be involved in human diseases such as non-insulin-dependent diabetes mellitus, cardiovascular and neurodegenerative diseases (23,24). Transcription factors regulated by GSK3β include Axin (25,26), β-catenin (27), c-Myc (28,29), NFκB (30), p53 (31,32) and Notch receptors (33,34). GSK3β phosphorylation of Notch2 inhibits transcription of the Notch target gene Hes1. Wnt signaling inhibits GSK3β, and since overexpression of Wnt upregulates Hes1, it has been suggested that Notch phosphorylation by GSK3β regulates cross-talk between the Notch and Wnt pathways (33).
In this study we have investigated how GSK3β regulates MAML1 activity. We found that GSK3β inhibits MAML1 transcriptional activity by directly targeting the N-terminal domain of MAML1. The MAML1 N-terminus is also crucial for interactions with Notch (2), p53 (13), MEF2C (14), and p300 (16,19), so we hypothesize that the N-terminus of MAML1 might be involved in controlling a possible competition for MAML1 by different signaling pathways. We recently reported that MAML1 increases global acetylation of histones (20), and in this paper we show that the GSK3 inhibitor SB41 further enhances MAML1-dependent acetylation of histone H3 in the cell. We also found that GSK3β interacts strongly with MAML1 regardless of its activity status, but that GSK3β must be active to inhibit MAML1 activity. Finally, we found that MAML1 translocates GSK3β to nuclear bodies, and this requires the full-length MAML1 protein.
MATERIALS AND METHODS
Plasmids
The expression plasmids pGEX-MAML1 1–1016, 1–300, 309–625, 499–804, 701–1016, pBIND-Notch1 ICD (19), pBIND-MAML1 1–1016 and 1–300 (20), have been described previously. cDNAs encoding MAML1 residues 1–127 and 1–200 were amplified with PCR and subcloned into pGEX-4T-3. pCDNA-HA-GSK3β WT was a gift from Dr A. Bigas. pCDNA-HA-GSK3β-S9A (Addgene plasmid 14754) and pCDNA-HA-GSK3β-K85R (Addgene plasmid 14755) were generously provided by Dr J. Woodgett.
Purification of proteins and protein interaction assay
GST-tagged proteins were expressed in the Escherichia coli strain BL21 and purified with glutathione-Sepharose 4B (Amersham Biosciences) following manufacturer's; protocol. In GST-MAML1 interaction assays, ∼2 µg of purified GST-tagged MAML1 proteins, bound to 20 µl of glutathione-Sepharose beads, was incubated with whole-cell extract from HEK-293 cells transfected with pCDNA-GSK3β lysed in RIPA buffer (25 mM Tris–HCl pH 7.6, 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS) and diluted 3 times with buffer A (50 mM Tris–HCl, pH 7.5, 150 mM KCl, 10% glycerol, 0.1% NP-40, 1 mM DTT and 1 mM PMSF) (Figure 3B) or incubated with HIS-GSK3β (Invitrogen) and FLAG-Notch1 ICD (as indicated in Figures 1E, 3C and 5D), expressed in Sf9 (Spodoptera frugiperda) cells via baculovirus and purified as described in ref. (17). After centrifugation of the suspension, the supernatant was removed and the beads were washed five times with 500 µl of buffer A. The proteins were eluted from the beads, resolved by SDS–PAGE and analyzed by immunoblot with a monoclonal mouse antibody recognizing GSK3β (BD Transduction Laboratories). GST-GSK3β bound to glutathione-Sepharose beads was incubated with FLAG-MAML1 1–1016 expressed in Sf9 cells via baculovirus and purified as described in ref. (17).
Figure 3.
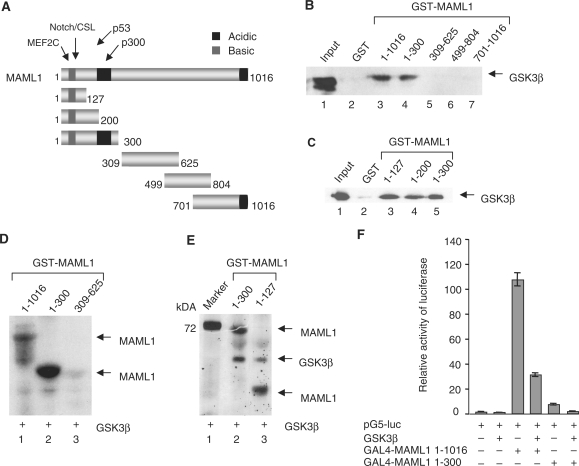
GSK3β directly interacts with the MAML1 N-terminus. (A) Schematic diagram of the MAML1 protein. GST-tagged MAML1 proteins as indicated, were incubated with whole-cell extract from HEK-293 cells transfected with pCDNA-GSK3β (B) or recombinant affinity-purified GSK3β (C), and MAML1-interacting GSK3β detected with immunoblot. (D) Recombinant HIS-GSK3β and GST-MAML1 proteins were affinity-purified and incubated with [γ-32P]ATP. Proteins were separated by SDS–PAGE and visualized by autoradiography. (E) GST-MAML1 proteins were incubated with HIS-GSK3β in the presence of cold ATP. The kinase reactions were separated by SDS–PAGE and stained with Pro-Q Diamond phosphoprotein staining kit. (F) Vectors expressing GSK3β and were cotransfected with a luciferase reporter containing five GAL4 binding sites into U2OS cells.
Figure 1.
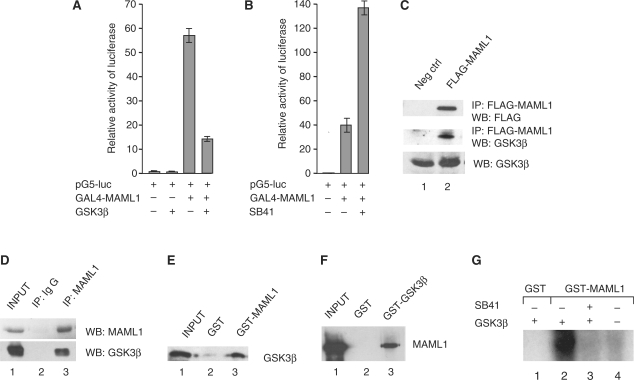
GSK3β decreases MAML1 activity. (A) HEK-293 cells were cotransfected with a luciferase reporter containing five GAL4 binding sites and vectors expressing GAL4-MAML1 and GSK3β as indicated. (B) GAL4-MAML1 was cotransfected into HEK-293 cells with the luciferase reporter in the absence or presence of SB41. (C) HEK-293 cells were transfected with a vector expressing FLAG-MAML1, MAML1 was affinity-purified with M2-agarose, and proteins separated by SDS–PAGE. Immunoblot detected MAML1, and GSK3β protein that interacted with MAML1. (D) Endogenous MAML1 protein was immunoprecipitated from C33A cells with an antibody recognizing the N-terminus of MAML1. Proteins were separated by SDS–PAGE and detection of MAML1, and MAML1-interacting proteins, was monitored by immunoblot with antibodies recognizing MAML1 and GSK3β. (E) GST-MAML1 and GST coupled to glutathione-Sepharose beads were incubated with affinity-purified HIS-GSK3β, and MAML1-interacting GSK3β was monitored by immunoblot. The input represents 10% of GSK3β used in the binding reaction. (F) GST-GSK3β and GST coupled to glutathione-Sepharose beads were incubated with affinity-purified FLAG-MAML1, and MAML1 binding to GSK3β was monitored by immunoblot. The input represents 10% of the MAML1 used in the binding reaction. (G) GSK3β phosphorylates MAML1 in vitro. Recombinant affinity-purified proteins HIS-GSK3β, GST-MAML1 or GST, were incubated with [γ-32P]ATP, in the presence or absence of SB41, proteins were separated by SDS–PAGE and visualized by autoradiography.
Figure 5.
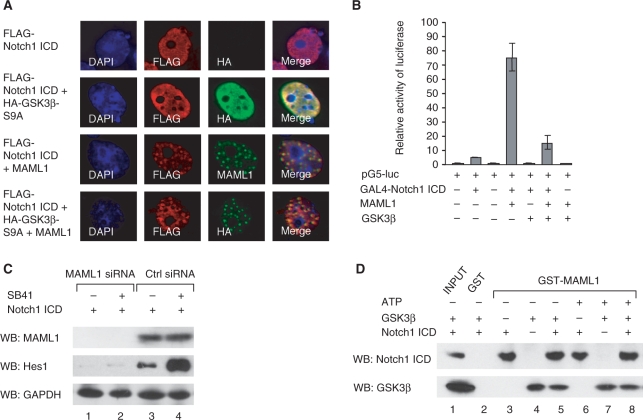
The MAML1-Notch1 ICD interaction is not abolished by GSK3β. (A) Vectors expressing FLAG-Notch1 ICD, HA-GSK3β-S9A and MAML1 1–1016 were cotransfected into Cos7 cells, and after 24 h cells were immunostained with antibodies recognizing the FLAG- or HA-tags or MAML1. (B) HEK-293 cells were cotransfected with a luciferase reporter containing five GAL4 binding sites and vectors expressing GAL4-Notch1 ICD, MAML1 and GSK3β. (C) C33A cells were transfected with Notch1 ICD and siRNA MAML1 or control (ctrl) siRNA, and incubated in the presence or absence of SB41. Expression of MAML1, GAPDH and the Notch target gene Hes1 was analyzed with immunoblot with antibodies recognizing MAML1, Hes1 and GAPDH. (D) GST-MAML1 and GST were incubated, in the presence or absence of ATP, with affinity-purified HIS-GSK3β and FLAG-Notch1 ICD proteins as indicated, and MAML1-interacting proteins were detected with immunoblot.
In vitro kinase assay
GST-MAML1 proteins were incubated with recombinant HIS-GSK3β (purified from Sf9 cells) in buffer B (50 mM Tris–HCl, pH 7.5, 100 mM KCl, 10% glycerol, 0.1 mg/ml BSA, 1 mM DTT, 1 mM PMSF, Phosphatase Inhibitor Cocktail 2 (Sigma)) containing 100 µM cold ATP with or without 5 µCi [γ-32P]ATP at 30°C for 30 min. The kinase reactions were separated by SDS–PAGE and analyzed with autoradiography or staining with Pro-Q Diamond phosphoprotein staining kit (Invitrogen P-33301) following manufacturer's; protocol.
Transient transfections and immunoprecipitation
HEK-293 cells or U2OS cells were transiently transfected in 24-well plates with 20 ng Notch1 ICD, 300 ng GSK3β, 100 ng MAML1 and 100 ng of the reporter plasmid pG5-luc. After 48 h, cells were lysed in 250 µl Roche lysis buffer and the levels of luciferase were measured. The bars in Figures 1A, B, 2A, 3F and 5B represent mean values obtained from at least three independent experiments. For siRNA transfections, we used DharmaFECT Duo Transfection Reagent, and predesigned MAML1 SMARTpool set of four siRNA, from Dharmacon. Protein expression was analyzed by western blotting using antibodies recognizing MAML1 (Santa Cruz Biotechnology), Hes1 (Santa Cruz Biotechnology) and GAPDH (Ambion). For immunoprecipitation experiments, HEK-293 cells were transfected with MAML1 using TransIT-LT1 (Mirus). The cells were harvested 48 h after transfection, and lysed in RIPA buffer and affinity-purification of MAML1 was performed with M2-agarose beads (Sigma) (Figure 1C) or MAML1 antibody (Figure 2B). Immunoprecipitated proteins were analyzed by western blotting using the following antibodies: MAML1 (Bethyl laboratories), FLAG (Sigma) and GSK3β (BD Transduction Laboratories). In addition, C33A cells were lysed in RIPA buffer and endogenous MAML1 immunoprecipitated with an antibody recognizing MAML1 N-terminus (Santa Cruz Biotechnology). Immunoprecipitated proteins were analyzed by western blotting using antibodies recognizing MAML1 (Santa Cruz Biotechnology) and GSK3β (BD Transduction Laboratories).
Figure 2.
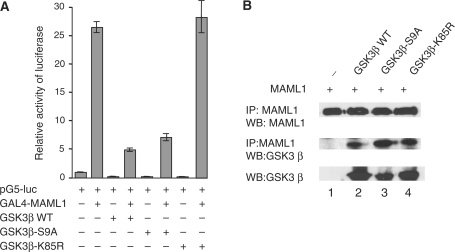
Active GSK3β regulates MAML1 transcription. (A) Vectors expressing GAL4-MAML1 and HA-GSK3β as WT, S9A or K85R, were cotransfected into HEK-293 cells with a luciferase reporter. (B) Vectors expressing MAML1 and GSK3β were cotransfected into HEK-293 cells as indicated, whole-cell extracts were prepared, and MAML1 protein immunoprecipitated. Proteins were separated by SDS–PAGE and detection of MAML1, and MAML1-interacting proteins, was monitored by immunoblot with antibodies recognizing MAML1 and GSK3β.
In vivo acetylation assay
HEK-293 cells stably expressing FLAG-MAML1 (f-MAML1 cell line) (20) and control HEK-293 cells were cultured in the absence or presence of SB41 (e.g. SB415286) for 24 h. The cells were lysed in RIPA lysis buffer and samples were analyzed by western blotting using the following antibodies: MAML1 (Bethyl Laboratories), acetylated histone H3 (Ac-H3) (Upstate Biotechnology), and Tubulin (Santa Cruz Biotechnology).
Immunostaining
Cos7 cells, 60–80% confluent, were transfected with vectors as indicated in the figures using TransIT-LT1 transfection reagent (Mirus) After 24 h, cells were fixed with 4% paraformaldehyde, washed with PBS and permeabilized with 1% Triton X-100 (Sigma) in PBS at RT. Slides were washed and blocked with 3% BSA (Sigma) in PBS for an hour at RT. After PBS wash, the slides were incubated with primary antibodies diluted in 3% BSA with 0.2% Triton X-100 in PBS, overnight at 4°C. Next day, slides were washed four times and incubated with FITC-conjugated rabbit or Cy3-conjugated mouse antibodies (Jackson ImmunoResearch Laboratories) diluted in PBS for 2 h at RT. Slides were washed four times and incubated with 0.1 µg/ml DAPI (Sigma) in PBS for 20 s. After three washes with PBS, slides were mounted with Immuno-Mount medium (Thermo) and analyzed with fluorescence microscope coupled with AxioCam digital camera (MRm), using Axio Vision software (Zeiss).
RESULTS
GSK3β decreases MAML1 transcriptional activity
To investigate if GSK3β regulates MAML1 activity, we transfected a GAL4 reporter gene with GAL4-MAML1 and GSK3β. As shown in Figure 1A, GAL4-MAML1 activated the reporter gene more than 50-fold, and expressing GSK3β together with MAML1 decreased MAML1 activity almost 4-fold. In the presence of the GSK3 inhibitor SB41, MAML1 transcriptional activation of the reporter gene (40-fold) was enhanced ∼3.5-fold (Figure 1B). Thus, our data suggest that GSK3β might target a coactivator, such as MAML1, in its regulation of gene expression. We next investigated if MAML1 and GSK3β associate in cells. HEK-293 cells were transfected with a vector expressing FLAG-MAML1, whole-cell extracts were prepared, and MAML1 protein affinity-purified with M2-agarose. As shown in Figure 1C, a significant amount of endogenous GSK3β co-immunoprecipitated with MAML1 (compare lanes 1 and 2). In addition, endogenous MAML1 protein was immunoprecipitated from C33A cells with an antibody recognizing the N-terminus MAML1 (Figure 1D). Endogenous GSK3β co-immunoprecipitated with MAML1 but not with the IgG control (compare lanes 2 and 3).
In order to examine if MAML1 directly and independently interacts with GSK3β, GST-tagged MAML1 was incubated with affinity-purified HIS-GSK3β. GSK3β significantly interacted with the GST-MAML1 full-length protein, but not with the GST negative control (Figure 1E, lanes 2 and 3). Consistently, we found that GST-GSK3β interacted with affinity-purified FLAG-MAML1 (Figure 1F, lane 3). In addition, we performed an in vitro kinase assay in which GST-MAML1 was incubated with HIS-GSK3β and [γ-32P]ATP in the presence or absence of the GSK3 inhibitor SB41 (Figure 1G). GSK3β strongly phosphorylated MAML1 in vitro (lane 2), and this was inhibited by SB41 (lane 3). Thus, the in vitro phosphorylation data indicated that MAML1 directly interacts with GSK3β.
GSK3β needs to be active to inhibit MAML1 activity
Phosphorylation of serine 9 in GSK3β has been shown to inactivate GSK3β and the lysine residue K85 is essential for GSK3β kinase activity. To explore if GSK3β needs to be in an active state to inhibit MAML1 activity, we cotransfected a GAL4 reporter gene and GAL4-MAML1 with GSK3β wild-type (WT), and a vector expressing GSK3β with serine 9 mutated to an alanine (GSK3β-S9A), which renders GSK3β constitutively active. We also used a construct expressing GSK3β with lysine 85 mutated to an arginine (GSK3β-K85R), which renders GSK3β kinase-dead. As shown in Figure 2A, GAL4-MAML1 increased expression of the reporter gene ∼26-fold, and both GSK3β WT and GSK3β-S9A inhibited this activity 5-fold. However, coexpressing GAL4-MAML1 with the inactive GSK3β-K85R did not significantly affect MAML1 activity, since the levels of transcription (28-fold), were similar to expressing GAL-MAML1 alone.
To determine if the activity status of GSK3β affects its interaction with MAML1 in cell culture, GAL4-MAML1 was cotransfected into HEK-293 cells with vectors expressing GSK3β WT, GSK3β-S9A or GSK3β-K85R. Whole-cell extracts were prepared, and MAML1 protein immunoprecipitated with an antibody recognizing the MAML1 C-terminus. MAML1, and GSK3β proteins interacting with MAML1, were detected by western blot, using antibodies against MAML1 and GSK3β. As shown in Figure 2B, the GSK3β antibody recognized all GSK3β proteins equally well (lanes 2–4). Immunoprecipitated MAML1 (lanes 1–4) interacted equally strongly with GSK3β WT, GSK3β-S9A and GSK3β-K85R (lanes 2–4).
The MAML1 N-terminus is a target of GSK3β
The MAML1 protein comprises various domains, and has acidic and basic regions (Figure 3A). We examined which domain(s) in MAML1 interact with GSK3β, and GST-tagged MAML1 proteins were incubated with whole-cell extract from HEK-293 cells transfected with HA-GSK3β. Figure 3B shows that GSK3β significantly interacted with GST-MAML1 full-length protein (lane 3) and MAML1 amino acids 1–300 (lane 4), but not with amino acids 309–625, 499–804, 701–1016 (lanes 5–7) or the GST negative control (lane 2). We continued to map GSK3β binding to MAML1 1–300 by incubating GST-tagged MAML1 domains 1–300, 1–200 and 1–127 with recombinant affinity-purified GSK3β. The data in Figure 3C show that GSK3β interacts equally well with these MAML1 truncations, suggesting that region 1–127 contains amino acids important for MAML1 binding to GSK3β. In addition, we performed an in vitro kinase assay and incubated GST-MAML1 1–1016, 1–300 and 309–625 with affinity-purified HIS-GSK3β and [γ-32P]ATP. GSK3β strongly phosphorylated MAML1 1–1016 and 1–300 (Figure 3D, lanes 1 and 2). However, GSK3β did not significantly phosphorylate MAML1 309–625 (lane 3), 499–804 or 701–1016 (data not shown), which are the regions in MAML1 that do not interact with GSK3β. GSK3β was then incubated with MAML1 1–300 and 1–127 in the presence of cold ATP, and phosphorylation of MAML1 determined with Pro-Q Diamond phosphoprotein staining. As shown in Figure 3E, GSK3β phosphorylated both MAML1 1–300 and 1–127. This indicates that MAML1 1–127 not only contains amino acids involved in GSK3β binding, but also comprises amino acid(s) that can be targeted for GSK3β phosphorylation.
To investigate if GSK3β inhibits the activity of the MAML1 N-terminus in cellular assays, we cotransfected cells with a GAL4 reporter gene, GAL4-MAML1 1–1016 or 1–300, and HA-GSK3β. As shown in Figure 3F, GSK3β decreased the transcriptional activity of both MAML1 1–1016 and 1–300 ∼4-fold.
MAML1 directs GSK3β to nuclear bodies
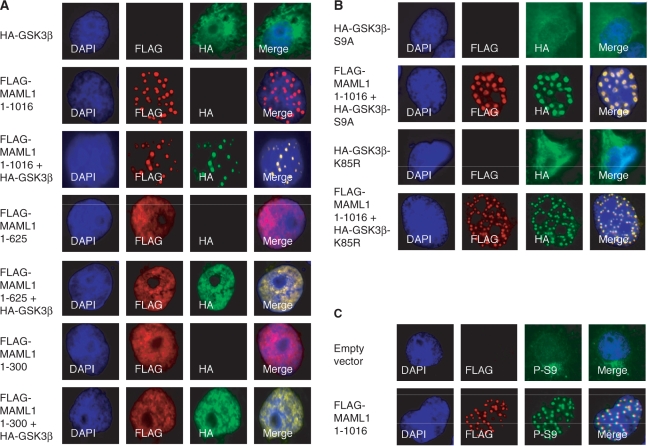
The MAML1 full-length protein has previously been shown to direct MAML1-interacting proteins such as Notch (2), p300 (16,20) and MEF2C (14) to nuclear bodies. In order to elucidate if MAML1 translocates GSK3β to nuclear bodies, Cos7 cells were cotransfected with HA-GSK3β and FLAG-MAML1 vectors and immunostained with antibodies recognizing the HA- and FLAG-tags (Figure 4A). Consistent with previously published data (16,20) MAML1 1–1016 was present in nuclear bodies (rows 2 and 3), while MAML1 1–625 and 1–300 were present in the nucleus but not in nuclear bodies (rows 4–7). In the absence of overexpressed MAML1, GSK3β seemed to be present mainly in the cytoplasm, but also appeared in the nucleus (row 1). When cells were cotransfected with GSK3β and MAML1 1–1016, MAML1 directed GSK3β to nuclear bodies (row 3). In cells transfected with GSK3β and MAML1 1–625 (rows 4 and 5) or MAML1 1–300 (rows 6 and 7), GSK3β was translocated to the nucleus but not present in nuclear bodies.
Figure 4.
MAML1 directs GSK3β to nuclear bodies. (A) Vectors expressing FLAG-MAML1 domains as indicated, and HA-GSK3β were cotransfected into Cos7 cells, and after 24 h the cells were immunostained with antibodies recognizing the FLAG- and HA-tags. (B) Vectors expressing HA-GSK3β-S9A, HA-GSK3β-K85R and FLAG-MAML1 were cotransfected into Cos7 cells, and cells were immunostained after 24 h with antibodies recognizing the FLAG- and HA-tags. (C) Cos7 cells were transfected with FLAG-MAML1 (or empty vector) and immunostained after 24 h with antibodies recognizing the FLAG-tag, and endogenous GSK3β phosphorylated at serine 9 (P-S9).
To further explore if MAML1 translocates both active and inactive forms of GSK3β to nuclear bodies, Cos7 cells were transfected with vectors expressing HA-GSK3β-S9A or HA-GSK3β-K85R and FLAG-MAML1. In the absence of transfected MAML1, GSK3β was present mainly in the cytoplasm, regardless of its activity status (Figure 4B, rows 1 and 3). FLAG-MAML1 strongly directed both HA-GSK3β-S9A and HA-GSK3β-K85R to nuclear bodies, and colocalized with these proteins (rows 2 and 4). To investigate if endogenous GSK3β was translocated to nuclear bodies by MAML1, Cos7 cells were transfected with FLAG-MAML1 or empty vector. Immunostaining with an antibody recognizing phosphorylated serine 9 in GSK (P-S9) showed that, in the absence of transfected MAML1, GSK3β was mainly present in the cytoplasm (Figure 4C, row 1). However, in the presence of transfected MAML1, endogenous GSK3β recognized by the P-S9 antibody was directed to nuclear bodies (row 2).
GSK3β does not inhibit the MAML1-Notch1 ICD interaction
Since GSK3β has been reported to potentiate or inhibit the activity of Notch proteins (Discussion), we decided to investigate how GSK3β affects MAML1-dependent Notch transcription. First, we transfected Cos7 cells with vectors expressing FLAG-Notch1 ICD, HA-GSK3β-S9A and MAML1, to see if Notch, MAML1 and active GSK3β colocalize in cells. In the absence of transfected MAML1, Notch1 ICD was widely distributed in the nucleus (Figure 5A, rows 1 and 2). When GSK3β-S9A was cotransfected with Notch1 ICD, GSK3β-S9A was directed to the nucleus (compare Figure 4B, row 1 to Figure 5A, row 2). Expressing MAML1 together with either Notch1 ICD or GSK3β-S9A directed Notch1 ICD (Figure 5A, row 3) and GSK3β-S9A (Figure 4B, row 2) to nuclear bodies. In Cos7 cells cotransfected with MAML1, Notch1 ICD and active GSK3β, all colocalized to these nuclear bodies (Figure 5, row 4).
We next cotransfected HEK-293 cells with a plasmid containing five GAL4 sites upstream of a luciferase gene and vectors expressing GAL4-Notch1 ICD, MAML1 1–1016 and GSK3β. GAL4-Notch1 ICD activated the reporter gene 5-fold, and the level of transactivation by Notch decreased almost 5-fold when cotransfected with GSK3β (Figure 5B). When we cotransfected MAML1 together with Notch, the levels of transcription were enhanced more than 15-fold, and expressing GSK3β together with Notch and MAML1 decreased the Notch activity ∼5-fold. Thus, we conclude that in our cell culture assay, GSK3β inhibits Notch1 ICD activity to the same extent in both the presence and absence of overexpressed MAML1. The GSK3 inhibitor LiCl has previously been reported to increase the mRNA levels of the Notch target gene Hes1 (33). Consistently, we found that expression of Hes1 protein was significantly potentiated in C33A cells cultured with SB41 (Figure 5C, compare lanes 3 and 4). When MAML1 expression was eliminated by MAML1 specific siRNA treatment, C33A cells expressed equally low levels of Hes1 in the absence or presence of SB41 (lanes 1 and 2), indicating that SB41-dependent increase in Hes1 protein levels might be dependent on the presence of MAML1.
We wanted to determine if GSK3β affects the interaction between MAML1 and Notch, and incubated GST-MAML1 with affinity-purified HIS-GSK3β and FLAG-Notch1 ICD, expressed in Sf9 cells via baculovirus. Figure 5D shows that GST-MAML1 interacted strongly, both in the absence or presence of ATP, with Notch1 ICD (lanes 3 and 6) and with GSK3β (lanes 4 and 7). GST-MAML1 interacted equally well with Notch and GSK when these three proteins were incubated together (compare lanes 5 and 8 to lanes 3–4 and 6–7). Furthermore, phosphorylation of MAML1 by GSK3β (in the presence of ATP) did not seem to affect MAML1 binding to GSK3β or Notch (compare lane 8 to lane 5).
GSK3 inhibits MAML1-dependent global histone acetylation
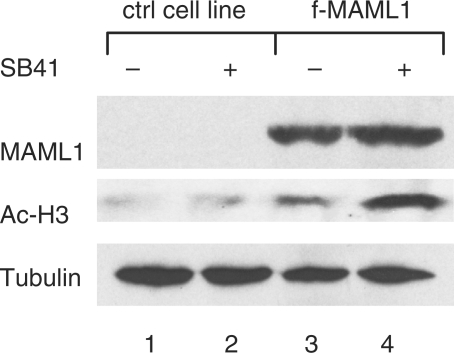
We previously reported that MAML1 increases global acetylation of histones in cell culture assay (20). To further explore if GSK3-mediated inhibition of MAML1 activity affects global acetylation of histones, we cultured HEK-293 cells and an HEK-293 cell line that stably expresses FLAG-MAML1 (f-MAML1), with the GSK3 inhibitor SB41. Consistent with our previous report, the data in Figure 6 show that histone acetylation increased in f-MAML1 cells (lanes 3 and 4), compared to HEK-293 control cells (lanes 1 and 2). SB41 increased acetylation of histone H3 significantly more in f-MAML1 cells compared to HEK-293 control cells (lanes 2 and 4). The expression level of tubulin, which is not an MAML1 target, was similar in both cell lines (lanes 1–4). Thus, our data indicated that GSK3 might be involved in regulating MAML1-dependent global acetylation of histones.
Figure 6.
GSK3β inhibits MAML1-dependent global histone acetylation. f-MAML1 or HEK-293 cells (ctrl cell line), were incubated in the presence or absence of SB41. The levels of expressed MAML1, acetylated histone H3 (Ac-H3), and tubulin, were monitored with immunoblot.
DISCUSSION
GSK3β, originally identified as a kinase that phosphorylates glycogen synthase, has been shown to phosphorylate, and regulate the activity of, many diverse proteins involved in several signaling pathways such as β-catenin, p53 and Notch (27,31–34). In this study we showed that GSK3β directly interacts with and decreases the activity of MAML1, which is a coactivator of Notch, p53 and β-catenin, (1–3,13,15). MAML1 interacted equally well with both the active and inactive states of GSK3β in a cellular assay, but GSK3β must be active to decrease MAML1 transcriptional activity. Thus, our data suggest that GSK3β might affect diverse signaling systems by targeting a coactivator involved in these pathways.
Inhibition of MAML1 activity by GSK3β required the N-terminus of MAML1, supporting the importance for this region, since the MAML1 N-terminus is reported to interact with Notch, p53, and MEF2C, and to play an important role in the transcription activity of these factors. Moreover, the MAML1 N-terminus has been shown to interact with histones (19) and p300 (16,17), and the MAML1 interaction with p300 increases p300 autoacetylation and HAT activity (although this requires the full-length MAML1 protein) (20). We therefore hypothesize that the N-terminus of MAML1 is involved in controlling the function of MAML1 in various signaling pathways, which might include the action of GSK3β. This does not necessarily include a physical competition for MAML1 between MAML1-interacting proteins. We found that GSK3β and Notch1 ICD do not seem to compete for MAML1 binding in cell culture assay or in vitro binding assay, and GSK3β phosphorylation of MAML1 and Notch1 ICD does not appear to affect the interaction between these proteins (Figure 5). Although GSK3β, MAML1 and Notch all colocalize in the same nuclear bodies, and these proteins do not seem to compete for binding to each other, it remains to be determined if GSK3β targets Notch and MAML1 individually, or targets a Notch-MAML1 complex, to regulate expression of genes.
MAML1 has been suggested to be phosphorylated (35), although no specific kinase has been shown to use MAML1 as a substrate. In this paper, we show that GSK3β phosphorylates amino acids within MAML1 1–300, in vitro. However, it will be crucial to identify amino acids in MAML1 that are targets for GSK3β phosphorylation, and to show that GSK3β can phosphorylate MAML1 in vivo. Since GSK3β prefers prior phosphorylation of its substrates, MAML1 is likely to be phosphorylated in vivo by another kinase. CDK8 is a kinase that is recruited by MAML1 to phosphorylate Notch, which facilitates Notch proteosomal degradation (18), so CDK8 could be involved in phosphorylating MAML1. So far, phosphorylation of Notch proteins has been correlated with Notch nuclear translocation (36,37), cellular transformation (38,39), and both activation (34) and inhibition (33) of Notch activity. A previous study demonstrated that GSK3β phosphorylates Notch1 ICD in vitro, Notch1 signaling is reduced in GSK3β deficient fibroblasts, and inhibition of endogenous GSK3β reduces the stability of Notch (34). Another report describes GSK3β phosphorylation of Notch2 ICD as negatively regulating Notch transcriptional activity (33). In addition, it was recently reported that Notch1 ICD can be negatively regulated by the GSK3α isoform (40). This study focused on GSK3β, and it remains to be explored if GSK3α plays a role in regulating MAML1 activity.
We found that MAML1 translocates GSK3β to nuclear bodies in the cell, and the GSK3β translocation requires the MAML1 full-length protein. Fryer and colleagues (16), and our previous work (20), showed that MAML1 full-length protein induces translocation of p300 to nuclear bodies, and that MAML1 protein contains a C-terminal region that is important for in vivo transcriptional activation. The MAML1 C-terminal amino acids 625–1016 appear to be important for MAML1 translocation of MAML1-interacting proteins to nuclear bodies, since MAML1 1–625 does not translocate p300 (20) or GSK3β (Figure 4) to nuclear bodies. Nuclear bodies are found in the nuclei of various cells, and are suggested to be involved in various cellular functions, such as transcriptional regulation and chromatin organization, post-translational modifications of proteins and identification and storage of proteins. In our cell culture assay MAML1 translocates both inactive and active GSK3β to nuclear bodies, and we speculate that nuclear bodies might act as an intracellular storage pool of GSK3β protein. In the absence of cotransfected MAML1 or Notch, GSK3β is mainly present in the cytoplasm. We showed that constitutively active GSK3β-S9A protein, can be translocated by Notch1 ICD to the nucleus (Figure 5), and this is in agreement with a previous study showing that Notch2 ICD translocates GSK3β WT to the nucleus (33). Interestingly, while MAML1 1–1016 translocates GSK3β to nuclear bodies, MAML1 1–625 and 1–300 translocate GSK3β to the nucleus, indicating that perhaps two regions in MAML1 are involved in translocating proteins in the cell. The N-terminus of MAML1 (aa 1–300) appears to comprise a region that directs translocation of cytoplasmic GSK3β to the nucleus, while the MAML1 C-terminus (aa 625–1016) has a region that is crucial for translocation to nuclear bodies.
We found that the specific GSK3 inhibitor SB41 significantly increases MAML1 activity in cellular cultures, indicating that MAML1 is most likely inhibited by endogenous GSK3. Furthermore, GSK3β appeared to interfere with MAML1-dependent global acetylation of histones. Cell immunostaining revealed that endogenous GSK3β and MAML1 associate in nuclear bodies, and we also found that GSK3β and MAML1 interact in cells when both proteins are present at physiological concentrations. Future studies will focus on elucidating the MAML1 mechanism for transporting GSK3β to nuclear bodies, and the GSK3β function in these cellular structures.
FUNDING
Swedish Research Council, the Swedish Cancer Society and the Swedish Children's; Cancer Foundation grants to A.E.W; fellowship from the Swedish Children's; Cancer Foundation to M.J.L. Funding for open access charge: The Swedish Cancer Society.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENT
Dr A. Bigas and Dr J. Woodgett are acknowledged for the gifts of expression plasmids described in the ‘Materials and methods’ section.
REFERENCES
- 1.Petcherski AG, Kimble J. Mastermind is a putative activator for Notch. Curr. Biol. 2000;10:R471–R473. doi: 10.1016/s0960-9822(00)00577-7. [DOI] [PubMed] [Google Scholar]
- 2.Wu L, Aster JC, Blacklow SC, Lake R, Artavanis-Tsakonas S, Griffin JD. MAML1, a human homolog of Drosophila mastermind, is a transcriptional coactivator for NOTCH receptors. Nat. Genet. 2000;26:484–489. doi: 10.1038/82644. [DOI] [PubMed] [Google Scholar]
- 3.Kitagawa M, Oyama T, Kawashima T, Yedvobnick B, Kumar A, Matsuno K, Harigaya K. A human protein with sequence similarity to Drosophila mastermind coordinates the nuclear form of notch and a CSL protein to build a transcriptional activator complex on target promoters. Mol. Cell Biol. 2001;21:4337–4346. doi: 10.1128/MCB.21.13.4337-4346.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lin SE, Oyama T, Nagase T, Harigaya K, Kitagawa M. Identification of new human mastermind proteins defines a family that consists of positive regulators for notch signaling. J. Biol. Chem. 2002;277:50612–50620. doi: 10.1074/jbc.M209529200. [DOI] [PubMed] [Google Scholar]
- 5.Wu L, Sun T, Kobayashi K, Gao P, Griffin JD. Identification of a family of mastermind-like transcriptional coactivators for mammalian notch receptors. Mol. Cell Biol. 2002;22:7688–7700. doi: 10.1128/MCB.22.21.7688-7700.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: cell fate control and signal integration in development. Science. 1999;284:770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- 7.Borggrefe T, Oswald F. The Notch signaling pathway: transcriptional regulation at Notch target genes. Cell Mol. Life. Sci. 2009 doi: 10.1007/s00018-009-8668-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bray SJ. Notch signaling: a simple pathway becomes complex. Nat. Rev. Mol. Cell Biol. 2006;7:678–689. doi: 10.1038/nrm2009. [DOI] [PubMed] [Google Scholar]
- 9.Talora C, Campese AF, Bellavia D, Felli MP, Vacca A, Gulino A, Screpanti I. Notch signaling and diseases: an evolutionary journey from a simple beginning to complex outcomes. Biochim. Biophys. Acta. 2008;1782:489–497. doi: 10.1016/j.bbadis.2008.06.008. [DOI] [PubMed] [Google Scholar]
- 10.Fortini ME, Artavanis-Tsakonas S. The suppressor of hairless protein participates in notch receptor signaling. Cell. 1994;79:273–282. doi: 10.1016/0092-8674(94)90196-1. [DOI] [PubMed] [Google Scholar]
- 11.Kurooka H, Honjo T. Functional interaction between the mouse notch1 intracellular region and histone acetyltransferases PCAF and GCN5. J. Biol. Chem. 2000;275:17211–17220. doi: 10.1074/jbc.M000909200. [DOI] [PubMed] [Google Scholar]
- 12.Oswald F, Tauber B, Dobner T, Bourteele S, Kostezka U, Adler G, Liptay S, Schmid RM. p300 acts as a transcriptional coactivator for mammalian Notch1. Mol. Cell Biol. 2001;21:7761–7774. doi: 10.1128/MCB.21.22.7761-7774.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhao Y, Katzman RB, Delmolino LM, Bhat I, Zhang Y, Gurumurthy CB, Germaniuk-Kurowska A, Reddi HV, Solomon A, Zeng MS, et al. The notch regulator MAML1 interacts with p53 and functions as a coactivator. J. Biol. Chem. 2007;282:11969–11981. doi: 10.1074/jbc.M608974200. [DOI] [PubMed] [Google Scholar]
- 14.Shen H, McElhinny AS, Cao Y, Gao P, Liu J, Bronson R, Griffin JD, Wu L. The Notch coactivator, MAML1, functions as a novel coactivator for MEF2C-mediated transcription and is required for normal myogenesis. Genes Dev. 2006;20:675–688. doi: 10.1101/gad.1383706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alves-Guerra MC, Ronchini C, Capobianco AJ. Mastermind-like 1 Is a specific coactivator of beta-catenin transcription activation and is essential for colon carcinoma cell survival. Cancer Res. 2007;67:8690–8698. doi: 10.1158/0008-5472.CAN-07-1720. [DOI] [PubMed] [Google Scholar]
- 16.Fryer CJ, Lamar E, Turbachova I, Kintner C, Jones KA. Mastermind mediates chromatin-specific transcription and turnover of the Notch enhancer complex. Genes Dev. 2002;16:1397–1411. doi: 10.1101/gad.991602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wallberg AE, Pedersen K, Lendahl U, Roeder RG. p300 and PCAF act cooperatively to mediate transcriptional activation from chromatin templates by notch intracellular domains in vitro. Mol. Cell Biol. 2002;22:7812–7819. doi: 10.1128/MCB.22.22.7812-7819.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fryer CJ, White JB, Jones KA. Mastermind recruits CycC:CDK8 to phosphorylate the Notch ICD and coordinate activation with turnover. Mol. Cell. 2004;16:509–520. doi: 10.1016/j.molcel.2004.10.014. [DOI] [PubMed] [Google Scholar]
- 19.Saint Just Ribeiro M, Hansson ML, Wallberg AE. A proline repeat domain in the Notch coactivator MAML1 is important for the p300-mediated acetylation of MAML1. Biochem. J. 2007;404:289–298. doi: 10.1042/BJ20061900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hansson ML, Popko-Scibor AE, Saint Just Ribeiro M, Dancy BM, Lindberg MJ, Cole PA, Wallberg AE. The transcriptional coactivator MAML1 regulates p300 autoacetylation and HAT activity. Nucleic Acids Res. 2009;37:2996–3006. doi: 10.1093/nar/gkp163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Frame S, Cohen P. GSK3 takes centre stage more than 20 years after its discovery. Biochem. J. 2001;359:1–16. doi: 10.1042/0264-6021:3590001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kockeritz L, Doble B, Patel S, Woodgett JR. Glycogen synthase kinase-3 – an overview of an over-achieving protein kinase. Curr. Drug Targets. 2006;7:1377–1388. doi: 10.2174/1389450110607011377. [DOI] [PubMed] [Google Scholar]
- 23.Force T, Woodgett JR. Unique and overlapping functions of GSK-3 isoforms in cell differentiation and proliferation and cardiovascular development. J. Biol. Chem. 2009;284:9643–9647. doi: 10.1074/jbc.R800077200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cohen P, Frame S. The renaissance of GSK3. Nat. Rev. Mol. Cell Biol. 2001;2:769–776. doi: 10.1038/35096075. [DOI] [PubMed] [Google Scholar]
- 25.Ikeda S, Kishida S, Yamamoto H, Murai H, Koyama S, Kikuchi A. Axin, a negative regulator of the Wnt signaling pathway, forms a complex with GSK-3beta and beta-catenin and promotes GSK-3beta-dependent phosphorylation of beta-catenin. EMBO J. 1998;17:1371–1384. doi: 10.1093/emboj/17.5.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yamamoto H, Kishida S, Kishida M, Ikeda S, Takada S, Kikuchi A. Phosphorylation of axin, a Wnt signal negative regulator, by glycogen synthase kinase-3beta regulates its stability. J. Biol. Chem. 1999;274:10681–10684. doi: 10.1074/jbc.274.16.10681. [DOI] [PubMed] [Google Scholar]
- 27.Yost C, Torres M, Miller JR, Huang E, Kimelman D, Moon RT. The axis-inducing activity, stability, and subcellular distribution of beta-catenin is regulated in Xenopus embryos by glycogen synthase kinase 3. Genes Dev. 1996;10:1443–1454. doi: 10.1101/gad.10.12.1443. [DOI] [PubMed] [Google Scholar]
- 28.Pulverer BJ, Fisher C, Vousden K, Littlewood T, Evan G, Woodgett JR. Site-specific modulation of c-Myc cotransformation by residues phosphorylated in vivo. Oncogene. 1994;9:59–70. [PubMed] [Google Scholar]
- 29.Sears R, Nuckolls F, Haura E, Taya Y, Tamai K, Nevins JR. Multiple Ras-dependent phosphorylation pathways regulate Myc protein stability. Genes Dev. 2000;14:2501–2514. doi: 10.1101/gad.836800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hoeflich KP, Luo J, Rubie EA, Tsao MS, Jin O, Woodgett JR. Requirement for glycogen synthase kinase-3beta in cell survival and NF-kappaB activation. Nature. 2000;406:86–90. doi: 10.1038/35017574. [DOI] [PubMed] [Google Scholar]
- 31.Watcharasit P, Bijur GN, Song L, Zhu J, Chen X, Jope RS. Glycogen synthase kinase-3beta (GSK-3beta) binds to and promotes the actions of p53. J. Biol. Chem. 2003;278:48872–48879. doi: 10.1074/jbc.M305870200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Turenne GA, Price BD. Glycogen synthase kinase3 beta phosphorylates serine 33 of p53 and activates p53's; transcriptional activity. BMC Cell Biol. 2001;2:12. doi: 10.1186/1471-2121-2-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Espinosa L, Ingles-Esteve J, Aguilera C, Bigas A. Phosphorylation by glycogen synthase kinase-3 beta down-regulates Notch activity, a link for Notch and Wnt pathways. J. Biol. Chem. 2003;278:32227–32235. doi: 10.1074/jbc.M304001200. [DOI] [PubMed] [Google Scholar]
- 34.Foltz DR, Santiago MC, Berechid BE, Nye JS. Glycogen synthase kinase-3beta modulates notch signaling and stability. Curr. Biol. 2002;12:1006–1011. doi: 10.1016/s0960-9822(02)00888-6. [DOI] [PubMed] [Google Scholar]
- 35.Jeffries S, Robbins DJ, Capobianco AJ. Characterization of a high-molecular-weight Notch complex in the nucleus of Notch(ic)-transformed RKE cells and in a human T-cell leukemia cell line. Mol. Cell Biol. 2002;22:3927–3941. doi: 10.1128/MCB.22.11.3927-3941.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Redmond L, Oh SR, Hicks C, Weinmaster G, Ghosh A. Nuclear Notch1 signaling and the regulation of dendritic development. Nat. Neurosci. 2000;3:30–40. doi: 10.1038/71104. [DOI] [PubMed] [Google Scholar]
- 37.Shimizu K, Chiba S, Hosoya N, Kumano K, Saito T, Kurokawa M, Kanda Y, Hamada Y, Hirai H. Binding of Delta1, Jagged1, and Jagged2 to Notch2 rapidly induces cleavage, nuclear translocation, and hyperphosphorylation of Notch2. Mol. Cell Biol. 2000;20:6913–6922. doi: 10.1128/mcb.20.18.6913-6922.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chiang MY, Xu ML, Histen G, Shestova O, Roy M, Nam Y, Blacklow SC, Sacks DB, Pear WS, Aster JC. Identification of a conserved negative regulatory sequence that influences the leukemogenic activity of NOTCH1. Mol. Cell Biol. 2006;26:6261–6271. doi: 10.1128/MCB.02478-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ronchini C, Capobianco AJ. Notch(ic)-ER chimeras display hormone-dependent transformation, nuclear accumulation, phosphorylation and CBF1 activation. Oncogene. 2000;19:3914–3924. doi: 10.1038/sj.onc.1203719. [DOI] [PubMed] [Google Scholar]
- 40.Jin YH, Kim H, Oh M, Ki H, Kim K. Regulation of Notch1/NICD and Hes1 expressions by GSK-3alpha/beta. Mol. Cells. 2009;27:15–19. doi: 10.1007/s10059-009-0001-7. [DOI] [PubMed] [Google Scholar]