Abstract
Among four types of bacterial restriction enzymes that cleave a foreign DNA depending on its methylation status, type I enzymes composed of three subunits are interesting because of their unique DNA cleavage and translocation mechanisms performed by the restriction subunit (HsdR). The elucidated N-terminal fragment structure of a putative HsdR subunit from Vibrio vulnificus YJ016 reveals three globular domains. The nucleolytic core within an N-terminal nuclease domain (NTD) is composed of one basic and three acidic residues, which include a metal-binding site. An ATP hydrolase (ATPase) site at the interface of two RecA-like domains (RDs) is located close to the probable DNA-binding site for translocation, which is far from the NTD nucleolytic core. Comparison of relative domain arrangements with other functionally related ATP and/or DNA complex structures suggests a possible translocation and restriction mechanism of the HsdR subunit. Furthermore, careful analysis of its sequence and structure implies that a linker helix connecting two RDs and an extended region within the nuclease domain may play a central role in switching the DNA translocation into the restriction activity.
INTRODUCTION
DNA imported into microorganisms is removed by restriction–modification (RM) systems (restriction enzymes) in host cells and, depending on their composition, cofactor requirements and target DNA recognition, RM systems are classified into four types: I–IV (1). Of these, type I RM systems are unique in their DNA translocation and restriction mechanisms as well as their DNA recognitions. They are hetero-oligomeric proteins encoded by three closely linked genes: a specificity subunit (HsdS or S) for recognizing a DNA sequence, a methylation subunit (HsdM or M) for methylating the recognized target bases, and a restriction subunit (HsdR or R) for the translocation and random cleavage of non-methylated DNA (2–3). They show diverse catalytic activities, including methyltransferase (MTase), ATP hydrolase (ATPase), DNA translocation and restriction activities.
The HsdS subunit recognizes two described sequences, which are asymmetric and separated by a spacer of 6–8 bp. Upon identification of specific DNA sequences containing a hemi-methylation, a methyl group is transferred from S-adenosylmethionine to the non-methylated adenine base in the remnant strand (4). When neither of two cognate bases on both strands is methylated, type I systems translocate onto and cut the DNA upon encountering an obstacle, at a site located at about 1000 bp from their recognized sequences. Type I systems are further divided into five subtypes (IA, IB, IC, ID and IE) according to the cross-hybridization of genes, antibody cross-reactivity and sequence conservation (5).
HsdM and HsdS subunits are capable of forming an independent active trimeric M2S1 MTase (6,7). However, a proper pentameric M2S1R2 assembly of all three subunits is requisite for the DNA cleavage and ATPase-related processive translocation activities of the HsdR subunit (8–10). According to the limited proteolysis, sequence and mutation studies of HsdR subunits such as EcoKI, EcoAI and EcoR124I, which are representatives of subtype IA, IB and IC HsdR proteins (2,11,12), respectively, the N-terminal polypeptide region includes a motif for an endonuclease activity, such as a PD − (D/E) × K sequence, whereas the C-terminal region forms a helix-rich domain that might serve as a contacting core for the MTase sub-complex. The intervening polypeptide region shows close sequence similarity to the RDs of helicases and includes a highly conserved DE×× sequence motif responsible for ATPase, which is directly related with the DNA translocation activity (11,13).
In contrast to the abundant biochemical studies since the first discovery of type I RM system, structural information has been limited. Two crystal structures of putative HsdS subunits from Methanococcus jannaschii (14) and Mycoplasma genitalium (15) have been reported. In succession, two crystal structures of HsdM subunits have been deposited in the Protein Data Bank (PDB) (PDB ID 2OKC and 2AR0 at http://www.rcsb.org/pdb). The recent solution structure of EcoKI MTase in complex with a DNA mimicking protein Ocr provided a more detailed structural model for the reaction mechanism of MTase activity (16). Structural studies of the HsdR subunit have also been published. A structural model for the EcoR124I HsdR subunit was predicted based on molecular modeling and small-angle-scattering data (17), although the study did not attempt to describe the detailed mechanisms of diverse activities at the molecular level. Its fragment crystal structure was reported very recently (18) and the overall structure included a proposed helical bundle domain at the C-terminal region. However, the presence of several disordered regions, such as the catalytic residues responsible for restriction (12) and the Q×××Y motif suggested to be involved in restriction (19), has made it difficult to provide satisfactory explanations for the diverse mechanisms of type I RM systems. Therefore, understanding of its diverse mechanisms remains obscure.
In order to add structural information and provide a molecular background on the multi-functional type I systems, we elucidated the crystal structure of the N-terminal fragment (∼590 amino acids) of a putative HsdR subunit (817 amino acids) from V. vulnificus YJ016 (HsdR_Vv). The crystal structure clearly reveals the catalytic sites for nuclease and ATPase activities and suggests a possible translocation mechanism of the HsdR subunit. The detailed structural features and its molecular mechanisms are discussed.
MATERIALS AND METHODS
Cloning, expression and purification of HsdR_Vv
The cloning and purification steps have been described in detail elsewhere (20). Briefly, the cloned full length HsdR_Vv (gi_37678449) was transformed into E. coli B834(DE3) cells and the expressed native protein homogeneously purified by following sequential chromatographic steps. The seleno-l-methionine (Se-Met) protein was prepared using the same procedures as those for the native protein.
Crystallization, data collection and structure determination
For crystallization, the purified native HsdR_Vv protein was concentrated to 5 mg/ml in a buffer consisting of 20 mM Tris–HCl at pH 7.5 and 100 mM NaCl. Native crystals suitable for diffraction experiments were obtained within 14 days using a hanging-drop vapor-diffusion method at 22°C by mixing 1 µl each of protein solution and reservoir solutions and equilibrating against 200 µl of reservoir solution, consisting of 8–10% (w/v) polyethylene glycol 3350, 0.15 M NH4Cl, 0.1 M 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid at pH 7.5 and 2 mM β-mercaptoethanol. Se-Met substituted protein crystals were obtained under the same crystallizing conditions as the native crystals. For data collection, the crystals were immersed briefly in a reservoir solution supplemented with 10% (v/v) glycerol and immediately placed in a 100 K nitrogen-gas stream. Single wavelength anomalous dispersion (SAD) data with a Se-Met crystal were collected at the MAX4A beamline of the Pohang Accelerator Laboratory (PAL) at a wavelength of 0.97940 Å. The data were indexed and scaled with the HKL-2000 suite (21). The crystal belonged to the orthorhombic space group P212121 with unit-cell dimensions of a = 71.01, b = 89.04 and c = 113.66 Å. Seven Se sites out of the expected eleven in the asymmetric unit were identified at 3.0 Å resolution using the program SOLVE (22). The electron density was further improved by density modification using the program RESOLVE (23), resulting in automatic modeling of 50% of the modeled residues. Further model building was performed manually using the programs, WinCoot (24) and O (25), and refinement performed with CCP4 refmac5 (26) and CNS (27). The data statistics are summarized in Table 1 and the quality of the model was analyzed with WinCoot (24) and PROCHECK (28).
Table 1.
Data collection and refinement statistics
| Parameters | High resolution | Se_Peak |
|---|---|---|
| Synchrotron | 4A (MX4A), PAL | 4A (MX4A), PAL |
| Wavelength (Å) | 0.9796 | 0.9794 |
| Space group | P212121 | |
| Cell parameters (Å) | a = 71.24, b = 88.78, c = 114.23 | a = 71.01, b = 89.04, c = 113.66 |
| Resolution (Å) | 50.0–2.50 (2.59–2.50) | 50.0–2.70 (2.80–2.70) |
| Completeness (%) | 99.1 (97.5) | 99.8 (99.6) |
| Redundancy | 5.6 (4.2) | 6.8 (6.3) |
| Rsyma (%) | 9.7 (32.0) | 9.0 (42.3) |
| Reflections, total/unique | 142 948/25 725 | 133 697/20 521 |
| I/Sigma(I) | 18.6 (3.7) | 21.1 (3.5) |
| FOMb, solve/resolve (50–3.0 Å) | 0.27/0.53 | |
| Rfactorc (%)/Rfreed (%) | 21.8/24.2 | |
| No. of atoms, protein/water | 4176/345 | |
| B factors (Å2), protein/water | 43.65/45.38 | |
| RMSDs, bonds (Å)/angles (°) | 0.008/1.60 | |
| Geometry (%) | ||
| Most favored | 92.6 | |
| Additionally allowed | 6.7 | |
| Generously allowed | 0.7 |
Values in parentheses are for the highest-resolution shell. FOM, figure of merit; rmsds, root-mean-square-deviations.
aRsym=Σhkl Σj|Ij – <I>|/ΣhklΣjIj, where <I> is the mean intensity of reflection hkl.
bFigure of merit = |ΣP(α)eiα/ΣP(α)|, where P(α) is the phase probability distribution and α is the phase (50.0–3.0 Å).
cRfactor= Σhkl||Fobs|–|Fcalc||/Σhkl|Fobs|; where Fobs and Fcalc are the observed and calculated structure factor amplitudes, respectively, for the reflections hkl included in the refinement.
dRfree is the same as Rfactor but calculated over a randomly selected fraction (10%) of the reflection data not included in the refinement.
RESULTS AND DISCUSSION
Overall structure
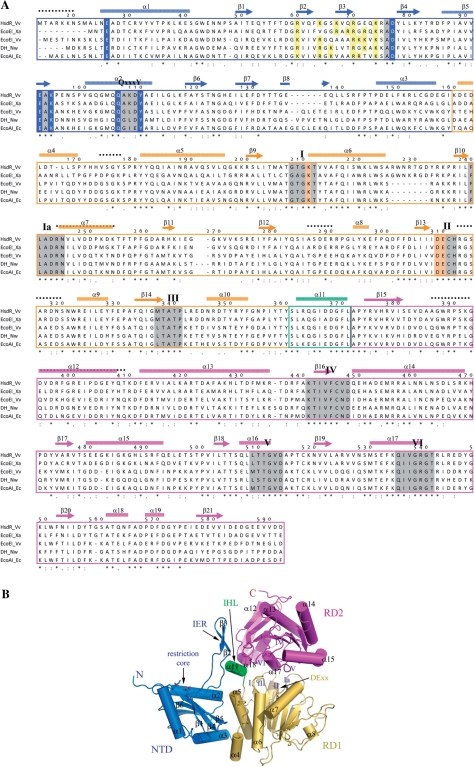
The putative HsdR_Vv subunit is composed of 817 amino acids with a calculated theoretical molecular weight of ∼95 kDa. Comparison with the other HsdR subunits shows a close sequence similarity to EcoAI, suggesting that HsdR_Vv may belong to the subtype IB (Figure 1A). Like other HsdRs, HsdR_Vv also has a conserved sequence motif of PD − (D/E) × K at the N-terminus and several conserved helicase-forming sequence motifs including the DECH, which was suggested to be responsible for the ATPase and DNA translocation activities (Figure 1A).
Figure 1.
Structure of HsdR_Vv. (A) Sequence comparison of HsdRs. Only the N-terminal fragment (Met14-Asp592) is displayed. The cylinders (α) and arrows (β) above the aligned sequences stand for the helix and strand, respectively. The numbering scheme follows the amino acid sequence of HsdR_Vv. Each domain for NTD, RD1 and RD2 is differentiated by colors. The catalytic residues at the NTD and the ATPase sites of RD1 are presented in blue and bright orange colored backgrounds, respectively. The conserved helicase-forming sequence motifs within the two RDs and the Q×××Y motif are presented in grey and green backgrounds, respectively. The regions of weak electron density in the present crystal structure are displayed by continuous black-dotted lines above the aligned sequences. Identical residues are marked by ‘asterisk’ and conserved residues by ‘:’ or ‘.’. Abbreviations used here: DH for DEAD-box containing helicase, Vv for V. vulnificus YJ016, Nw for Nitrobacter winogradskyi, Xa for Xanthobacter autotrophicus and Eco for E. coli. (B) Overall shape of the N-terminal fragment of HsdR_Vv structure. Each domain is displayed by ribbon diagrams and labeled using the same colors as in (A). The critical residues responsible for the restriction (NTD) and ATPase activities (RD1) are drawn by stick models. The spatial position of the conserved helicase-forming sequence motifs on both RDs are marked by Roman figures (I–VI). The figures, except for Figure 1A, were prepared by the PyMol Molecular graphics program of Delano Scientifics.
The crystal structure of HsdR_Vv was determined by SAD analysis using a Se-Met substituted protein crystal and refined to 2.5 Å resolution (Table 1). Even though the full length protein was cloned and the expressed, purified protein had shown a molecular weight of >90 kDa on SDS–PAGE (data not shown), only the N-terminal fragment (∼600 amino acids) of the HsdR_Vv protein was modeled in the refined structure. The HsdR_Vv crystal was examined by SDS–PAGE, which showed a ∼60 kDa fragment (data not shown), suggesting cleavage of the protein during the crystallization trials. One protein molecule was present in the asymmetric unit and analysis of the modeled residues by WinCoot (24) and PROCHECK (28) showed that all modeled residues were found in valid regions of the Ramachandran plot (Table 1).
Overall, the N-terminal fragment of HsdR_Vv comprises three globular domains: an N-terminal nuclease domain (NTD, Ala21-Ile159) and two continuous globular RecA-like domains (RD1 for Lys160-Tyr360 and RD2 for Ala372-Val590). An internal helical linker (IHL, Ser361-Leu371) formed by a two-turn helical structure α11 connects the RD1 to RD2 domains (Figure 1).
The NTD nuclease domain
About the N-terminal 160 residues of HsdR_Vv constitute a single NTD (blue in Figure 1). However, the first 20 residues at the extreme N-terminus (Met1-Ala20) were disordered in the present structure. Therefore, the current NTD structure is effectively composed of ∼140 residues (Figure 1A).
The NTD could be separated into two structural parts: a globular part and an internal extended region (IER). In the globular part, one β-sheet composed of six β-strands (β1 and β4–β8) is enclosed by three helices at both sides and the top, respectively. The polypeptide IER (Thr55-Arg75) forms a short anti-parallel β-sheet by two β-strands (β2 and β3), which is connected to the globular part through long loop regions (Figure 1). However, this IER does not show specific interactions with the globular part of the NTD, except for one region, a Q×××Y sequence motif, as described below.
The structural homolog search using DALI server shows that the globular part of ∼100 residues has a close relationship to the nuclease-forming domains, such as the holiday junction resolvase, tRNA endonuclease and EcoRV type II restriction enzymes with a Z-score of ∼5 and root-mean-square deviations (rmsd) of less than 4.0 Å (29–31). However, the structure-based sequence identity is very low among the aligned residues, for example, with a maximum of 16%. The NTD (Asn13-Met272) of the EcoR124I HsdR subunit, one of the most closely functionally related proteins to HsdR_Vv, has higher sequence identity (17%), but shows larger deviations (rmsd of 4.6 Å) among the superposed Cα atoms (Supplementary Figure S1) than the type II enzymes of EcoRV and NgoMIV.
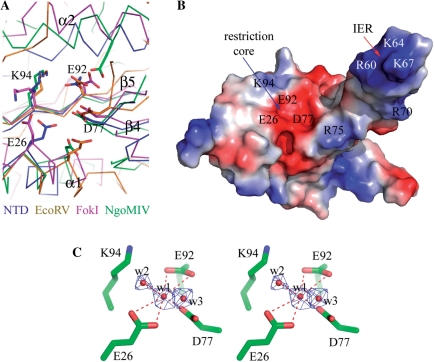
The nuclease determining PD – (D/E) × K sequence motif in the NTD is located on the two strands of β4 and β5 (grey background in Figure 1A). Structure comparison with the structurally and catalytically related nuclease cores of EcoRV (31) and NgoMIV (32) type II restriction enzymes and FokI nuclease (33) also suggests a well-defined restriction site in HsdR_Vv (Figure 2), containing three conserved residues of Asp77, Glu92 and Lys94 from the PD − (D/E) × K sequence motif and another conserved Glu26 residue among the type I enzymes at the extreme N-terminus (Figure 1A). Among them, the former three residues (Asp77, Glu92 and Lys94) are consistent with those confirmed by a mutagenesis study in the subtype IB EcoAI protein (12). On the other hand, Glu33 in EcoR124I, the corresponding fourth nucleolytic residue (Glu26) in HsdR_Vv, is far from the other three catalytic residues (Asp151, Glu165 and Lys167), because the structure is partially disordered here (Supplementary Figure S1) (18). Therefore, the catalytic residues of the NTD in EcoR124I might not be constituted for the instantaneous cleavage reaction.
Figure 2.
Catalytic site of the NTD. (A) The NTD is superposed with other restriction endonucleases. The Cα tracings of NgoMIV, EcoRV, FokI and HsdR_Vv (NTD) are differentiated by colors. The catalytic residues are displayed by stick models. For clarity, only the HsdR_Vv residues are labeled. (B) Surface potential presentation of the NTD with similar orientation to (A). The positively and negatively charged surfaces are colored in blue and red, respectively. The key residues for DNA binding and subsequent restriction are indicated on the surface. (C) Stereo-representation of a catalytic site. The catalytic residues for nuclease activity are displayed with stick models and three water molecules bound at the catalytic site by red dots. The 2Fo-Fc density (blue) for the water molecules at the restriction site were contoured at 2.0σ. The hydrogen bonds among the protein side chain atoms and water molecules are displayed by red-dotted lines.
A magnesium ion is often requisite for DNA cleavage (32). In the active site of the NgoMIV in complex with DNA, one of two Mg ions is hexa-coordinated by two protein atoms, three water molecules and a substrate DNA atom. In the nucleolytic site of NTD, three water molecules are observed. One tentatively modeled water molecule (w1) is also coordinated with six oxygen atoms from Glu26, Asp77, Glu92 and another water molecule (w2) (Figure 2C) and occupies a structurally equivalent position of one of two Mg ions in the NgoMIV structure (Figure 2), suggesting that w1 might be a catalytic Mg ion at the nucleolytic site. However, this putative Mg ion at the NTD was not observed in the EcoR124I structure (18).
A mutagenic study of EcoR124I has shown that the conserved Q × × × Y motif at the NTD was confirmed to participate in restriction (34). However, this motif was disordered in the EcoR124I structure (Supplementary Figure S1) (18), but it is located close to the four putative nucleolytic residues in the present structure (Figure 2). Its two conserved residues (Gln106 and Tyr110) do not directly interact with any of the four catalytic residues. Instead, the –OH of Tyr110 interacts with the peptide carbonyl oxygen of Arg75. Interestingly enough, this residue connects the IER to the globular part of the NTD, implying another role of the Q × × × Y motif in the restriction activity of type I enzymes, as discussed below.
Collectively, the single nucleolytic site with a putative Mg ion is directly exposed to the solvent accessible region, but has a very shallow cleft (Figures 1 and 2), which may not allow transient tight interactions, if any, with the bulky DNA substrate for the cleavage reaction at either a single or double strand.
Two RD domains
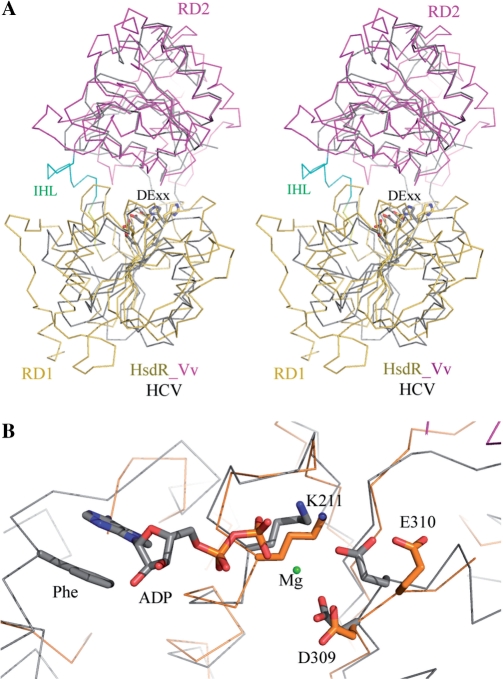
The polypeptide region of Lys160-Tyr360 in HsdR_Vv forms a single globular RD (RD1, gold in Figure 1) and has four conserved sequence motifs (I, Ia, II and III). RD1 is composed of a 6-parallel β-sheet (β9–β14), which is flanked by a three-helical bundle structure (α4, α5 and α6) at one side and by three helices (α7, α8 and α9) at the other (Figure 3A). The four peptide regions within RD1 have rather a weak electron density: residues of Val174-Tyr177, Arg244-Lys254, Ala286-Arg291 and Arg313-Asn321 (the black dotted lines in Figure 1A). A structural similarity search using the DALI server shows that its overall folding is closely related with the RDs frequently observed in DNA repair systems and helicases, such as RAD25, UvrABC and SNF2/RAD54 family proteins (34–36) (Figure 3A). Four conserved sequence motifs are located at the domain interface with the RD2 (Figure 1).
Figure 3.
Comparison of RDs. (A) The RD1 (gold) and RD2 (magenta) of HsdR_Vv are superposed with HCV NS3 helicase (HCV, grey). The residues of the DECH sequence (DE××) for ATP hydrolysis are displayed with stick models. (B) Close-up view of ATP-binding site. The catalytic residues for ATP hydrolysis are displayed with stick models and Mg ion with a green dot and occupy positions structurally equivalent to HCV helicase. In other functionally related enzymes, the aromatic ring of a protein side chain (Phe) stacks with the base of adenine nucleotide.
The RD2 (Ala372-Val590, magenta in Figure 1) also forms another single globular structure. Seven-parallel-strands (β15–β21) form a pleated β-sheet that is sandwiched by several helices: two long-kinked helices (α13 and α14) at one side and five short helices (α15–α19) at the other. The latter forms a large cleft at the domain interface along with the RD1 (Figure 1B). The polypeptide region of Ala388-Lys410 in RD2 has very weak electron density (the black-dotted lines in Figure 1A). Despite the low sequence homology and the difference in the β-strand orientations between the two RD domains, the RD2 architecture is similar to that of RD1 with an rmsd of 3.5 Å among the superposed main-chain atoms. Thereby, the RD2 also shows close structural relationship to helicases and DNA repair systems (36–39) (Figure 3A). Three conserved sequence motifs (IV, V and VI) within the RD2 are located close to the cleft formed with the RD1 (Figure 1B).
ATPase site
Motor proteins use ATPase activity to produce mechanical power for translocating an ss-DNA or ds-DNA. They commonly consist of at least two structurally conserved RDs, whose interface contains an ATP-binding pocket formed by several conserved helicase-forming sequence motifs I–VI (34,40,41).
As in EcoR124I (18), HsdR_Vv has several conserved regions in the middle of the primary sequences, which form two RDs in the present structure (Figure 1). Despite the disordered regions within these two RDs, most of the conserved sequence motifs are defined around the interface of the two RDs. Furthermore, the conserved DECH sequence (motif II), responsible for the Mg ion and ATP binding and for the subsequent ATP hydrolysis, occupies structurally equivalent positions to those of other DE × × helicases (Figure 3). This suggests that HsdR_Vv has a similar ATP binding and a similar ATP hydrolysis mechanism of HsdR_Vv to those of the other two RD-containing enzymes (42). However, a minute difference may exist in the ATP-binding mode between the HsdR_Vv and other proteins. In other related enzymes (42), an aromatic residue (Phe on Figure 3B) in the ATP-binding pocket stacks with the bound adenine base, but the corresponding aromatic residue is absent in the current HsdR_Vv structure and also in the primary sequence of subtype IB HsdR proteins (Figure 1A). Nor was any aromatic stacking observed between the adenine base and protein in the ATP complex structure of EcoR124I (18). The conserved RYYQ sequence motif among subtype IB HsdRs is, however, on the α5 helix and a non-conserved Tyr177 is located near the modeled adenine base in the HsdR_Vv fragment structure (Figure 1A). Because the flexible, partially disordered region (Val174-Ser178) is directly connected to this RYYQ motif and includes the Tyr177 residue, we cannot totally exclude the possibility that the RYYQ motif and/or Tyr177 may be ordered upon ATP binding, resulting in the similar aromatic interaction with the adenine base.
The IHL α11 plays a central role in domain assembly
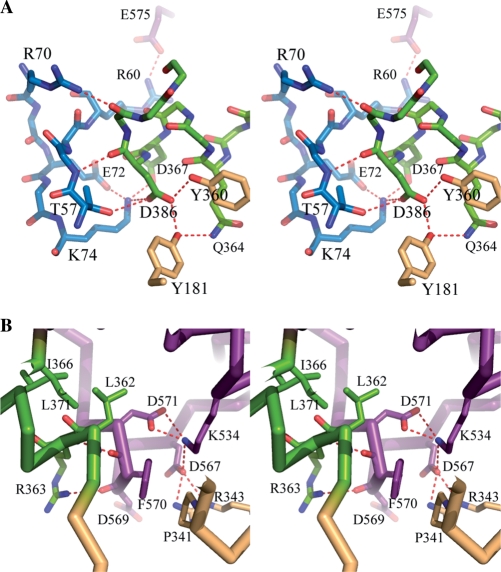
NTD interacts with two RDs through a large number of intertwined hydrophobic residues on the α3 of the NTD and the α4 and α5 helices of the RD1 (Figure 1B). In addition, Asp85 in the β4–β5 loop of NTD interacts with the peptidyl nitrogen atom of Tyr171 and the NE2 atom of His173 in the α4–α5 loop of the RD1. The IER within the NTD also forms several polar interactions with the IHL α11 (Ser361-Leu371) and the RD2 (Figure 4A). Arg60 and Arg70 interact ionically with Glu575 and Glu545 of the RD2. Another positively charged residue Lys74 interacts with two acidic residues (Asp367 and Asp368) on the IHL. Polar interaction is not unique here. Residues of Val61, Phe63 and Val68 of the NTD participate in hydrophobic interactions with the hydrophobic moieties of Pro373, Arg375, Leu550, Trp551 and Glu575 of the RD2.
Figure 4.
Interaction at the interface of two RDs. (A) Stereo-representation of polar interaction at the linker region with three domains. The residues are displayed by stick models and hydrogen bonds by continuous red-dotted lines. (B) Stereo-representation of RD1 and RD2 near the linker helix α11. The hydrophobic residues on the linker helix contribute to the domain assembly at the interface.
As implicated above, the IHL has highly conserved amino acid sequences among the subtype IB HsdR proteins and connects the RD1 to the RD2 (Figure 1). In the overall fragment structure of HsdR_Vv, the IHL is located at the intersecting region of three domains and plays a central role in their assembly (Figure 1B). Besides the aforementioned interaction with the NTD through the IER, the IHL interacts with both RDs through complicated polar networks (Figure 4). The side chain atoms of Gln364 form a hydrogen bond with the –OH of the conserved Tyr181, which in turn interacts with the side chain atoms of the conserved Asp368 that simultaneously has polar interactions with those of the conserved Tyr360 and Lys74. The latter residue also interacts ionically with the negatively charged residues of Asp367 and Asp368 (Figure 4A). The non-polar atoms of the conserved Leu362 and Ile366 are intertwined with those of Thr209, Ala206, Tyr360, Phe370, Leu371, Tyr374, Val537, Thr541, Phe552 and Ile554 (Figure 4B).
A short peptide region (Pro568-Asp571) of the RD2 forms a one-turn helix, α18 and interacts with the IHL through its main chain atoms. In addition to the polar interactions formed between the peptidyl oxygen atom of Asp569 and the NH1 atom of Arg363, the peptidyl oxygen atom of Phe570 in the RD2 is trapped with two peptidyl nitrogen atoms of Leu362 and Arg363 at the helical cap of α11 (Figure 4B).
Near the IHL of the two-RD interface, another complicated polar interaction between both RDs is observed: the Asp571 carboxyl group forms an ionic interaction with the Lys534 that interacts with the Asp567 side chain, which has a polar interaction with Arg343 at the same time. Ser531 is located within the distance able to form a hydrogen bond with three oxygen atoms of Thr340, Asn346 and Asp345. The Asp345 makes another hydrogen bond with the Ser531 peptidyl nitrogen (Figure 4B).
DNA binding and cleavage at the restriction site
For cleavage reactions, substrate DNA may be bound to the catalytic core or a transient interaction can occur between the enzyme and the DNA substrate (32,33).
Like the EcoR124I, the N-terminal fragment of HsdR_Vv has a single nuclease domain, which contains a sole nucleolytic core (Figure 2). Conserved catalytic residues (Glu26, Asp77, Glu92 and Lys94) occupy positions structurally equivalent to those of other related nucleolytic enzymes (31–33). However, the restriction site is exposed directly to the solvent region (Figures 1B and 2) and has a shallow surface with several negatively charged residues (Glu26, Asp77 and Glu92), which may not allow favorable interactions with the negatively charged ds-DNA for cleavage reaction. Therefore, other regions may be involved in DNA binding at the restriction site, for example, the IER (Thr55-Arg75) and/or α1 helix within the NTD or the C-terminal helical bundle structure that was missing in our structure but closely located to the NTD in the EcoR124I structure (Supplementary Figure S1) (18). Interestingly, the IER has several basic residues that are conserved among subtype IB HsdR proteins (the yellow background in Figure 1A), which possibly accommodate transient non-specific interactions with the ds-DNA backbone, if any. In the present apo-structure, the IER does not interact with the NTD, but with the RD2 and the linker helix α11 (Figures 1 and 4). However, there are indications of a local rearrangement occurring around the nucleolytic site upon DNA binding. First, the EcoR124I structure shows several disordered regions around the restriction site including a Q × × × Y motif and a catalytic residue Glu33 (Supplementary Figure S1) (18). Second, the replacement of Gln and Tyr residues in the conserved Q × × × Y motif with the respective Ala residue or both with Ala residues dramatically reduced the nuclease activity (19). Third, the Q × × × Y region in the EcoR124I structure belongs to one α-helix that is partially disordered (Supplementary Figure S1) (18), which appears to participate in the DNA restriction activity through the direct interaction with DNA, not with nucleolytic residues. Fourth, the corresponding region in HsdR_Vv, which is located on the rigid secondary structure (α2), interacts with the main-chain atom (peptide carbonyl oxygen of Arg75) at the beginning of the IER using the –OH of Tyr110, but does not form any direct interaction to the nucleolytic residues. For the IER region to take part in the restriction activity, therefore, a local structural change might be invoked on the NTD surface, probably from the domain rearrangement upon collisions of two type I systems (Figure 5, as discussed below). A modeling study of the DNA complex at the NTD also suggests intriguing results in which a β-sheet of the IER is fitted into the DNA major groove (Figure 5B). Similar β-sheet insertion into the DNA major groove upon DNA binding was observed in the EcoRV–DNA complex structure (31). On the other hand, the α1 helix of an abnormal secondary structure at the beginning fits into the DNA major groove at the opposite site of the β4–β5 sheet, by restoring the normal α-helix conformation observed frequently in other DNA complex structures of nucleases (Figure 2A) (29,33).
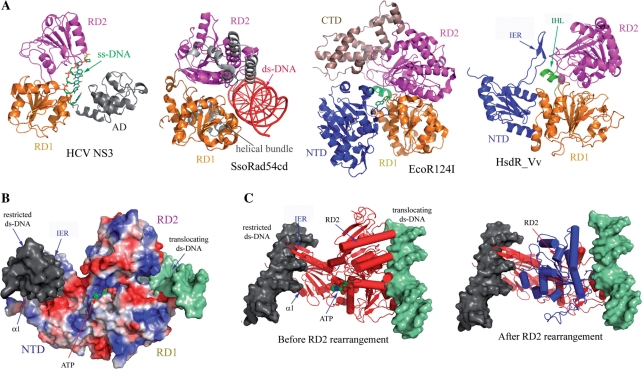
Figure 5.
Proposed translocation mechanism of HsdR_Vv. (A) HsdR_Vv is compared with ss-DNA translocator (HCV NS3), ds-DNA translocator (SsoRad54cd) and type I EcoR124I translocator (EcoR124I). The C-terminal helical bundle domain of EcoR124I was labeled by CTD. Note the remarkable structural difference in the relative spatial orientation of RD2 in HCV helicase and SsoRad54cd, which might imply the translocation mechanism of HsdR_Vv. (B) Modeling of ATP and two ds-DNAs at the NTD and at the surface along the two RDs. The N-terminal fragment structure of HsdR_Vv is represented by the surface potential map. The restricted (grey surface) and translocating (shallow green surface) ds-DNAs of HsdR_Vv were modeled by superposing the respective DNA complex structure of NgoMIV type II enzyme and SsoRad54cd ds-DNA translocator with the fragment structure of HsdR_Vv. The modeled ATP is displayed by the space filling model. In this model, the α1 helix of the NTD was straightened up to fit into the DNA groove as in the NgoMIV-DNA complex structure, while the IER (extended region) within the NTD was rotated towards the ds-DNA at the nucleolytic site, thereby locating the two stranded β-sheet into the DNA major groove. (C) Relative reorientation of RD2 in HsdR_Vv upon ATP and ds-DNA binding to RDs. The RD2 in the present structure is superposed well with the ss-DNA translocator HCV NS3 (before RD2 rearrangement), while RD2 is rearranged (blue in ‘After RD2 rearrangement’) when it is compared with the ds-translocator SsoRad54cd (After RD2 rearrangement).
The collision of two HsdR motor proteins or the intervention of a translocation reaction by an unusual DNA structure (12) appears to be the most promising mechanism for the cleavage initiation in the type I system. Supposedly, there is no dimer formation of the HsdR subunit in solution and this NTD might not constitute a dimeric structure either. Therefore, a single-strand cut of a ds-DNA substrate appears to be more plausible than double strand breaks by a monomeric HsdR subunit. The previous report that two single-strand cutting HsdR subunits are necessary for a ds-DNA break (43) supports this structural feature of HsdR proteins described here and in EcoR124I. Otherwise, the single NTD might flip between two orientations, cleaving first one DNA strand and then the other, as suggested in the FokI endonuclease that has a single NTD and cleaves both strands (33). Even the dimeric state observed in type I EcoKI enzyme (44), where each single neighboring HsdR subunit may introduce a single cut on a DNA strand, will provide a much easier background for understanding the ds-DNA restriction by a single NTD in each HsdR subunit.
Implications for the DNA translocation and restriction initiation
In HCV NS3 and PcrA helicases that perform translocations on DNA by unwinding ds-DNA, two RDs and an additional domain (AD) grip the bases of the separated ss-DNA (Figure 5A) (41,42). In contrast, SWI2/SNF2 enzymes that remodel chromatin or other DNA–protein complexes by translocating on ds-DNA have not shown DNA unwinding activity but have instead generated superhelical torsion into DNA (36). The crystal structure of the catalytic domain of Sulfolobus solfataricus Rad54 homolog (SsoRad54cd) (36), a protein of the SWI2/SNF family, shows the absence of this AD. In both cases, a nucleic acid at the translocation site binds to the surface that encompasses two RDs (Figure 5A).
Besides the two separate base recognition sites, another biophysical property of type I systems is fast DNA translocation on ds-DNA or ss-DNA, in which M2S1 MTase binds to a specific DNA sequence and recognizes the methylation status of the target nucleotides. When neither recognized bases is methylated, two HsdR subunits are recruited and each continuously pulls the ds-DNA toward the MTase sub-complex until it collides with the neighboring type I system (12,45). Sequence analysis has suggested that the HsdR subunit might possess a helicase activity; however, recent studies in the EcoR124I system have shown the absence of such activity (46). Instead, supercoiling of ds-DNA was generated during the DNA translocation (47). As shown in both the fragment structures of EcoR124I (18) and HsdR_Vv here, there is no AD that is responsible for gripping ss-DNA in HCV NS3 and PcrA helicases, either (41,42). Therefore, HsdR motor proteins are likely to translocate on DNA, as in the case of SsoRad54cd that translocates on ds-DNA using the different spatial orientations of RDs (36).
Consideration of the outlined biochemical and structural features may allow the respective DNA-binding model at the nucleolytic core and the translocation site of HsdR_Vv, which induces an abnormal DNA topology if there is a direct connection between the two DNA fragments at both modeled sites (Figure 5B). To derive the translocation mechanism of the HsdR subunit, two RDs of the HsdR_Vv fragment structure were superposed with those of the ds-DNA-bound SsoRad54cd and other ATP-bound motor proteins (Figure 5). The relative orientation of the RD2 in the HsdR_Vv protein is well superposed with the ATP complex structure, suggesting that the domain assembly of two RDs in the present structure is similar to the ATP-binding form (Figures 3A and 5A). However, the RD2 in SsoRad54cd was rotated by more than 180° while fixing the RD1 structure (Figure 5). Similarly in the SsoRad54cd motor protein (36), we suggest that the rearrangement of RD2 observed in this apo-structure may be related with the ds-DNA translocation and can subsequently induce several structural differences, including the IER, which is bound to both RD domains by forming several polar and non-polar interactions (Figure 4). Specifically, the loop region (Arg542-Trp551) of RD2, which has two positively charged residues (Arg542 and Lys549) and occupies the space of the IER region upon reorientation, might conflict with several positively charged residues (Arg60, Lys64, Lys67 and Arg70) on the IER region, resulting in the reorientation of the IER region observed in the current structure to the nuclease core site. Although the missing C-terminal helical bundle structure was suggested as an interaction site with the MTase sub-complex (2,11,12), and as possibly being involved in DNA binding at the restriction site (18), the possibility that the C-terminal region might partially joins the translocation activity could not be totally ignored since decreased translocation rate was observed due to the loss of the nuclease activity in the HsdR subunit of EcoR124I system (48). In the same context, a portion of the MTase sub-complex may have taken part in the translocation activity, since the translocation activity of the HsdR subunit is fulfilled only after all three subunits are properly arranged (7–9). Of course, the proper assembly of all subunits could invoke a completely different domain arrangement observed in the HsdR structures alone and the domain rearrangement suggested here.
There are, however, several intrinsic differences in characteristics between the HsdR and SsoRad54cd motor proteins. HsdR subunits have an additional NTD and C-terminal region, compared with SsoRad54cd. The NTD is essential to substrate DNA cleavage and is located far from the putative DNA-binding site for translocation in the fragment structures of HsdR (Figure 5). On the other hand, the missing C-terminal region in the present structure forms a helical bundle structure in EcoR124I and was suggested to be involved in DNA interaction at the restriction site, along with the NTD (18). Another difference can be found in the amino acid compositions of the RD1s. Compared with that of HsdR_Vv, the RD1 of SsoRad54cd has an extra polypeptide region, which forms a three-helical bundle structure and binds to the DNA groove (the grey structure in the SsoRad54cd of Figure 5A). Finally, the translocation mechanisms appear to differ, following the DNA in SsoRad54cd (36) or pulling the DNA in HsdR while fixed on the recognition sequence by the MTase sub-complex (9). Therefore, the HsdR structures in complex with other subunits and/or DNA will provide a clearer background for understanding the mechanisms of DNA cleavage and translocation activity of this complicated system.
ACCESSION NUMBER
Coordinates and structure factors have been deposited in the PDB with the accession number 3H1T and the RCSB ID rcsb052584.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Korea Research Foundation Grant by Korean Government (MOEHRD) (KRF-2007-331-C00209); partially by 21C Frontier Microbial Genomics and Application Center Program, Ministry of Science and Technology, Republic of Korea; partially by grant from BioGreen 21 Program (Code 20080701034001), Rural Development Administration, Republic of Korea. Funding for open access charge: Rural Development Administration, Republic of Korea.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Drs K. H. Kim, K. J. Kim and Y. J. Kim, Pohang Light Source, for helping with the data collection.
REFERENCES
- 1.Murray NE. Type I restriction systems: sophisticated molecular machines (a legacy of Bertani and Weigle) Microbiol. Mol. Biol. Rev. 2000;64:412–434. doi: 10.1128/mmbr.64.2.412-434.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dryden DTF, Murray NE, Rao DN. Nucleoside triphosphate-dependent restriction enzymes. Nucleic Acids Res. 2001;29:3728–3741. doi: 10.1093/nar/29.18.3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sistla S, Rao DN. S-Adenosyl-L-methionine–dependent restriction enzymes. Crit. Rev. Biochem. Mol. Biol. 2004;39:1–19. doi: 10.1080/10409230490440532. [DOI] [PubMed] [Google Scholar]
- 4.Wilson GG, Murray NE. Restriction and modification systems. Annu. Rev. Genet. 1991;25:585–627. doi: 10.1146/annurev.ge.25.120191.003101. [DOI] [PubMed] [Google Scholar]
- 5.Chin V, Valinluck V, Magaki S, Ryu J. KpnBI is the prototype of a new family (IE) of bacterial type I restriction-modification system. Nucleic Acids Res. 2004;32:e138. doi: 10.1093/nar/gnh134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Taylor IPJ, Firman K, Kneale G. Purification and biochemical characterisation of the EcoR124 type I modification methylase. Nucleic Acids Res. 1992;20:179–186. doi: 10.1093/nar/20.2.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Janscak P, Bickle TA. The DNA recognition subunit of the type IB restriction-modification enzyme EcoAI tolerates circular permutation of its polypeptide chain. J. Mol. Biol. 1998;284:937–948. doi: 10.1006/jmbi.1998.2250. [DOI] [PubMed] [Google Scholar]
- 8.Firman K, Szczelkun MD. Measuring motion on DNA by the type I restriction endonuclease EcoR1241 using triplex displacement. EMBO J. 2000;19:2094–2102. doi: 10.1093/emboj/19.9.2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Seidel R, van Noort J, van der Scheer C, Bloom JG, Dekker NH, Dutta CF, Blundell A, Robinson T, Firman K, Dekker C. Real-time observation of DNA translocation by the type I restriction modification enzyme EcoR124I. Nat. Struct. Mol. Biol. 2004;11:838–843. doi: 10.1038/nsmb816. [DOI] [PubMed] [Google Scholar]
- 10.Dryden DT, Cooper LP, Murray NE. Purification and Characterization of the methyltransferase from the Type 1 Restriction and Modification System of Escherichia coli K12. J. Biol. Chem. 1993;268:13228–13236. [PubMed] [Google Scholar]
- 11.Davies GP, Martin I, Sturrock SS, Cronshaw A, Murray NE, Dryden DT. On the structure and operation of type I DNA restriction enzymes. J. Mol. Biol. 1999;292:565–579. doi: 10.1006/jmbi.1999.2908. [DOI] [PubMed] [Google Scholar]
- 12.Janscak P, MacWilliams MP, Sandmeier U, Nagaraja V. DNA translocation blockage, a general mechanism of cleavage site selection by type I restriction enzymes. EMBO J. 1999;18:2638–2647. doi: 10.1093/emboj/18.9.2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bird LE, Subramanya HS, Wigley DB. Helicases: a unifying structural theme? Curr. Opin. Struct. Biol. 8:14–18. doi: 10.1016/s0959-440x(98)80004-3. [DOI] [PubMed] [Google Scholar]
- 14.Kim JS, DeGiovanni A, Jancarik J, Adams PD, Yokota H, Kim R, Kim SH. Crystal structure of DNA sequence specificity subunit of a type I restriction-modification enzyme and its functional implications. Proc. Natl Acad. Sci. USA. 2005;102:3248–3253. doi: 10.1073/pnas.0409851102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Calisto RM, Pich OQ, Pinol J, Fita I, Querol E, Carpena X. Crystal structure of a putative type I restriction–modification S subunit from Mycoplasma genitalium. J. Mol. Biol. 2005;351:749–762. doi: 10.1016/j.jmb.2005.06.050. [DOI] [PubMed] [Google Scholar]
- 16.Kennaway CK, Obarska-Kosinska A, White JH, Tuszynska I, Cooper LP, Bujnicki JM, Trinick J, Dryden DT. The structure of M.EcoKI Type I DNA methyltransferase with a DNA mimic antirestriction protein. Nucleic Acids Res. 2009;37:762–770. doi: 10.1093/nar/gkn988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Obarska-Kosinska A, Taylor JE, Callow P, Orlowski J, Bujnicki JM, Kneale GG. HsdR subunit of the type I restriction-modification enzyme EcoR124I: biophysical characterisation and structural modeling. J. Mol. Biol. 2008;376:438–452. doi: 10.1016/j.jmb.2007.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lapkouski M, Panjikar S, Janscak P, Smatanova I, Carey J, Ettrich R, Csefalvay E. Structure of the motor subunit of type I restriction-modification complex EcoR124I. Nat. Struct. Mol. Biol. 2009;16:94–95. doi: 10.1038/nsmb.1523. [DOI] [PubMed] [Google Scholar]
- 19.Sisakova E, Stanley LK, Weiserova M, Szczelkun MD. A RecB-family nuclease motif in the Type I restriction endonuclease EcoR124I. Nucleic Acids Res. 2008;36:3939–3949. doi: 10.1093/nar/gkn333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nguyen TU, Nishi K, Park SY, Choi JW, Lee HJ, Kim JS. Crystallization and preliminary X-ray diffraction analysis of HsdR subunit of a putative type I restriction enzyme from Vibrio vulnificus YJ016. Acta Crystallogr. Sect. F. 2008;64:926–928. doi: 10.1107/S1744309108027516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Otwinowski Z, Minor W. Procession of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 22.Terwilliger TC, Berendzen J. Automated MAD and MIR structure solution. Acta Crystallogr. D Biol. Crystallogr. 1999;55:849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Terwilliger TC. Maximum-likelihood density modification. Acta Crystallogr. D Biol. Crystallogr. 2000;56:965–972. doi: 10.1107/S0907444900005072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 25.Jones TA, Zou ZY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A. 1991;47:110–117. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 26.Winn MD, Isupov MN, Murshudov GN. Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta Crystallogr. D Biol. Crystallogr. 2001;57:122–133. doi: 10.1107/s0907444900014736. [DOI] [PubMed] [Google Scholar]
- 27.Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 28.Laskowski R, MacArthur M, Hutchinson E, Thorton J. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 1993;26:283–291. [Google Scholar]
- 29.Bond CS, Kvaratskhelia M, Richard D, White MF, Hunter WN. Structure of Hjc, a Holliday junction resolvase, from Sulfolobus solfataricus. Proc. Natl Acad. Sci. USA. 2001;98:5509–5514. doi: 10.1073/pnas.091613398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li H, Trotta CR, Abelson J. Crystal structure and evolution of a transfer RNA splicing enzyme. Science. 1998;280:279–284. doi: 10.1126/science.280.5361.279. [DOI] [PubMed] [Google Scholar]
- 31.Horton NC, Perona JJ. Recognition of flanking DNA sequences by EcoRV endonuclease involves alternative patterns of water-mediated contacts. J. Biol. Chem. 1998;273:21721–21729. doi: 10.1074/jbc.273.34.21721. [DOI] [PubMed] [Google Scholar]
- 32.Deibert M, Grazulis S, Sasnauskas G, Siksnys V, Huber R. Structure of the tetrameric restriction endonuclease NgoMIV in complex with cleaved DNA. Nat. Struct. Biol. 2000;7:792–799. doi: 10.1038/79032. [DOI] [PubMed] [Google Scholar]
- 33.Wah DA, Hirsch JA, Dorner LF, Schildkraut I, Aggarwal AK. Structure of the multimodular endonuclease FokI bound to DNA. Nature. 1997;388:97–100. doi: 10.1038/40446. [DOI] [PubMed] [Google Scholar]
- 34.Eryilmaz J, Ceschini S, Ryan J, Geddes S, Waters TR, Barret TE. Structural insights into the cryptic DNA-dependent ATPase activity of UvrB. J. Mol. Biol. 2006;357:62–72. doi: 10.1016/j.jmb.2005.12.059. [DOI] [PubMed] [Google Scholar]
- 35.Fan L, Arvai A, Cooper PK, Iwai S, Hanaoka F, Tainer JA. Conserved XPB core structure and motifs for DNA unwinding: implications for pathway selection of transcription or excision repair. Mol. Cell. 2006;22:27–37. doi: 10.1016/j.molcel.2006.02.017. [DOI] [PubMed] [Google Scholar]
- 36.Durr H, Korner C, Muller M, Hickmann V, Hopfner KP. X-ray structures of the Sulfolobus solfataricus SWI2/SNF2 ATPase core and its complex with DNA. Cell. 2005;121:363–373. doi: 10.1016/j.cell.2005.03.026. [DOI] [PubMed] [Google Scholar]
- 37.Machius M, Henry L, Palnitkar M, Deisenhofer J. Crystal structure of the DNA nucleotide excision repair enzyme UvrB from Thermus thermophilus. Proc. Natl Acad. Sci. USA. 1999;96:11717–11722. doi: 10.1073/pnas.96.21.11717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Andersen CB, Ballut FI, Johansen JS, Chamieh H, Nielsen KH, Oliveira CLP, Pedersen JS, Seraphin B, Hir HL, Andersen GR. Structure of the exon junction core complex with a trapped DEAD-box ATPase bound to RNA. Science. 2006;313:1968–1972. doi: 10.1126/science.1131981. [DOI] [PubMed] [Google Scholar]
- 39.Richards JD, Johnson KA, Liu H, McRobbie AM, McMahon S, Oke M, Carter L, Naismith JH, White MF. Structure of the DNA repair helicase Hel308 reveals DNA binding and autoinhibitory domains. J. Biol. Chem. 2008;283:5118–5126. doi: 10.1074/jbc.M707548200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim JL, Morgenstern KA, Griffith JP, Dwyer MD, Thomson JA, Murcko MA, Lin C, Caron PR. Hepatitis C virus NS3 RNA helicase domain with a bound oligonucleotide: the crystal structure provides insights into the mode of unwinding. Structure. 1998;6:89–100. doi: 10.1016/s0969-2126(98)00010-0. [DOI] [PubMed] [Google Scholar]
- 41.Jindrova E, Schmid-Nuoffer S, Hamburger F, Janscak P, Bickle TA. On the DNA cleavage mechanism of type I restriction enzymes. Nucleic Acids Res. 2005;33:1760–1766. doi: 10.1093/nar/gki322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shi H, Cordin O, Minder CM, Linder P, Xu RM. Crystal structure of the human ATP-dependent splicing and export factor UAP56. Proc. Natl Acad. Sci. USA. 2004;101:17628–17633. doi: 10.1073/pnas.0408172101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Studier FW, Bandyopadhyay PK. Model for how type-I restriction enzymes select cleavage sites in DNA. Proc. Natl Acad. Sci. USA. 1988;85:4677–4681. doi: 10.1073/pnas.85.13.4677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Berge T, Ellis DJ, Dryden DT, Edwardson JM, Henderson RM. Translocation-independent dimerization of the EcoKI endonuclease visualized by atomic force microscopy. Biophys. J. 2000;79:479–484. doi: 10.1016/S0006-3495(00)76309-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Velankar SS, Soultanas P, Dillingham MS, Subramanya HS, Wigley DB. Crystal structures of complexes of PcrA DNA helicase with a DNA substrate indicate an inchworm mechanism. Cell. 1999;97:75–98. doi: 10.1016/s0092-8674(00)80716-3. [DOI] [PubMed] [Google Scholar]
- 46.Stanley LK, Seidel R, van der Scheer C, Dekker NH, Szczelkun MD, Dekker C. When a helicase is not a helicase: dsDNA tracking by the motor protein EcoR124I. EMBO J. 2006;25:2230–2239. doi: 10.1038/sj.emboj.7601104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Janscak P, Bickle TA. DNA supercoiling during ATP-dependent DNA translocation by the Type I restriction enzyme EcoAI. J. Mol. Biol. 2000;295:1089–1099. doi: 10.1006/jmbi.1999.3414. [DOI] [PubMed] [Google Scholar]
- 48.Sisáková E, Weiserová M, Dekker C, Seidel R, Szczelkun MD. The interrelationship of helicase and nuclease domains during DNA translocation by the molecular motor EcoR124I. J. Mol. Biol. 2008;384:1273–1286. doi: 10.1016/j.jmb.2008.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.