Abstract
The transcription factor RUNX1 (AML1) is an important regulator of haematopoiesis, and an important fusion partner in leukaemic translocations. High-affinity DNA binding by RUNX1 requires the interaction of the RUNX1 Runt-Homology-Domain (RHD) with the core-binding factor β protein (CBFβ). To generate novel reagents for in vitro and in vivo studies of RUNX1 function, we have selected high-affinity RNA aptamers against a recombinant RHD–CBFβ complex. Selection yielded two sequence families, each dominated by a single consensus sequence. Aptamers from each family disrupt DNA binding by the RUNX1 protein in vitro and compete with sequence-specific dsDNA binding. Minimal, high-affinity (∼100–160 nM) active aptamer fragments 28 and 30 nts in length, consisting of simple short stem-loop structures, were then identified. These bind to the RHD subunit and disrupt its interaction with CBFβ, which is consistent with reduced DNA affinity in the presence of aptamer. These aptamers represent new reagents that target a novel surface on the RHD required to stabilize the recombinant RHD–CBFβ complex and thus will further aid exploring the functions of this key transcription factor.
INTRODUCTION
The transcription factor RUNX1 (AML1) is one of the most important regulators of haematopoiesis and is involved in the regulation of transcription of a range of blood cell-specific genes. It is expressed at high levels in haematopoietic stem cells and cells committed to all blood cell lineages, including myeloid precursors (1,2). Analysis of RUNX1-deficient mice showed that they do not generate definitive haematopoietic cells and the embryos die at around day 12 of development (3). The region most conserved in RUNX1 proteins is the 128 amino acid Runt-Homology-Domain (RHD), which is located at the N-terminus of the protein and is responsible for binding to the consensus DNA sequence PyGPyGGTPy (Py = pyrimidine) (4,5). The RHD allows RUNX1 to heterodimerize with the core-binding factor beta (CBFβ) protein (4) to form a complex that binds more tightly to its DNA target (6). This interaction is vital for RUNX1 function, as shown by the finding that mice carrying a targeted mutation of the CBFβ gene display the same phenotype as RUNX1-null mice (6). RUNX1 also interacts with a number of other proteins, including other transcription factors, as well as co-activators and co-repressors. The latter includes histone acetyltransferases, such as CBP, p300 and MOZ and repressor molecules such as Sin3A (7–9), for review see (1). These proteins interact with a number of different domains C-terminal of the RHD. RUNX1 can thus function both as an activator and a repressor and these activities are context dependent.
In addition to its role in the regulation of normal haematopoiesis, the RUNX1 gene is an important proto-oncogene. Chromosomal translocations affecting this gene are a recurring feature in acute leukaemias with t(8;21), t(16;21) and t(12;21) being three of the most frequently observed (1,8). Both of these translocations retain the DNA-binding domain of RUNX1 and are therefore still able to bind to DNA but have lost the ability to be properly regulated, leading to a reprogramming of the chromatin structure of target genes and the deregulation of gene expression (10–12). The fusion proteins also maintain the capacity to interact with CBFβ and this interaction contributes to their activities (13). The interactions of the RHD domain of RUNX1 with DNA and CBFβ are therefore potential targets for therapeutic intervention.
A novel, promising class of compounds with potential as both research tools and therapeutics are aptamers-structured polynucleotide sequences that can be isolated by in vitro selection from randomized oligonucleotide libraries (14–17). Aptamers have distinct advantages over antibodies as potential therapeutics and diagnostics as they are significantly smaller, can be isolated rapidly in vitro and modified to include chromophores, fluorophores, radiolabels or novel chemical groups. In addition, aptamers do not carry the secondary functional signals of antibodies, such as complement fixation, and do not elicit a significant immune response (17,18). The first aptamer-based drugs are beginning to appear in the clinic (19) [reviewed in (20)].
Besides their value as novel therapeutic agents, aptamers can be selected against defined protein target surfaces, facilitating the study of inter-molecular interactions and their sites of action. Indeed, RNA aptamers have been generated against a number of transcription factors and shown to interfere with a range of molecular interactions both in vitro and in vivo (21–25). In a similar effort, we have isolated high-affinity aptamers that alter the affinity of RUNX1 for DNA and investigated their effects on DNA binding and CBF complex formation. Analysis of selected aptamers revealed two dominant sequence families, each of which disrupts the binding of RUNX1 proteins to DNA. We have identified minimal short stem-loop sequences from these consensus sequences which retain binding activity (Kds ∼ 100–160 nM). These short structures compete with DNA binding by the RHD–CBFβ complex by binding the RHD subunit disrupting the interaction with CBFβ. This competition between DNA- and RNA-binding activities raises the possibility that RNA binding might be involved in regulating the function of this complex in vivo.
MATERIALS AND METHODS
Biotinylation and immobilization of RHD–CBFβ
The recombinant RHD–CBFβ protein complex was partially biotinylated with ∼20-fold molar excess of EZ-Link® Sulfo-NHS-LC-LC-biotin (Pierce), according to the manufacturer’s protocol. The biotinylated samples were then immobilized on 1 μm MyOne™ streptavidin-coated Dynabeads® (Invitrogen), according to the manufacturer’s protocol.
In vitro transcription
The initial dsDNA SELEX library consists of ∼1015 sequences, each containing an N50 random region, flanked by two fixed primer regions; one of which carries the T7 RNA polymerase promoter. All RNAs were prepared in 50 µl reactions containing a final concentration of 40 mM HEPES NaOH pH 7.6, 26 mM MgCl2, 2 mM spermidine, 40 mM DTT, 2.5 mM each ATP, GTP, 2′-fluoro-UTP, 2′-fluoro-CTP, 5 µl dsDNA template, 100 U T7 RNA polymerase, 0.5 U Yeast Inorganic Pyrophosphatase (Sigma). This was thoroughly mixed then incubated at 37°C for 3 h. To improve RNA yields T7 R&DNA™ polymerase (Epicentre Biotechnologies) was used in all transcription reactions containing 2′-fluoro-modified nucleotides. Template DNA was then removed by addition of 6 µl DNase 1 buffer (to final concentration of 40 mM Tris–HCl pH 8, 10 mM MgSO4, 1 mM CaCl2) and 4 U RQ1 DNase (Promega), then incubating at 37°C for 20 min. The DNase was inactivated by heating the reaction to 65°C for 5 min. The RNA was then purified using RNAClean™ resin following the manufacturer’s protocol (Agencourt).
In vitro selection
The purified RNA pool was counter selected in every round by pre-incubation with 200 μg of underivatized MyOne™ Dynabeads® (Invitrogen) in 100 µl selection buffer (10 mM HEPES NaOH pH 7.4, 100 mM NaCl) at 37°C for 5 min. The beads were then separated and the unbound RNA was removed and added to 200 μg Dynabeads, saturated with biotinylated RHD–CBFβ. After incubation at 37°C for 20 min, the beads were separated and gently washed six times to remove any unbound RNA. The beads were then suspended in 50 µl of selection buffer and incubated (37°C, 5 min) to allow any weakly bound RNA to dissociate. The beads were again separated and unbound material was removed. Aptamers were then eluted by heating the beads to 95°C for 10 min in 30 µl RNase-free water. Eluted RNAs were reverse transcribed and polymerase chain reaction (PCR) amplified as described below. The stringency of selection was gradually increased by reducing the amount of RHD–CBFβ beads from 300 to 50 µg of total protein, reducing the incubation time (from 20 to 5 min) and increasing the dissociation time (from 5 to 20 min).
RT–PCR
Twenty microlitre RT–PCR mix was added to the eluted RNA to give a final concentration of 50 mM Tris–HCl pH 8.5, 30 mM KCl, 8 mM MgCl2, 2 µM reverse primer (5′-GGGGTCATAGTATCCTAGTTG-3′), 1 mM each dNTP, 20 U Transcriptor™ reverse transcriptase (Roche). This was incubated at 52°C for 2 h before adding PCR mix. PCR amplification was carried out in 70 µl reactions containing 20 mM Tris–HCl pH 8.4, 50 mM KCl, 1.5 mM MgCl2, 200 µM dNTP mix and 500 nM forward primer (5′-CCCAAGCTTAATACGACTCACTATAGGGAGCTCCAGAAGATAAATTACAGG-3′). Five hundred nanomolar reverse primer and 2.5 U of Taq DNA polymerase (Invitrogen) were then added before cycling. This PCR product was used as the template in the subsequent round of selection. A Biomek 2000 automated workstation (Beckman Coulter) was used to carry out 10 rounds of in vitro selection.
RNA/DNA synthesis
RNA for use in EMSAs was transcribed using in vitro transcription reactions (as described earlier), spiked with 2.5 mCi ml−1 guanosine 5′-[α-32P] triphosphate (GE Healthcare). As in the selections, T7 R&DNA™ polymerase was used to improve aptamer yields. After incubation (37°C, 3 h) RNA samples were DNase 1 treated and purified by acidified phenol/chloroform extraction, then ethanol precipitation. Samples were then run out on a 10% (w/v) denaturing polyacrylamide gel (19 : 1 bis ratio, 1× TBE, 7 M urea) and gel purified by standard electroelution protocols.
Minimal aptamer sequences were purchased from Dharmacon and resuspended in nuclease-free water to a final concentration of 100 µM. 5′-fluorescein-labelled minimal sequences were prepared and purified using standard protocols (26), whilst their ssDNA equivalents and RUNX1 probe fragments were purchased from MWG.
Cell culture and reagents
Kasumi-1 cells (CRL-2724) were obtained from the American Type Culture Collection (ATCC), VA, USA and were cultured in RPMI (Sigma-Aldrich, UK) containing 10% (v/v) foetal calf serum, 10 U ml−1 penicillin–streptomycin, 2 mM l-glutamine (Gibco, UK). HL60 cells (CCL-240) were from the ATCC and were cultured in DMEM (Sigma-Aldrich) supplemented with 10% (v/v) foetal calf serum, 10 U ml−1 penicillin–streptomycin and 2 mM l-glutamine. The goat anti-RUNX1 antibodies C19 (sc-8564x) and N20 (sc-8564x) were purchased from Santa Cruz Biotechnology, CA, USA.
Preparation of nuclear extracts
Cells (2 × 107) were washed in ice-cold PBS containing 1× complete mini protease inhibitors (Roche) and lysed in a mild lysis buffer (0.4% (v/v) NP40, 10 mM Tris–HCl pH 7.3, 10 mM KCl, 1.5 mM MgCl2, 0.5 mM PMSF, 0.5 mM β-mercaptoethanol) containing 1× complete mini protease inhibitors. Following centrifugation for 5 min at 16 000g to separate the nuclei from the cytosolic fraction, the nuclei were lysed with high-salt buffer (0.4 M NaCl, 20 mM HEPES NaOH pH 7.9, 1 mM EDTA, 1 mM DTT, 1 mM PMSF and 1× complete mini protease inhibitors).
Electrophoretic mobility shift assays
Electrophoretic mobility shift assays (EMSAs) were performed using either 32P-body-labelled aptamers or end-labelled, double-stranded (ds) synthetic oligonucleotides harbouring a RUNX1-binding site-(5′-TTTCCGCCCACACAGGCTG-3′) (11). Recombinant RHD–CBFβ complex or nuclear extracts were incubated for 15 min at room temperature, together with competitors and/or antibodies, in 10% (v/v) glycerol, 10 mM Tris–HCl pH 7.5, 5 mM HEPES NaOH pH 7.9, 50 mM KCl, 80 mM NaCl, 1.2 mM EDTA, 0.1 mg ml−1 poly-dIdC, 2.2 mM DTT and 300 mg ml−1 BSA. Following this the 32P-labelled aptamer or dsDNA probe was added and the incubation continued for a further 15 min. The samples were analysed on 5% (w/v) polyacrylamide (37.5:1 acylamide:bis acrylamide) gels containing 1× TBE.
Aptamer sequencing
To prepare dsDNA with ‘A’-overhangs, PCR were performed using 0.05 U ml−1 Taq DNA polymerase (Promega) in a reaction also containing 1× PCR buffer, 1.25 mM MgCl2, 125 µM dNTPs, 500 nM primer 1 and 500 nM primer 2. The PCR products were cloned into pCR2.1 TA-cloning vector (Invitrogen) as per manufacturer’s instructions. Individual clones were sequenced using M13-21 primer (5′-TGTAAAACGACGGCCAGT-3′).
Enzymatic secondary structure probing
Individual aptamer clones were selected and transcribed as described above. 2′-F-modified nucleotides were replaced with regular 2′-OH nucleotides. Regular T7 RNA polymerase (Roche) was also used. Five microgram of purified RNA was dephosphorylated and 5′-end-labelled with [γ-32P]-ATP following standard protocols. This was then phenol/chloroform extracted, ethanol precipitated and resuspended in 50 µl RNase free water. One microlitre samples were digested with various concentrations of RNase A, T1, V1 (Ambion) or S1 nuclease (Invitrogen) following manufacturers’ protocols. Digestion products were run on a pre-warmed, 20% (w/v) denaturing SequaGel™ (National Diagnostics). A radiolabelled Decade Marker™ (Ambion) and an alkaline hydrolysis ladder were also run. Digestion products were imaged by autoradiography.
Nuclease protection assays
Individual aptamers were labelled and digested with RNase A or T1 as described above. Samples were also digested in the presence of a 2–16-fold molar excess of RHD–CBFβ. As previously, digestion products were run on a 20% (w/v) SequaGel™ and visualized by autoradiography.
Analytical ultracentrifugation
Samples for analysis by sedimentation velocity were prepared in 20 mM HEPES (pH 6.6), 300 mM NaCl, 10 mM DTT and 1 mM EDTA and were equilibrated for 1 h at room temperature. About 0.42 ml samples were centrifuged in 1.2 cm path-length two-sector aluminium centrepiece cells, assembled with sapphire windows, in either an eight-place An-50 Ti or a four-place An-60 Ti analytical rotor running in an Optima XL-I analytical ultracentrifuge (Beckman Instruments, Inc., Palo Alto, California) at 50 000 or 60 000 r.p.m., respectively, at 20°C. Changes in solute concentration were detected by Rayleigh interference and absorbance scans. Samples containing protein and aptamer were measured simultaneously at 260 and 280 nm.
Data analysis
The results were analysed by whole boundary profile analysis using the program Sedfit v 11.3 (27). Sample buoyancy parameters, buffer densities and viscosities were calculated using the program Sednterp (28). The partial specific volume (v-bar) of proteins was calculated from amino acid composition using Sednterp (28). For protein–RNA complexes a weight-average v-bar was calculated, using a default RNA v-bar of 0.45 ml g-1. For samples in the presence of denaturants the v-bar was calculated as for a loosely conjugated component, using Sednterp. The time series of radial absorbance scans was fitted to the Continuous c(s) model in Sedfit (27). After optimizing meniscus position and fitting limits, the best-fit frictional ratio (f/f0) was determined by iterative least squares fitting. The weight-average MW of peaks in the s-value distribution was displayed on the plot and the sedimentation coefficient and MW values determined by integrating each peak. The MW determined depends on the buoyancy parameters calculated, and on the frictional ratio value determined.
Fluorescence anisotropy titrations
The affinities of the minimal RNA aptamers, Apt28A and Apt45A, their ssDNA equivalents and a dsDNA encompassing a RUNX1-binding site was determined using fluorescence anisotropy. Experiments were performed on a Spex Fluorolog Tau spectrofluorometer (HORIBA Jobin Yvon) controlled by Datamax software. The excitation and emission wavelengths were set to 495 and 520 nm, respectively. Slit widths were set to 5 mm and all titrations were carried out at 20°C. The detector integration time was 5 s and three readings were averaged. Changes in fluorescence anisotropy were calculated using the following equation:
wherer= fluorescence anisotropy
F|| = fluorescence intensity parallel to the excitation plane
F┴= fluorescence intensity perpendicular to the excitation plane
The data were fitted to a sigmoidal growth logistic model using the following equation:
where A1 refers to the initial value, A2 refers to the final value, x0 refers to the mid-point and p is a value relating to the slope, of the transition. Errors refer the standard deviation of three titrations. Data fitting was carried out using OriginPro 7.5 software.
Titrations of RHD–CBFβ were into solutions (160 nM) of RNA or ssDNA oligonucleotides encompassing 28A and 45A, or the RUNX1 DNA in 10 mM HEPES, 100 mM NaCl pH 7.6 at room temperature. The RNA aptamer fragments and their DNA equivalents were labelled with fluorescein at their 5′-ends. The RUNX1-binding site was annealed from two oligonucleotides and was dye labelled on the top strand. All labelled DNAs were purchased from MWG. During titrations, the reaction volume did not increase by more than 5%. Each sample was mixed with a pipette and left to equilibrate for 15 min before data acquisition. The relative standard error was <4% for all measurements.
The sequences of the labelled oligonucleotides was as follows:
Apt28A: 5′ Fluorescein-AUUACCGGCGAGUUUUGAACCGCGUAAU-3′
Apt45A: 5′ Fluorescein-GGUCCAGGCGCGUUAGCAAAACCGCGGAUC-3′
DNA Apt28A: 5′ Fluorescein-ATTACCGGCGAGTTTTGAACCGCGTAAT-3′
DNA Apt45A: 5′ Fluorescein-GGTCCAGGCGCGTTAGCAAAACCGCGGATC-3′
RUNX1 top strand: 5′ Fluorescein-TTTCCGCCCACACAGGCTG-3′
RUNX1 bottom strand: 5′-CAGCCTGTGTGGGCGGAAA-3′
Unlabelled minimal aptamer sequences Apt28A and Apt45A were identical except lacking the 5′ dye.
RESULTS
Selection and characterization of aptamers against the RUNX1–CBFβ complex
The aptamer selection target was recombinant RUNX1 RHD and its interaction partner CBFβ. This protein dimer has previously been characterized structurally and its complex with a high-affinity dsDNA oligonucleotide has been determined (5). The protein was purified from Escherichia coli as described previously (29) and then prepared for automated aptamer selection by immobilization on streptavidin-coated microspheres following brief biotinylation using EZ-link® biotin (Pierce), which modifies lysine side chains (30,31). Western blotting with anti-biotin antibodies suggested that both components of the protein dimer were biotinylated (data not shown). These microspheres were then used as the target for 10 rounds of in vitro RNA aptamer selection with an N50 degenerate library as described in the ‘Materials and Methods’ section. PCR reactions at the end of various selection rounds showed increasing amounts of DNA product. Since these reactions were restricted to 10 cycles to avoid artefacts, they were not saturated so this result is consistent with recovery of increasing amounts of anti-target aptamers throughout the SELEX process.
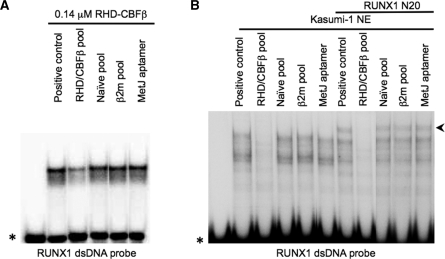
The selected anti-RHD–CBFβ aptamer pool was then tested for its ability to alter sequence-specific DNA binding by the unmodified RHD–CBFβ using EMSAs. The protein was pre-incubated with the test aptamer pool before addition of a 32P-labelled ds oligonucleotide probe encompassing a RUNX1-binding site (11,32). The mixture was allowed to equilibrate before being separated on a native polyacrylamide gel. In addition to the selected pool a number of negative control RNAs were also assayed. These were the naïve library used for the current selections, an individual aptamer from the same library but selected against the bacterial transcription factor MetJ (33) or an aptamer pool derived from an unrelated library and selected against the amyloid forming protein β2m (30). These controls allowed us to show that any effects were due to the selected region of the anti-RHD–CBFβ pool, and were independent of the fixed flanking regions. This aptamer pool caused detectable inhibition of RHD–CBFβ binding to the RUNX1 probe (Figure 1A), whereas none of the negative controls showed any effect. In order to confirm that similar effects would occur with native RUNX1 protein, the EMSA assay was repeated using nuclear extracts from Kasumi-1 cells which are known to express RUNX1 (34) (Figure 1B). The nuclear extract shows a series of retarded bands, presumably because RUNX1 exists in various isoforms and can be a component of larger protein complexes. The presence of RUNX1 in the shifted bands was confirmed by supershifting the complex with a polyclonal antibody directed against the N-terminal region of RUNX1. The formation of these complexes was completely ablated by the anti-RHD–CBFβ 10th round pool and unaffected by any of the negative control RNAs, consistent with the idea that the pool inhibits specific DNA binding by RUNX1. EMSAs were then performed using radioactively labelled aptamer pool and the recombinant RHD–CBFβ protein. These showed formation of an RNA–protein complex (data not shown), confirming that anti-RHD–CBFβ aptamers had been selected.
Figure 1.
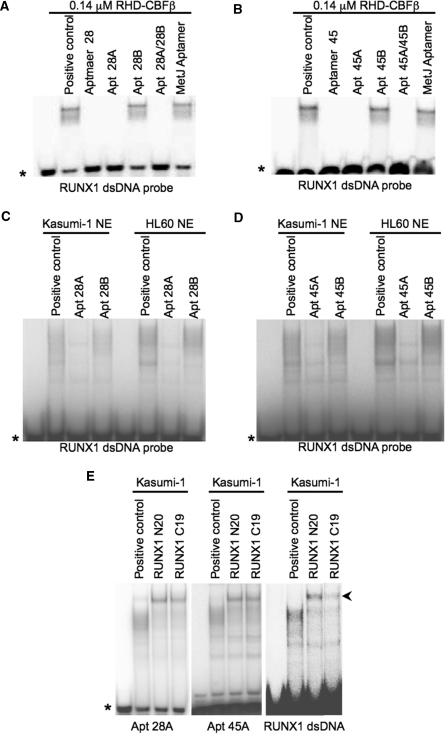
Aptamers selected against recombinant RHD–CBFβ prevent RUNX1 DNA binding. Recombinant RHD–CBFβ at the concentration indicated (A) or 10 µg of total protein from a Kasumi-1 nuclear extract (B) were incubated without (positive control) or with 1 µM concentrations of the following RNAs: the 10th round aptamer pool selected against RHD–CBFβ; the naïve pool used in this selection; the anti-β2m pool or the anti-MetJ aptamer. A 32P-end-labelled RUNX1 dsDNA probe (28) was then added and the sample equilibrated for 15 min before electrophoresis. In (B) when the anti-RUNX1 antibody N20 was added to an identical group of samples at the same time as the aptamers, a proportion of the retarded species was supershifted. The asterisk indicates the positions of the free DNA probe and the arrowhead indicates the RUNX1 supershift.
Isolation of high-affinity individual aptamer sequences
In order to isolate individual aptamer sequences that bind the RHD–CBFβ complex with high affinity, aptamer–protein complexes were isolated from an EMSA gel. Protein-bound RNAs were eluted, reverse transcribed into DNA, cloned and sequenced. All sequences between the expected flanking sequences were ∼50 nucleotides in length (Figure 2). The sequences clustered into two sequence families, each of which contained a single dominant sequence that was represented 14 and 15 times, respectively. The majority of the remaining sequences were also closely related, but contained insertions, deletions or sequence variations. This degree of sequence identity suggested that each aptamer family was directed against a single epitope.
Figure 2.
Aptamer sequence alignments and consensus sequences. The random regions of all the sequenced aptamers were aligned using the multiple alignment program ‘Genebee’ (http://www.genebee.msu.su/services/malign_reduced.html). Of the 58 aptamer sequences, 32 fall into the first family and 17 into the second. One aptamer (aptamer 55) appears in both. The remaining 10 ‘orphan’ sequences do not match any part of these alignments. The consensus sequence, generated using the WebLogo program (http://weblogo.berkeley.edu/logo.cgi), is shown below each family.
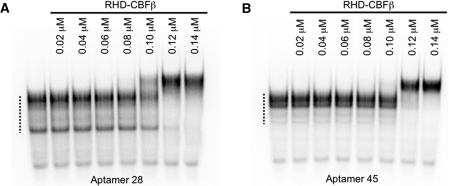
A representative aptamer from each family, 28 and 45, was chosen for further characterization. The affinities of these individual anti-RHD–CBFβ aptamers for their target were estimated by EMSAs. As shown in Figure 3, aptamers 28 and 45 bind to RHD–CBFβ with apparent affinities of ∼100 nM, as judged by the point at which 50% input RNA was shifted. No binding to the anti-MetJ aptamer occurred under these conditions (data not shown). EMSA assays with unmodified versions of aptamers 28 and 45, i.e. without the 2′-fluoro-modified pyrimidines showed that both versions of the aptamers have similar affinities for RHD–CBFβ (data not shown), suggesting that these modified nucleotides do not contribute to binding affinity and/or the structure of the epitope-binding domain within the aptamer. This is perhaps a surprising observation but other well characterized aptamers are known with similar properties. For instance the aptamer that led to MacugenTM was substantially modified post-selection, to give the final drug molecule (19). All but two nucleotides of the original aptamer were modified, without significant effect on the aptamer-binding properties. To our knowledge this may be the first instance were aptamer sequences selected with modified nucleotides are also active in their absence and this may indicate a role for RNA binding by the RHD–CBFβ in vivo.
Figure 3.
Anti-RHD–CBFβ aptamers 28 and 45 bind with high affinity. EMSAs were performed with 32P-labelled aptamers (5000 cpm) incubated without (first lanes) or with RHD–CBFβ at the concentrations shown (A and B). The dotted line on the left indicates the positions of the unbound aptamer.
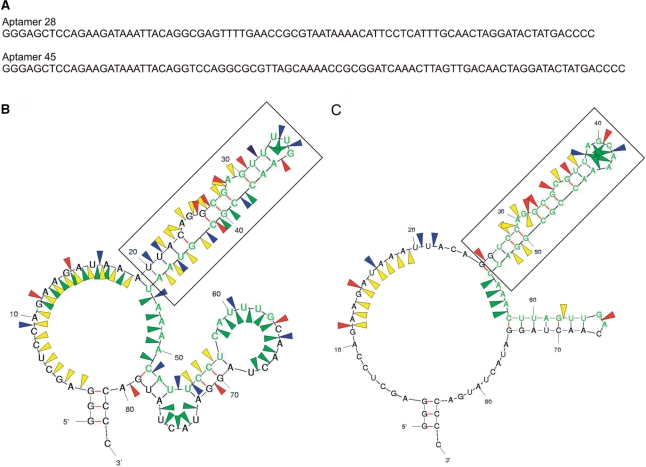
Although aptamer 28 and 45 transcripts are single bands on denaturing gels, they migrate on native gels as a series of discrete bands, suggesting that they have the ability to equilibrate between several secondary/tertiary structures. To test which of the possible folds interacted with RHD–CBFβ, their sequences were first analysed using the program Mfold, which yielded a single secondary structure prediction for aptamer 45 and three possible structures for aptamer 28. We then used enzymatic solution structure probing to identify the most likely solution secondary structure of aptamers without modified pyrimidines (30) (Figure 4 and Supplementary Figure S1A and B) and nuclease protection assays to identify the aptamer residues contacting RHD–CBFβ (Supplementary Figure S1C and D). The resultant protection profiles demonstrate that the 5′ stem-loop structures in both aptamers are likely to be the principal epitope-binding domains (Figure 4B and C, boxed regions).
Figure 4.
Sequence and secondary structures of representative aptamers. The RNA sequences of aptamers 28 and 45 (A), were analysed using ‘Mfold’. The resultant secondary structures that are most consistent with the enzymatic structure probing (Supplementary Figure S1A and B) are shown for both aptamers 28 and 45 (B and C, respectively). The selected sequence region is shown in green. Nuclease cleavage sites are indicated by blue, red, green and yellow arrows, for the C- and U-specific RNase A, the G-specific RNase T1, the single-strand-specific nuclease S1, and ds/helical-specific nuclease V1, respectively. Regions shown to be protected from nuclease attack by RHD–CBFβ are shown in the boxed regions (Supplementary Figure S1C and D), which includes the conserved secondary structure motif (see Discussion) running from C23–G42 and C29–G50 for aptamers 28 and 45, respectively.
Aptamers and DNA bind to similar sites on the RUNX1–CBFβ complex
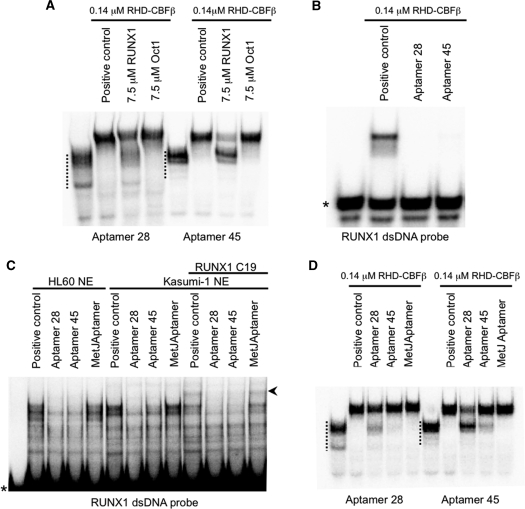
In order to determine how aptamers 28 and 45 inhibit RHD–CBFβ DNA binding, a series of competition experiments between the aptamers and the dsRUNX1 DNA probe were carried out. Figure 5A shows that both aptamers bound to the RHD–CBFβ complex in the absence of DNA but binding was significantly reduced in the presence of a molar excess of the RUNX1 DNA probe. This effect was not seen with a negative control dsDNA probe containing a binding site for the transcription factor Oct1 (Oct1 dsDNA). The converse experiment showed that the specific 32P-labelled-dsDNA probe was incapable of disrupting the complexes formed between the aptamers and RHD–CBFβ when the aptamers were present in molar excess (Figure 5B). These results confirm that the pyrimidine-modified aptamers interfere with formation of DNA–RHD–CBFβ complexes and imply that the protein has similar affinities for both RNA and dsDNA targets, as was later confirmed (Figure 7). Similar effects were also seen when unmodified aptamers were used (data not shown).
Figure 5.
Aptamers 28 and 45 can prevent DNA binding of RUNX1 by binding to similar sites. (A) RHD–CBFβ was incubated with either buffer alone (positive control) or unlabelled dsDNA probes containing RUNX1 or Oct1 consensus-binding sites. 32P-labelled aptamers 28 or 45 (1000 cpm) were then added and EMSAs performed. The RUNX1 DNAs compete with aptamer binding but the Oct1 sequence does not. (B) RHD–CBFβ was pre-incubated with unlabelled aptamers 28, 45 or buffer (positive control) followed by addition of 32P-end-labelled RUNX1 dsDNA probe (10 000 cpm) followed by EMSAs. Both aptamers compete with binding to the DNA. (C) As in (B), Kasumi-1 or HL60 nuclear extracts were pre-incubated with unlabelled aptamers 28, 45, the anti-MetJ aptamer or a buffer alone (positive control) followed by addition RUNX1 probe (10 000 cpm) and EMSA. In some of the experiments a polyclonal antibody against the C-terminal region of RUNX1, C19, was added at the same time as the aptamers. The asterisk indicates the positions of the free DNA probe and the arrowhead indicates the RUNX1 supershift. Both aptamers compete with DNA binding by the extracts but the anti-MetJ aptamer does not. (D) RHD–CBFβ was incubated with buffer blank (positive control) or unlabelled aptamers 28, 45 or the anti-MetJ aptamer. The complexes were then challenged by addition of 32P-labelled aptamers 28 or 45 (1000 cpm) and EMSAs performed. Both aptamers show direct competition as expected, with aptamer 28 having the higher affinity. The positions of unshifted aptamers in these gels are indicated by the dotted lines.
Figure 7.
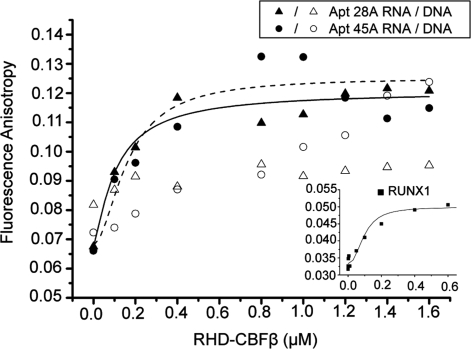
Affinities of the minimal aptamer domains and dsDNA RUNX1 probe for RHD–CBFβ. Plots of fluorescence anisotropy versus RHD–CBFβ concentration are shown for titrations against Apt28A, Apt45A and their equivalent ssDNAs, as well as the dsDNA RUNX1 probe (inset). Curves represent fits to the binding data, as described in the text. Anisotropy and solution conditions, including the sequences of the oligonucleotides are described in ‘Materials and Methods’ section.
In order to confirm these effects on the native RUNX1 protein, competition assays were performed using nuclear extracts from the RUNX1-expressing cells, Kasumi-1 and HL60 (Figure 5C). There was clear inhibition of transcription factor complex formation by the individual aptamers, whereas there was no effect using the anti-MetJ control aptamer. These data confirm that the aptamers are able to block binding of the RUNX1 protein to DNA, suggesting that either they bind to a site that overlaps the DNA-binding domain or somehow disrupt it. We next tested whether the aptamers bind to independent sites on the RHD–CBFβ complex. Figure 5D shows that aptamers 28 and 45 compete with each other, as well as themselves, suggesting that they bind to sites on the protein complex which are very close to each other if not identical.
In order to determine whether the RHD–CBFβ footprints on aptamers 28 and 45 identified functional, minimal length RNAs, we assayed unmodified, synthetic RNA oligonucleotides encompassing the 5′ and 3′ stem-loops from each aptamer (Figure 4). Oligonucleotides ‘Apt 28A’ and ‘Apt 45A’ comprise the 5′ stem-loops, which were strongly protected by RHD–CBFβ and ‘Apt 28B’ and ‘Apt 45B’ comprise the non-protected 3′ stem-loops (Figure 4 and Supplementary Figure S1C and D). Figure 6 shows that the 5′ stem-loops in each case were as effective as the full length, modified aptamers in blocking DNA binding of both purified RHD–CBFβ protein and RUNX1 in nuclear extracts. Incubation with the 3′ stem-loops had no effect and no synergy of inhibition was seen when they were combined with the 5′ stem-loops in the same reaction. The obvious conclusion is that the 5′ stem-loops of 28 and 30 nts (for aptamers 28 and 45, respectively), contain all the sequence/structure information required to bind the RUNX1/RHD–CBFβ proteins.
Figure 6.
The 5′ stem-loops of aptamers 28 and 45 bind to the RHD of RUNX1 to block DNA binding. (A) and (B) RHD–CBFβ was incubated with 2.5 µM unlabelled aptamers 28 or 45, the anti-MetJ aptamer or buffer alone (positive control), or the minimal aptamer fragments Apt28A, Apt45A, Apt28B or Apt45B and then 32P-labelled RUNX1 dsDNA probe (10 000 cpm) added and EMSAs performed. The full length aptamers as well as the ‘A’ fragments, but not the anti-MetJ aptamer or ‘B’ fragments, were equally effective at competing with binding to the RUNX1 probe. (C) and (D) nuclear extracts from Kasumi-1 or HL60 cells were pre-incubated with the putative minimal aptamer fragments Apt28A and Apt28B or Apt45A and Apt45B prior to challenge with RUNX1 probe, as in (A). Only the ‘A’ fragments show inhibition of DNA binding. (E) In order to identify which subunit within the RHD–CBFβ dimer the minimal aptamer fragments bind, 32P-end-labelled Apt28A and Apt45A were incubated with Kasumi-1 nuclear extracts (positive control) and complexes were supershifted with antibodies N20 or C19. The retarded minimal aptamer species supershift with both antibodies confirming the presence of RHD in the aptamer complexes. A similar EMSA with RUNX1 dsDNA is shown alongside to confirm the activity of the antibodies used. Asterisks indicate the positions of free oligonucleotides and the arrowheads the positions of the RUNX1 supershifts.
Aptamers inhibit RHD–CBFβ DNA binding by high-affinity binding to the RHD
The experiments described above are consistent with the aptamers exerting their effects by binding to the DNA-binding domain of the RHD–CBFβ complex (Figure 5). However, an additional possibility was that they could disrupt the interaction between subunits, which would also lower DNA affinity (35). To examine these possibilities we analysed the aptamer–RHD–CBFβ protein complexes by sedimentation velocity in an analytical ultracentrifuge (36). The data (Table 1) show that the RHD–CBFβ dimer remained intact after biotinylation but was disrupted in the presence of either Apt28A or Apt45A, which each seemed to make 1 : 1 complexes with one of the two subunits. The resolution of the sedimentation data was not sufficient to determine which subunit of the RHD–CBFβ dimer was bound. However, antibodies directed against N- and C-terminal regions of RUNX1 caused supershifting of the aptamer–protein complex (Figure 6E), demonstrating that aptamers bound to the RHD subunit. These results suggest that the inhibitory action of the aptamers is at least partially due to disruption of the RHD–CBFβ dimer, formation of which increases DNA affinity by the RHD.
Table 1.
Sedimentation velocity analysis of aptamer/RHD-CBFβ complex formation
| Sample | Concentration | Expected Mass (Da) | Observed Sed Vel Massa (Da) | Comment |
|---|---|---|---|---|
| RHD–CBFβ | 250 nM | 32 514 | 36 000 | |
| Biotinylated RHD–CBFβ | 250 nM | 32 514b | 35 600 | Biotinylation does not disrupt the protein dimer |
| Apt28A | 500 nM | 8939 | 7600 | |
| Apt28B | 500 nM | 9150 | 10 000 | |
| Apt45A | 500 nM | 9665 | 10 600 | |
| Apt45B | 500 nM | 5411 | Not done | |
| RHD–CBFβ +Apt28A | Protein 250 nM | (RHD–CBFβ):Apt28A =41 453 | Single peak at 23 300c | There is no evidence that the Apt28A binds to the dimeric target, rather it is making a 1 : 1 complex with one of the monomers |
| Aptamer 500 nM | (RHD):Apt28A =25 660 | |||
| (CBFβ):Apt28A =24 732 | ||||
| RHD–CBFβ +Apt28B | Protein 250 nM | (RHD–CBFβ):Apt28B =41 664 | Peaks at 33 000 and 9000 | Apt28B does not appear to bind to the protein |
| Aptamer 500 nM | (RHD):Apt28B =25 871 | |||
| (CBFβ):Apt28B =24 943 | ||||
| RHD–CBFβ +Apt45A | Protein 250 nM | (RHD–CBFβ):Apt45A =42 179 | Single peak at 25 000c | There is no evidence that the Apt45A binds to the dimeric target, rather it is making a 1 : 1 complex with one of the monomers |
| Aptamer 500 nM | (RHD):Apt45A =26 386 | |||
| (CBFβ):Apt45A =25 458 | ||||
| RHD–CBFβ +Apt45B | Protein 250 nM | (RHD–CBFβ):Apt45B =37 925 | Peaks at 31 100 and 5500 | Apt45B does not appear to bind to the protein |
| Aptamer 500 nM | (RHD):Apt45B =22 132 | |||
| (CBFβ):Apt45B =21 204 |
a±10%.
bPlus mass of EZ-link.
cBroad peak at higher s values was also present, consistent with aggregating material.
Finally, in order to understand the competition observed between RNA aptamer and DNA binding, we determined the equilibrium dissociation constants (Kds) for RHD–CBFβ dimer binding to the RUNX1 dsDNA probe and Apt28A and Apt45A. To confirm that the action of the aptamers was RNA specific, rather than a non-specific effect of adding polynucleotides, we also assayed ssDNA versions of both minimal aptamer sequences. Affinities were determined using fluorescence anisotropy with fluorescein end-labelled oligonucleotides, as described in the Materials and Methods section. The results are shown in Figure 7. The anisotropy plots show binding curves for the dsDNA RUNX1, Apt28A and Apt45A targets that yield Kds of 95 ± 15; 108 ± 34 and 162 ± 102 nM, respectively. In contrast, the ssDNA version of Apt28A showed no anisotropy change over the protein concentration range tested, whilst the Apt45A equivalent showed a linear increase across this range, consistent with non-specific binding. These Kd values are entirely consistent with the competition results of Figure 5A and B.
DISCUSSION
The experiments described provide novel insights into the structural and functional features of the RHD–CBFβ complex. The high-affinity RNA aptamers selected against the RHD–CBFβ complex were composed of two major sequence families that both bind to the RHD domain of the selection target (Figure 2). RNA aptamers from both of these sequence families contain a functional stem-loop with a similar secondary structure, encompassing a base-paired region interrupted on the 5′ leg by a two base bulge (A.G) followed by 3 bp and a single base insertion (A or C, depending on the aptamer) (Figure 3). Binding of both aptamers, including the minimized fragments encompassing just the stem-loop motifs, occurs to a similar, presumably identical, site on the RHD. Aptamer–RHD complex formation disrupts the RHD–CBFβ dimer which would be expected to lower its DNA affinity (Table 1). It is not clear whether the epitope on RHD bound also overlaps with its DNA-binding domain, although the efficient ablation of DNA binding would be consistent with both these effects. There are multiple examples for transcription factors that use the same protein domain to specifically bind both RNA and DNA sequences to regulate gene expression, e.g. Xenopus laevis TFIIIA, TRA-1, bicoid, p53 and STAT1 (37). X-ray crystallographic analysis of an RNA–protein complex of a high-affinity aptamer selected against the NF-κB p50 homodimer revealed that two RNA molecules bind independently to the p50 N-terminal immunoglobulin-like domains (24). Each p50 monomer uses the same surface to recognize the distorted RNA major groove of the aptamer as is observed in the NF-κB DNA/p50 complex. Interestingly, the nucleic acid binding interfaces of p53, STAT1, NF-κB and RUNX1 are strikingly similar (37). The DNA-binding domains of these proteins belong to the p53 family of transcription factors that share a common DNA-binding immunoglobulin fold. Although speculative, this raises the possibility that specific cellular RNAs may target RUNX1 to mediate a further level of regulatory control. The idea that there might be dual roles for transcription factors in RNA and DNA binding has been reinforced recently by the observation that ribosomal protein Rps3 is an RNA-binding KH domain component in NF-κB complexes that mediates selective gene regulation (38).
Given these precedents and the fact that the RNA aptamers described here efficiently compete with the cognate RUNX1 DNA in vitro, it seems reasonable to suggest that the aptamer is binding the p53-like immunoglobulin fold DNA-binding domain of the RHD. One hypothesis is that the RNA aptamers could act as molecular DNA mimics, adopting three-dimensional structures with similar physico-chemical properties to cognate RHD DNA. However, the aptamers lack extended base-paired matches to the RUNX1 DNA consensus and only function as ribo-oligonucleotides (Figure 7). This suggests that they are novel recognition partners with the ability to both bind RHD and displace the CBFβ subunit.
Experiments performed by others demonstrated that the binding of small molecular weight compounds sharing a common structural feature, namely a 2-aminothiazole attached to an aromatic ring, can cause disruption of the RHD–CBFβ complex (39). These potential drugs bind to a site on the CBFβ subunit that is displaced from the interface with RUNX1 DNA. Our data suggest that we have isolated aptamer inhibitors with a different mode of action, i.e. with a primary binding target located on the RHD domain and a significantly higher affinity (Kd ∼ 100 nM versus >10 µM) than the small molecular weight ligands. One possibility is that the aptamers bind to the RHD DNA-binding motif in such a way that they stabilize its low-affinity conformation for CBFβ, a type of negative RNA allostery. Whilst our experiments have uncovered a novel effect of oligonucleotide binding to RHD, understanding the consequences for the CBFβ interaction can only be resolved by structural studies of the aptamer–RHD complexes and these are underway.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
This work was funded by Leukemia Research (UK). Aptamer selection facilities within Leeds have been supported by funding from the UK Medical Research Council and The Wellcome Trust. Funding for open access charge: Leukemia Research.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Mr Ben Mantle for technical assistance throughout this work.
REFERENCES
- 1.Mikhail FM, Sinha KK, Saunthararajah Y, Nucifora G. Normal and transforming functions of RUNX1: a perspective. J. Cell Physiol. 2006;207:582–593. doi: 10.1002/jcp.20538. [DOI] [PubMed] [Google Scholar]
- 2.Speck NA. Core binding factor and its role in normal hematopoietic development. Curr. Opin. Hematol. 2001;8:192–196. doi: 10.1097/00062752-200107000-00002. [DOI] [PubMed] [Google Scholar]
- 3.Okuda T, vanDeursen J, Hiebert SW, Grosveld G, Downing JR. AML1, the target of multiple chromosomal translocations in human leukemia, is essential for normal fetal liver hematopoiesis. Cell. 1996;84:321–330. doi: 10.1016/s0092-8674(00)80986-1. [DOI] [PubMed] [Google Scholar]
- 4.de Bruijn M, Speck NA. Core-binding factors in hematopoiesis and immune function. Oncogene. 2004;23:4238–4248. doi: 10.1038/sj.onc.1207763. [DOI] [PubMed] [Google Scholar]
- 5.Bravo J, Li Z, Speck NA, Warren AJ. The leukemia-associated AML1 (Runx1)-CBF beta complex functions as a DNA-induced molecular clamp. Nat. Struct. Biol. 2001;8:371–378. doi: 10.1038/86264. [DOI] [PubMed] [Google Scholar]
- 6.Wang Q, Stacy T, Miller JD, Lewis AF, Gu T-L, Huang X, Bushweller JH, Bories J-C, Alt FW, Ryan G, et al. The CBF[beta] subunit is essential for CBF[alpha]2 (AML1) function in vivo. Cell. 1996;87:697. doi: 10.1016/s0092-8674(00)81389-6. [DOI] [PubMed] [Google Scholar]
- 7.Kitabayashi I, Yokoyama A, Shimizu K, Ohki M. Interaction and functional cooperation of the leukemia-associated factors AML1 and p300 in myeloid cell differentiation. EMBO J. 1998;17:2994–3004. doi: 10.1093/emboj/17.11.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lutterbach B, Westendorf JJ, Linggi B, Isaac S, Seto E, Hiebert SW. A mechanism of repression by acute myeloid leukemia-1, the target of multiple chromosomal translocations in acute leukemia. J. Biol. Chem. 2000;275:651–656. doi: 10.1074/jbc.275.1.651. [DOI] [PubMed] [Google Scholar]
- 9.Petrovick HS, Hiebert SW, Friedman AD, Hetherington CJ, Tenen DG, Zhang DE. Multiple functional domains of AML1: PU.1 and C/EBP alpha synergize with different regions of AML1. Mol. Cell Biol. 1998;18:3915–3925. doi: 10.1128/mcb.18.7.3915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Di Croce L. Chromatin modifying activity of leukaemia associated fusion proteins. Hum. Mol. Genet. 2005;14:R77–R84. doi: 10.1093/hmg/ddi109. [DOI] [PubMed] [Google Scholar]
- 11.Follows GA, Tagoh H, Lefevre P, Hodge D, Morgan GJ, Bonifer C. Epigenetic consequences of AML1-ETO action at the human c-FMS locus. EMBO J. 2003;22:2798–2809. doi: 10.1093/emboj/cdg250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Follows GA, Tagoh H, Richards SJ, Melnik S, Dickinson H, de Wynter E, Lefevre P, Morgan GJ, Bonifer C. c-FMS chromatin structure and expression in normal and leukaemic myelopoiesis. Oncogene. 2005;24:3643–3651. doi: 10.1038/sj.onc.1208655. [DOI] [PubMed] [Google Scholar]
- 13.Roudaia L, Cheney MD, Manuylova E, Chen W, Morrow M, Park S, Lee CT, Kaur P, Williams O, Bushweller JH, et al. CBF beta is critical for AML1-ETO and TEL-AML1 activity. Blood. 2009;113:3070–3079. doi: 10.1182/blood-2008-03-147207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ellington AD, Szostak J. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 15.Robertson DL, Joyce GF. Selection in vitro of an RNA enzyme that specifically cleaves single-stranded-DNA. Nature. 1990;344:467–468. doi: 10.1038/344467a0. [DOI] [PubMed] [Google Scholar]
- 16.Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment – RNA ligands to bacteriophage-T4 DNA-polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 17.White RR, Sullenger BA, Rusconi CP. Developing aptamers into therapeutics J. Clin. Invest. 2000;106:929934. doi: 10.1172/JCI11325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wlotzka B, Leva S, Eschgfäller B, Burmeister J, Kleinjung F, Kaduk C, Muhn P, Hess-Stumpp H, Klussmann S. In vitro properties of an Anti-GnRH Spiegelmer: An Example of oligonucleotide-based therapeutic substance class. Proc. Natl Acad. Sci. USA. 2002;99:8898–8902. doi: 10.1073/pnas.132067399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ng EWM, Shima DT, Calias P, Emmett T, Cunningham J, Guyer DR, Adamis AP. Pegaptanib, a targeted anti-VEGF aptamer for ocular vascular disease. Nat. Rev. Drug Disc. 2006;5:123–132. doi: 10.1038/nrd1955. [DOI] [PubMed] [Google Scholar]
- 20.Bunka DHJ, Stockley PG. Aptamers come of age – at last. Nat. Rev. Microbiol. 2006;4:588–596. doi: 10.1038/nrmicro1458. [DOI] [PubMed] [Google Scholar]
- 21.Park MW, Choi KH, Jeong S. Inhibition of the DNA binding by the TCF-1 binding RNA aptamer. Biochem. Biophys. Res. Commun. 2005;330:11–17. doi: 10.1016/j.bbrc.2005.02.119. [DOI] [PubMed] [Google Scholar]
- 22.Shi H, Fan XC, Sevilimedu A, Lis JT. RNA aptamers directed to discrete functional sites on a single protein structural domain. Proc. Natl Acad. Sci. USA. 2007;104:3742–3746. doi: 10.1073/pnas.0607805104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhao XC, Shi H, Sevilimedu A, Liachko N, Nelson HCM, Lis JT. An RNA aptamer that interferes with the DNA binding of the HSF transcription activator. Nucleic Acids Res. 2006;34:3755–3761. doi: 10.1093/nar/gkl470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang DB, Vu D, Cassiday LA, Zimmerman JM, Maher LJ, Ghosh G. Crystal structure of NF-kappa B (p50)(2) complexed to a high-affinity RNA aptamer. Proc. Natl Acad. Sci. USA. 2003;100:9268–9273. doi: 10.1073/pnas.1632011100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reiter NJ, Maher LJI, Butcher SE. DNA mimicry by a high-affinity anti-NF-{kappa}B RNA aptamer. Nucleic Acids Res. 2008;36:1227–1236. doi: 10.1093/nar/gkm1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Murray JB, Collier AK, Arnold JRP. A general purification procedure for chemically synthesized oligoribonucleotides. Anal. Biochem. 1994;218:177–184. doi: 10.1006/abio.1994.1157. [DOI] [PubMed] [Google Scholar]
- 27.Schuck P. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and Lamm equation modeling. Biophys. J. 2000;78:1606–1619. doi: 10.1016/S0006-3495(00)76713-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Laue T, Shah B, Ridgeway T, Pelletier S. Analytical Ultracentrifugation in Biochemistry and Polymer Science. Cambridge: Royal Society of Chemistry; 1992. [Google Scholar]
- 29.Warren AJ, Bravo J, Williams RL, Rabbitts TH. Structural basis for the heterodimeric interaction between the acute leukaemia-associated transcription factors AML1 and CBF beta. EMBO J. 2000;19:3004–3015. doi: 10.1093/emboj/19.12.3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bunka DHJ, Mantle BJ, Morten IJ, Tennent GA, Radford SE, Stockley PG. Production and characterization of RNA aptamers specific for amyloid fibril epitopes. J. Biol. Chem. 2007;282:34500–34509. doi: 10.1074/jbc.M703679200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ellingham M, Bunka DHJ, Rowlands DJ, Stonehouse NJ. Selection and characterization of RNA aptamers to the RNA-dependent RNA polymerase from foot-and-mouth disease virus. RNA. 2006;12:1970–1979. doi: 10.1261/rna.161006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Himes SR, Cronau S, Mulford C, Hume DA. The Runx1 transcription factor controls CSF-1-dependent and -independent growth and survival of macrophages. Oncogene. 2005;24:5278–5286. doi: 10.1038/sj.onc.1208657. [DOI] [PubMed] [Google Scholar]
- 33.McGregor A, Murray JB, Adams CJ, Stockley PG, Connolly BA. Secondary structure mapping of an RNA ligand that has high affinity for the MetJ repressor protein and interference modification analysis of the protein-RNA complex. J. Biol. Chem. 1999;274:2255–2262. doi: 10.1074/jbc.274.4.2255. [DOI] [PubMed] [Google Scholar]
- 34.Asou H, Tashiro S, Hamamoto K, Otsuji A, Kita K, Kamada N. Establishment of a human acute myeloid-leukemia cell-line (Kasumi-1) with 8-21 chromosome-translocation. Blood. 1991;77:2031–2036. [PubMed] [Google Scholar]
- 35.Tahirov TH, Inoue-Bungo T, Morii H, Fujikawa A, Sasaki M, Kimura K, Shiina M, Sato K, Kumasaka T, Yamamoto M, et al. Structural analyses of DNA recognition by the AML1/Runx-1 Runt domain and its allosteric control by CBFbeta. Cell. 2001;104:755–767. doi: 10.1016/s0092-8674(01)00271-9. [DOI] [PubMed] [Google Scholar]
- 36.Cassiday LA, Lebruska LL, Benson LM, Naylor S, Owen WG, Maher LJ. Binding stoichiometry of an RNA aptamer and its transcription factor target. Anal. Biochem. 2002;306:290–297. doi: 10.1006/abio.2002.5710. [DOI] [PubMed] [Google Scholar]
- 37.Cassiday LA, Maher LJ. Having it both ways: transcription factors that bind DNA and RNA. Nucleic Acids Res. 2002;30:4118–4126. doi: 10.1093/nar/gkf512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gorczynski MJ, Grembecka J, Zhou YP, Kong Y, Roudaia L, Douvas MG, Newman M, Bielnicka I, Baber G, Corpora T, et al. Allosteric inhibition of the protein-protein interaction between the leukemia-associated proteins Runx1 and CBF beta. Chem. Biol. 2007;14:1186–1197. doi: 10.1016/j.chembiol.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 39.Schneider D, Tuerk C, Gold L. Selection of high-affinity RNA ligands to the bacteriophage-R17 coat protein. J. Mol. Biol. 1992;228:862–869. doi: 10.1016/0022-2836(92)90870-p. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.