Abstract
Plasmids harbored by Staphylococcus aureus are a major contributor to the spread of bacterial multi-drug resistance. Plasmid conjugation and partition are critical to the dissemination and inheritance of such plasmids. Here, we demonstrate that the ArtA protein encoded by the S. aureus multi-resistance plasmid pSK41 is a global transcriptional regulator of pSK41 genes, including those involved in conjugation and segregation. ArtA shows no sequence homology to any structurally characterized DNA-binding protein. To elucidate the mechanism by which it specifically recognizes its DNA site, we obtained the structure of ArtA bound to its cognate operator, ACATGACATG. The structure reveals that ArtA is representative of a new family of ribbon–helix–helix (RHH) DNA-binding proteins that contain extended, N-terminal basic motifs. Strikingly, unlike most well-studied RHH proteins ArtA binds its cognate operators as a dimer. However, we demonstrate that it is also able to recognize an atypical operator site by binding as a dimer-of-dimers and the extended N-terminal regions of ArtA were shown to be essential for this dimer-of-dimer binding mode. Thus, these data indicate that ArtA is a master regulator of genes critical for both horizontal and vertical transmission of pSK41 and that it can recognize DNA utilizing alternate binding modes.
INTRODUCTION
The emergence of multi-drug resistant bacteria has now become a global threat to human health. Indeed, more people in the United States now die from multi-drug resistant forms of Staphylococcus aureus than AIDS (1–4). Multi-drug resistance determinants in S. aureus are found chromosomally and on plasmids (5). The largest staphylococcal plasmids are the conjugative multi-resistance plasmids, which are typified by the prototype pSK41 (6). Members of the pSK41 family of plasmids encode a wide array of resistance phenotypes; for example, pLW1043 confers resistance to five different classes of anti-microbial agents. The conjugation system of these plasmids makes them efficient vehicles of horizontal transfer, and they therefore represent important mediators of multi-drug resistance transmission between bacterial strains, and hence a significant medical threat.
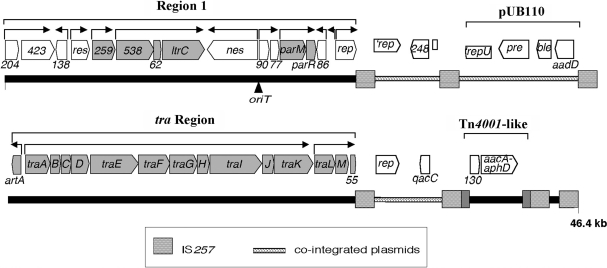
pSK41-like plasmids show a high degree of structural and sequence similarity such that a conserved plasmid backbone can be recognized, into which distinct DNA segments encoding various resistance genes have been integrated, often mediated by the activities of the insertion element IS257 that consequently flanks many of these segments (5). In pSK41, resistance segments divide the backbone into two segments; viz., the transfer (tra) region that contains genes associated with conjugative transfer, and Region 1 that contains genes involved plasmid replication and maintenance, encoding the replication initiation protein, a resolvase and partitioning proteins, as well as the conjugative nickase (6,8–10). The artA gene is encoded at one end of the tra region, divergently transcribed from the other tra genes. The 7 kDa product of the identical homolog from the pSK41-like plasmid pGO1, TrsN, has been shown to bind to the three tra region promoters, PtrsN, PtrsA and PtrsL, and to repress transcription of the latter (11). The equivalent promoters in pSK41 each contain the sequence CATGACA overlapping their −35 sequences (12), and three promoters with this feature were subsequently identified within Region 1 (6), including the promoter responsible for transcription of the plasmid’s parMR type II partitioning system (10). These observations suggested that ArtA might act as a key regulator that coordinates transcription of most pSK41 backbone genes.
The regulation of genes required for conjugative transfer and partition/segregation is critical to plasmid maintenance (13). ArtA shows no sequence homology to any structurally characterized DNA-binding protein and thus how it binds DNA and regulates transcription is unknown. We have therefore carried out cellular, biochemical and X-ray crystallographic studies to determine the role of ArtA in transcriptional regulation of pSK41 genes in vivo and to elucidate its structural mechanism of DNA binding. These data indicate that ArtA is a global regulator of genes critical for pSK41 transmission and that the ArtA protein utilizes different modes for binding consensus versus atypical DNA operator sites.
MATERIALS AND METHODS
Bacterial strains, growth conditions and plasmids
Bacterial strains and plasmids used in this study are listed in Table 1. Bacterial strains were grown at 37°C in LB or on plates containing LB medium and 1.5% w/v Oxoid agar, unless otherwise stated. S. aureus electrocompetent cells were prepared using B2 medium as described earlier (14). When required, media was supplemented with ampicillin (Ap) 100 µg/ml, cadmium chloride (CdCl2) 0.1 or 5 µM, neomycin (Nm) 15 µg/ml, tetracycline (Tc) 10 µg/ml and trimethoprim (Tm) 250 µg/ml.
Table 1.
Bacterial strains and plasmids
 |
DNA manipulations
Plasmid DNA was isolated from Escherichia coli using the alkaline lysis method (15) or the Quantum Prep plasmid miniprep kit (Bio-Rad). Cloning in E. coli was performed using standard methods and restriction enzymes, calf alkaline phosphatase and T4 DNA ligase were purchased from New England Biolabs. DNA fragments were PCR-amplified using ‘Taq’ (New England Biolabs) or ‘Pfu’ Turbo DNA polymerase (Stratagene). Automated DNA sequencing was performed by the Australian Genome Research Facility (University of Queensland, Australia).
CAT Assays
Chloramphenicol acetyl transferase (CAT) assays based on the method of Shaw (16) were adapted to microplate format as described earlier (8). Lysostaphin, acetyl Coenzyme A and 5-5′-dithio-bis[2-nitrobenzoic acid] were purchased from Sigma Aldrich and bovine serum albumin from New England Biolabs. CAT units are expressed as nanomoles of chloramphenicol acetylated per milligram of protein per minute at 37°C and are the average of at least three independent assays.
Primer extension
Total RNA was extracted using Trizol reagent (Gibco-BRL) from exponential-phase cultures of S. aureus RN4220 containing the appropriate plasmid. Glass beads (100 µm; Sigma) in combination with a bead beater (Bio 101) were used for cell lysis. Primer extension was performed as previously described (8) using M-MuLV reverse transcriptase (New England Biolabs) and sequencing ladders were prepared with the SequiTherm EXCEL II DNA sequencing kit (Epicentre Technologies).
Protein overexpression and purification
The pSK41 artA coding region was PCR-amplified and digested with EcoRI and PstI, and cloned into the respective sites of the expression vector pTTQ18RGSH6. An RGSH6 tag was fused to the C-terminal end to facilitate purification of ArtA. The fusion construct, pSK6825, was checked by DNA sequencing and used to transform E. coli BL21(DE3). Recombinant ArtA protein was purified using Ni-NTA chromatography. Pure ArtA was eluted with native elution buffer (50 mM Tris–HCl pH 7.0, 300 mM NaCl, 300 mM imidazole) and aliquots of each purification step were analyzed by SDS-PAGE. ArtA was then re-buffered into DNA-binding buffer (10 mM Tris–HCl pH 7.5, 10 mM MgCl2, 100 mM NaCl, 0.2 mM DTT, 10% glycerol), using a Sephadex PD10 column (GE Healthcare).
DNA-binding experiments
The pSK41 backbone promoter regions were PCR-amplified and end-labeled using [γ32P]ATP (GE Healthcare) and T4 polynucleotide kinase (New England Biolabs). The end-labeled promoter fragments were purified using the illustra™ DNA and Gel Purification Kit (GE Healthcare), eluted in water and stored at −20°C. Electrophoretic mobility shift assays (EMSAs) were performed by incubating the end‐labeled fragments (6000 c.p.m.) with 2 µg of poly[dI-dC] (Sigma Aldrich) and increasing amounts of purified ArtA in DNA-binding buffer (10 mM Tris–HCl pH 7.5, 10 mM MgCl2, 100 mM NaCl, 0.2 mM DTT, 10% glycerol). Binding reactions (20 µl total volume) were incubated for 30 min at room temperature and analyzed using 4% polyacrylamide gels and 0.5 X TBE buffering.
DNase I footprinting was performed using end-labeled promoter fragments (one primer end-labeled prior to PCR-amplification), which were incubated with increasing amounts of purified ArtA using the EMSA conditions described above. The volume of each reaction was brought to 200 µl with DNase I buffer (10 mM Tris–HCl pH 8.0, 5 mM MgCl2, 1 mM CaCl2, 100 mM KCl, 2 mM DTT, 50 µg/ml BSA, 2 µg/ml salmon sperm DNA). DNase I, at a concentration pre-determined to nick ∼50% of the DNA once, was added in 20 µl of DNase I buffer and digestion was allowed to proceed for 2 min at room temperature before the addition of 700 µL of DNase I stop solution (92% ethanol, 3 M sodium acetate, 10 µg/ml salmon sperm DNA). DNA samples were ethanol precipitated and analyzed using denaturing 8% polyacrylamide sequencing gels. Sequencing ladders were prepared using the SequiTherm EXCEL II DNA sequencing kit (Epicentre Technologies).
Protein expression and purification for crystallization
For crystallization, ArtA was produced using a different expression construct, as well diffracting crystals could not be obtained with a His-tagged ArtA protein. To produce this construct, the artA gene was cloned into pET-15b (Novagen) using NdeI and XhoI restriction sites, producing a protein containing a cleavable N-terminal His-tag. The protein was purified using Ni2+-NTA chromatography and then dialyzed into digestion buffer (20 mM Tris–HCl pH 8.0, 500 mM NaCl) for 5–6 h. Thrombin cleavage (GE Healthcare Biosciences) was then carried out overnight at room temperature to remove the N-terminal His-tag. Finally, gel filtration was applied to remove residual thrombin. ArtA was buffer exchanged and concentrated to 30 mg/ml in 20 mM Tris–HCl pH 8.0, 50 mM NaCl.
Crystallization
For crystallization, a 12 bp dsDNA (5′-GACATGACATGT-3′ and 5′-CACATGTCATGT-3′, Oligos Etc.) was annealed by heating the DNA to 95°C for 5 min, followed by slow cooling at room temperature. The best crystals were obtained using a complex composed of a molar ratio of one ArtA dimer to one DNA duplex. Crystals were grown by hanging-drop vapor diffusion using 5% (w/v) PEG 2000, 100 mM Acetate pH 4.60 as a crystallization solution.
Data collection, structure determination and refinement
Because ArtA contains only one methionine at its N-terminus, the structure was solved by bromo-MAD phasing using crystals containing bromouracil substitutions for thymines in the DNA operator site (5′-GACAXGACAXGT-3′ and 5′-CACAXGTCAXGT-3′, where X represents 5-bromo-deoxyuridine). The crystals contained an ArtA dimer and DNA duplex in the crystallographic asymmetric unit (ASU) and all four bromine sites were located with SOLVE (17), resulting in a figure of merit of 0.58 to 2.80 Å resolution. Model building was carried out using the graphics programs O and Coot (18,19). After multiple rounds of refinement in CNS (20) and re-building, using the high-resolution native data, the Rfactor/Rfree converged to 23.9/26.4% at 2.35 Å resolution. The refinement statistics are summarized in Table 2. The coordinates and structure factors have been deposited with RCSB Protein Data Bank.
Table 2.
Crystallographic data for ArtA–DNA complex
| Bromo-Uracil MAD data | |||
| Energy (keV) | 13 474.5/ peak | 13 471.0/ inflection | 13 800.0/ remote |
| Resolution (Å) | 60.86–2.80 | 60.86–2.80 | 60.86–2.80 |
| Overall Rsym(%)a | 6.5 (26.4)b | 6.5 (26.2) | 6.9 (29.3) |
| Overall I/σ(I) | 7.8 (2.7) | 7.8 (3.2) | 7.6 (2.4) |
| No. of total reflections | 17 763 | 18 131 | 19 789 |
| No. of unique reflections | 4109 | 4129 | 4567 |
| Multiplicity | 4.3 | 4.4 | 4.3 |
| Overall figure of Meritc | 0.580 | ||
| Refinement statistics | |||
| Resolution (Å) | 36.0–2.35 | ||
| Overall Rsym(%)a | 7.4 (25.8) | ||
| Overall I/σ(I) | 7.8 (2.8) | ||
| No. of total reflections | 21 181 | ||
| No. of unique reflections | 10 661 | ||
| Complete (%) | 93.1 (93.0) | ||
| Rwork/Rfree (%)d | 23.9/26.4 | ||
| RMSD | |||
| Bond angles (°) | 1.35 | ||
| Bond lengths (A) | 0.008 | ||
| Ramachandran analysis | |||
| Most favored (%) | 88.4 | ||
| Additional allowed (%) | 10.5 | ||
| Generously allowed (%) | 1.1 | ||
| Disallowed (%) | 0.0 | ||
aRsym=ΣΣ|Ihkl–Ihkl(j)|/ΣIhkl, where Ihkl(j) is observed intensity and Ihkl is the final average value of intensity.
bValues in parentheses are for the highest resolution shell.
cFigure of Merit = <|ΣP(α)eIα/ΣP(α)|>, where α is the phase and P(α) is the phase probability distribution.
dRwork = Σ||Fobs|–|Fcalc||/Σ|Fobs| and Rfree = Σ||Fobs|–Fcalc||/Σ|Fobs|; where all reflections belong to a test set of 5% randomly selected data.
Fluorescence polarization assays
Fluorescence polarization (FP) assays of ArtA–DNA binding were performed using a Pavera Beacon Fluorescence Polarization system (21). All oligonucleotides used in the assays were 5′-fluorescein labeled. For each assay, increasing concentrations of ArtA were titrated into the binding mixture containing 2 nM DNA in 20 mM Tris–HCl pH 7.5, 50 mM NaCl. The excitation and emission wavelengths were 490 and 530 nm, respectively. All data were processed in Kaleidagraph and fit with the equation P = {(Pbound – Pfree)[Protein]/(Kd + [Protein])} + Pfree, where P is the polarization magnitude at a given protein concentration, Pfree is the initial polarization of the free oligonucleotide and Pbound is the maximum polarization when the oligonucleotide is saturated by ArtA. Non-linear least squares analysis was applied to determine Pbound, and Kd. In all assays, poly[dI-dC] (5 µg/ml, Sigma Aldrich) was used as a non-specific binding competitor. FP stoichiometry experiments were carried out by titrating ArtA into a solution with 20 mM Tris–HCl pH 7.5, 50 mM NaCl, 2 nM fluoresceinated oligonucleotide and 25-fold excess non-fluoresceinated oligonucleotide. In all titrations, 1 μg/ml poly(dI-dC) was added as a non-specific DNA competitor. All titration curves were fitted by Kaleidagraph.
RESULTS
The ArtA regulon
To establish which pSK41 backbone promoters (Figure 1) are regulated by ArtA, the three pSK41 tra promoters and ten Region 1 promoters were amplified from pSK41 by PCR and cloned upstream of the promoterless cat reporter gene of the promoter-probe vector pSK5483 (8), which facilitates measurement of CAT activity as an indicator of promoter strength in S. aureus. CAT assays were performed on whole-cell lysates prepared from S. aureus RN4220 cells harboring individual reporter constructs co-resident with the ArtA expression plasmid pSK7701 or the corresponding control vector pSK7700. Thus in this assay, ArtA responsiveness was indicated by significantly reduced CAT activity from a given pSK41 promoter when co-resident with pSK7701, in comparison to that obtained in the presence of pSK7700.
Figure 1.
Genetic map of pSK41 (6). Promoters of the pSK41 backbone (Region 1 and tra region) are denoted by arrows, with arrowheads indicating the direction of transcription. ArtA-regulated genes are colored grey. Resistance genes shown are aacA–aphD and aadD (aminoglycoside resistance), ble (bleomycin resistance) and qacC (antiseptic/disinfectant resistance). Other loci of known function include nes (conjugative nickase), oriT (origin of conjugative transfer), parM and parR (partitioning), rep (replication initiation) and res (multi-mer resolution). Also indicated are the locations of cointegrated plasmids, including pUB110, copies of IS257 and a Tn4001-like transposon.
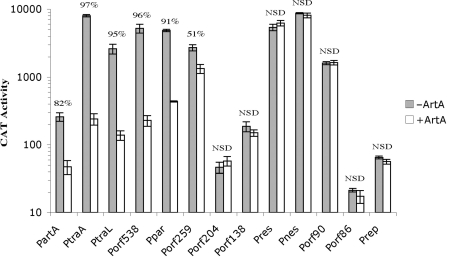
As shown in Figure 2, six of the promoters tested, PartA, PtraA, PtraL, Porf538, Porf259 and Ppar, were significantly repressed in cells expressing the ArtA protein. The promoters of the two tra region operons, PtraA and PtraL, were each repressed by >90%, as were Porf538 and Ppar from Region 1. PartA was auto-regulated (82% repression) and Porf259 was the least sensitive ArtA-regulated promoter (51% repression). Six other Region 1 promoters were not regulated by ArtA, including those for the replication initiation gene, rep, the resolvase, res and the conjugative nickase, nes. Additionally, since ArtA had no affect on transcription from Prep, it is likewise expected to have no influence on the promoter for the rep anti-sense regulator RNAI (8), which was present in the Prep-cat reporter plasmid and hence should have revealed any such regulation; this could not be tested directly because PrnaI-cat constructs appear to be non-viable.
Figure 2.
CAT activity (n mol of chloramphenicol acetylated per milligram per minute) of the pSK41 backbone promoter reporter constructs in the absence/presence of ArtA. The mean CAT activity of several replicates is shown with error bars denoting the standard deviation. The percentage decrease in CAT activity in the presence of ArtA is shown for each promoter region. NSD, no significant difference.
Primer extension mapping confirmed the existence of transcript start points (TSPs) for each of the ArtA-regulated promoters; TSPs for Prep, and Pres have been reported previously (8,9). The present studies employed RNA isolated from S. aureus cells harboring pSK41, and additionally for promoters within Region 1, from cells containing the pSK41 derivative pSK5093 that lacks the tra region, and hence artA, due to a deletion resulting from homologous recombination between flanking IS257 elements (6). pSK41-derived RNA facilitated the detection of appropriately located TSPs for Porf259, Porf538 and PtraA (Supplementary Figure S1; Figure 3). Consistent with artA derepression, pSK5093-derived RNA additionally enabled detection of Ppar, and yielded more intense signals for both Porf259 and Por538. TSPs for PartA and PtraL were similarly detected in the absence of artA by using RNA isolated from cells harboring the relevant Ptra-cat fusion constructs employed above (pSK7759 and pSK7761, respectively).
Figure 3.
Alignment of the ArtA-regulated pSK41 backbone promoter sequences. The −10 and −35 promoter regions are highlighted in grey and the consensus sequences are shown above. ArtA-protected regions determined by footprint analysis are underlined. Arrows indicate the transcriptional start points and dots in the sequence denote gaps.
Delineation of ArtA operators
In EMSAs, purified ArtA protein was shown to bind specifically to DNA fragments containing each of the promoters found to be repressed by ArtA in CAT assays, but not those containing the ArtA-insensitive promoters (data not shown). DNase I footprinting was undertaken to localize ArtA binding at each promoter. These studies revealed that the region protected by ArtA encompassed CATGACA sequences that overlay the −35 sequence of each regulated promoter (Supplementary Figure S2; Figure 3).
Overall structure of ArtA–DNA complex
ArtA shows no sequence homology to any structurally characterized DNA-binding protein. Thus to elucidate the mechanism by which ArtA binds its cognate DNA site, we next crystallized and determined the structure of ArtA bound to a 12-mer DNA duplex containing the ArtA consensus site (top strand; 5′-GACATGACATGT-3′, consensus shown in bold) (Figure 4A). The structure was solved by multiple wavelength anomalous diffraction (MAD) using DNA in which thymines were substituted with 5-Bromo-uracil (see ‘Materials and Methods’ section). There are two ArtA molecules and one 12-mer DNA duplex in the ASU. The structure includes 11 bp of the DNA duplex and residues 7–59 of each subunit and has been refined to an Rwork/Rfree of 23.9/26.4% to 2.35 Å resolution (Table 2).
Figure 4.
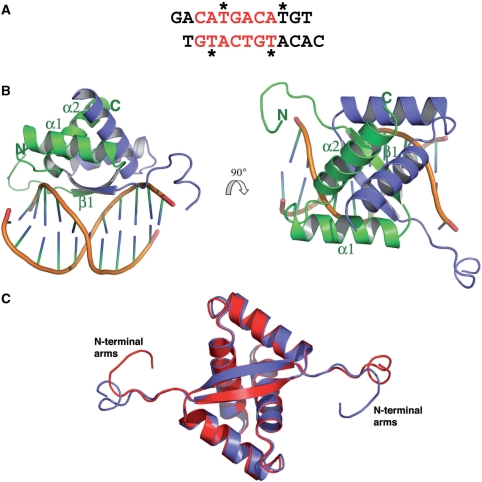
(A) DNA sequence used in crystallization of the ArtA–DNA complex. The ArtA consensus site is colored red and the positions of the thymines that were substituted with 5-bromouracil for MAD phasing are indicated by asterisks. (B) Overall structure of ArtA, one subunit of the dimer is colored in green and the other in purple. Shown are two views of the complex related by a 90° rotation. (C) Superimposition of the ArtA dimer subunits showing the conserved nature of the RHH-fold and the distinct conformations adopted by the N-terminal arms. This figure and Figures 5A–C and Figure 8 were made using PyMOL (7).
The structure reveals that ArtA belongs to the ribbon–helix–helix (RHH) family of DNA-binding proteins and displays the topology β1–α1–α2 (β1; residues 17–23, α1; residues 26–38, α2; residues 43–59) (Figure 4B). Structural homology searches revealed that the ArtA RHH unit shows the strongest structural similarity with the transcriptional repressor CopG protein; the ArtA and CopG structures superimpose with a root mean squared deviation (RMSD) of 1.57 Å for 44 corresponding Cα atoms (22). Two ArtA subunits tightly associate in the structure to form the functional RHH2 unit. The hydrophobic core of ArtA is extensive and is formed by residues V17, L19 and L21 from the β strand, residues M26, I31, I32, Y34 from α1 and residues L43, I50, L51, L55, I58 from α2.
While the RHH units of each ArtA monomer adopt essentially identical conformations, as underscored by the RMSD of 0.31 Å for superimposition of Cα atoms of residues 17–59, the N-terminal residues 7–16 adopt distinct conformations in each subunit and extend outward towards the DNA (Figure 4C). Indeed, residues from this N-terminal arm provide some interactions with the DNA phosphate backbone (shown below). However, all DNA–nucleobase interactions are provided by residues located on the anti-parallel β strands. Notably, specification of the ArtA DNA consensus sequence is mediated by one ArtA dimer (Figure 5A–D). This reveals a significant distinction between ArtA and most other structurally characterized RHH proteins, which bind DNA as dimer-of-dimers (23).
Figure 5.
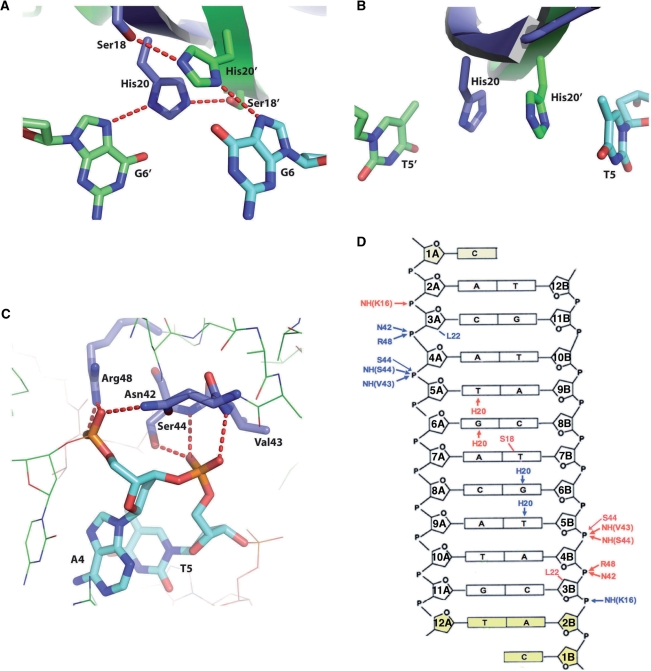
ArtA–DNA interactions. (A) Close up of hydrogen bond interactions between H20 and guanine 6. (B) Stacking interactions between H20 and thymine 5. (C) Interactions between the RHH-loop region and the DNA phosphate backbone. (D) Schematic diagram showing the interactions between ArtA and the DNA site. Residues from different subunits of ArtA are colored in blue (chain A) and red (chain B), respectively. Hydrogen bond and van der Waals contacts are indicated by arrows and lines, respectively, between the residue and nucleotide. Nucleotides not visible in the crystal structure are colored yellow.
Operator recognition
In most RHH proteins, three residues in the ribbon (β-strand) mediate specific interactions with bases in the DNA major groove (8,23–32). In ArtA, the corresponding residues, S18, H20 and L22, participate in either base specific or phosphate contacts. The L22 side chain makes hydrophobic contacts with DNA ribose groups. In addition, the carbonyl group of L22 forms hydrogen bonds with the Nε of R48′ (where ‘ ′ ’ indicates other subunit of the dimer), which positions the R48 side chain optimally for interaction with the phosphate backbone. Finally, the amide nitrogen of L22 interacts with same phosphate group indirectly via a water mediated contact. Interestingly, the majority of base contacts are made by one ArtA residue, H20, from each subunit. The environment of the DNA likely influences the side chain H20 pKa as we find that ArtA is able to bind its consensus site with equal affinities at pH values ranging from 4.6 to 8.5 (Supplementary Figure S3). The H20 side chain participates in hydrogen bonds, hydrophobic and stacking interactions. Specifically, Nδ of H20 interacts with the N7 of guanine 6 (Figure 5A). The H20 imidazole ring also stacks with thymine 7, where the distance between methyl group of thymine and imidazole ring of H20 is ∼4 Å (Figure 5B). The Nε of H20 hydrogen bonds with Oγ of S18′ from its dimer mate and this interaction positions the S18 side chain Cβ from one subunit to make hydrophobic contacts with the methyl group of the central thymine base. Notably, this contact from S18 is the only base interaction not provided by H20 (Figure 5D).
An important structural characteristic of RHH family proteins is a conserved loop motif G-X-S/T/N between α2 and α2, which makes contacts with the DNA phosphate backbone and helps dock the RHH onto the DNA (Figure 5C). These interactions involve direct contacts from amide nitrogens and side chains near the N-terminus of α2 as well as a positive contribution from the helical dipole of α2, which points directly towards the phosphate backbone. In ArtA, the G-X-S/T/N motif is slightly elongated and has a different residue content than other structurally characterized RHH. Nonetheless, it retains a structure similar to other RHH proteins and the N-terminus of its α2 makes numerous interactions with the DNA phosphate backbone. The Nδ of N42 in the loop interacts with O2P of Adenine 4 and two nitrogens of main-chain amides of V43 and S44 contact phosphate backbone atoms O2P and O1P of thymine 5 (Figure 5C). Finally, R48 forms salt bridges with the O1P moiety of Adenine 4.
ArtA-bound DNA conformation
The ArtA-bound DNA is primarily B form in conformation. For example, the average twist of the ArtA-complexed DNA is ∼33.4 Å compared with 34.3 Å for B-DNA. The central consensus site specified by ArtA is not significantly bent (33). However, conformational alterations caused by ArtA binding include major groove widening near the bound H20 residues, whereby the DNA major groove width is 15.0 Å compared with 11.7 Å in B-DNA. The minor groove is correspondingly compressed to a width of 4.2 Å, compared with 5.7 Å in B-DNA. This DNA distortion in groove width may play some role in transcriptional regulation by affecting binding of the σ factor (see below).
FP analysis of ArtA-operator binding
As noted, ArtA is somewhat unique among RHH proteins in that it binds the cognate TGACA site located in the promoters it regulates as a dimer. However, in addition to its RHH motif, ArtA contains a 16 residue N-terminal arm, parts of which cannot be identified in electron density maps. Interestingly, this extended region (MNNNEENSVFFGKKKK) lies close to the phosphate backbone in the structure and contains four consecutive lysine residues, K13 through K16, indicating that it may play a role in DNA binding. Indeed, the amide nitrogens of the 2-fold related K16 residues make contacts to the phosphate backbone while the lysine side chains make electrostatic interactions with the DNA. The remaining lysine residues appear too far from the DNA to contribute directly to nucleotide binding. However, the DNA we used for crystallization was a 12-mer oligonucleotide containing the minimal ArtA consensus binding site and thus, conceivably, might not be long enough to provide phosphates for interaction with lysine residues in the extended arm. This prompted us to ask whether this region might play a role in binding longer DNA sites. To address this possibility, a truncation mutant, Δ14ArtA, was made in which the first 14 amino acids were removed. FP studies were then carried out to analyze the DNA-binding activities of the wild-type ArtA and the truncation mutant Δ14ArtA. Three oligonucleotides of different length (11-, 16- and 22-mer) were designed based on the consensus binding site in the ArtA-regulated promoters to assay DNA binding to the minimal versus longer DNA sites. The results are summarized in Table 3. These studies revealed no differences in the binding affinities of the 16- and 22-mer DNA sites for ArtA and Δ14ArtA and a very minor, or 2-fold decrease, in wild-type ArtA and Δ14 ArtA binding to the 11-mer compared with the 16- and 22-mer. The slight reduction in DNA binding of ArtA to the 11-mer compared with the longer sites may indicate that residues other than those in the N-terminal arms are providing minor contributions to DNA binding to longer sites, likely to the phosphate backbone. However, 2-fold differences in binding are on the order of one to a few weak interactions and indicate that the binding to the short site is essentially the same as to longer sites. In any case, the data clearly show that the N-terminal arms are not important for high-affinity binding to these single site operators.
Table 3.
KdS of ArtA and Δl4ArtA binding to operator DNA sites
| Oligonucleotides | ArtA-Kd(nM) | Δ14ArtA-Kd (nM) |
|---|---|---|
| ACATGACATGT | 127 + 5 | 222 + 8 |
| ACATGACATGACATGT | 60 + 3 | 58 + 3 |
| CAAACATGATATGACATGTAAT | 62 + 6 | 58 + 5 |
| TTGTCATGACATGTCATGTGTAA (PartA) | 28 + 8 | 41 + 9 |
| TTACATGACATGACATGTAATAC (PtraA) | 42 + 9 | 52 + 6 |
| TGCATTACATGACATGACATGTAAT (PtraL) | 23 + 5 | 39 + 5 |
| ACATGACAGGT (Porf259) | 790 + 34 | – |
| TAAACATTGCATAACATGACAGGT (Porf259) | – | – |
| ACACGACATCA (Porf538) | – | – |
| AAATGACACGT (Porf538) | – | – |
| ACACGACATGAAATGACACGT (Porf538) | 58+5 | – |
| ACACTAAATGAAATGACACGT (Mutant 1) | – | – |
| ACACGACATGAAATTAAACGT (Mutant 2) | – | – |
Note: ‘–’ indicates that no measurable binding was detected.
Interestingly, like Ppar, the promoters for PtraL and PtraA contain two consecutive TGACA motifs that multiple ArtA molecules might bind. However, modeling shows that two ArtA dimers cannot dock simultaneously onto DNA-containing contiguous TGACA repeats such as in Ppar, PtraL and PtraA without steric clash. These combined data are consistent with the idea that dimeric ArtA is the functional DNA-binding unit for these promoter operators. Moreover, these data also indicate that, in these cases, the long N-terminal arms of ArtA do not contribute to DNA binding by either contacting the DNA or participating in protein–protein interactions that aid DNA binding. Indeed, FP experiments examining binding to the full length promoter operators of Ppar, PartA, PtraA and PtraL showed no difference in binding affinity between the wildtype and Δ14 mutant of ArtA and the binding affinities were essentially the same as obtained for the single ArtA consensus site (Table 3).
The lower binding affinity of ArtA for its operator in Porf259 (790 nM compared with 127 nM for the optimal site) may be explained by the fact that it contains a single base difference compared with the consensus ArtA-binding site (in position 9 in which the T is changed to G). The phosphate group of this nucleotide interacts extensively with N cap of α2 and these contacts may be influenced by nucleotide identity (Figure 5D). Notably, the reduced affinity of ArtA for Porf259 is consistent with the finding that Porf259 was the least sensitive of the ArtA-regulated promoters (Figure 2). The ArtA DNase I footprint data obtained for each of the six ArtA-regulated promoters (Supplementary Figure S2; Figure 3) are in good agreement with our FP data. However, the protected regions determined here are markedly smaller than those described previously for the ArtA homolog, TrsN (11). The larger footprints observed in that study, however, are likely attributable to the much larger TrsN fusion protein, which contained a glutathione S-transferase affinity tag.
Interestingly, one operator bound by ArtA, Porf538, presented a paradox. Although this site is regulated by ArtA, an obvious consensus site within the promoter is unclear. Two possible matches that are close to the consensus were identified (Figure 3; Table 3). Individually, neither of these sites showed detectable binding as measured by FP. However, the 21-mer oligonucleotide, which covers both sites was found to bind wild-type ArtA saturably, with a Kd of 58 nM. Because this site contains two possible binding motifs (Table 3) but neither is sufficient for DNA binding, we reasoned that this site might bind two ArtA dimers. Indeed, modeling revealed that two ArtA dimers would bind on the same side of the DNA duplex on this operator (Supplementary Figure S4). This binding mode shows no steric clash and critically, the N-terminal arm of one subunit is juxtaposed next to the adjacent molecule in the neighboring dimer, suggesting that ArtA may utilize its N-terminal arms in protein–protein interactions when binding to Porf538. If this is the case, the ArtA truncation mutant should display weakened binding to this site. Indeed, FP experiments showed that removal of the N-terminal arms essentially abrogated binding to the 21-mer Porf538 site (Figure 6). To further test the Porf538 dimer-of-dimer model, two Porf538 mutants were designed (Mutant 1: ACACTAAATGAAATGACACGT; Mutant 2: ACACGACATGAAATTAAACGT). In each of these oligonucleotides, the two key bases involved in H20 recognition (G and C) were mutated (G to T and C to A). As predicted, none of these mutant oligonucleotides were bound by ArtA, consistent with the previous result that ArtA cannot bind either 12-mer oligonucleotide with only one binding site derived from Porf538. In contrast, as noted, previous FP experiments revealed that both Δ14 ArtA and wild-type ArtA bound the full length promoter regions of Ppar, PartA, PtraA and PtraL with the same affinity as the single ArtA consensus site, consistent with a single ArtA dimer binding to these promotors.
Figure 6.
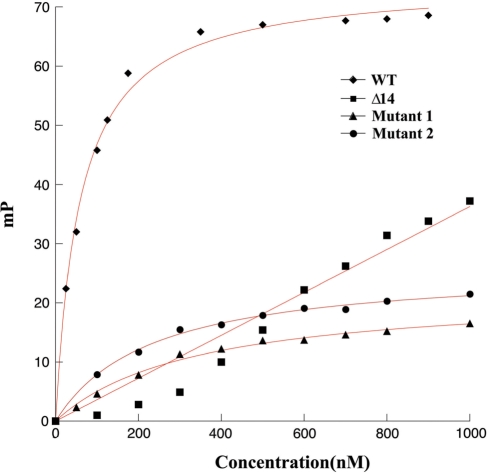
FP binding isotherms for wild type ArtA and Δ14ArtA binding to the 21-mer DNA sites from Porf538. Filled diamond represents the curve for wild-type ArtA binding to Porf538. Filled squares for Δ14ArtA binding to Porf538. Filled triangles for wild-type ArtA binding to the oligonucleotide mutant 1 Porf538 site. Filled circles for wild-type ArtA binding to the oligonucleotide mutant 2 Porf538 site. FP units (mP, millipolarization) and ArtA concentrations are along the y- and x-axis, respectively.
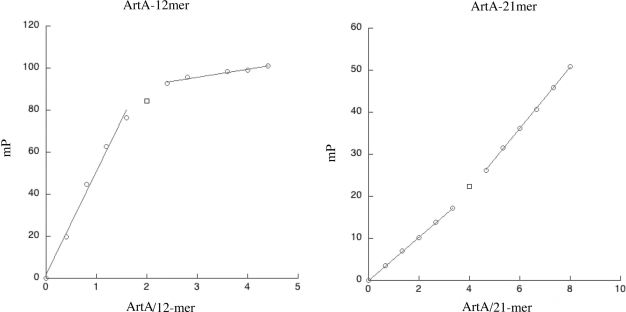
Finally, we utilized FP to directly ascertain the stoichiometry of ArtA binding to the Porf538 site and to the consensus operator site. These studies, which showed that two ArtA subunits (i.e. a ArtA dimer) bound the consensus operator site, while Porf538 is bound by four ArtA subunits, are consistent with the model that ArtA binds Porf538 as a dimer-of-dimers and the consensus sites as only a dimer (Figure 7). While the inflection point on the ArtA-consensus site curve is typical of most DNA-binding proteins in that, after all the DNA sites are saturated, the curve is primarily flat, it is interesting that although the inflection point in the ArtA–Porf538 curve is clear and indicates saturation of the specific sites by four ArtA subunits, addition of more ArtA protein in this case does not lead to a flat curve but an additional increase. This suggests that after the specific sites are saturated, more ArtA molecules non-specifically interact with either the protein or the DNA. Thus, the combined data show that ArtA binds DNA utilizing different modes of binding. In the case of the Ppar, PartA, PtraA, PtraL and Porf259 operators, ArtA binds as a dimer and there are no protein–protein interactions involved in this binding outside the contacts between subunits in the dimer. In contrast, ArtA binds to the atypical Porf538 operator as a dimer-of-dimers and utilizes, in addition to its RHH, its extended N-terminal arms, presumably for mediating dimer–dimer contacts.
Figure 7.
Stoichiometry of ArtA–DNA binding by FP. (A) Titration curve of ArtA into the consensus 12-mer used in crystallization resulted in a molar ratio of ArtA subunit to DNA duplex of ∼2. (B) ArtA titrated into the 21-mer DNA sites containing Porf538 resulted in a molar ratio of ArtA subunit to DNA duplex of ∼4.
ArtA transcription repression mechanism
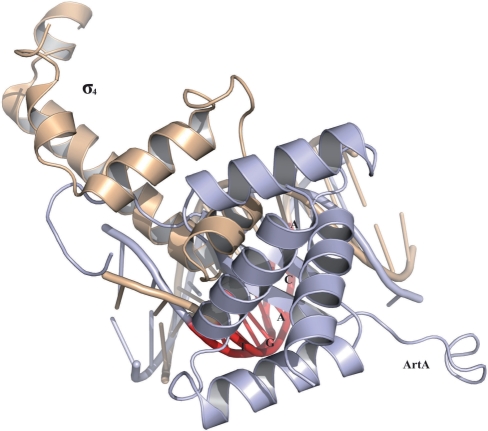
Notably, the ArtA-binding sites within the Ppar, PartA, PtraA and PtraL promoters overlap the −35 motifs, while the ArtA-binding site within the Porf259 promoter is located slightly downstream of the −35 position. Finally, the ArtA-binding site within the Porf538 promoter, which we find ArtA binds as a dimer-of-dimers, extends over the entire −35 box. Thus, all the ArtA-binding sites overlap or impinge on the −35 boxes of the promoters that it regulates. This suggested that ArtA may repress transcription by preventing binding of the sigma factor. Regions 4 of sigma factors, which specify binding to the −35 boxes of the promoters, are very conserved among bacteria (34−36). The structure of the Thermus aquaticus sigma Region 4–DNA complex has been solved and can serve as a model for the S. aureus sigma Region 4–DNA complex as the two share 79% sequence similarity and importantly, all the residues involved in −35 box binding are identical between the two proteins (36). The nucleotides within the −35 box in the promoters bound by ArtA are ATGACA instead of the typical −35 TTGACA. Superposition of the T. aquaticus Sigma A Region 4–DNA complex onto ArtA–DNA complex using the conserved TGACA −35 box region as a guide shows explicitly that binding of ArtA would completely block the binding of Region 4 of S. aureus sigma factor A at the −35 box (Figure 8). Furthermore, the DNA distortion induced by ArtA binding would also hinder the accessibility of the sigma factor. Thus, these data indicate that ArtA represses transcription by physically blocking access of the promoter to the sigma factor and by altering the DNA conformation such that sigma binding would be unfavorable.
Figure 8.
Overlay of σ4-DNA (coordinates 1KU7) and ArtA–DNA complexes indicating that ArtA binding would prevent σ4 binding to the −35 promoter site. The phosphate backbone of the TGACA DNA sites (−35 site) of the σ4-DNA (wheatish) and ArtA–DNA (light blue) complexes were superimposed. For reference the −35 TGACA site is colored red.
DISCUSSION
We have shown here that ArtA represses six promoters in Region 1 and the tra region of pSK41. All but PartA itself and Porf259 are likely to direct transcription of operons. As a consequence, ArtA is expected to regulate the expression of 21 out of a total of 30 coding sequences contained within the pSK41 backbone (Figure 1). It should be noted that the only operon promoter not regulated by ArtA, Porf204, corresponds to an example of an IS257-hybrid promoter where the −35 sequence is located within the terminal inverted repeat of the upstream IS257 that is partnered to a fortuitously located −10 sequence present in the flanking sequence. Such IS257-hybrid promoters have previously been shown to direct transcription of resistance genes (37,38). It was hypothesized that the acquisitions of IS257 elements and associated resistance genes are recent evolutionary events and that Region 1 and the tra region were previously contiguous in a pSK41 ancestor (6). If correct, it is likely that transcription of orf204 and orf423 would have initiated at PtraL, and hence would also have been under the control of ArtA. With regard to this, it is worth noting that pSK41-like plasmids have now been identified where the Region 1 and tra region termini evident in pSK41 are indeed contiguous. In the mupirocin resistance plasmid pV030–8 (GenBank entry EU366902), the truncated remnant present at the end of the pSK41 tra region, orf55 (Figure 1), corresponds to a 242 codon ORF, which is immediately followed by a 207 codon ORF and then orf204 and orf423 homologues. Therefore, in pV030–8 PtraL probably directs transcription of a six gene operon. Transposon mutagenesis and complementation studies of pGO1 indicate that trsL and/or trsM are required for conjugative transfer (39), so these co-transcribed genes may likewise be associated with this function. Although the majority of ArtA-regulated genes are involved in conjugative transfer, ArtA also participates in the repression of Ppar, which transcribes the functional parMR type II partitioning system. This operon is also auto-regulated by the centromere binding protein ParR, which like ArtA, contains a RHH DNA-binding fold (10). Thus, transcription of the par operon is subject to two levels of control, mediated by two different proteins that utilize RHH folds specific for distinct DNA-binding sites.
The RHH DNA-binding motif is a common DNA-binding motif found in prokaryotes (23). Indeed, ∼2000 RHH-domain containing protein sequences have been identified. To date, >15 structures have been determined of RHH proteins in their apo or DNA-bound forms. A characteristic feature of most RHH proteins is that they recognize their DNA sites by forming dimer-of-dimers. This increases specificity in DNA binding through the formation of DNA base contacts from two dimeric modules as a single RHH module can only specify ∼6 nt in one major groove. Indeed, several RHH proteins have been shown to bind cooperatively to extended DNA segments to form superstructures on DNA. This was first demonstrated for the RHH protein CopG (40). Subsequently, such cooperative binding by the pSK41 ParR protein was shown to lead to the formation of a specific superhelical partition complex that is critical for plasmid DNA segegation by the type II ParR-ParM proteins (10). Therefore, it is interesting that ArtA appears to display altered ability in how it binds DNA in that it functionally represses most of its promoters by binding as a dimer, but can also bind a non-canonical DNA operator as a dimer-of-dimers.
The formation of ArtA dimer-of-dimers was shown to be dependent on its N-terminal arm regions. Long N-terminal regions have been found in the RHH proteins MetJ, ParR and Omega repressor (10,24,25). However, in these proteins, the N-terminal arms play different roles than they do in ArtA. Specifically, the N-terminal regions of MetJ fold back and interact with Helix 2 and are involved in binding to the MetJ corepressor, S-adenosylmethionine and the N-terminal regions of ParR and the Omega repressor are known to interact with their respective partition NTPase partner proteins, ParM and Delta, respectively (10,25,26). Thus, the utilization of N-terminal arms to mediate RHH dimer-of-dimer interactions is so far unique to ArtA. The formation of dimer-of-dimers by other RHH proteins has been shown to involve different regions and sometimes domains of each protein. Examples include Arc (27), MetJ (24), NikR (28), FitAB (29) and ParR (10). In these cases, the binding sites for each dimer are normally arranged as inverted repeats. In contrast, the ArtA-binding sites in Porf538 are arranged as direct repeats. One further example of a RHH protein that appears to function as a dimer in DNA binding is the E. coli proline utilization protein A (PutA). However, the only other RHH protein that may function similarly to ArtA in being capable of binding as a dimer or dimer-of-dimers is the F plasmid TraY protein (41). Although the structure of TraY has not yet been solved, the protein sequence contains two direct repeats that are predicted to each contain a RHH-fold. If this is the case, TraY forms a monomeric RHH2-fold. Biochemical studies have suggested that it can bind either as a monomer (mimicking a RHH dimer) or a dimer (mimicking a RHH dimer of dimers). Whether these proposed modes of binding are relevant in vivo, however, has not yet been determined.
Although the RHH proteins, Mnt repressor and ParR contain histidines in their ribbon, ArtA is the first RHH protein observed to utilize histidine as its primary sequence specifying residue. In the ParR–DNA structure, the side chain of ParR residue H8 faces the hydrophobic core rather than the DNA and although biochemical data suggest that Mnt uses H6 for DNA contacts, there are no structures available for a Mnt–DNA complex, so this remains to be determined (10,32). ArtA is notable in that not only does it utilize H20 in base contacts, but, in fact, it also relies on this residue for mediating almost all its base specifying interactions. The histidine side chain is somewhat unique in that it has the ability to form hydrophobic contacts, stacking interactions and multiple hydrogen bonds all of which can result in a specific interaction with a given ligand depending on the binding context. In ArtA, the H20 residues make stacking interactions with the central five nucleotides. Stacking interactions between aromatic residues and nucleobases have recently been suggested to play important roles in specific binding of proteins to their DNA targets (42,43). In addition, the H20 residues also make specific hydrophobic interactions with the methyl groups of thymine 4A and thymine 6B. Finally, the 2-fold related H20s also hydrogen bond with guanine 5A and guanine 7B. As a result, the ArtA H20 residues alone are able to almost entirely mediate specific operator binding.
Our FP and footprinting studies show that ArtA binds operator sites that overlap or are next to the −35 box of each promoter suggesting that ArtA may function to repress transcription by physically blocking binding of Sigma A to these promoters. Modeling shows clearly that ArtA and Sigma A cannot bind the −35 region simultaneously. Moreover, ArtA also alters the DNA structure to one that is not favorably bound by Sigma A, Region 4. Thus, together these findings provide a mechanism for ArtA-mediated transcription repression.
The data presented indicates that ArtA coordinately regulates the expression of most cognate pSK41 coding sequences via transcriptional repression. The genes controlled include plasmid housekeeping functions such as plasmid partitioning and conjugation, as well as genes for which functions are yet to be ascribed. ArtA presumably sets a basal level of activity from its target promoters, which may be subject to additional levels of control, as in the case of Ppar. However, the possibility that the expression or activity of ArtA itself might be subject to exogenous control cannot be excluded. Tight control of plasmid gene expression is expected to enhance evolutionary fitness by reducing the burden of plasmid carriage on the host cell. As such, the ArtA regulatory system, which is highly conserved across the pSK41 plasmid family, likely contributes to the capacity of these clinically significant plasmids to confer multiple and diverse antimicrobial resistance phenotypes.
ACCESSION NUMBER
3GXQ.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
M.D. Anderson Trust Fellowship and the National Institutes of Health (grant GM068453 to M.A.S.); University of Sydney R&D Grant (to N.F., M.H.B. and R.A.S.); National Health and Medical Research Council of Australia Project (grant 457454 to N.F., M.A.S., S.M.K., S.O.J. and R.A.S.). Funding for open access charge: National Institutes of Health.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the Advanced Light Source (ALS) and their support staff. The ALS is supported by the Director, Office of Science, Office of Basic Energy Sciences and Material Science Division of the US Department of Energy at the Lawrence Berkeley National Laboratory.
REFERENCES
- 1.Goetghebeur M, Landry PA, Han D, Vicente C. Methicillin-resistant Staphylococcus aureus: a public health issue with economic consequences. Can. J. Infect Dis. Med. Microbiol. 2007;18:27–34. doi: 10.1155/2007/253947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Deurenberg RH, Vink C, Kalenic S, Friedrich AW, Bruggeman CA, Stobberingh EE. The molecular evolution of methicillin-resistant Staphylococcus aureus. Clin. Microbiol. Infect. 2007;13:222–235. doi: 10.1111/j.1469-0691.2006.01573.x. [DOI] [PubMed] [Google Scholar]
- 3.Clements A, Halton K, Graves N, Pettitt A, Morton A, Looke D, Whitby M. Overcrowding and understaffing in modern health-care systems: key determinants in meticillin-resistant Staphylococcus aureus transmission. Lancet Infect. Dis. 2008;8:427–434. doi: 10.1016/S1473-3099(08)70151-8. [DOI] [PubMed] [Google Scholar]
- 4.Navarro MB, Huttner B, Harbarth S. Methicillin-resistant Staphylococcus aureus control in the 21st century: beyond the acute care hospital. Curr. Opin. Infect. Dis. 2008;21:372–379. doi: 10.1097/QCO.0b013e3283013add. [DOI] [PubMed] [Google Scholar]
- 5.Firth N, Skurray RA. The Staphylococcus-genetics: accessory elements and genetic exchange. In: Fischetti VA, Novick RP, Ferretti JJ, Portnoy DA, Rood JI, editors. Gram-Positive Pathogens. 2nd edn. Washington DC: American Society for Microbiology; 2006. pp. 413–426. [Google Scholar]
- 6.Berg T, Firth N, Apisiridej S, Hettiaratchi A, Leelaporn A, Skurray RA. Complete nucleotide sequence of pSK41: evolution of staphylococcal conjugative multiresistance plasmids. J. Bacteriol. 1998;180:4350–4359. doi: 10.1128/jb.180.17.4350-4359.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Delano WL. San Carlos, CA: Delano Scientific; 2002. The PyMOL molecular graphics system. [Google Scholar]
- 8.Kwong SM, Skurray RA, Firth N. Staphylococcus aureus multiresistance plasmid pSK41: analysis of the replication region, initiator protein binding and antisense RNA regulation. Mol. Microbiol. 2004;51:497–509. doi: 10.1046/j.1365-2958.2003.03843.x. [DOI] [PubMed] [Google Scholar]
- 9.LeBard RJ, Jensen SO, Arnaiz IA, Skurray RA, Firth N. A multimer resolution system contributes to segregational stability of the prototypical staphylococcal conjugative multiresistance plasmid pSK41. FEMS Microbiol. Lett. 2008;284:58–67. doi: 10.1111/j.1574-6968.2008.01190.x. [DOI] [PubMed] [Google Scholar]
- 10.Schumacher MA, Glover TC, Brzoska AJ, Jensen SO, Dunham TD, Skurray RA, Firth N. Segrosome structure revealed by a complex of ParR with centromere DNA. Nature. 2007;450:1268–1271. doi: 10.1038/nature06392. [DOI] [PubMed] [Google Scholar]
- 11.Sharma VK, Johnston JL, Morton TM, Archer GL. Transcriptional regulation by TrsN of conjugative transfer genes on staphylococcal plasmid pGO1. J. Bacteriol. 1994;176:3445–3454. doi: 10.1128/jb.176.12.3445-3454.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Firth N, Ridgway KP, Byrne ME, Fink PD, Johnson L, Paulsen IT, Skurray RA. Analysis of a transfer region from the staphylococcal conjugative plasmid pSK41. Gene. 1993;136:13–25. doi: 10.1016/0378-1119(93)90442-6. [DOI] [PubMed] [Google Scholar]
- 13.Thomas CM. Transcription regulatory circuits in bacterial plasmids. Biochem. Soc. Trans. 2006;34:1072–1074. doi: 10.1042/BST0341072. [DOI] [PubMed] [Google Scholar]
- 14.Schenk S, Laddaga RA. Improved methods for electroporation of Staphylococcus aureus. FEMS Microbiol. Lett. 1992;73:133–138. doi: 10.1016/0378-1097(92)90596-g. [DOI] [PubMed] [Google Scholar]
- 15.Birnboim HC, Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979;7:1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shaw WV. Chloramphenicol acethyltransferase from chloramphenicol-resistant bacteria. Methods Enzymol. 1975;43:737–355. doi: 10.1016/0076-6879(75)43141-x. [DOI] [PubMed] [Google Scholar]
- 17.Terwilliger TC, Berendzen J. Automated MAD and MIR structure solution. Acta Crystallogr. D Biol. Crystallogr. 1999;55:849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A. 1991;47(Pt 2):110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 19.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 20.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse‐Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 21.Lundblad JR, Laurance M, Goodman RH. Fluorescence polarization analysis of protein-DNA and protein-protein interactions. Mol. Endocrinol. 1996;10:607–612. doi: 10.1210/mend.10.6.8776720. [DOI] [PubMed] [Google Scholar]
- 22.Gomis-Ruth FX, Sola M, Acebo P, Parraga A, Guasch A, Eritja R, Gonzalez A, Espinosa M, del Solar G, Coll M. The structure of plasmid-encoded transcriptional repressor CopG unliganded and bound to its operator. EMBO J. 1998;17:7404–7415. doi: 10.1093/emboj/17.24.7404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schreiter ER, Drennan CL. Ribbon–helix–helix transcription factors: variations on a theme. Nat. Rev. Microbiol. 2007;5:710–720. doi: 10.1038/nrmicro1717. [DOI] [PubMed] [Google Scholar]
- 24.Somers WS, Phillips SE. Crystal structure of the met repressor-operator complex at 2.8 A resolution reveals DNA recognition by beta-strands. Nature. 1992;359:387–393. doi: 10.1038/359387a0. [DOI] [PubMed] [Google Scholar]
- 25.Weihofen WA, Cicek A, Pratto F, Alonso JC, Saenger W. Structures of omega repressors bound to direct and inverted DNA repeats explain modulation of transcription. Nucleic Acids Res. 2006;34:1450–1458. doi: 10.1093/nar/gkl015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dmowski M, Sitkiewicz I, Ceglowski P. Characterization of a novel partition system encoded by the delta and omega genes from the streptococcal plasmid pSM19035. J. Bacteriol. 2006;188:4362–4372. doi: 10.1128/JB.01922-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Raumann BE, Rould MA, Pabo CO, Sauer RT. DNA recognition by beta-sheets in the Arc repressor-operator crystal structure. Nature. 1994;367:754–757. doi: 10.1038/367754a0. [DOI] [PubMed] [Google Scholar]
- 28.Schreiter ER, Sintchak MD, Guo Y, Chivers PT, Sauer RT, Drennan CL. Crystal structure of the nickel-responsive transcription factor NikR. Nat. Struct. Biol. 2003;10:794–799. doi: 10.1038/nsb985. [DOI] [PubMed] [Google Scholar]
- 29.Mattison K, Wilbur JS, So M, Brennan RG. Structure of FitAB from Neisseria gonorrhoeae bound to DNA reveals a tetramer of toxin-antitoxin heterodimers containing pin domains and ribbon-helix-helix motifs. J. Biol. Chem. 2006;281:37942–37951. doi: 10.1074/jbc.M605198200. [DOI] [PubMed] [Google Scholar]
- 30.Zhou Y, Larson JD, Bottoms CA, Arturo EC, Henzl MT, Jenkins JL, Nix JC, Becker DF, Tanner JJ. Structural basis of the transcriptional regulation of the proline utilization regulon by multifunctional PutA. J. Mol. Biol. 2008;381:174–188. doi: 10.1016/j.jmb.2008.05.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gu D, Zhou Y, Kallhoff V, Baban B, Tanner JJ, Becker DF. Identification and characterization of the DNA-binding domain of the multifunctional PutA flavoenzyme. J. Biol. Chem. 2004;279:31171–31176. doi: 10.1074/jbc.M403701200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Knight KL, Sauer RT. Biochemical and genetic analysis of operator contacts made by residues within the beta-sheet DNA binding motif of Mnt repressor. EMBO J. 1992;11:215–223. doi: 10.1002/j.1460-2075.1992.tb05044.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ravishanker G, Swaminathan S, Beveridge DL, Lavery R, Sklenar H. Conformational and helicoidal analysis of 30 PS of molecular dynamics on the d(CGCGAATTCGCG) double helix: ‘curves’, dials and windows. J. Biomol. Struct. Dyn. 1998;6:669–699. doi: 10.1080/07391102.1989.10507729. [DOI] [PubMed] [Google Scholar]
- 34.Helmann JD, Chamberlin MJ. Structure and function of bacterial sigma factors. Annu. Rev. Biochem. 1988;57:839–872. doi: 10.1146/annurev.bi.57.070188.004203. [DOI] [PubMed] [Google Scholar]
- 35.Gruber TM, Bryant DA. Molecular systematic studies of eubacteria, using sigma70-type sigma factors of group 1 and group 2. J. Bacteriol. 1997;179:1734–1747. doi: 10.1128/jb.179.5.1734-1747.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Campbell EA, Celenov M, Sun JL, Olson CA, Weinman O, Trester-Zeditz ML, Darst SA. Structure of the bacterial RNA polymerase promoter specificity sigma subunit. Mol. Cell. 2002;9:527–539. doi: 10.1016/s1097-2765(02)00470-7. [DOI] [PubMed] [Google Scholar]
- 37.Leelaporn A, Firth N, Paulsen IT, Skurray RA. IS257-mediated cointegration in the evolution of a family of staphylococcal trimethoprim resistance plasmids. J. Bacteriol. 1996;178:6070–6073. doi: 10.1128/jb.178.20.6070-6073.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Simpson AE, Firth N, Skurray RA. An IS257-derived hybrid promoter directs transcription of a tetA(K) tetracycline resistance gene in the Staphylococcus aureus chromosomal mec region. J. Bacteriol. 2000;182:3345–3352. doi: 10.1128/jb.182.12.3345-3352.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Morton TM, Eaton DM, Johnston Jl, Archer GL. DNA sequence and units of transcription of the conjugative transfer gene complex (trs) of Staphylococcus aureus plasmid pGO1. J. Bacteriol. 1993;175:4436–4447. doi: 10.1128/jb.175.14.4436-4447.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Costa M, Sola M, del Solar G, Eritja R, Hernandez-Arriaga AM, Espinosa M, Gomis-Ruth FX, Coll M. Plasmid transcriptional repressor CopG oligomerizes to render helical superstructures unbound and in complexes with oligonucleotides. J. Mol. Biol. 2001;310:403–417. doi: 10.1006/jmbi.2001.4760. [DOI] [PubMed] [Google Scholar]
- 41.Nelson WC, Matson SW. The F plasmid traY gene product binds DNA as a monomer or a dimer: structural and functional implications. Mol. Microbiol. 1996;20:1179–1187. doi: 10.1111/j.1365-2958.1996.tb02638.x. [DOI] [PubMed] [Google Scholar]
- 42.Tao F, Goswami J, Bernasek SL. Competition and coadsorption of di-acids and carboxylic acid solvents on HOPG. J. Phy. Chem. B. 2006;110:19562–19569. doi: 10.1021/jp063923a. [DOI] [PubMed] [Google Scholar]
- 43.Lesley R, Rutledge LSCV, Stacey DW. Characterization of the stacking interactions between DNA or RNA nucleobases and the aromatic amino acids. Chem. Phys. Lett. 2007;444:167–175. [Google Scholar]
- 44.Kreiswirth BN, Lofdahl S, Betley MJ, O’Reilly M, Schlievert PM, Bergdoll MS, Novick RP. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature. 1983;305:709–712. doi: 10.1038/305709a0. [DOI] [PubMed] [Google Scholar]
- 45.Stark MJ. Multicopy expression vectors carrying the lac repressor gene for regulated high-level expression of genes in Escherichia coli. Gene. 1987;51:255–267. doi: 10.1016/0378-1119(87)90314-3. [DOI] [PubMed] [Google Scholar]
- 46.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 47.Novick RP, Richmond MH. Nature and interactions of the genetic elements governing penicillinase synthesis in Staphylococcus aureus. J. Bacteriol. 1965;90:467–480. doi: 10.1128/jb.90.2.467-480.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Firth N, Apisiridej S, Berg T, O’Rourke BA, Curnock S, Dyke KGH, Skurray RA. Replication of staphylococcal multiresistance plasmids. J. Bacteriol. 2000;182:2170–2178. doi: 10.1128/jb.182.8.2170-2178.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kwong SM, Skurray RA, Firth N. Replication control of staphylococcal multiresistance plasmid pSK41: an antisense RNA mediates dual-level regulation of Rep expression. J. Bacteriol. 2006;188:4404–4412. doi: 10.1128/JB.00030-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kwong SM, Lim R, LeBard RJ, Skurray RA, Firth N. Analysis of the pSK1 replicon, a prototype from the staphylococcal multiresistance plasmid family. Microbiol. 2008;154:3084–3094. doi: 10.1099/mic.0.2008/017418-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.