Abstract
Double strand breaks (DSB) are severe DNA lesions, and if not properly repaired, may lead to cell death or cancer. While there is considerable data on the repair of simple DSB (sDSB) by non-homologous end-joining (NHEJ), little is known about the repair of complex DSBs (cDSB), namely breaks with a nearby modification, which precludes ligation without prior processing. To study the mechanism of cDSB repair we developed a plasmid-based shuttle assay for the repair of a defined site-specific cDSB in cultured mammalian cells. Using this assay we found that repair efficiency and accuracy of a cDSB with an abasic site in a 5′ overhang was reduced compared with a sDSB. Translesion DNA synthesis (TLS) across the abasic site located at the break prevented loss of DNA sequences, but was highly mutagenic also at the template base next to the abasic site. Similar to sDSB repair, cDSB repair was totally dependent on XrccIV, and altered in the absence of Ku80. In contrast, Artemis appears to be specifically involved in cDSB repair. These results may indicate that mammalian cells have a damage control strategy, whereby severe deletions are prevented at the expense of the less deleterious point mutations during NHEJ.
INTRODUCTION
Double strand breaks (DSBs) are among the most detrimental types of DNA damage, and if not properly repaired, can lead to cell death or cancerous transformation (1). DSB are repaired in mammalian cells primarily by non-homologous end joining (NHEJ), in which the two broken parts of the chromosomes are ligated (2–4). The core components in NHEJ include the Ku80/70 heterodimer, which protects DNA ends from degradation, and recruits other components of the repair machinery (5–8). The DNA-dependent protein kinase catalytic subunit (DNA-PKcs) joins Ku to form the DNA-PK holoenzyme, which connects the two termini of the break, and phosphorylates proteins involved in DNA damage response. Ligation is carried out by the DNA ligase IV complex, composed of DNA Ligase IV, XRCC IV and Cernunnos/XLF all of which are essential for NHEJ (8–10). In addition to the core components of NHEJ, other enzymes participate in DSB repair mainly through DNA end processing prior to ligation. This includes the nuclease Artemis, which is activated by DNA-PK and involved in the repair of some DSBs (6,11,12), and DNA polymerases µ and λ, both implicated in DNA end-processing prior to ligation (6,13–16).
Most studies on NHEJ addressed simple DSB (sDSB), namely DNA breaks which did not include chemical modification of the base, sugar or phosphate moieties near the break point. However, some DNA damaging agents, most notably ionizing radiation, cause clusters of DNA damage, which often lead to modification in the vicinity of a DSB (17). For example, Winter and his colleagues have reported that abasic sites as well as other oxidized nucleotide derivatives reside in the vicinity of a DSB that was formed by ionizing radiation (18,19). The presence of modified bases in the vicinity of the DSB can interfere with its repair, and may therefore require special processing. Indeed, it has been observed that some DSB have slow repair kinetics in vivo and in vitro, suggesting that their chemical structure hampers their repair (11,20). Despite their importance little is known on how cells cope with such complex DSB (cDSB), and specifically on how they are processed by the DSB repair machinery. Here we present a novel plasmid-based assay for the repair of a model cDSB in mammalian cells. We found that: (i) the presence of a single synthetic abasic site on a short 5′-overhang on one side of a DSB reduces the efficiency of DSB repair; (ii) translesion DNA synthesis across the abasic site enables NHEJ with minimal loss of DNA bases; (iii) remarkably, full-length TLS at the DNA terminus is highly mutagenic at the nucleotide following the abasic site, and it violates the A-rule at the abasic site; (iv) XrccIV and Ku80 are involved in the repair of both sDSB and cDSB, whereas Artemis is involved specifically in the repair of cDSB in this assay system.
MATERIALS AND METHODS
Cell cultures and media
The human cell lines H1299 and PC3 were derived from a large cell lung carcinoma and a prostate carcinoma, respectively. They were cultured in RPMI 1640 supplemented with 10% FBS (GIBCO/BRL). 48BR normal and Artemis-deficient CJ179, telomerase-immortalized fibroblasts (11,20) were grown in MEM medium with 10% FBS. Human Artemis deficient Guetel cells and their isogenic control Guetel/DA4 are skin fibroblasts transformed with SV40 and immortalized with telomerase (21). They were grown in RPMI 1640 10% FCS. The SV40-transformed NBS1 fibroblasts and their complemented controls (22) were grown in DMEM media supplemented with 10% fetal bovine serum, 2 mM Glutamine, 100 units/ml penicillin and 100 µg/ml streptomycin. The CHO cell lines K1 (wild-type) and Xrs5 (Ku80−/−) were purchased from ATCC, and XR-1 (Xrcc4−/−) from Coriell cell repositories. All CHO cells were grown either in MEMα or HAM medium with 10% FCS. All the cells were incubated at 37°C in a 5% CO2 atmosphere.
DNA substrates
Construction of the linear plasmids with complex and simple DSB
The modified-linear plasmid LP41 was constructed by ligating a short duplex oligonucleotide carrying a site-specific synthetic abasic site to a restriction nuclease-cleaved plasmid (Figure 1s). The short duplex oligonucleotide was prepared by annealing oligonucleotides 5′-AXCAGACCTGCGTGTACCG-3′ (X is the abasic site) and 5′-ACACGCAGGTCTG-3′ (200 pmol each) in 20 µl of a solution containing 10 mM Tris–HCl (pH 7.5), 1 mM EDTA, and 150 mM NaCl by heating to 70°C for 10 min, and then cooling to room temperature over a period of 2–3 h. The control non-modified linear plasmid LP40 was prepared in a similar way, except that the ligation was to a control short duplex oligonucleotide prepared by annealing onligonucleotides 5′-AACAGACCTGCGTGTACCG-3′ and 5′-ACACGCAGGTCTG-3′. The vector was the 3000 bp BstXI-BglI fragment of plasmid pSKSL (23), to which EcoRV site was introduced (pSKSL-EcoRV-Bgl1) (Figure 1s). The vector was obtained by cleavage of the plasmid with BstXI and BglI (see Figure 1sA), followed by fractionation on a 0.8% agarose gel, and purification by electroelution using the BIOTRAP* device (Schleicher & Schuell).
Preparative ligation of the insert to the vector was carried out in a 7.5 : 1 molar ratio, with 350 ng/µl vector DNA, and 1.5 unit/µl T4 ligase (New England Biolabs) at 16°C overnight. Next, the ligase was heat inactivated at 65°C for 10 min, and the efficiency of ligation was examined. This was done by digesting 1 µg of the linear plasmid with restriction nuclease Afe 1 for 3 h, followed by dephosphorelation, and then 5′ end labeling with 32P using polynucleotide kinase and [γ-32P] ATP. The fragments were then separated on 6% PAGE. The ligated DNA fragment migrates slower than the non-ligated one. Ligation efficiency was at least 90% as determined by phosphorimaging (Figure 2s). After confirming the ligation efficiency the linear plasmid was digested with EcoRV, which removed 10 nucleotides from one terminus, leaving a blunt end (Figures 1B and 1sC). Finally, the DNA was again gel purified. Plasmid pSA26 (3356 bp long; cmR) was previously described (23).
Figure 1.
Quantitative assay for the repair of cDSB in cultured mammalian cells. (A) Flow chart of the assay. Cells were transfected with a DNA mixture containing a modified linear plasmid with an abasic site on a 5′-overhang (LP41, kanR), a normalizing intact plasmid (pSA26, cmR) and a carrier plasmid (pUC18, ampR). In parallel, cells were transfected with a control mixture containing a non-modified linear plasmid with a 5′-overhang but no abasic site (LP40, kanR). Cells were incubated to allow repair, after which the plasmids were extracted and elctroporated into indicator E. coli cells. Finally, the bacteria cells were seeded in parallel on LB plates containing kanamycin or chloramphenicol. The relative repair of the cDSB relative to the sDSB was deduced from the colonies count, as described in the text. LP40 and LP41 descendent plasmids were recovered from the bacteria colonies, and subjected to DNA sequence analysis at the vicinity of the original break point. (B) The break points of of the linear plasmids carrying a cDSB (LP41) and a sDSB (LP40).
NHEJ assay for cDSB in cultured cells
The assay for the repair of cDSB included the following steps (Figure 1): (i) Transfection of the mammalian cells with a plasmid mixture containing the modified linear plasmid (LP41; kanR), the internal control plasmid pSA26 (cmR), and the carrier plasmid pUC18. As a control, a parallel transfection was performed with the non-modified linear plasmid, LP40, pSA26 and pUC18. (ii) Extraction of the plasmids from the mammalian cells. (iii) Transformation of an Escherichia coli indicator strain with the plasmid mixture. (iv) Deduction of the extent of cDSB repair from the number of transformants. More specifically, mammalian cells were co-transfected with a DNA mixture containing 150–350 ng of linear plasmid, 3–24 ng of the internal control plasmid pSA26, and 10 µg of the carrier plasmid pUC18. Transfection of H1299 cells was done using the JetPEI (Polyplus transfection), whereas other cells were transfected by electroporation using the Nucleofector system (Amaxa; Koln, Germany) with Nucleofector kits T (for CHO cells) and V and R (for other human cells). The cells were incubated in their appropriate medium for 16–24 h at 37°C in a 5% CO2 atmosphere. At the end of the incubation the cells were collected, washed three times with PBS and their plasmid content was extracted by using a plasmid purification kit based on alkaline lysis (Wizard miniprep, Promega). The recovered plasmids were electroporated into 150 µl of competent E. coli JM109recA cells, followed by incubation in 1 ml SOC medium at 37°C for 1 h, and then plating in parallel on LB-agar plates containing kanamycin (50 µg/ml) or chloramphenicol (30 µg/ml). Relative repair of the cDSB was calculated based on the number of colonies, as described below. When desired, plasmids were extracted from kanR colonies originating from several independent transfection experiments, and subjected to DNA sequence analysis at the vicinity of the break point.
RESULTS
Outline of the assay for cDSB repair in cultured cells
The cDSB repair assay measures the ability of cells in culture to circularize a transfected linearized plasmid that carries on one end a two-nucleotides 5′ overhang with a site-specific abasic site next to the terminal nucleotide, whereas the other end is blunt (LP41, kanR; Figure 1). In order to enable quantification, the mammalian cells were co-transfected with a plasmid mixture containing the modified linearized plasmid (LP41, kanR), an intact plasmid as an internal normalizing reference (pSA26; cmR), and a carrier plasmid (pUC18). As a control, cells were transfected in parallel with a similar mixture, wherein the modified linearized plasmid was replaced with a non-modified linearized control plasmid (LP40 kanR). The measurement of the linear plasmid repair was done in a subsequent step, in which the plasmid mixture was isolated from the mammalian cells, and used to transform an indicator E. coli strain. The transformants were plated in parallel on LB plates containing kanamycin, to select for cells harboring repaired plasmids that had originally carried a DSB, or chloramphnicol, to select for cells carrying the normalizing intact plasmid (Figure 1A). Linear plasmids poorly transform E. coli, and therefore the E. coli colonies observed had been formed by a circular (repaired) plasmid molecule. To further decrease the background transformation of linear plasmids, isolation of the plasmid content of the mammalian cells was done under mild alkaline conditions followed by neutralization, which cause denaturation of linear, but not covalently closed circular plasmids.
The efficiency of DSB repair in mammalian cells was calculated based on the number of E. coli transformants. First, the number of LP41 kanR transformants was divided by the number of pSA26 cmR transformants, to yield a normalized DSB repair efficiency for the cDSB. The same was done for the pair of LP40 and pSA26 transformants, to obtain the normalized DSB repair efficiency for the sDSB. We then divided the two to get the relative repair efficiency of the cDSB compared with sDSB. Importantly, none of the plasmids can be replicated within the mammalian cells, hence number of E. coli transformants correlates to the extent of repair within the mammalian cells.
A single abasic site near a DSB reduces both the efficiency and accuracy of DSB repair
To examine whether linearized plasmids with a cDSB are repaired in human cells, we performed a series of experiments with the human lung cancer cell line H1299. As can be seen in Table 1, which presents the results of 7 independent experiments, the number of kanR and cmR E. coli tranformants obtained after extraction of the plasmids mixture from the human cells varied considerably among experiments. However, the relative repair of cDSB (relative to the sDSB) was remarkably reproducible, yielding a value of 20 ± 2%. Thus, the assay yields quantitative and reproducible measurements of the repair of the cDSB relative to the sDSB. The value obtained indicates that the presence of a single modification near the break points significantly decreased its repair by 5-fold relative to a sDSB.
Table 1.
Repair of complex DSB (Substrate LP41) relative to simple DSB (Substrate LP40) in human H1299 cells
| Experiment no. | Number of colonies |
KanR/CmR | Relative repaira (%) | ||
|---|---|---|---|---|---|
| KanR | CmR | ||||
| No passageb | LP41 | 0 | 3200 | <3 × 10−4 | NA |
| LP40 | 0 | 2272 | <5 × 10−4 | ||
| 1 | LP41 | 182 | 834 | 0.22 | 21 |
| LP40 | 1860 | 1782 | 1.04 | ||
| 2 | LP41 | 94 | 1448 | 0.065 | 16 |
| LP40 | 147 | 352 | 0.41 | ||
| 3 | LP41 | 359 | 283 | 1.27 | 20 |
| LP40 | 416 | 65 | 6.4 | ||
| 4 | LP41 | 62 | 100 | 0.62 | 19 |
| LP40 | 285 | 88 | 3.2 | ||
| 5 | LP41 | 233 | 311 | 0.75 | 17 |
| LP40 | 325 | 72 | 4.5 | ||
| 6 | LP41 | 584 | 896 | 0.65 | 22 |
| LP40 | 2784 | 962 | 2.9 | ||
| 7 | LP41 | 90 | 638 | 0.14 | 22 |
| LP40 | 361 | 572 | 0.63 | ||
| Average relative repair: | 20 ± 2 | ||||
Human H1299 cells were transfected with a mixture of linear plasmid LP41 with an abasic site in a 5′ overhang, the normalizing intact plasmid pSA26, and the carrier plasmid pUC18. A parallel transfection was conducted with a mixture of the control linear plasmid LP40, which had no abasic site, along with the control and carrier plasmids. After 16 h the plasmids were extracted using an alkaline procedure, and used to transform E. coli recA cells, which were then seeded in parallel on LB plates containing kanamycin (to select for repaired LP41 or LP40 plasmids) or chloramphenicol (to select for the normalizing plasmid pSA26). The repair of cDSB relative to sDSB was calculated by dividing the kanR/cmR colonies ratio obtained for LP41, by the ratio obtained for LP40. See text for details.
aRelative repair of cDSB relative to sDSB.
bThe plasmid mixture was used to directly transform the E. coli recA cells without prior passage through the H1299 human cells.
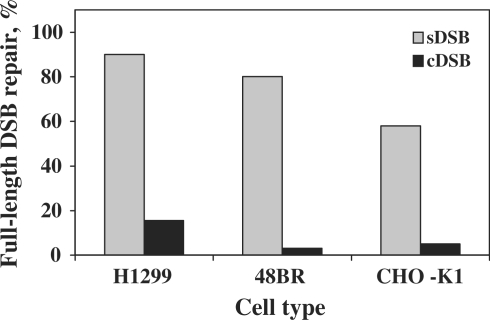
To analyze the accuracy of DSB repair we performed DNA sequence analysis of repaired descendents of the two types of linearized plasmids. As can be seen in Figure 2, 90% (45/50 events) of the sDSB (substrate LP40) were accurately repaired in H1299 cells, with restoration of the original DNA sequence. Largely accurate repair of sDSB was observed also in human 48BR cells (80%), and to lesser extent in the CHO K1 cells (57%; Figure 2). The picture dramatically changed for cDSB (substrate LP41). In H1299 cells only 16% (19/123 events) of the repair events preserved the correct length of the plasmid, whereas 84% contained either deletions or insertions. Similarly, in human 48BR and CHO K1 cells the original DNA length was rarely preserved, and the vast majority of events included deletions and insertions (>96 and 95%, respectively; Figure 2).
Figure 2.
Repair of sDSB and cDSB without loss of DNA sequences. The proportion of DSB repair events with no deletions or insertions was determined for sDSB and cDSB in the indicated cell lines. In human H1299 cells full-length repair accounted for 45/50 all sequenced sDSB repair isolates and 19/123 cDSB repair isolates; in human 48BR cells the respective fractions were 20/25 and 0/25, and the Chinese hamster ovary cells CHO-K1 13/22 and 1/19. The data was obtained from experiments performed as described in the legends to Table 1. The sequences for each cell line were obtained from at least three different experiments.
Spectrum of repair events of cDSB in human cells
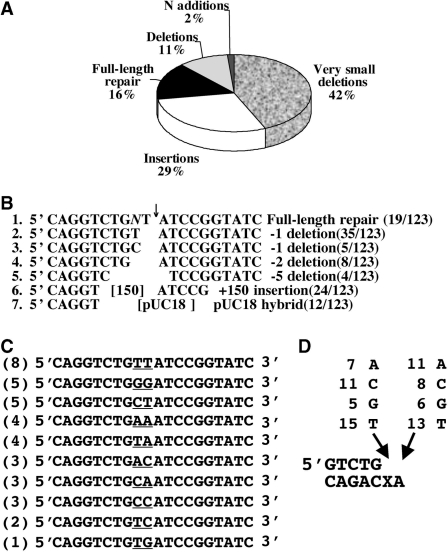
DNA sequence analysis of 123 plasmid isolates that have resulted from the repair of cDSB in human H1299 cells revealed a wide variety of mutations types (Figure 3). The most frequent class of events comprised very small deletions of up to 5 bp, and accounted for 43% of all repair events. Of these, eight contained −2 deletions, indicating precise elimination of the 5′ overhang in plasmid LP41 (line 4 in Figure 3B). Minus 1 deletions and full-length restoration comprised 39 and 19 events, respectively (Figure 3B). These events are the signature of the activity of a DNA polymerase(s) that skipped over the lesion and replicated the next template nucleotide, or performed full-length TLS, respectively (see below). Another mechanism that contributed to formation of deletions is microhomology-directed end joining (MHEJ). According to this mechanism the ligation of the two ends is mediated by short homologies of 1–4 bases, and therefore the DNA is chewed until the microhomology is found (line 5 in Figure 3B). Ten out of thirteen of larger deletion events are consistent with such a mechanism. Insertions constituted 29% of all the repair events (36/123) and could be divided into two subclasses: (i) insertion of 50–200 bp DNA fragments originating most likely from bovine genomic DNA that is present in the serum of the cells growth medium (24 out of 36 insertion events); (ii) hybrids with pUC18 that had served as carrier DNA in the assay.
Figure 3.
Spectrum of mutations at repaired sites of cDSB in human H1299 cells. (A) DNA sequence analysis was performed for 123 repair events of cDSB, and the results were classified according to repair scenario. (B) Examples of DNA sequences at the repaired cDSB. The numbers in parenthesis show the occurrence of each event out of 123 repair events. (C) DNA sequence analysis of plasmids with cDSB that had been repaired without any loss of nucleotides, suggestive of a mechanism of TLS-assisted NHEJ. The number of occurrence of each sequence is presented in the parentheses. The nucleotides inserted opposite the original 5′-overhang are underlined. (D) Summary of the identity and occurrence of the nucleotides inserted opposite and 3′ to the abasic site during the NHEJ of the linear plasmids with the cDSB.
To examine whether treatment of the cells by DNA damaging agents might affect the repair of cDSB, we repeated the experiments with γ-irradiated human H1299 cells. We observed no effect of the radiation on the relative efficiency, nor the accuracy of cDSB repair (data not shown). This is consistent with the lack of effect of pre-irradiating cells on another plasmid-based repair assay used in our laboratory, which measures translesion DNA synthesis (24). It may indicate that pre-induction of the DNA damage response does not affect NHEJ of cDSB, or else that the transfection process itself, being stressful to the cells, induces the relevant genes (25).
Full-length TLS across an abasic sites near a DSB lacks template sequence instruction
Those cDSB whose repair resulted restoration of full DNA length, a +1 addition, or a −1 deletion opposite the abasic site, must have occurred via a mechanism in which the abasic site was tolerated rather than removed. Since the substrate contained a two-nucleotide 5′ overhang on one side, and a blunt end on the other side, the simplest mechanism would have been TLS by a DNA polymerase, to generate a blunt end that is sealable. A total of 129 out of 232 repair events (56%) in H1299 cells, which involved loss of one (86 events) or no (38 events) nucleotides, or gain of one nucleotide (five events) are likely to have occurred by such a mechanism. The DNA sequences of the 38 full-length TLS products are shown in Figure 3C and D. Each of the four possible nucleotides was found at the position corresponding to the abasic site, suggesting that bypass synthesis across this site occurred without any particular specificity (Figure 3C and D). This is different from the preferential insertion of a dAMP opposite an abasic site observed by most DNA polymerases (the so-called A-rule; see ‘Discussion’ section). Surprisingly, the nucleotide next to the abasic site was highly mutated too. Only in 13 out of 38 cases (34%) was the correct nucleotide T inserted opposite the template A (Figure 3C and D). The majority of the insertion events at this site were mutagenic, with no strong bias for a specific nucleotide. This suggests that the polymerase(s) that perform the bypass synthesis acts in a manner that is template independent. However, the precise length synthesized (2 nt) suggests that if such a mechanism had operated, it is template length-dependent.
TLS prevents sequence losses during repair of a cDSB in various cell lines
To examine whether the TLS observed at the cDSB in H1299 cells is a general phenomenon, we analyzed DNA sequence changes caused during the repair of cDSB in several other cell lines. Table 2 presents the results of such experiments performed with the human prostate carcinoma cell line PC3, an SV40-transformed fibroblast cell line from an NBS1 patient, and the latter stably complemented with the NBS1 gene. In each cell line a significant fraction of repaired plasmids included the signature of TLS, primarily of full-length products and −1 deletions. One case of a single nucleotide insertion, which led to a net one-nucleotide increase in sequence length was observed too (Table 2), and is likely to result from TLS activity. The fraction of TLS events varied: 56% in H1299 cells, 53% in PC3 cells, 21% in NBS1– cells, and 17% in NBS1+ cells. The average for the 4 cell lines was 37%, of which about a one-third were full-length TLS events and the rest primarily −1 deletions, consistent with skipping across the abasic site.
Table 2.
Abundance of TLS-assisted NHEJ events in various cell lines
| Cell line | TLS event | DNA sequence | Occurrence |
|---|---|---|---|
| PC3 | Full-length | 5′CAGGTCTGTT↓ATCCGGTATC | 2 |
| Skipping | 5′CAGGTCTG-T ATCCGGTATC | 5 | |
| Skipping | 5′CAGGTCTG-A ATCCGGTATC | 1 | |
| Skipping | 5′CAGGTCTG-C ATCCGGTATC | 1 | |
| N addition | 5′CAGGTCTGGGGATCCGGTATC | 1 | |
| Total TLS events: | 10/19 (53%) | ||
| NBS1+ | Full length | 5′CAGGTCTGCA↓ATCCGGTATC | 2 |
| Full length | 5′CAGGTCTGTT ATCCGGTATC | 1 | |
| Skipping | 5′CAGGTCTG-T ATCCGGTATC | 2 | |
| Total TLS events: | 5/29 (17%) | ||
| NBS1− | Full length | 5′CAGGTCTGTA↓ATCCGGTATC | 2 |
| Full length | 5′CAGGTCTGAT ATCCGGTATC | 1 | |
| Skipping | 5′CAGGTCTG-T ATCCGGTATC | 2 | |
| Total TLS events: | 5/24 (21%) | ||
| H1299a | Full length | 5′CAGGTCTGNM↓ATCCGGTATC | 38 |
| Skipping | 5′CAGGTCTG-T ATCCGGTATC | 79 | |
| Skipping | 5′CAGGTCTG-N ATCCGGTATC | 7 | |
| N addition | 5′CAGGTCTGGGGATCCGGTATC | 5 | |
| Total TLS events: | 129/232 (56%) |
NHEJ experiments were conducted with the indicated cell lines, after which descendants of plasmids with a cDSB were isolated and subjected to DNA sequence analysis.
aThe data for H1299 cells was taken from Figure 3, from sequence data obtained with pre-irradiated H1299 cells which were described in the text (data not shown). The detailed sequence changes in the 38 NHEJ events associated with full-length TLS (schematically presented as NM) are presented in Figure 3C and D.
The repair of linear plasmids is dependent on XrccIV and altered in the absence of Ku80
In order to examine whether the repair of linear plasmids in our system depends on the known component of the NHEJ machinery, we performed the in vivo assay in XrccIV-deficient CHO XR1 cells, which are impaired in NHEJ, and sensitive to ionizing radiation (26,27). As a control we used the parent CHO K1 cells. As can be seen in Table 3, in K1 cells the numbers of kanR colonies, originating from repaired linearized substrates, was similar to that obtained in other cell types, yielding a relative repair of 56% for the cDSB relative to the sDSB. In the XR1 XrccIV -deficient cells the results were totally different. The number of cmR colonies was normal (hundreds/plate), however very few kanR colonies were observed (typically 0–6) for both the sDSB or cDSB substrates, indicating that these cells were severely deficient in the repair of both sDSB and cDSB (Table 3).
Table 3.
Repair of complex DSB (substrate LP41) relative to simple DSB (substrate LP40) in hamster and human cells defective in NHEJ genes
| Cell type/vector | Number of colonies (DNA amounta) |
Relative repair b % | |
|---|---|---|---|
| KanR | CmR | ||
| CHO K1 (wild-type) | |||
| LP41 | 203 (350) | 253 (24) | 56.1 ± 7.0 |
| LP40 | 393 (350) | 260 (24) | |
| CHO XR1 (XrccIV−/−) | |||
| LP41 | 6 (350) | 636 (24) | NA |
| LP40 | 2 (350) | 364 (24) | |
| CHO XRS5 (Ku80−/−) | |||
| LP41 | 106 (200) | 1228 (24) | 51.0 ± 4.9 |
| LP40 | 203 (200) | 1122 (24) | |
| Human Guetel/DA4 (Artemis+) | |||
| LP41 | 106 (200) | 1072 (12) | 35.3 ± 9.6 |
| LP40 | 278 (200) | 784 (12) | |
| Human Guetel (Artemis–) | |||
| LP41 | 54 (200) | 1164 (12) | 18. 5 ± 3.4 |
| LP40 | 242 (200) | 1084 (12) | |
| Human 48BR (Artemis+) | |||
| LP41 | 28 (300) | 666 (24) | 15 ± 4.9 |
| LP40 | 59 (300) | 205 (24) | |
| Human CJ179 (Artemis–) | |||
| LP41 | 28 (300) | 237 (24) | 7 ± 2 |
| LP40 | 108 (300) | 80 (24) | |
NHEJ assays were performed with the indicated cell lines as described in the legend to Table 1 and under ‘Materials and Methods’ section. The colonies numbers shown represent a typical experiment, whereas the relative repair represents the average obtained from three to six experiments.
aThe numbers in the parentheses show the amount of DNA (in nanograms) used to transfect the mammalian cells.
bRelative repair of cDSB relative to sDSB.
We examined the repair of sDSB and cDSB also in Ku80-deficient CHO XRS5 cells. Unlike in the XrccIV-deficient cells, DSB repair did occur in the Ku80-deficient cells, and the relative repair of the cDSB was similar to that observed in the Ku80-proficient K1 cells (Table 3). DNA sequence analysis of plasmids that underwent DSB repair showed that in Ku80-proficient cells, most repair events at sDSB did not cause any nucleotides loss at either the protruding or the blunt ends of the break (Figures 4A and 3s). The pattern of repair of cDSB was different: While about half of the blunt ends (53%) were preserved, most protruding ends were degraded (95% P = 0.001; Figures 4A and 3s), indicating that the blunt end was selectively protected compared with the complex protruding end. This selective protection was lost in the Ku80−/− cells, where sequences were lost from both sides of the cDSB, as well as the sDSB (Figures 4B and 3s). Analysis of the number of deletions larger than 10 bp showed a significant increase in Ku80−/− cells compared with Ku80+/+ cells for both sDSB (from 18 to 73% P = 0.0008) and cDSB (from 26 to 79% P = 0.003) (Figures 4 and 3s). Thus, although Ku80 does not affect the relative efficiency of repair of the cDSB, it does protect its ends from nucleolytic degradation.
Figure 4.
Accuracy of sDSB and cDSB repair in Ku80+/+and Ku80−/− cells. Occurrence of accurate and inaccurate repair events in CHO K1 Ku80+/+ cells (A) and Xrs5 Ku80−/− cells (B). The percentage of each type of repair event was calculated out of the total number of repair events including hybrids with pUC18, based on sequences presented in Figure 3s.
Deficiency in the Artemis nuclease modestly reduces the relative repair efficiency of cDSB
Since most of the cDSB repair events involved a nuclease activity, we examined whether the Artemis nuclease was involved. As can be seen in Table 3, the relative repair of cDSB was reduced by nearly 2-fold in Artemis-deficient cells compared with Artemis-complemented cells (18 ± 3 and 35 ± 10%, respectively; Table 3). DNA sequence analysis of linear plasmids with cDSB that underwent repair revealed similar patterns of repair events in Artemis-proficient and deficient cells (Figure 4s). However, the mean size of deletions larger than 2 bp was 78% longer in Artemis-proficient cells compared with Artemis-deficient cells (52.4 versus 29.4 bp, respectively; Figure 4s). Similar effects were obtained when another normal human cell (48BR) was compared with the Artemis-deficient cell line CJ179: The relative repair in the Artemis-deficient cells was 2-fold lower than in the normal cells (Table 3), and the average size of deletions larger than 2 bp was 58% longer in Artemis-proficient cells (42.2 bp) compared with Artemis-deficient cells (26.7 bp) (Figure 5s).
DISCUSSION
The presence of chemical modifications near DSB may interfere with mechanisms that function to seal the break, such as NHEJ. As a result, both the efficiency and the accuracy of the process may diminish. Obviously, nucleolytic processing can in principle eliminate chemically damaged DNA regions, and generate simple and ligatable ends, however, this will cause loss of DNA sequences near the break point. Although NHEJ is generally considered to be an error-prone repair process, mechanisms that minimize the mutagenic outcome of NHEJ and/or its severity may exist in mammalian cells. Indeed, several studies have shown that NHEJ is capable of aligning, annealing and patching partially complementary overhangs, thereby minimizing, or even avoiding loss of DNA sequences (6,13–16). However, the effect of chemical modifications on such processes is just beginning to emerge (28,29).
For such a purpose a plasmid assay system offers significant advantages, since it enables engineering of the DNA ends, including the incorporation of damaged nucleotides. Being a model assay system it is not expected to fully mimic the chromosomal repair of cDSB, but its relevance is indicated by its response to proteins known to be involved in chromosomal DSB repair, and its power is the ability to report on individual repair events of cDSB at a single nucleotide resolution in a large variety of mammalian cells. In the past, several predictions that were made by plasmid-based assays were later confirmed by chromosomal-based assays. For example the use of microhomology directed repair in NHEJ (15,30,31) and the end protection from degradation by the Ku complex (5,32).
DSB repair in our assay system was totally dependent on the XrccIV protein for both cDSB and sDSB. This is consistent with the critical role of the XrccIV-LigIV in NHEJ (26,27). As for Ku80, we observed similar extents of repair in Ku80-deficient and proficient cells, consistent with previous results with plasmid and chromosomal assays (5,26,32). However, degradation of the DNA ends was more extensive in Ku80-deficient cells, and the fraction of deletions longer than 10 bp significantly increased, consistent with the loss of the protection usually endowed by the Ku proteins. Interestingly, Ku80 suppressed the potentially deleterious repair-associated deletions also in cDSB repair, when accurate repair was not possible. Recently, it was reported that a DNA ligase IV-independent alternative end joining (AEJ) pathway is robust in mammalian cells (33–35). This pathway depends to great extent on long microhomology DNA termini, and therefore relies on the sequence context. It is possible that in some sequence contexts AEJ can repair cDSB with high efficiency but with a great risk to genomic stability. Interestingly, human cells deficient in the Artemis nuclease were specifically defective in the repair of cDSB. This was associated with shorter deletions compared with Artemis-complemented cells (Figures 4s and 5s).
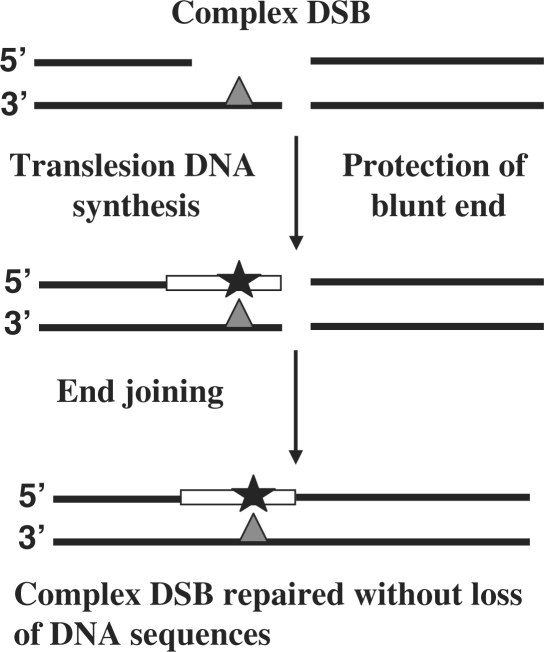
The DNA substrates that we have used carried DSB that were blunt on one side, and had a short two-nucleotides 5′ overhang on the other side. Such a configuration enables DNA synthesis to convert the protruding terminus into a blunt end, and subsequent ligation could lead to DSB repair with a minimal or no loss of DNA sequences (Figure 5). In human cells most of the repair events of sDSB indeed involved template-directed DNA synthesis, clearly indicating that the repair process that preserves the original DNA length was favored over the one that causes loss of DNA sequences, consistent with previous results (30). The presence of a single abasic site in the overhang caused a significant 5-fold decrease in the efficiency of DSB repair compared with the sDSB in H1299 cells. While abasic sites inhibit DNA synthesis in at least some mammalian cells, the significant decrease in repair efficiency was surprising, since a simple nucleolytic cleavage of the problematic overhang and beyond, might have easily solved the problem, and enabled ligation. The reason for this significant effect of a single abasic site on the relative repair of the DSB is not clear, however it might hint on the presence of molecular protection against loss of sequences from the end.
Figure 5.
Model for TLS-assisted NHEJ in mammalian cells. TLS across a DNA lesion located on a 5′ overhang in one side of a cDSB leads to the formation of a blunt end. If the other side of the DSB is blunt, this will enable repair of the cDSB without loss of nucleotides. See text for details.
With the abasic site present in the 5′ overhang, 56% of the DSB repair events in H1299 cells occurred without loss of DNA sequences or with a minimal one nucleotide deletion or addition at the site corresponding to the lesion. These sequences outcomes are hallmarks of TLS, with the main events being full-length bypass, or skipping over the lesion bypass. A similar picture was observed also for the prostate cancer cell line PC3, in which 53% of DSB repair events could be attributed to TLS-assisted NHEJ, whereas in the SV40-immortalized fibroblasts examined TLS was involved in 17–21% of NHEJ repair events (Table 2). Still, in some cell lines we observed only a small fraction of TLS. This may reflect a limitation of our plasmid assay, in which the DNA ends are likely to be less protected from accidental nucleolytic degradation compared with chromosomal cDSB.
Among the TLS events, skipping over the lesion was generally more abundant than full-length TLS. This is different from TLS across an abasic site opposite a gap, where the fraction of –1 deletions was smaller than full-length bypass (36). This difference might be attributed to the substrates difference, since a higher DNA flexibility of a single-stranded overhang compared with a short ssDNA segment embedded in a duplex DNA might facilitate misalignment and skipping by the polymerase. Alternatively, this may reflect the activity of the DNA polymerase(s) involved. In most skipping events, the template A 5′ to the abasic site was accurately copied. However, there were events in which skipping was accompanied by a mutation in the next nucleotide, which is generally consistent with the mutations caused at that site during TLS without skipping. Currently, there are no effective methods to study the repair of chromosomal cDSB at high resolution. However, it is possible that in chromosomes, where flexibility of the DNA is more restricted, the dominant TLS event during NHEJ of cDSB will be full-length bypass. In any case, the outcome of these events is a point mutation (base substitution, −1 deletion, or +1 addition), which is considerably less severe than long deletions.
An abasic site is an inherently miscoding lesion, and therefore in theory, any of the four nucleotides could be inserted opposite it by DNA polymerases. However, it is well documented that many prokaryotic and mammalian purified DNA polymerases tend to insert dAMP opposite abasic sites (the so-called ‘A-rule’) (37–39). Moreover, it was shown both in our laboratory and by others that dAMP is preferentially inserted opposite abasic sites in human cells (36,40), including H1299 cells used in this study (23). This implies that when operating near a cDSB, TLS in H1299 cells violates the A-rule. Interestingly, nucleotide insertion opposite the terminal nucleotide of the overhang appears to have occurred at random, without template instruction. Taken together with the events opposite the abasic site, it appears that TLS at the cDSB occurred via template-directed but sequence-independent polymerization activity. Several mammalian DNA polymerases were shown to be able to bypass an abasic site (38,39,41–44); however, we do not know yet which DNA polymerase carries out the TLS step during the NHEJ events monitored in this study. A potential candidate is DNA polymerase µ, which was implicated to be involved in NHEJ, and was shown by us to possess a template-dependent, but sequence-independent activity during TLS across a synthetic basic site (45). However, an abasic site is a strongly blocking lesion, which is bypassed slowly in human cells, and likely to require more than one polymerase (36). Specifically, in vivo experiments have shown that polζ (36) as well as an aphidiclin-sensitive polymerase (23) are involved, and in vitro experiments have implicated also REV1 (46) and polθ (47). The high mutability of TLS across and near an abasic site may be relevant to somatic hypermutation in the immune system, where abasic sites generated by the combined action of AID and UDG play a major role (48).
In summary, using a DNA substrate with a single abasic site on a 5′ overhang had a surprisingly big inhibitory effect on NHEJ in human cells, and uncovered a TLS-assisted NHEJ mechanism, that allowed repair of cDSB with diminished loss of DNA sequences, albeit with the concomitant formation of point mutations. To our knowledge this is the first demonstration of TLS-assisted NHEJ in mammalian cells. Further studies are needed in order to elucidate the mechanism and regulation of this process, which might represent a damage control strategy, whereby severe deletions are prevented at the expense of less deleterious point mutations.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Flight Attendant Medical Research Institute, Florida, USA grant; and the Israel Science Foundation grant (no 564/04 and 1136/08). Funding for open access charge: Flight Attendant Medical Research Institute, Florida, USA.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMETS
Z.L. is the incumbent of the Maxwell Ellis Professorial Chair in Biomedical Research. The authors thank Yosef Shiloh (Tel Aviv University, Israel) for NBS1 and complemented cells,
REFERENCES
- 1.Friedberg EC, Walker GC, Siede W, Wood RD, Schultz RA, Ellenberger T. DNA Repair and Mutagenesis. 2nd edn. Washington, DC: ASM Press; 2006. [Google Scholar]
- 2.Bassing CH, Alt FW. The cellular response to general and programmed DNA double strand breaks. DNA Repair (Amst) 2004;3:781–796. doi: 10.1016/j.dnarep.2004.06.001. [DOI] [PubMed] [Google Scholar]
- 3.Lieber MR, Lu H, Gu J, Schwarz K. Flexibility in the order of action and in the enzymology of the nuclease, polymerases, and ligase of vertebrate non-homologous DNA end joining: relevance to cancer, aging, and the immune system. Cell Res. 2008;18:125–133. doi: 10.1038/cr.2007.108. [DOI] [PubMed] [Google Scholar]
- 4.Weterings E, Chen DJ. The endless tale of non-homologous end-joining. Cell Res. 2008;18:114–124. doi: 10.1038/cr.2008.3. [DOI] [PubMed] [Google Scholar]
- 5.Liang F, Jasin M. Ku80-deficient cells exhibit excess degradation of extrachromosomal DNA. J. Biol. Chem. 1996;271:14405–14411. doi: 10.1074/jbc.271.24.14405. [DOI] [PubMed] [Google Scholar]
- 6.Ma Y, Lu H, Tippin B, Goodman MF, Shimazaki N, Koiwai O, Hsieh CL, Schwarz K, Lieber MR. A biochemically defined system for mammalian nonhomologous DNA end joining. Mol. Cell. 2004;16:701–713. doi: 10.1016/j.molcel.2004.11.017. [DOI] [PubMed] [Google Scholar]
- 7.Nick McElhinny SA, Snowden CM, McCarville J, Ramsden DA. Ku recruits the XRCC4-ligase IV complex to DNA ends. Mol. Cell Biol. 2000;20:2996–3003. doi: 10.1128/mcb.20.9.2996-3003.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li Z, Otevrel T, Gao Y, Cheng HL, Seed B, Stamato TD, Taccioli GE, Alt FW. The XRCC4 gene encodes a novel protein involved in DNA double-strand break repair and V(D)J recombination. Cell. 1995;83:1079–1089. doi: 10.1016/0092-8674(95)90135-3. [DOI] [PubMed] [Google Scholar]
- 9.Ahnesorg P, Smith P, Jackson SP. XLF interacts with the XRCC4-DNA ligase IV complex to promote DNA nonhomologous end-joining. Cell. 2006;124:301–313. doi: 10.1016/j.cell.2005.12.031. [DOI] [PubMed] [Google Scholar]
- 10.Buck D, Malivert L, de Chasseval R, Barraud A, Fondanèche MC, Sanal O, Plebani A, Stéphan JL, Hufnagel M, le Deist F, et al. Cernunnos, a novel nonhomologous end-joining factor, is mutated in human immunodeficiency with microcephaly. Cell. 2006;124:287–299. doi: 10.1016/j.cell.2005.12.030. [DOI] [PubMed] [Google Scholar]
- 11.Riballo E, Kuhne M, Rief N, Doherty A, Smith GC, Recio MJ, Reis C, Dahm K, Fricke A, Krempler A, et al. A pathway of double-strand break rejoining dependent upon ATM, Artemis, and proteins locating to gamma-H2AX foci. Mol. Cell. 2004;16:715–724. doi: 10.1016/j.molcel.2004.10.029. [DOI] [PubMed] [Google Scholar]
- 12.Moshous D, Callebaut I, de Chasseval R, Corneo B, Cavazzana-Calvo M, Le Deist F, Tezcan I, Sanal O, Bertrand Y, Philippe N, et al. Artemis, a novel DNA double-strand break repair/V(D)J recombination protein, is mutated in human severe combined immune deficiency. Cell. 2001;105:177–186. doi: 10.1016/s0092-8674(01)00309-9. [DOI] [PubMed] [Google Scholar]
- 13.Mahajan KN, Nick McElhinny SA, Mitchell BS, Ramsden DA. Association of DNA polymerase mu (pol mu) with Ku and ligase IV: role for pol mu in end-joining double-strand break repair. Mol. Cell Biol. 2002;22:5194–5202. doi: 10.1128/MCB.22.14.5194-5202.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee JW, Blanco L, Zhou T, Garcia-Diaz M, Bebenek K, Kunkel TA, Wang Z, Povirk LF. Implication of DNA polymerase lambda in alignment-based gap filling for nonhomologous DNA end joining in human nuclear extracts. J. Biol. Chem. 2004;279:805–811. doi: 10.1074/jbc.M307913200. [DOI] [PubMed] [Google Scholar]
- 15.Capp JP, Boudsocq F, Bertrand P, Laroche-Clary A, Pourquier P, Lopez BS, Cazaux C, Hoffmann JS, Canitrot Y. The DNA polymerase lambda is required for the repair of non-compatible DNA double strand breaks by NHEJ in mammalian cells. Nucleic Acids Res. 2006;34:2998–3007. doi: 10.1093/nar/gkl380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Capp JP, Boudsocq F, Besnard AG, Lopez BS, Cazaux C, Hoffmann JS, Canitrot Y. Involvement of DNA polymerase mu in the repair of a specific subset of DNA double-strand breaks in mammalian cells. Nucleic Acids Res. 2007;35:3551–3560. doi: 10.1093/nar/gkm243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ward JF. Complexity of damage produced by ionizing radiation. Cold Spring Harbor Symp. Quant. Biol. 2000;65:377–382. doi: 10.1101/sqb.2000.65.377. [DOI] [PubMed] [Google Scholar]
- 18.Datta K, Neumann RD, Winters TA. Characterization of complex apurinic/apyrimidinic-site clustering associated with an authentic site-specific radiation-induced DNA double-strand break. Proc. Natl Acad. Sci. USA. 2005;102:10569–10574. doi: 10.1073/pnas.0503975102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Datta K, Jaruga P, Dizdaroglu M, Neumann RD, Winters TA. Molecular analysis of base damage clustering associated with a site-specific radiation-induced DNA double-strand break. Radiation Res. 2006;166:767–781. doi: 10.1667/RR0628.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nikjoo H, O'N;eill P, Terrissol M, Goodhead DT. Quantitative modelling of DNA damage using Monte Carlo track structure method. Radiat. Environ. Biophys. 1999;38:31–38. doi: 10.1007/s004110050135. [DOI] [PubMed] [Google Scholar]
- 21.Poinsignon C, de Chasseval R, Soubeyrand S, Moshous D, Fischer A, Hache RJG, de Villartay JP. Phosphorylation of Artemis following irradiation-induced DNA damage. Eur. J. Immunol. 2004;34:3146–3155. doi: 10.1002/eji.200425455. [DOI] [PubMed] [Google Scholar]
- 22.Tauchi H, Kobayashi J, Morishima K, Matsuura S, Nakamura A, Shiraishi T, Ito E, Masnada D, Delia D, Komatsu K. The forkhead-associated domain of NBS1 is essential for nuclear foci formation after irradiation but not essential for hRAD50·hMRE11·NBS1 complex DNA repair activity. J. Biol. Chem. 2001;276:12–15. doi: 10.1074/jbc.C000578200. [DOI] [PubMed] [Google Scholar]
- 23.Avkin S, Adar S, Blander G, Livneh Z. Quantitative measurement of translesion replication in human cells: evidence for bypass of abasic sites by a replicative DNA polymerase. Proc. Natl Acad. Sci. USA. 2002;99:3764–3769. doi: 10.1073/pnas.062038699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Avkin S, Sevilya Z, Toube L, Geacintov NE, Chaney SG, Oren M, Livneh Z. p53 and p21 regulate error-prone DNA repair to yield a lower mutation load. Mol. Cell. 2006;22:407–413. doi: 10.1016/j.molcel.2006.03.022. [DOI] [PubMed] [Google Scholar]
- 25.Renzing J, Lane DP. p53-dependent growth arrest following calcium phosphate-mediated transfection of murine fibroblasts. Oncogene. 1995;10:1865–1868. [PubMed] [Google Scholar]
- 26.Schulte-Uentrop L, El-Awady RA, Schliecker L, Willers H, Dahm-Daphi J. Distinct roles of XRCC4 and Ku80 in non-homologous end-joining of endonuclease- and ionizing radiation-induced DNA double-strand breaks. Nucleic Acids Res. 2008;36:2561–2569. doi: 10.1093/nar/gkn094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Giaccia AJ, MacLaren RA, Denko N, Nicolaou D, Stamato TD. Increased sensitivity to killing by restriction enzymes in the XR-1 DNA double-strand break repair-deficient mutant. Mutat. Res. 1990;236:67–76. doi: 10.1016/0921-8777(90)90034-3. [DOI] [PubMed] [Google Scholar]
- 28.Chen S, Inamdar KV, Pfeiffer P, Feldmann E, Hannah MF, Yu Y, Lee JW, Zhou T, Lees-Miller SP, Povirk LF. Accurate in vitro end joining of a DNA double strand break with partially cohesive 3′-overhangs and 3′-phosphoglycolate termini: effect of Ku on repair fidelity. J Biol. Chem. 2001;276:24323–24330. doi: 10.1074/jbc.M010544200. [DOI] [PubMed] [Google Scholar]
- 29.Zhou RZ, Blanco L, Garcia-Diaz M, Bebenek K, Kunkel TA, Povirk LF. Tolerance for 8-oxoguanine but not thymine glycol in alignment-based gap filling of partially complementary double-strand break ends by DNA polymerase lambda in human nuclear extracts. Nucleic Acids Res. 2008;36:2895–2905. doi: 10.1093/nar/gkn126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roth DB, Wilson JH. Nonhomologous recombination in mammalian cells: role for short sequence homologies in the joining reaction. Mol. Cell Biol. 1986;6:4295–4304. doi: 10.1128/mcb.6.12.4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hazkani-Covo E, Covo S. Numt-mediated double-strand break repair mitigates deletions during primate genome evolution. PLoS Genet. 2008;4:e1000237. doi: 10.1371/journal.pgen.1000237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guirouilh-Barbat J, Huck S, Bertrand P, Pirzio L, Desmaze C, Sabatier L, Lopez BS. Impact of the KU80 pathway on NHEJ-induced genome rearrangements in mammalian cells. Mol. Cell. 2004;14:611–623. doi: 10.1016/j.molcel.2004.05.008. [DOI] [PubMed] [Google Scholar]
- 33.Yan CT, Boboila C, Souza EK, Franco S, Hickernell TR, Murphy M, Gumaste S, Geyer M, Zarrin AA, Manis JP, et al. IgH class switching and translocations use a robust non-classical end-joining pathway. Nature. 2007;449:478–482. doi: 10.1038/nature06020. [DOI] [PubMed] [Google Scholar]
- 34.Corneo B, Wendland RL, Deriano L, Cui X, Klein IA, Wong SY, Arnal S, Holub AJ, Weller GR, Pancake BA, et al. Rag mutations reveal robust alternative end joining. Nature. 2007;449:483–486. doi: 10.1038/nature06168. [DOI] [PubMed] [Google Scholar]
- 35.Soulas-Sprauel P, Le Guyader G, Rivera-Munoz P, Abramowski V, Olivier-Martin C, Goujet-Zalc C, Charneau P, de Villartay JP. Role for DNA repair factor XRCC4 in immunoglobulin class switch recombination. J. Exp. Med. 2007;204:1717–1727. doi: 10.1084/jem.20070255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shachar S, Ziv O, Avkin S, Adar S, Wittschieben J, Reisner T, Chaney SG, Friedberg EC, Wang Z, Carell T, et al. Two-polymerase mechanisms dictate error-free and error-prone translesion DNA synthesis in mammals. EMBO J. 2009;28:383–393. doi: 10.1038/emboj.2008.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Strauss BS. The ‘A rule’ of mutagen specificity: a consequence of DNA polymerase bypass of non-instructional lesions? Bioessays. 1991;13:79–84. doi: 10.1002/bies.950130206. [DOI] [PubMed] [Google Scholar]
- 38.Daube SS, Arad G, Livneh Z. Translesion replication by DNA polymerase beta is modulated by sequence context and stimulated by fork-like flap structures in DNA. Biochemistry. 2000;39:397–405. doi: 10.1021/bi991443m. [DOI] [PubMed] [Google Scholar]
- 39.Daube SS, Tomer G, Livneh Z. Translestion replication by DNA polymerase delta depends on processivity accessory proteins and differs in specificity from DNA polymerase beta. Biochemistry. 2000;39:348–355. doi: 10.1021/bi9917784. [DOI] [PubMed] [Google Scholar]
- 40.Takeshita M, Eisenberg W. Mechanism of mutation on DNA templates containing synthetic abasic sites: study with double strand vector. Nucleic Acids Res. 1994;22:1897–1902. doi: 10.1093/nar/22.10.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ohashi E, Ogi T, Kusumoto R, Iwai S, Masutani C, Hanaoka F, Ohmori H. Error-prone bypass of certain DNA lesions by the human DNA polymerase kappa. Genes Dev. 2000;14:1589–1594. [PMC free article] [PubMed] [Google Scholar]
- 42.Efrati E, Tocco G, Eritja R, Wilson SH, Goodman MF. Abasic translesion synthesis by DNA polymerase b violates the “A-rule”. J. Biol. Chem. 1997;272:2559–2569. doi: 10.1074/jbc.272.4.2559. [DOI] [PubMed] [Google Scholar]
- 43.Maga G, Shevelev I, Ramadan K, Spadari S, Hubscher U. DNA polymerase theta purified from human cells is a high-fidelity enzyme. J. Mol. Biol. 2002;319:359–369. doi: 10.1016/S0022-2836(02)00325-X. [DOI] [PubMed] [Google Scholar]
- 44.Maga G, Villani G, Ramadan K, Shevelev I, Le Gac NT, Blanco L, Blanca G, Spadari S, Hubscher U. Human DNA polymerase lambda functionally and physically interacts with proliferating cell nuclear antigen in normal and translesion DNA synthesis. J. Biol. Chem. 2002;277:48434–48440. doi: 10.1074/jbc.M206889200. [DOI] [PubMed] [Google Scholar]
- 45.Covo S, Blanco L, Livneh Z. Lesion bypass by human DNA polymerase mu reveals a template-dependent sequence-independent nucleotidyl transferase activity. J. Biol. Chem. 2004;279:859–865. doi: 10.1074/jbc.M310447200. [DOI] [PubMed] [Google Scholar]
- 46.Lin W, Xin H, Zhang Y, Wu X, Yuan F, Wang Z. The human REV1 gene codes for a DNA template-dependent dCMP transferase. Nucleic Acids Res. 1999;27:4468–4475. doi: 10.1093/nar/27.22.4468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Seki M, Masutani C, Yang LW, Schuffert A, Iwai S, Bahar I, Wood RD. High-efficiency bypass of DNA damage by human DNA polymerase Q. EMBO J. 2004;23:4484–4494. doi: 10.1038/sj.emboj.7600424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Di Noia JM, Neuberger MS. Molecular mechanisms of antibody somatic hypermutation. Annu. Rev. Biolchem. 2007;76:1–22. doi: 10.1146/annurev.biochem.76.061705.090740. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.