Abstract
The aim of the present study was to analyse the molecular mechanisms involved in the Interleukin-6 (IL-6) silencing in pancreatic adenocarcinoma cell lines. Our results demonstrate that TNF-α, a major IL-6 inducer, is able to induce IL-6 only in three out of six cell lines examined. 5-aza-2′-deoxycytidine (DAC), but not trichostatin A (TSA), activates the expression of IL-6 in all cell lines, indicating that DNA methylation, but not histone deacetylation, plays an essential role in IL-6 silencing. Indeed, the IL-6 upstream region shows a methylation status that correlates with IL-6 expression and binds MeCP2 and H3meK9 only in the non-expressing cell lines. Our results suggest that critical methylations located from positions –666 to –426 relative to the transcription start site of IL-6 may act as binding sites for MeCP2.
INTRODUCTION
Interleukin-6 (IL-6) is a cytokine produced constitutively and/or under specific stimuli by almost all nucleated cells (1). Its biological role includes a wide range of activities, such as the regulation of the immune response, the acute-phase reaction, cell proliferation and differentiation, apoptosis and cell trafficking (1,2). All these activities may have a direct correlation to the pathogenesis of several diseases, including various types of cancers (3–5).
Pancreatic adenocarcinoma is a very aggressive cancer, characterized by a high frequency of gene mutations (TP53, K-RAS, P16 and DPC4) (6) and alterations in the expression of secondary messengers involved in cancer pathogenesis (1). In pancreatic cancer, IL-6 deregulation has been associated with carcinogenesis, cell proliferation, angiogenesis, alteration in cell adhesion molecules and chemotaxis, and reduction of sensitivity to apoptosis (1,7,8).
Transcriptional activation of IL-6 has been extensively studied (9–12), whereas the mechanisms involved in its repression remain largely unclarified (13–19).
Epigenetic events, such as DNA methylation and histone modifications, play critical roles in gene silencing, which is a key event in both carcinogenesis and drug resistance (20,21). DNA methylation leads to transcriptional repression either by directly interfering with the binding of transcription factors or by the involvement of proteins that specifically recognize methylated CpGs (mCpGs) (22), the prototype of which is the methyl-CpG-binding protein 2 (MeCP2). Two independent, not mutually exclusive, mechanisms have been claimed for MeCP2 activities (23). In the enzymatic model, MeCP2 specifically targets mCpGs and determines repressive heterochromatin by interacting with the co-repressor mSin3A, which is part of a large complex containing histone deacetylases (HDACs) (24,25). MeCP2 can also recruit histone methyltransferase that methylates local H3 lysine 9 strengthening the repressive state (26). In this model, repression may be partially or totally relieved by the HDAC inhibitor trichostatin A (TSA). The structural model, which is not affected by HDAC inhibitors, postulates the ability of MeCP2 to repress gene transcription by mechanisms of chromatin condensation independent of histone acetylation (23).
In the present work, we have studied the molecular mechanisms leading to IL-6 repression in three non-expressing and three IL-6-expressing pancreatic adenocarcinoma cell lines. We demonstrate that in all cell lines the DNA methyltransferase inhibitor 5-aza-2′-deoxycytidine (DAC) induces IL-6 transcription, while the HDAC inhibitor TSA does not significantly modify IL-6 expression. Furthermore, we report that the IL-6 upstream region shows a methylation status that correlates with IL-6 expression and binds MeCP2 and H3meK9 only in the non-expressing cell lines.
MATERIALS AND METHODS
Reagents
TSA and DAC were purchased from Sigma (Sigma–Aldrich), solubilized in DMSO and stored at −80°C until use. TNF-α (PeproTech, Calbiochem) was solubilized in PBS/1% BSA. Sodium bisulphite was obtained from Sigma–Aldrich.
Cell lines
Six human pancreatic adenocarcinoma cell lines, PaCa44, CFPAC1, PT45P1, PC, HPAFII and Panc1 [Moore et al. (6) for genetic characterization and primary tissue source], were grown in RPMI 1640 supplemented with 10% FBS and 50 µg/ml gentamicin sulphate (BioWhittaker), and were incubated at 37°C with 5% CO2. All cell lines were routinely screened for mycoplasma contamination by PCR analysis.
RNA extraction and northern blot analysis
Total RNA from 5 × 106 cells was extracted by TRIzol Reagent (Invitrogen) according to the manufacturer’s instructions. Cells were treated with TNF-α (5 ng/ml) for 1 h. Ten micrograms of denaturated RNA was separated by electrophoresis in 1% agarose gel containing 5% formaldehyde and 20 mM 3-[N-morpholino]propanesulphonic acid (MOPS) and was transferred onto nylon membranes (Hybond-N, Amersham-Pharmacia Biotech.) in 10× SSC. The membranes were hybridized overnight at 65°C with a 32P-labelled IL-6 cDNA probe (DECAprimeII kit, Ambion) in a solution containing 7% SDS, 1% BSA, 1 mM EDTA and 50% NaH2PO4. Filters were cold washed twice with a solution containing 5% SDS, 0.5% BSA, 1 mM EDTA and 4% NaH2PO4, then twice at 65°C with a pre-warmed solution containing 1% SDS, 1 mM EDTA and 4% NaH2PO4. Specific mRNAs were detected and quantified by scanning the filters in the PhosphorImager (Molecular Dynamics) using ImageQuant software (ACHS).
IL-6 enzyme-linked immunosorbent assay
Cells were seeded in 96-well microtiter plates (4 × 103 cells/well) and treated for 24 h with TNF-α (5 ng/ml). Culture supernatants were assayed for IL-6 production by PREDICTATM kit (Genzyme diagnostic) according to the manufacturer’s instructions.
Nuclear extracts and electrophoretic mobility shift assay
Nuclear extracts were prepared according to Osborn et al. (27) from 5 × 106 cells treated with TNF-α (5 ng/ml) for 30 min and/or DAC (2.5 µM) for 72 h and/or TSA (300 nM) for the last 8 h. Protein concentration was measured by the Coomassie Protein Assay Reagent (Pierce), using BSA as a standard. Binding reactions were carried out using 10 µg of nuclear protein extracts in a binding buffer containing 20 mM Hepes (pH 7.5), 50 mM KCl, 0.5 mM DTT, 0.1 mM EDTA, 10% glycerol and 2 µg Poly(dI-dC) (Amersham). Radiolabelled probe (0.02 pmol; sp.act 3 × 106 cpm/pmol) was added last to each reaction mix, and samples were incubated at room temperature for 30 minutes. In competition assays, a 200-fold molar excess of cold double-stranded oligonucleotides was added to the reaction mix. Samples were run on 5% (30 : 1.2) native polyacrylamide gel in 0.5× TBE, dried, detected and quantified by scanning the filters in the PhosphorImager (Molecular Dynamics) using ImageQuant software (ACHS). The oligonucleotide sequences used were 5′-AATGTGGGATTTTCCCATG-3′ for IL-6-NF-κB and 5′-CTCAACCCCCAATAAATAT-3′ for IL-6-C/EBP. The formation of specific complexes was previously established by using specific antibodies.
Plasmid constructs and transient transfections
The reporter constructs p4xNF-κB-LUC and p4xC/EBP-LUC were obtained by cloning four tandem repeats of NF-κB or C/EBP binding sites of the IL-6 promoter into the pGL Promoter vector (Promega). The pIL-6(−592) construct was obtained by inserting the MscI/XhoI fragment of the human IL-6 promoter into the pGL Basic vector (Promega). All plasmids were amplified in dam− bacterial strains. 2.5 × 105 cells/well were seeded in 6-well plates. Twenty-four hours later, transfections were carried out with FuGENE 6 Transfection Reagent (Roche) keeping a transfection reagent:DNA ratio of 3:2. TNF-α (5 ng/ml) stimulation was carried out 18 h later. Twenty-four hours after transfection, reporter gene activity was determined by Luciferase Assay System (Promega) according to the manufacturer’s instructions.
When DAC treatment was performed, cells were seeded in a six-well plate at a density of 1.5 × 105 cells/well and DAC (2.5 µM) was added after 24 h. Forty-eight hours later, the pIL-6(−592) vector was transfected with FuGENE 6 Transfection Reagent. Twenty-four hours after transfection, reporter gene activity was determined by Luciferase Assay System.
In each transfection, 0.2 μg of pSVβgal was added, and the luciferase activity was normalized with respect to the β-galactosidase activity.
RT–PCR
Total RNA was extracted as described above. One microgram of total RNA was reverse-transcribed by First Strand cDNA Synthesis (Invitrogen) in a final volume of 20 μl. One microlitre of cDNA was used for PCR amplification. The sequences of the primers were as follows: IL6 forward 5′-CTTCTCCACAAGCGCCTTCG-3′; IL6 reverse 5′-TACTCTTGTTACATGTCTCC-3′; β-ACTIN forward 5′-ACCAACTGGGACGACATGGAGAA-3′; β-ACTIN reverse 5′-TGGTGGTGAAGCTGTAGCC-3′. PCR conditions included 25–40 cycles (depending on cell lines and genes) at 94°C for 1 min, 58°C for 1 min and 72°C for 30 s, and a final extension at 72°C for 4 min. β-ACTIN was used as an internal standard to evaluate the relative expression levels of IL-6. PCR products were separated by electrophoresis in 1.5% agarose gel and stained with ethidium bromide.
DNA methylation analysis
The DNA methylation status was analysed by bisulphite genomic sequencing (BGS). Genomic DNA was extracted from cultured cells by Wizard Genomic DNA Purification Kit (Promega) and used for bisulphite treatment performed with EZ DNA Methylation-Gold kit (Zymo Research, Orange, California) according to the manufacturer’s instructions.
Fifty nanograms of each modified DNA was amplified by PCR from −1150 to +50 with respect to the IL-6 transcriptional start site.
PCR products were separated by electrophoresis in 1.5% agarose gel stained with ethidium bromide, excised, cleaned by Qiagen Gel Extraction Kit (Qiagen) and used for pyrosequencing analysis. The pyrosequencing service was offered by EpigenDx Inc (Worcester, Massachusetts). The assay design package including primer sequences and PCR conditions is available on request.
Chromatin immunoprecipitation assays.
Chromatin immunoprecipitation (ChIP) assays were performed using the EpiQuik TM Chromatin Immunoprecipitation Kit (Epigentek) according to the manufacturer’s instructions. Briefly, DMSO (control), TNF-α-, DAC- or TNF-α/DAC-treated cells were fixed using 1% formaldehyde at room temperature for 10 min. Cells were washed twice in ice-cold PBS, resuspended in SDS lysis buffer and sonicated until crosslinked chromatin was sheared to an average DNA fragment molecular length of 300–500 bp. Five per cent of the sonicated lysate was used to quantify the total amount of DNA present in different samples before immunoprecipitation (inputs). Chromatin preparations were immunoprecipitated using antibodies anti-RNA polymerase II (PolII), anti-histone H4 acetylated (H4Ac, Upstate Biotechnology), anti–dimethylhistone H3 (Lys9) (H3meK9, Upstate Biotechnology), anti-methyl-CpG-binding protein 2 (MeCP2, Abcam) and a non-specific antibody (IgG). Non-immunoprecipitated samples (NoAb) were used as negative controls. Precipitated complexes were bound to Protein A Agarose, washed and then eluted in 1% SDS/0.1 M NaHCO3. Crosslinking between DNA and proteins was reversed by heating the samples at 65°C for 15 min, followed by Proteinase K digestion at 65°C for 1.5 h. After cleaning on spin columns, DNA was eluted in 20 μl of 10 µM Tris–EDTA. Four microlitres of ChIP-DNA was analysed using Brilliant® SYBR® Green QPCR Master Mix (Stratagene, La Jolla, CA, USA) and the Stratagene Mx3005P™ QPCR System. qPCR was performed in triplicate with primers for IL-6, β-ACTIN and the human chromosome 16 centromere region (16CEN).
The sequences of the primers used for β-ACTIN and 16CEN were: β-ACTIN For 5′-CTGCGCATAGCAGACATACAA-3′ and β-ACTIN Rev 5′-CTGGGCTTGAGAGGTAGAGTG-3′; 16CEN For 5′-GTCTCTTTCTTGTTTTTAAGCTGGG-3′ and 16CEN Rev 5′-TGAGCTCATTGAGACATTTGG-3′. The sequences of the six primer pairs used for IL-6 were: IL6-1 For 5′-GGGCTTCTGAACCAGCTTGA-3′ and IL6-1 Rev 5′-CAGGCACGGCTCTAGGCTC-3′; IL6-2 For 5′-AAGATGCCACAAGGTCCTCCT-3′ and IL6-2 Rev 5′-CCACTTGGTTCAGGGCAGA-3′; IL6-3 For 5′-CAGCAGCCAACCTCCTCTAAGT-3′ and IL6-3 Rev 5′-CAAGGCGTCTCCAGGTGG-3′; IL6-4 For 5′-AGGATGGCCAGGCAGTTCTA-3′ and IL6-4 Rev 5′-AAGCCTGGGATTATGAAGAAGGTA-3′; IL6-5 For 5′-TGCATGACTTCAGCTTTACTC-3′ and IL6-5 Rev 5′-GCAGAACCACTCTTCCTTTAC-3′; IL6-6 For 5′-ACCGGGAACGAAAGAGAAGC-3′ and IL6-6 Rev 5′-CTGGCAGTTCCAGGGCTAAG-3′. The following cycling conditions were used: 10 min at 95°C, 40 cycles at 95°C for 30 s, 65°C for 1 min and 72°C for 30 s. The average of cycle threshold (CT) of each triplicate was analysed according to the 2(−ΔΔCt) method. For each cell line in each condition, the result of the NoAb sample was subtracted from those of the IgG, PolII, H4Ac, MeCP2 and H3MeK9 samples. The resulting data were divided by their corresponding input DNA and finally plotted in a scale in which the final value of IgG was arbitrarily set to 1.
RESULTS
IL-6 expression in human pancreatic adenocarcinoma cell lines
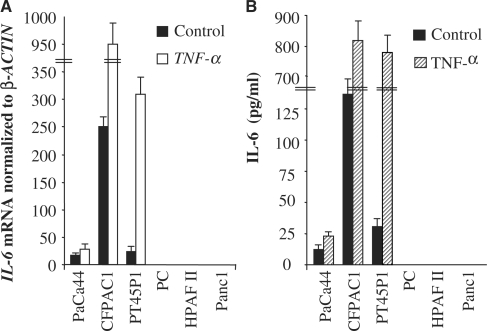
We analysed IL-6 expression in six human pancreatic adenocarcinoma cell lines treated with TNF-α for 1 h. The presence of IL-6 mRNA was evaluated by northern blot analysis. Figure 1A shows that mRNA IL-6 was constitutively expressed in three of the cell lines examined (PaCa44, CFPAC1 and PT45P1), and its level was increased in response to TNF-α. PC, HPAFII and Panc1 cells exhibited neither basal nor TNF-α-induced expression of IL-6 (Figure 1A). Consistent with the data of gene expression, IL-6 was constitutively present in PaCa44, CFPAC1 and PT45P1 cell lines and significantly increased after 24 h of TNF-α treatment (Figure 1B). In contrast, no IL-6 was detected in PC, HPAFII and Panc1 cells both in the absence and in the presence of TNF-α.
Figure 1.
IL-6 expression in human pancreatic adenocarcinoma cell lines. (A) Cells were incubated in the absence or presence of TNF-α for 1 h. IL-6 induction was evaluated by northern blot analysis performed as described in ‘Materials and Methods’ section. The expression of β-ACTIN was used as an internal control. (B) Cells were incubated in the absence or presence of TNF-α for 24 h. Culture supernatants from untreated and treated cells were assayed for IL-6 production as described in ‘Materials and Methods’ section. The results are the means ± SE of three independent experiments.
NF-κB and C/EBP DNA binding activity and their functional role
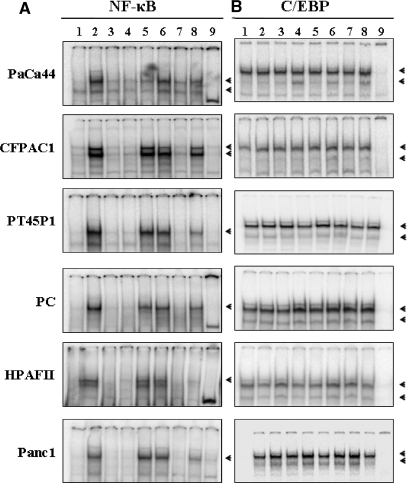
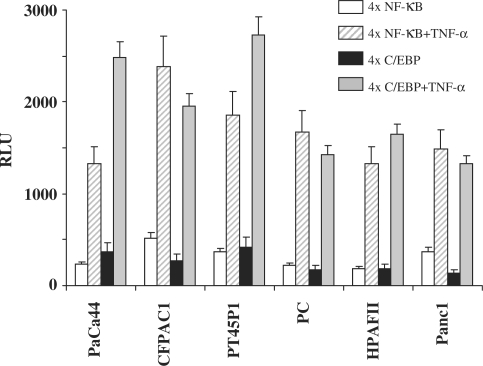
To verify whether the absence of IL-6 production was due to alterations in the transcriptional machinery, we analysed the DNA binding activities of NF-κB and C/EBP, the main transcription factors for IL-6 promoter activation. Electrophoretic mobility shift assays (EMSAs) performed with the IL-6-NF-κB oligonucleotide as a probe showed the formation of specific and TNF-α-inducible DNA-protein complexes in all cell lines (Figure 2A: lanes 1, 2 and 9). EMSAs performed with the IL-6-C/EBP oligonucleotide showed specific DNA-protein complexes constitutive and non-inducible by TNF-α (Figure 2B: lanes 1, 2 and 9). To determine whether NF-κB and C/EBP were functionally active, we carried out transfection assays with the reporter constructs p4xNF-κB-LUC and p4xC/EBP-LUC, containing four tandem repeats of the NF-κB or C/EBP binding sites of the IL-6 promoter. In all cell lines, transfections with both constructs produced a strong activation of the luciferase gene following TNF-α induction (Figure 3), indicating that either in non-expressing or in IL-6-expressing cell lines TNF-α signalling and NF-κB and C/EBP transcriptional activities were similarly functional.
Figure 2.
NF-κB and C/EBP DNA binding activity. Cells were treated as described in ‘Materials and Methods’ section and cell extracts were analysed by EMSAs using (A) the IL-6-NF-κB or (B) the IL-6-C/EBP oligonucleotides, as probes. Lanes: (1) uninduced; (2) TNF-α; (3) TSA; (4) DAC; (5) TNF-α + TSA; (6) TNF-α + DAC; (7) TSA + DAC; (8) TNF-α + TSA + DAC; (9) specific competitor. The arrows indicate specific complexes.
Figure 3.
NF-κB and C/EBP transcriptional activity. Cell lines were transfected with the plasmids p4xNF-κB-LUC and p4xC/EBP-LUC obtained by cloning four tandem repeats of NF-κB or C/EBP binding sites of the IL-6 promoter into the pGL Promoter vector. TNF-α (5 ng/ml) stimulation was carried out 18 h later. Twenty-four hours after transfection, luciferase gene activity was normalized with respect to the β-galactosidase activity (RLU). The results are the means ± SE of three independent experiments.
IL-6 regulation by DAC and/or TSA treatments
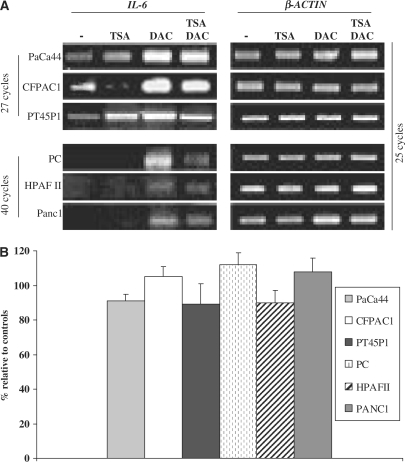
To study the involvement of epigenetic events in the IL-6 silencing in pancreatic cell lines, we examined IL-6 mRNA expression by RT–PCR analyses following treatments with DAC and/or TSA. Cells were treated with DAC (2.5 µM) for 72 h and/or TSA (300 nM) for the last 8 h. The time of TSA and DAC treatments was chosen on the basis of our published results (28,29) and preliminary experiments aimed to obtain the highest IL-6 mRNA level and the most direct effects of TSA and DAC. Figure 4A shows that TSA did not significantly modify IL-6 expression, while DAC was able to induce IL-6 mRNA both in non-expressing and in IL-6-expressing cell lines. The association TSA/DAC did not further increase the level of DAC-induced IL-6 mRNA. It is worth to note that a very different number of PCR cycles between the two groups of cell lines was necessary to obtain a similar signal. Enzyme-linked immunosorbent assay (ELISA) confirmed that DAC, but not TSA, enables PC, HPAFII and Panc1 cell lines to secrete detectable levels of IL-6 (data not shown). To rule out the possibility that DAC indirectly induces IL-6, we performed transfection experiments with the unmethylated pIL-6(−592) luciferase reporter plasmid in the absence or presence of DAC. No significant difference in luciferase activity between untreated and DAC-treated cells was observed (Figure 4B).
Figure 4.
IL-6 expression following treatment with DAC or TSA. (A) Cells were treated with DAC (2.5 µM) for 72 h and/or TSA (300 nM) for the last 8 h. Cell extracts and RT–PCR analyses were performed as described in ‘Materials and Methods’ section. The expression of β-ACTIN was used as an internal control. A representative experiment is reported. (B) Cell lines were transfected with the pIL-6(–592) plasmid. Twenty-four hours after transfection, cells were treated with DAC (2.5 µM) for 24 h. Luciferase gene activity was normalized with respect to the β-galactosidase activity and expressed as percentage relative to controls. The results are the means ± SE of three independent experiments.
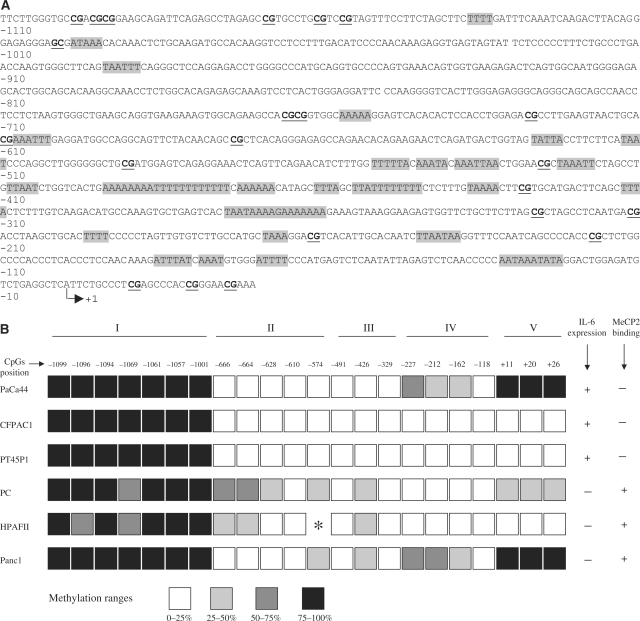
Methylation pattern of the IL-6 upstream region
To verify whether DAC activates IL-6 expression by epigenetic mechanisms, we examined by BGS (bisulphite genomic sequencing) the methylation profile of the IL-6 upstream region spanning from positions −1099 to +26 and containing 22 CpGs (Figure 5A). Figure 5B shows that the CpG methylation pattern correlated with IL-6 expression (Figure 1). To evidence the relation between the levels of methylation and gene expression, we clustered the CpGs into five groups (I, II, III, IV and V) and we summarized the methylation levels in four ranges, as indicated in the figure. Group I was highly methylated in all cell lines, while groups II and III presented different sites of methylation exclusively in the non-expressing cell lines; remarkably, only position −426 was methylated in all three non-expressing cell lines. Groups IV and V were methylated in two non-expressing cell lines, PC and PANC1, and in Paca44, the least IL-6-expressing cell line. These results strongly suggest that the IL-6-inducing activity of DAC in the non-expressing cell lines depends on directly removing specific mCpGs located between positions −666 and −426 and responsible for transcriptional repression.
Figure 5.
Schematic map of the methylation status of the IL-6 upstream region. (A) Sequence of the IL-6 upstream region. CpGs are underlined and (A/T) > 4 motifs are marked with shaded boxes. (B) Numbers at the top of the squares represent the position of 22 consecutive CpGs located from −1099 to +26 respect to IL-6 transcriptional start site. CpGs are clustered into five groups (I, II, III, IV and V) and methylation levels are represented by four ranges, as indicated at the bottom of the figure. Asterisk indicates the presence of a G to T mutation.
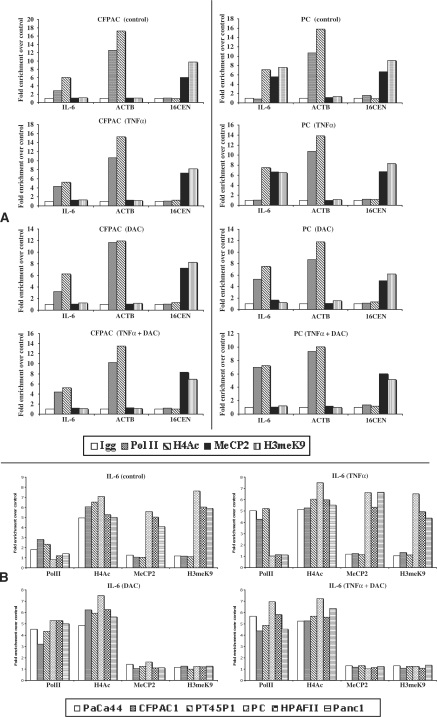
ChIP analysis of the IL-6 upstream region
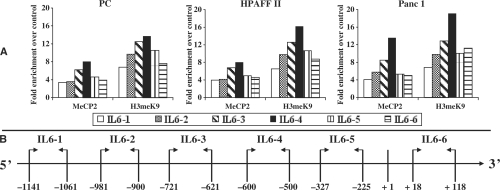
To verify whether the differentially methylated IL-6 upstream region could bind MeCP2 and/or H3meK9, we performed ChIP analysis. Figure 6A shows the pattern of binding of RNA polymerase II, acetylated histone H4, methylated histone H3 and MeCP2 on the upstream region of IL-6 and β-ACTIN and on 16CEN, in an IL-6-expressing (CFPAC1) and a non-expressing (PC) cell line. While in CFPAC1 cells the pattern of binding on IL-6 was similar to that of β-ACTIN, in PC cells, it corresponded to that of 16CEN in control and TNF-α-treated cells. Interestingly, following DAC or TNF-α/DAC treatments, the pattern of binding on IL-6 became similar to that of β-ACTIN also in PC cells. Results comparable to those shown for each representative cell line were obtained with the other cell lines (data not shown). Figure 6B shows the pattern of binding of RNA polymerase II, acetylated histone H4, methylated histone H3 and MeCP2 on the upstream region of IL-6 in all cell lines following treatment with TNF-α and/or DAC. H3meK9 and MeCP2 were present only in the three non-expressing cell lines and disappeared after DAC treatment, while H4Ac was present in all cell lines in the absence or presence of treatments. To finely identify the sequence that allows the recruitment of MeCP2/H3meK9 on the 5′-flanking region of IL-6, we performed ChIP assays with a more systematic quantitative RT–PCR analysis using six sets of primers located from −1141 to +118. Figure 7 shows that the peak of MeCP2/H3meK9 binding was obtained with the primer set amplifying the region located from −600 to −500. This finding indicates that in IL-6 non-expressing cell lines MeCP2/H3meK9 binds at the 5′-flanking region of IL-6 around positions −600/−500. Together with the observation that DAC is able to induce IL-6 mRNA (Figure 4) and prevent MeCP2/H3meK9 binding (Figure 6B), these data strongly suggest that IL-6 repression in pancreatic adenocarcinoma cell lines is mediated by the binding of MeCP2 and H3meK9 to the methylated CpG located from positions −666 to −426 of IL-6.
Figure 6.
ChIP analysis. ChIP, qPCR and data analyses were performed as reported in ‘Materials and Methods’ section. DNA from each cell line in each condition (control, TNF-α, DAC TNF-α/DAC) was immunoprecipitated in the absence or presence of the antibodies against PolII, H4Ac, H3meK9, MeCP2 and a non-specific antibody (IgG). qPCR for each ChIP-DNA sample was performed for IL-6, β-ACTIN and 16CEN. Results are reported as fold enrichment of immunoprecipitated DNA from each sample relative to the DNA immunoprecipitated with the non-specific antibody (control), and were plotted in a scale in which the final value of IgG was arbitrarily set to 1. (A) ChIP analysis of IL-6, β-ACTIN and 16CEN in two representative cell lines, an IL-6-expressing (CFPAC1) and a non-expressing (PC) cell line. (B) ChIP analysis of IL-6 in all six cell lines performed with IL6-5 primer pairs.
Figure 7.
Systematic ChIP analysis of the IL-6 5′-flanking region. ChIP, qPCR and data analyses were performed as reported in ‘Materials and Methods’ section. DNA from each cell line was immunoprecipitated in the absence or presence of the antibodies against MeCP2, H3meK9 and a non-specific antibody (IgG). qPCR for each ChIP-DNA sample was performed with six primer pairs spanning from −1141 to +118 relative to IL-6 transcriptional start site (+1). Results are reported as fold enrichment of immunoprecipitated DNA from each sample relative to the DNA immunoprecipitated with the non-specific antibody (control), and were plotted in a scale in which the final value of IgG was arbitrarily set to 1. (A) ChIP analysis of IL-6 in the three non-expressing cell lines. (B) Location of the PCR products analysed in A on the IL-6 flanking region.
DISCUSSION
In this study, we have investigated the molecular mechanisms involved in IL-6 silencing in pancreatic adenocarcinoma cell lines. Our results demonstrate that TNF-α is able to induce IL-6 only in three out of six cell lines examined, although all cell lines possess wild type IL-6 and functional transcriptional machinery. Furthermore, we show that either in non-expressing or in IL-6-expressing cell lines DAC, but not TSA, is able to induce IL-6, indicating that DNA methylation, but not histone deacetylation, plays an essential role in IL-6 silencing.
DAC might induce IL-6 by activating the expression of genes that in turn activate IL-6 transcription. To verify this hypothesis, we performed transfection experiments with the pIL-6(−592) luciferase reporter plasmid (Figure 4B). No significant difference in luciferase activity between untreated and DAC-treated cells was observed. Since the transfected plasmid was completely unmethylated, this result rules out the possibility that DAC acts by an indirect mechanism. Consistent with this hypothesis, we observed that neither DAC nor TSA significantly modified the binding activity of both NF-κB and C/EBP, the major transcription factors in IL-6 activation (Figure 2).
Quantitative pyrosequencing analysis shows that the CpG methylation status of the IL-6 upstream region in the six cell lines examined correlates with IL-6 expression. Methylation-dependent silencing of gene promoters generally depends on direct interfering with the binding of transcription factors and/or specific recognition of mCpGs by methyl-CpG-binding proteins that indirectly repress transcription (22,23). To determine whether one or both mechanisms were involved in IL-6 silencing of pancreatic adenocarcinoma cell lines, we analysed the DNA methylation profile of the IL-6 upstream region spanning from −1099 to +26. Positions −227, −212 and −162, which are close to the binding sites for transcription factors necessary for IL-6 activation, were methylated in a non-expressing cell line (Panc1) and in the least expressing cell line (PaCa44). This finding suggests that the reduced IL-6 induction by TNF-α in PaCa44 cells (Figure 1) may depend on the interference between transcription factors and methylated CpG, thus supporting a direct mechanism of repression. On the other hand, all IL-6 non-expressing cell lines showed mCpGs in a region comprised from positions −666 to −426, which does not contain transcription factor binding sites. This result suggested the occurrence of an indirect methyl-CpG-binding protein-mediated repression. Indeed, ChIP analysis at the IL-6 upstream region revealed, around positions −600/−500, the presence of both MeCP2 and H3meK9, which disappeared after DAC treatment, only in the non-expressing cell lines. The presence of H3meK9 in non-expressing cells is consistent with its role in the formation of a condensed and transcriptionally inactive chromatin (26,30). Altogether, our results strongly suggest that the major contribution to IL-6 silencing in pancreatic adenocarcinoma cell lines is due to the binding of MeCP2/H3meK9 to the methylated CpG located from positions −666 to −426 of IL-6.
Klose et al. (31) have recently reported that MeCP2 requires (A/T) >4 sequences adjacent to the methyl-CpGs for efficient DNA binding. In particular, three to eight/nine base pairs away from the methyl-CpG are necessary and sufficient for high-affinity MeCP2 binding. The analysis of the IL-6 upstream region shows several (A/T) >4 motifs (Figure 5A). Interestingly, only the sequences immediately downstream to the methyl-CpG at positions −666, −664 and −426 contain (A/T) >4 motifs corresponding to the required feature. This observation strongly supports the hypothesis that these methyl-CpGs, especially the one located at position −426 that is methylated in all three non-expressing cell lines, may act as binding sites for MeCP2. Nevertheless, we cannot rule out the possibility that MeCP2 binds unmethylated DNA of the IL-6 upstream region through the connection to faraway DNA sequences via still unknown mechanisms (32).
In the expressing cell lines, IL-6 induction by DAC, which is lower than that observed in the non-expressing cell lines, may depend on the demethylation of the IL-6 regions localized upstream to position −1001. We suppose that in these cell lines DAC treatment is able to improve the accessibility of the already transcriptionally active IL-6 promoter and to increase the constitutive IL-6 expression.
The methylation at any given site of the IL-6 region downstream to position −666 appears to be incomplete, as indicated by the different percentage of the methylation level (Figure 5B). This finding suggests allelic/cellular heterogeneity and is consistent with results already reported by other authors (33,34).
In conclusion, we have presented the first evidence of an MeCP2/H3meK9-mediated epigenetic mechanism leading to IL-6 silencing.
FUNDING
Fondazione Giorgio Zanotto, Verona, Italy; Associazione Italiana Ricerca sul Cancro, Milan, Italy; Italian Ministry of University and Research. Funding for open access charge: University of Verona.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENT
The authors thank Dr Ilaria Mazzon for her technical support and helpful comments.
REFERENCES
- 1.Feurino LW, Fisher WE, Bharadwaj U, Yao Q, Chen C, Li M. Current update of cytokines in pancreatic cancer: pathogenic mechanisms, clinical indication, and therapeutic values. Cancer Invest. 2006;24:696–703. doi: 10.1080/07357900600981398. [DOI] [PubMed] [Google Scholar]
- 2.Hodge DR, Hurt EM, Farrar WL. The role of IL-6 and STAT3 in inflammation and cancer. Eur. J. Cancer. 2005;41:2502–2512. doi: 10.1016/j.ejca.2005.08.016. [DOI] [PubMed] [Google Scholar]
- 3.Bellone G, Smirne C, Mauri FA, Tonel E, Carbone A, Buffolino A, Dughera L, Robecchi A, Pirisi M, Emanuelli G. Cytokine expression profile in human pancreatic carcinoma cells and in surgical specimens: implications for survival. Cancer Immunol. Immunother. 2006;55:684–698. doi: 10.1007/s00262-005-0047-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ishihara K, Hirano T. IL-6 in autoimmune disease and chronic inflammatory proliferative disease. Cytokine Growth Factor Rev. 2002;13:357–368. doi: 10.1016/s1359-6101(02)00027-8. [DOI] [PubMed] [Google Scholar]
- 5.Heinrich PC, Behrmann I, Haan S, Hermanns HM, Muller-Newen G, Schaper F. Principles of interleukin (IL)-6-type cytokine signalling and its regulation. Biochem. J. 2003;374:1–20. doi: 10.1042/BJ20030407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moore PS, Sipos B, Orlandini S, Sorio C, Real FX, Lemoine NR, Gress T, Bassi C, Kloppel G, Kalthoff H, et al. Genetic profile of 22 pancreatic carcinoma cell lines. Analysis of K-ras, p53, p16 and DPC4/Smad4. Virchows Arch. 2001;439:798–802. doi: 10.1007/s004280100474. [DOI] [PubMed] [Google Scholar]
- 7.Feurino LW, Zhang Y, Bharadwaj U, Zhang R, Li F, Fisher WE, Brunicardi FC, Chen C, Yao Q, Li M. IL-6 Stimulates Th2 Type Cytokine Secretion and Upregulates VEGF and NRP-1 Expression in Pancreatic Cancer Cells. Cancer Biol. Ther. 2007;6:1096–1100. doi: 10.4161/cbt.6.7.4328. [DOI] [PubMed] [Google Scholar]
- 8.ten Kate M, Hofland LJ, van Koetsveld PM, Jeekel J, van Eijck CH. Pro-inflammatory cytokines affect pancreatic carcinoma cell. Endothelial cell interactions. Jop. 2006;7:454–464. [PubMed] [Google Scholar]
- 9.Merola M, Blanchard B, Tovey MG. The kappa B enhancer of the human interleukin-6 promoter is necessary and sufficient to confer an IL-1 beta and TNF-alpha response in transfected human cell lines: requirement for members of the C/EBP family for activity. J. Interferon Cytokine Res. 1996;16:783–798. doi: 10.1089/jir.1996.16.783. [DOI] [PubMed] [Google Scholar]
- 10.Akira S, Kishimoto T. NF-IL6 and NF-kappa B in cytokine gene regulation. Adv. Immunol. 1997;65:1–46. [PubMed] [Google Scholar]
- 11.Vanden Berghe W, Vermeulen L, De Wilde G, De Bosscher K, Boone E, Haegeman G. Signal transduction by tumor necrosis factor and gene regulation of the inflammatory cytokine interleukin-6. Biochem. Pharmacol. 2000;60:1185–1195. doi: 10.1016/s0006-2952(00)00412-3. [DOI] [PubMed] [Google Scholar]
- 12.Faggioli L, Merola M, Hiscott J, Furia A, Monese R, Tovey M, Palmieri M. Molecular mechanisms regulating induction of interleukin-6 gene transcription by interferon-gamma. Eur. J. Immunol. 1997;27:3022–3030. doi: 10.1002/eji.1830271140. [DOI] [PubMed] [Google Scholar]
- 13.Plaisance S, Vanden Berghe W, Boone E, Fiers W, Haegeman G. Recombination signal sequence binding protein Jkappa is constitutively bound to the NF-kappaB site of the interleukin-6 promoter and acts as a negative regulatory factor. Mol. Cell Biol. 1997;17:3733–3743. doi: 10.1128/mcb.17.7.3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Armenante F, Merola M, Furia A, Palmieri M. Repression of the IL-6 gene is associated with hypermethylation. Biochem. Biophys. Res. Commun. 1999;258:644–647. doi: 10.1006/bbrc.1999.0566. [DOI] [PubMed] [Google Scholar]
- 15.Armenante F, Merola M, Furia A, Tovey M, Palmieri M. Interleukin-6 repression is associated with a distinctive chromatin structure of the gene. Nucleic Acids Res. 1999;27:4483–4490. doi: 10.1093/nar/27.22.4483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Palmieri M, Sasso MP, Monese R, Merola M, Faggioli L, Tovey M, Furia A. Interaction of the nuclear protein CBF1 with the kappaB site of the IL-6 gene promoter. Nucleic Acids Res. 1999;27:2785–2791. doi: 10.1093/nar/27.13.2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gerlo S, Haegeman G, Vanden Berghe W. Transcriptional regulation of autocrine IL-6 expression in multiple myeloma cells. Cell Signal. 2008;20:1489–1496. doi: 10.1016/j.cellsig.2008.04.004. [DOI] [PubMed] [Google Scholar]
- 18.Kannabiran C, Zeng X, Vales LD. The mammalian transcriptional repressor RBP (CBF1) regulates interleukin-6 gene expression. Mol. Cell Biol. 1997;17:1–9. doi: 10.1128/mcb.17.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vales LD, Friedl EM. Binding of C/EBP and RBP (CBF1) to overlapping sites regulates interleukin-6 gene expression. J. Biol. Chem. 2002;277:42438–42446. doi: 10.1074/jbc.M207363200. [DOI] [PubMed] [Google Scholar]
- 20.Ballestar E, Wolffe AP. Methyl-CpG-binding proteins. Targeting specific gene repression. Eur. J. Biochem. 2001;268:1–6. doi: 10.1046/j.1432-1327.2001.01869.x. [DOI] [PubMed] [Google Scholar]
- 21.Sansom OJ, Maddison K, Clarke AR. Mechanisms of disease: methyl-binding domain proteins as potential therapeutic targets in cancer. Nat. Clin. Pract. Oncol. 2007;4:305–315. doi: 10.1038/ncponc0812. [DOI] [PubMed] [Google Scholar]
- 22.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 23.Bowen NJ, Palmer MB, Wade PA. Chromosomal regulation by MeCP2: structural and enzymatic considerations. Cell Mol. Life Sci. 2004;61:2163–2167. doi: 10.1007/s00018-004-4177-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nan X, Campoy FJ, Bird A. MeCP2 is a transcriptional repressor with abundant binding sites in genomic chromatin. Cell. 1997;88:471–481. doi: 10.1016/s0092-8674(00)81887-5. [DOI] [PubMed] [Google Scholar]
- 25.Nan X, Ng HH, Johnson CA, Laherty CD, Turner BM, Eisenman RN, Bird A. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature. 1998;393:386–389. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- 26.Fuks F, Hurd PJ, Wolf D, Nan X, Bird AP, Kouzarides T. The methyl-CpG-binding protein MeCP2 links DNA methylation to histone methylation. J. Biol. Chem. 2003;278:4035–4040. doi: 10.1074/jbc.M210256200. [DOI] [PubMed] [Google Scholar]
- 27.Osborn L, Kunkel S, Nabel GJ. Tumor necrosis factor alpha and interleukin 1 stimulate the human immunodeficiency virus enhancer by activation of the nuclear factor kappa B. Proc. Natl Acad. Sci. USA. 1989;86:2336–2340. doi: 10.1073/pnas.86.7.2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moore PS, Barbi S, Donadelli M, Costanzo C, Bassi C, Palmieri M, Scarpa A. Gene expression profiling after treatment with the histone deacetylase inhibitor trichostatin A reveals altered expression of both pro- and anti-apoptotic genes in pancreatic adenocarcinoma cells. Biochim. Biophys. Acta. 2004;1693:167–176. doi: 10.1016/j.bbamcr.2004.07.001. [DOI] [PubMed] [Google Scholar]
- 29.Missiaglia E, Donadelli M, Palmieri M, Crnogorac-Jurcevic T, Scarpa A, Lemoine NR. Growth delay of human pancreatic cancer cells by methylase inhibitor 5-aza-2′-deoxycytidine treatment is associated with activation of the interferon signalling pathway. Oncogene. 2005;24:199–211. doi: 10.1038/sj.onc.1208018. [DOI] [PubMed] [Google Scholar]
- 30.Nakagawachi T, Soejima H, Urano T, Zhao W, Higashimoto K, Satoh Y, Matsukura S, Kudo S, Kitajima Y, Harada H, et al. Silencing effect of CpG island hypermethylation and histone modifications on O6-methylguanine-DNA methyltransferase (MGMT) gene expression in human cancer. Oncogene. 2003;22:8835–8844. doi: 10.1038/sj.onc.1207183. [DOI] [PubMed] [Google Scholar]
- 31.Klose RJ, Sarraf SA, Schmiedeberg L, McDermott SM, Stancheva I, Bird AP. DNA binding selectivity of MeCP2 due to a requirement for A/T sequences adjacent to methyl-CpG. Mol. Cell. 2005;19:667–678. doi: 10.1016/j.molcel.2005.07.021. [DOI] [PubMed] [Google Scholar]
- 32.Nikitina T, Shi X, Ghosh RP, Horowitz-Scherer RA, Hansen JC, Woodcock CL. Multiple modes of interaction between the methylated DNA binding protein MeCP2 and chromatin. Mol. Cell Biol. 2007;27:864–877. doi: 10.1128/MCB.01593-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Danam RP, Howell SR, Remack JS, Brent TP. Heterogeneous methylation of the O(6)-methylguanine-DNA methyltransferase promoter in immortalized IMR90 cell lines. Int. J. Oncol. 2001;18:1187–1193. doi: 10.3892/ijo.18.6.1187. [DOI] [PubMed] [Google Scholar]
- 34.Qian XC, Brent TP. Methylation hot spots in the 5′ flanking region denote silencing of the O6-methylguanine-DNA methyltransferase gene. Cancer Res. 1997;57:3672–3677. [PubMed] [Google Scholar]