Abstract
Accurate aminoacylation of tRNAs by the aminoacyl-tRNA synthetases (aaRSs) plays a critical role in protein translation. However, some of the aaRSs are missing in many microorganisms. Helicobacter pylori does not have a glutaminyl-tRNA synthetase (GlnRS) but has two divergent glutamyl-tRNA synthetases: GluRS1 and GluRS2. Like a canonical GluRS, GluRS1 aminoacylates tRNAGlu1 and tRNAGlu2. In contrast, GluRS2 only misacylates tRNAGln to form Glu-tRNAGln. It is not clear how GluRS2 achieves specific recognition of tRNAGln while rejecting the two H. pylori tRNAGlu isoacceptors. Here, we show that GluRS2 recognizes major identity elements clustered in the tRNAGln acceptor stem. Mutations in the tRNA anticodon or at the discriminator base had little to no impact on enzyme specificity and activity.
INTRODUCTION
In protein translation, each aminoacyl-tRNA synthetase (aaRS) recognizes and connects its cognate tRNA to its cognate amino acid (aa), forming a specific aminoacyl-tRNA (or isoacceptor set). These aminoacyl-tRNAs are then brought into the ribosome by elongation factor (EF-Tu) where they are used in protein translation. Intuitively, a complete set of 20 aaRSs is required with one enzyme matching each of the cognate 20 amino acids to the appropriate tRNA(s) (1). However, many microorganisms lack a full set of aaRSs. For example, Helicobacter pylori does not have glutaminyl-tRNA synthetase (GlnRS) or asparaginyl-tRNA synthetase (AsnRS) (2). In fact, many bacteria and archaea are missing one or both of these enzymes (3).
In the absence of GlnRS or AsnRS, Gln-tRNAGln and Asn-tRNAAsn are generated indirectly through similar two-step processes (4–11). Typically, non-discriminating glutamyl- and aspartyl-tRNA synthetases (ND-GluRS and ND-AspRS) misacylate tRNAGln and tRNAAsn, respectively, to generate Glu-tRNAGln and Asp-tRNAAsn (Reactions 1 and 3, respectively, misacylated tRNAs are shown in bold) (1,12). These enzymes are non-discriminatory because they still recognize and aminoacylate their cognate tRNAs to generate Glu-tRNAGlu and Asp-tRNAAsp (Reactions 2 and 4, respectively). Next, a glutamine-dependent amidotransferase (AdT) identifies and repairs Glu-tRNAGln and Asp-tRNAAsn to generate Gln-tRNAGln and Asn-tRNAAsn, respectively (AdT rxn not shown) (4,6,9).
-
(1)
ND-GluRS: Glu +ATP +tRNAGln → Glu-tRNAGln + AMP + PPi
-
(2)
ND-GluRS: Glu +ATP +tRNAGlu → Glu-tRNAGlu + AMP + PPi
-
(3)
ND-AspRS: Asp+ATP +tRNAAsn →Asp-tRNAAsn+ AMP + PPi
-
(4)
ND-AspRS: Asp +ATP+tRNAAsp→Asp-tRNAAsp+ AMP + PPi
Instead of a canonical ND-GluRS, H. pylori and a small subset of other bacteria utilize two paralogous GluRSs—GluRS1 and GluRS2. Helicobacter pylori GluRS1 is discriminatory and only generates Glu-tRNAGlu1 and Glu-tRNAGlu2 (H. pylori has two tRNAGlu isoacceptors; Figure 1). GluRS2 is consequently responsible for the biosynthesis of Glu-tRNAGln. Interestingly, although GluRS1 and GluRS2 are closely related, GluRS2 does not make Glu-tRNAGlu (13,14).
-
(5)
GluRS1: Glu + ATP + tRNAGlu → Glu-tRNAGlu + AMP + PPi
-
(6)
GluRS2: Glu + ATP + tRNAGln → Glu-tRNAGln + AMP + PPi
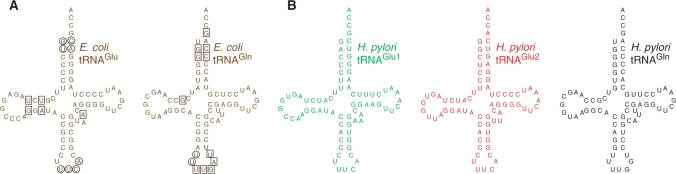
Figure 1.
Sequences and secondary structures of tRNAs. (A) The known major identity elements in E. coli tRNAGlu and tRNAGln are boxed (21–23); minor elements are circled. (B) Helicobacter pylori tRNAGlu1 is shown in green, H. pylori tRNAGlu2 is shown in red and H. pylori tRNAGln is in black.
The close evolutionary relationship between GluRS1 and GluRS2 and the unusual non-cognate tRNAGln specificity of GluRS2 led to the proposal that GluRS2 could represent an abortive or ongoing attempt by bacteria to evolve a bacterial GlnRS (14). (All known GlnRSs originated in eukarya (15–18) and the factors that have prevented the emergence and/or utilization of GlnRS in most bacteria are not well understood.) It has also been proposed that the divergence of GluRS1 and GluRS2 occurred to accommodate changes in the length of the tRNAGlu and tRNAGln D-stems (4 versus 3 base pairs, respectively) (13).
We are interested in understanding how GluRS2 diverged from GluRS1 to gain unique specificity for tRNAGln, while rejecting the two tRNAGlu isoacceptors tRNAGlu1 and tRNAGlu2. We have previously shown that a single point mutation in the anticodon-binding domain of GluRS2 converts this enzyme into one that only aminoacylates tRNAGlu1 instead of tRNAGln, demonstrating recognition of the tRNA anticodon by GluRS2. Unexpectedly, this G417T mutation did not induce aminoacylation activity towards tRNAGlu2, despite the fact that this tRNA has the same UUC anticodon (19). In order to identify the mechanisms used by GluRS2 to select tRNAGln and reject tRNAGlu2, here we have introduced varying degrees of tRNAGlu2 character into tRNAGln. Analysis of these tRNAs demonstrates that the anticodon loop and the discriminator base are not identity elements for GluRS2 aminoacylation of tRNAGln. Instead, the major identity elements are localized in the acceptor stem of tRNAGln. These results are put into an evolutionary context.
MATERIALS AND METHODS
Materials
Oligonucleotides were purchased from Invitrogen and used without further purification. The pCR 2.1 TOPO plasmid was also from Invitrogen. Pfu polymerase was purchased from Stratagene. Taq polymerase was from New England Biolabs. Radiolabeled glutamate (L-[3,4-3H]-glutamic acid) was purchased from Perkin Elmer. All buffers were filtered through a 0.22 µm filter prior to use. When appropriate, solutions were autoclaved. Unless otherwise stated, reagents were used without further purification. All gene constructs were verified by DNA sequencing of the entire gene insert.
Cloning of Hp tRNA variants
For each tRNA chimera, two partially overlapping primers were designed to reconstitute the entire tRNA gene with appended BamHI and EcoRI restriction sites onto the 5′- and 3′-ends of the gene, respectively (see Supplementary Table S1). Each primer pair was used in a template-independent polymerase chain reaction (PCR) with Pfu polymerase. Each PCR product was inserted into the pCR2.1 TOPO vector after incubation with Taq polymerase. The correct insert was verified by DNA sequencing and then sub-cloned into the BamHI and EcoRI sites of the pES300 vector to enable isopropyl β-d-1-thiogalactopyranoside (IPTG)-induced in vivo overtranscription of the cloned tRNA gene (14).
Mutations were introduced by QuikChange© mutagenesis according to the directions provided by Stratagene. Primer sequences are provided in Supplementary Table S1.
Overtranscription and purification of Hp tRNAs and tRNA chimeras
Each tRNA and chimera was overexpressed in the Escherichia coli strain MV1184 at 37°C in Luria Broth (500 ml LB) supplemented with ampicillin (100 µg/ml) and glucose (0.5% w/v). When the A600 nm was between 0.4 and 0.6, IPTG (to a final concentration of 1 mM) was added to induce production of the tRNA. Cells were pelleted by centrifugation 4–5 h after induction and stored at −80°C for future use. Each overproduced tRNA was purified by Nucleobond affinity chromatography (Clontech) as previously described (14). This procedure generates a mixture of E. coli tRNAs that is enriched with the encoded H. pylori tRNA of interest. In vivo tRNA production was conducted (instead of in vitro transcription) to allow for the introduction of a 2-thiouridine at position 34 of all three wild-type tRNAs and all mutations. This modification is essential for aminoacylation by GluRS (20). Thus, any tRNAs that lack this modification will have negligible activity in the experiments described below.
The concentration of each tRNA was determined using GluRS1 or GluRS2, depending on the substrate specificity of the tRNA, in standard aminoacylation assays (see below). Expression of tRNA variants that were not robust substrates for either GluRS1 or GluRS2 was verified by urea gel and northern blot (see next section). Final tRNA concentrations were highly variable and ranged from being too low to accurately quantify (<100 pmol/A260 nm) to ∼700 pmol/A260 nm (see Supplementary Table S3).
Northern blots of acceptor mutations and D stem/loop tRNAs
Northern blots were performed for tRNAs that could not be robustly quantified by aminoacylation. Total tRNA concentration was measured by A260 nm value; either 0.5 or 0.05 A260 nm aliquots were used for analysis of each tRNA. Each tRNA was diluted to 40 µl in 100 mM NaOAc, pH 5.0, 8 M urea, 0.05% (w/v) bromophenol blue and 0.05% (w/v) xylene cyanol; the sample was boiled for 5 min. Each sample (20 µl) was immediately loaded onto a 12% urea gel. Electrophoresis was performed for 75 min at 150 V. The tRNAs were transferred from the gel to an Immobilon-NY+ membrane (Millipore) using a Semi-Dry Blotting Unit (Fisher Biotech); transfer was conducted for 90 min at 320 mA. The membrane was baked for 90 min at 75°C and then subjected to overnight hybridization with a 32P-labeled oligonucleotide (TLH-14C: 5′ CTCGGAATGCCAGGACCAA 3′) selected to be specific for all mutant tRNAs (15). The membrane was washed four times with 20 ml of the following buffer: 450 mM NaCl, 90 mM Tris–HCl, pH 8.0, 6 mM Na2EDTA, 0.1% SDS w/v, before exposure to a storage phosphor screen (Molecular Dynamics). Bound radioactivity was visualized using a Typhoon 9210 (Amersham Biosciences).
Aminoacylation assays with GluRS1 and GluRS2
Helicobacter pylori GluRS1 and GluRS2 were purified to homogeneity as previously described (14). Aminoacylation reactions were conducted in 40 mM HEPES-OH, pH 7.5, 4 mM ATP, 8 mM MgCl2, 200 µM unlabelled Glu and 50 µCi 3H-Glu at 37°C. For assays aimed at measuring the expression level of different tRNAs, the experiments were performed for 90 min with 1 µM GluRS1 or GluRS2. For initial rate assays, 0.1 µM GluRS1 or GluRS2 was used with 10 µM enriched tRNA (concentration was estimated from A260 nm readings) and time points were taken at shorter intervals. The unit definition of GluRS1 is defined as the amount of enzyme that aminoacylates 0.1 pmol tRNAGlu1 per second; a unit of GluRS2 aminoacylates tRNAGln at a rate of 0.1 pmol per second (14). All assays were conducted in triplicate and the reported error measurements reflect standard deviation.
RESULTS
The tRNA acceptor stem is important for the tRNAGln specificity of GluRS2
There are two tRNAGlu isoacceptors in H. pylori—tRNAGlu1 and tRNAGlu2—and one tRNAGln (2) (Figure 1; the sequences of E. coli tRNAGlu and tRNAGln are given for comparison, with known identity elements marked by circles and squares) (21–23). While tRNAGlu1 and tRNAGlu2 share 78% sequence identity, H. pylori GluRS2 apparently uses different mechanisms to reject these two tRNAs. It has been shown that a single mutation in the GluRS2 anticodon-binding domain can switch this enzyme’s tRNA substrate specificity from tRNAGln to tRNAGlu1. However, this mutated GluRS2 failed to aminoacylate tRNAGlu2 (19). The main aim of the present work is to investigate the important elements that distinguish tRNAGlu2 from tRNAGln, with respect to H. pylori GluRS1 and GluRS2.
A series of four tRNA chimeras were designed according to different domains of the tRNA. These tRNAGln/Glu2 chimeras each contain ∼75% tRNAGln and ∼25% tRNAGlu2 character (Figures 2 and 3; Supplementary Table S2). Chimera 2 was further modified to contain the tRNAGlu2 variable loop in order to maintain stable tertiary structure (21). All chimeras, including chimera 3, retain the tRNAGln anticodon. Each tRNA was overtranscribed in vivo and purified by ion exchange chromatography, as previously described (14). Levels of overexpression were quantified by aminoacylation assays using excess GluRS1 and GluRS2, and results from the assay that produced the highest aminoacylation levels were used. (Note: The calculated expression levels for each tRNA are included in Supplementary Table S3.)
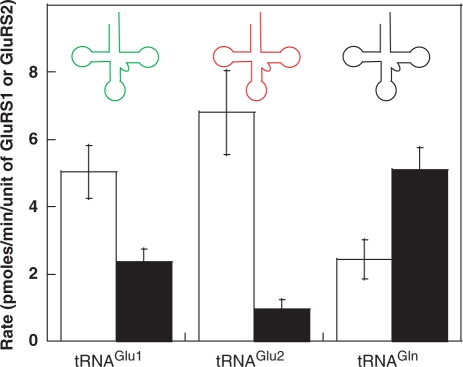
Figure 2.
Initial rates of aminoacylation for H. pylori tRNAGlu1 (green), tRNAGlu2 (red) and tRNAGln (black) using H. pylori GluRS1 (white bars) and GluRS2 (black bars). GluRS2 preferentially aminoacylates tRNAGln, whereas GluRS1 aminoacylates tRNAGlu1 and tRNAGlu2. The aminoacylation rate is determined by measuring the rate of formation of glutamyl-tRNAs in pmoles per unit of GluRS1 or GluRS2. Error bars represent standard deviation from triplicate assays.
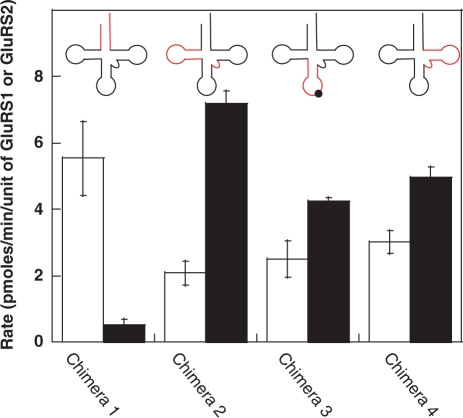
Figure 3.
Initial rates of aminoacylation for different tRNA chimeras using H. pylori GluRS1 (white bars) and GluRS2 (black bars). Each tRNA chimera represents the insertion of different regions of H. pylori tRNAGlu2 (red) into H. pylori tRNAGln (black). Chimera 1 is a better substrate for GluRS1 than GluRS2, while the remaining chimeras retain tRNAGln-like specificity and are substrates for GluRS2. The tRNAGln anticodon was retained in chimera 3. Error bars represent standard deviation from triplicate assays.
Each chimeric tRNA was assayed with GluRS1 and with GluRS2; for comparison, identical assays were conducted with the three wild-type H. pylori tRNAs. The results of these initial rate assays are shown in Figure 2 (for wild-type tRNAs) and in Figure 3 (for the chimeric tRNAs). (Note: the primary sequences of these chimeras and all other tRNA constructs are included in the Supplementary Table S2). Only Chimera 1, wherein the acceptor stem of tRNAGlu2 was transplanted into tRNAGln, has tRNAGlu2-like activity. Unlike the parent tRNAGln, Chimera 1 is a strong substrate for GluRS1 but not for GluRS2. In contrast, Chimeras 2, 3 and 4 all retain tRNAGln-like activity as they are predominantly aminoacylated by GluRS2 but not by GluRS1. These results argue that the key identity elements for GluRS2 recognition of tRNAGln are localized only within the acceptor stem of tRNAGln. In fact, the aminoacylation profiles of Chimeras 3 and 4 are virtually indistinguishable from that of tRNAGln. (Note: For tRNAGln, the low observed rate of GluRS1-catalyzed aminoacylation is the result of aminoacylation of contaminating E. coli tRNAs (14); thus, it is likely that the GluRS1 data for chimeras 2–4 is misleadingly high. Given the negligible impact that these chimeras had on GluRS2 activity, the role of contaminating tRNAs was not investigated further). Interestingly, Chimera 2 is actually a better substrate for GluRS2 than is tRNAGln. This result is seemingly in contradiction with the proposed role of the D-stem length in the emergence of GluRS2 (13); see below for further discussion and analysis.
Based on these results and the known identity elements for other tRNAGln aminoacylation systems (21–23), we chose to further dissect the acceptor stem, D-stem/loop, and anticodon stem/loop to more precisely define the role(s) of these regions and to confirm the unexpected results that neither the D-stem/loop nor the anticodon stem/loop are strong sources of identity for GluRS2 recognition of tRNAGln.
The role of the D-stem/loop
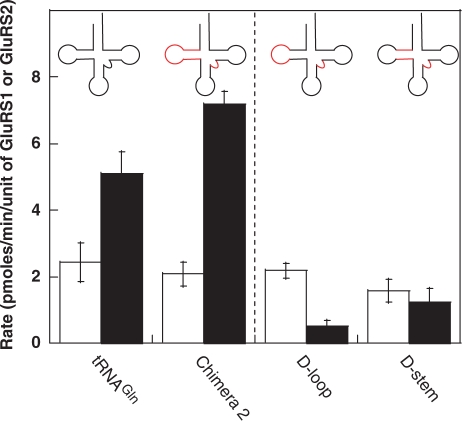
Transfer RNAGlu isoacceptors typically have an augmented D-stem, containing four base pairs instead of the three base pairs seen in tRNAGln. In E. coli tRNAGlu, this larger D-stem contains major identity elements that are recognized by the discriminating E. coli GluRS (Figure 1) (21). Moreover, it has also been proposed that the size of the tRNAGlu D-stem is an important feature for the divergence in tRNA specificity between GluRS1 and GluRS2; this hypothesis was partially based on the observation that Acidithiobacilus ferrooxidans GluRS1 aminoacylates one of its tRNAGln isoacceptors and this tRNAGln has a four base pair D-stem (13). Because of the apparent contradiction between these previous observations and our data showing that Chimera 2, which contains an engineered four base pair D-stem, is still tRNAGln-like in activity, we evaluated two additional D-stem/loop constructs. In the first construct, the D-loop of tRNAGlu2 was transplanted into tRNAGln; in the second, the tRNAGlu2 D-stem was introduced into tRNAGln. Neither of these tRNAs were robust substrates for either GluRS1 or GluRS2, a result that is strikingly different from both tRNAGln and Chimera 2 (Figure 4A). Interestingly, when the concentration of GluRS1 or GluRS2 and the length of the assay is increased (the conditions we use to quantify tRNA expression levels), both of these tRNAs can be aminoacylated by either GluRS1 or GluRS2 (see Supplementary Table S3). Consequently, neither the D-stem nor the D-loop contain major identity elements for GluRS1 or GluRS2. However, because both the D-stem and the D-loop tRNAs have diminished activity towards GluRS2, the possibility that this region contains minor identity elements cannot be ruled out.
Figure 4.
Investigation of the role of the D-stem and D-loop. Transfer RNAs were constructed to contain only the tRNAGlu2 D-stem (red) or D-Loop (red) in a tRNAGln (black) background. Neither mutant tRNA was a robust substrate for either GluRS1 or GluRS2. Data to the left of the dashed line are reproduced from Figures 1 and 2 for comparison. The white and black bars indicate data obtained from GluRS1 and GluRS2 assays, respectively. Error bars represent standard deviation from triplicate assays.
Mutagenesis of the anticodon loop
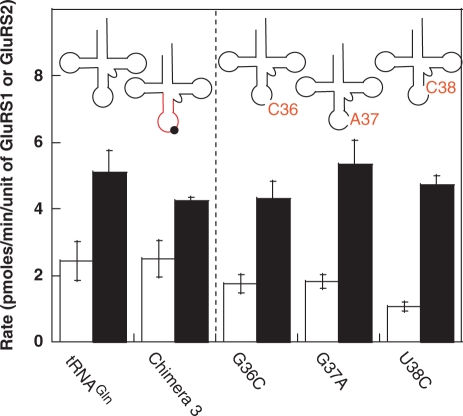
Next, we focused our attention on the anticodon stem/loop of tRNAGln. In Thermus thermophilus D-GluRS, a single mutation in the anticodon-binding domain switched this enzyme to an ND-GluRS with dual specificity for tRNAGlu and tRNAGln (24). And in H. pylori, a G417T mutation in the GluRS2 anticodon-binding domain was sufficient to introduce tRNAGlu1, but not tRNAGlu2, aminoacylation activity into GluRS2 (19). It has also been shown that both the anticodon of E. coli tRNAGlu (UUC) and E. coli tRNAGln (UUG) are important for E. coli GluRS and E. coli GlnRS recognition, respectively (21–23). Also, post-transcriptional modification of position 34 in the anticodon, to generate 5-methylaminomethyl 2-thiouridine (mnm5s2U34) enhances both aminoacylation activity and specificity (20). This modification is expected to be present in all three H. pylori tRNA isoacceptors (2) and in our different tRNAs (14). Because U34 is present in all three tRNAs, it was not evaluated. The H. pylori tRNAGlu1, tRNAGlu2, and tRNAGln anticodon loops vary at four positions (Figure 1). Among these four nucleotides, N36, N37, and N38 have been shown to be important identity elements for both E. coli GluRS and GlnRS (21–23). We individually evaluated each of these positions by mutating the nucleotide in tRNAGln into that of tRNAGlu2 (Figure 5). Consistent with the wild-type behavior of Chimera 3, mutagenesis at each of these positions had no effect on the substrate behavior of tRNAGln. These results show that the anticodon loop is not important for GluRS2 recognition, in contrast to patterns seen with other GluRSs and with many other tRNA/aaRS pairs (12,19,24,26,27).
Figure 5.
Impact of specific nucleotides in the anticodon loop on the initial aminoacylation rates of tRNAGln. The third nucleotide of the tRNAGln anticodon G36, G37 and U38, were individually mutated to the corresponding nucleotides in tRNAGlu2 (G36C, G37A, U38C) and assayed with both GluRS1 (white bars) and GluRS2 (black bars). These mutations did not impact the substrate specificity of tRNAGln. Data to the left of the dashed line are reproduced from Figures 1 and 2 for comparison. Error bars represent standard deviation from triplicate assays.
Mutagenesis of the tRNAGln acceptor stem and discriminator base
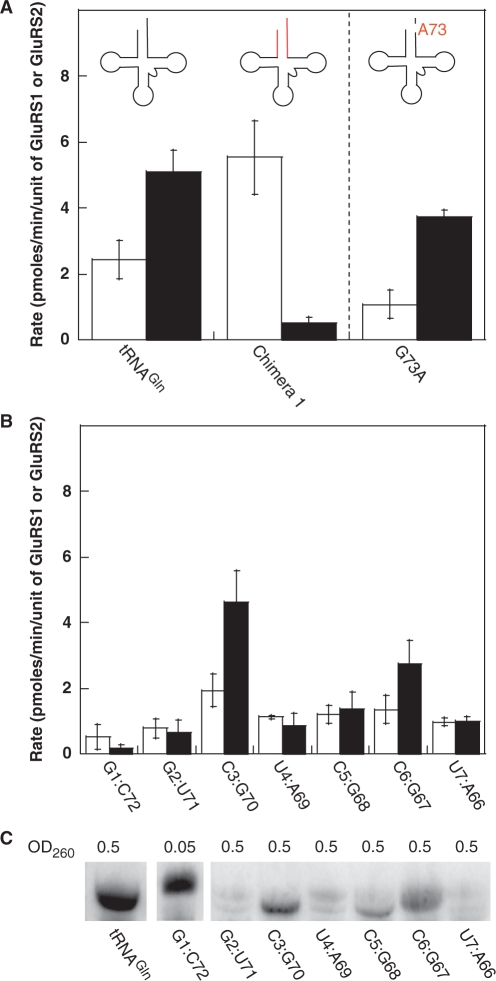
Finally, we turned our attention to the acceptor stem of tRNAGln, the region that holds the most promise based on the original survey of tRNA chimeras. We first examined the discriminator base (N73), another position that is often used by aaRSs to achieve tRNA specificity (27,28). The G73 discriminator base of E. coli tRNAGln is a major identity element for E. coli GlnRS (22,23). Interestingly, H. pylori tRNAGlu1 and tRNAGln have the same G73 discriminator base, despite being recognized by different GluRSs (GluRS1 and GluRS2, respectively); tRNAGlu2 has an A73 in this position. A G73A-mutant tRNAGln was evaluated to test the impact of the discriminator base on GluRS1 and GluRS2 aminoacylation. Unexpectedly, given the common importance of this position, the identity of the discriminator base is not important for GluRS2 recognition of tRNAGln (Figure 6A).
Figure 6.
Influence of the discriminator base and acceptor stem on the initial aminoacylation rate of tRNAGln. (A) The discriminator base of H. pylori tRNAGln was mutated to that of tRNAGlu2 (A G73A mutation). This change did not affect substrate specificity of this tRNA. Initial rates of aminoacylation with GluRS1 (white bars) versus GluRS2 (black bars). Error bars represent standard deviation. (B) Evaluation of single base pair mutations in the acceptor stem of H. pylori tRNAGln. Initial rates of aminoacylation with GluRS1 (white bars) versus GluRS2 (black bars). Each mutation is labeled according to the base pair inserted into tRNAGln (from tRNAGlu2). Error bars represent standard deviation. (C) Evaluation of the expression efficiency of each acceptor stem mutant tRNA by Northern blot. Transfer RNAGln and the G1:C72 mutant tRNA were strongly expressed. The remaining mutant tRNAs were expressed at lower and varying levels. The white bars separating the first three lanes are the result of digital removal of empty lanes in the gel. The image shown was adjusted with uniform contrast using ImageQuant TL (Amersham Biosciences, v. 2005). Another image of the blot, showing the last 6 lanes with darker contrast, is provided in the Supplementary Figure S1.
Next, we individually evaluated each base pair in the acceptor stem of tRNAGln, in order to better understand how GluRS1 recognizes Chimera 1. In other GluRS and GlnRS systems, residues within the acceptor stem are known identity elements (The first two base pairs of E. coli tRNAGlu with GluRS, and the second and third base pairs of E. coli tRNAGln with GlnRS) (21–23). To more precisely characterize the impact of the tRNA acceptor stem on H. pylori GluRS1 and GluRS2 aminoacylation activity, each acceptor stem base pair in tRNAGln was separately mutated to that found in tRNAGlu2; as above, each tRNA was assayed with both GluRS1 and GluRS2 (Figure 6B). Consistent with the analysis of Chimera 1, all but two of these mutations are poor substrates for GluRS2. The exceptions are the third and sixth base pairs (G3:C70 and U6:A67 in tRNAGln). Inversion of the G3:C70 base pair to C3:G70 had no effect on this tRNA’s specificity, whereas the C6:G67 mutation caused a slight decrease in the rate of aminoacylation by GluRS2 but did not induce GluRS1 recognition. In contrast, none of the remaining mutations were robust substrates for either GluRS1 or GluRS2.
In order to interpret these results, it was necessary to verify that these mutant tRNAs were overtranscribed and present in our assay mixtures. To this end, a sample of each tRNA was analyzed by northern blot (Figure 6C). Mutation at the first base pair (converting the U1:A72 in tRNAGln into the G1:C72 of tRNAGlu2), generated a tRNA that was overtranscribed in vivo at levels greater than that of wild-type tRNAGln. This high level of tRNA production and the lack of activity with GluRS2 clearly demonstrate that the G1:C72 base pair of tRNAGlu2 is a major antideterminant for GluRS2. The fifth base-pair mutation (G5:C68 inverted to C5:G68) was overtranscribed at moderate levels, confirming that this position is also important as an antideterminant in tRNAGlu2, preventing GluRS2 recognition. The second, fourth and seventh base pairs were not robustly overexpressed (Figure 6C and Supplementary Figure S1). Each of these tRNAs was assayed at the maximum possible concentration. Consequently, the poor aminoacylation activities with each of these mutations likely indicate that each of these positions is an important antideterminant that prevents tRNAGlu2 from being aminoacylated by GluRS2, however the possibility that tRNA expression levels were simply too low to observe aminoacylation activity cannot be ruled out.
Interestingly, while many of the acceptor stem positions in tRNAGlu2 are clearly important for rejection by GluRS2, no single base-pair mutation led to recognition by GluRS1. This observation is in sharp contrast to Chimera 1, which is a robust substrate for GluRS1. Clearly, some or all of these positions are tRNAGlu2 determinants for GluRS1, but they are only strong enough to induce recognition when combined. Perhaps, tRNAGln also contains an antideterminant for GluRS1 distal to the acceptor stem.
DISCUSSION
GluRS2 uses specialized mechanisms to recognize tRNAGln
These studies demonstrate that GluRS2 achieves its unique tRNAGln specificity, rejecting tRNAGlu2, solely by distinguishing between differences in the acceptor stems of these two tRNAs. It is surprising that neither the anticodon nor the discriminator base is important, as these positions are critical for tRNAGlu and tRNAGln aminoacylation in other systems (21–23,27). In E. coli tRNAGlu, identity elements are spread throughout the tRNA scaffold, with major determinants located in the augmented D-stem (Figure 1) (21). The identity elements of E. coli tRNAGln are mainly located in the two distal ends of the tRNA, at the discriminator base, the second and third base pairs in the acceptor stem, and the anticodon loop (Figure 1) (22,23). Furthermore, in T. thermophilus D-GluRS the size and identity of the amino acid that interacts with the third nucleotide of the anticodon plays an important role in discrimination between tRNAGlu and tRNAGln (24); this region is important for GluRS2 rejection of tRNAGlu1 as well (19). In contrast, here we show that GluRS2 rejects tRNAGlu2 by predominantly looking at only one region of the tRNA—the acceptor stem.
Although this work focused on the rejection of tRNAGlu2, the results also provide some insight into how GluRS2 rejects tRNAGlu1. As we have previously reported, a G417T mutant GluRS2 aminoacylates tRNAGlu1 but not tRNAGln or tRNAGlu2 (19). In light of the present work, this result is surprising because tRNAGlu1 contains the same G1:C72 base pair as tRNAGlu2, a strong antideterminant for GluRS2 (Figure 6A and B). Thus, it appears that the G417T mutation unmasks a role for the tRNAGlu1 anticodon that is sufficient to overcome the potency of the G1:C72 acceptor stem antideterminant. The combination of these results suggest that tRNAGlu1, unlike tRNAGlu2, contains distal antideterminants for GluRS2 that are located in both the acceptor stem and the anticodon. This observation is unexpected since tRNAGlu1 and tRNAGlu2 contain the same UUC anticodon, and it suggests subtle differences in the shapes of the two tRNAs. Mutagenesis experiments within the tRNAGlu1 framework are needed in order to truly understand the differences in how these tRNAs are rejected by GluRS2.
Our results indicate that the first acceptor stem base pair (U1:A72) is critically important for GluRS2s tRNA specificity—namely, the accurate recognition of tRNAGln and the rejection of tRNAGlu1 and tRNAGlu2, which both contain a G1:C72 base pair. The importance of this position is conserved throughout indirect aminoacylation. Like GluRS2, AdT, the amidotranferase that converts Glu-tRNAGln into Gln-tRNAGln, relies on the U1:A72 base pair for recognition of Glu-tRNAGln (29,30); the archaeal type AdT (Methanothermobacter thermautotrophicus GatDE) also relies on this position for recognition of tRNAGln (in this case, it is an A1:U72 base pair which is recognized by GatDE) (25). The archaeal GatCAB does not use the first base pair as a strong identity element (31).
Evolutionary implications
Substantial evidence has accumulated to suggest that progenitor tRNAs were smaller than their modern counterparts and consisted of either a single acceptor-stem microhelix or a minihelix comprised of the acceptor stem and TΨC-stem/loop (32–34). These smaller RNAs were putatively aminoacylated by ancestral aaRSs, comprised solely of catalytic domains (34,35). Acquisition of divergent anticodon-binding domains was a likely key step in the separation of GluRS and GlnRS (34,36). GluRS2 deviates from this picture, however, because it contains a GluRS-like anticodon-binding domain (13,14). Instead, this enzyme has capitalized on primordial mechanisms of tRNA recognition, in effect rendering the anticodon-binding domain useless, at least with respect to distinguishing between tRNAGln and tRNAGlu2.
A very recent report also demonstrated that the truncated catalytic domain of E. coli D-GluRS is capable of discriminating against tRNAGln, in favor of tRNAGlu, even in the presence of a GlnRS anticodon-binding domain (37). While this D-GluRS truncation was large enough to include recognition of known D-stem/loop identity determinants, this report further supports the evolutionary hypothesis that ancestral identity elements were recognized solely by the catalytic domain of GluRSs, as we see predominate here for GluRS2.
The data presented herein also show that the strongest determinants for H. pylori GluRS1 are localized within the acceptor stem of tRNAGlu. Chimera 1 was the only chimera to show robust aminoacylation activity with GluRS1. However, individual base pair mutations in the acceptor stem of tRNAGln were insufficient to induce GluRS1 recognition. Thus, tRNAGlu2 does not contain a single potent determinant for GluRS1, rather aminoacylation activity is induced by recognition of a set of identity elements apparently distributed throughout the acceptor stem. Thus, like GluRS2, GluRS1 uses ancestral mechanisms to recognize its tRNA substrates and to reject tRNAGln.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health (GM071480); Wayne State University. Funding for open access charge: National Institutes of Health.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Liangjun Zhou and Shirin Fatma for critical reading of the article and the two anonymous reviewers for their thoughtful and thorough comments on the article.
REFERENCES
- 1.Ibba M, Söll D. Aminoacyl-tRNA synthesis. Annu. Rev. Biochem. 2000;69:617–650. doi: 10.1146/annurev.biochem.69.1.617. [DOI] [PubMed] [Google Scholar]
- 2.Tomb JF, White O, Kerlavage AR, Clayton RA, Sutton GG, Fleischmann RD, Ketchum KA, Klenk HP, Gill S, Dougherty BA, et al. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature. 1997;388:539–547. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
- 3.Woese CR, Olsen GJ, Ibba M, Söll D. Aminoacyl-tRNA synthetases, the genetic code, and the evolutionary process. Microbiol. Mol. Biol. Rev. 2000;64:202–236. doi: 10.1128/mmbr.64.1.202-236.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schön A, Kannangara CG, Gough S, Söll D. Protein biosynthesis in organelles requires misaminoacylation of tRNA. Nature. 1988;331:187–190. doi: 10.1038/331187a0. [DOI] [PubMed] [Google Scholar]
- 5.Wilcox M. Gamma-phosphoryl ester of glu-tRNA-GLN as an intermediate in Bacillus subtilis glutaminyl-tRNA synthesis. Cold Spring Harb. Symp. Quant. Biol. 1969;34:521–528. doi: 10.1101/sqb.1969.034.01.059. [DOI] [PubMed] [Google Scholar]
- 6.Curnow AW, Hong K, Yuan R, Kim S, Martins O, Winkler W, Henkin TM, Söll D. Glu-tRNAGln amidotransferase: a novel heterotrimeric enzyme required for correct decoding of glutamine codons during translation. Proc. Natl Acad. Sci. USA. 1997;94:11819–11826. doi: 10.1073/pnas.94.22.11819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Curnow AW, Ibba M, Söll D. tRNA-dependent asparagine formation. Nature. 1996;382:589–590. doi: 10.1038/382589b0. [DOI] [PubMed] [Google Scholar]
- 8.Wilcox M. Gamma-glutamyl phosphate attached to glutamine-specific tRNA. A precursor of glutaminyl-tRNA in Bacillus subtilis. Eur. J. Biochem. 1969;11:405–412. doi: 10.1111/j.1432-1033.1969.tb00788.x. [DOI] [PubMed] [Google Scholar]
- 9.Becker HD, Kern D. Thermus thermophilus: a link in evolution of the tRNA-dependent amino acid amidation pathways. Proc. Natl Acad. Sci. USA. 1998;95:12832–12837. doi: 10.1073/pnas.95.22.12832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Becker HD, Min B, Jacobi C, Raczniak G, Pelaschier J, Roy H, Klein S, Kern D, Söll D. The heterotrimeric Thermus thermophilus Asp-tRNA(Asn) amidotransferase can also generate Gln-tRNA(Gln) FEBS Lett. 2000;476:140–144. doi: 10.1016/s0014-5793(00)01697-5. [DOI] [PubMed] [Google Scholar]
- 11.Becker HD, Reinbolt J, Kreutzer R, Giege R, Kern D. Existence of two distinct aspartyl-tRNA synthetases in Thermus thermophilus. Structural and biochemical properties of the two enzymes. Biochemistry. 1997;36:8785–8797. doi: 10.1021/bi970392v. [DOI] [PubMed] [Google Scholar]
- 12.Cathopoulis T, Chuawong P, Hendrickson TL. Novel tRNA aminoacylation mechanisms. Mol. Biosyst. 2007;3:408–418. doi: 10.1039/b618899k. [DOI] [PubMed] [Google Scholar]
- 13.Salazar JC, Ahel I, Orellana O, Tumbula-Hansen D, Krieger R, Daniels L, Söll D. Coevolution of an aminoacyl-tRNA synthetase with its tRNA substrates. Proc. Natl Acad. Sci. USA. 2003;100:13863–13868. doi: 10.1073/pnas.1936123100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Skouloubris S, Ribas de Pouplana L, De Reuse H, Hendrickson TL. A noncognate aminoacyl-tRNA synthetase that may resolve a missing link in protein evolution. Proc. Natl Acad. Sci. USA. 2003;100:11297–11302. doi: 10.1073/pnas.1932482100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brown JR, Doolittle WF. Gene descent, duplication, and horizontal transfer in the evolution of glutamyl- and glutaminyl-tRNA synthetases. J. Mol. Evol. 1999;49:485–495. doi: 10.1007/pl00006571. [DOI] [PubMed] [Google Scholar]
- 16.Lamour V, Quevillon S, Diriong S, N'G;uyen VC, Lipinski M, Mirande M. Evolution of the Glx-tRNA synthetase family: the glutaminyl enzyme as a case of horizontal gene transfer. Proc. Natl Acad. Sci. USA. 1994;91:8670–8674. doi: 10.1073/pnas.91.18.8670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wolf YI, Aravind L, Grishin NV, Koonin EV. Evolution of aminoacyl-tRNA synthetases–analysis of unique domain architectures and phylogenetic trees reveals a complex history of horizontal gene transfer events. Genome Res. 1999;9:689–710. [PubMed] [Google Scholar]
- 18.Doolittle RF, Handy J. Evolutionary anomalies among the aminoacyl-tRNA synthetases. Curr. Opin. Genet. Dev. 1998;8:630–636. doi: 10.1016/s0959-437x(98)80030-0. [DOI] [PubMed] [Google Scholar]
- 19.Lee J, Hendrickson TL. Divergent anticodon recognition in contrasting glutamyl-tRNA synthetases. J. Mol. Biol. 2004;344:1167–1174. doi: 10.1016/j.jmb.2004.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Madore E, Florentz C, Giege R, Sekine S, Yokoyama S, Lapointe J. Effect of modified nucleotides on Escherichia coli tRNAGlu structure and on its aminoacylation by glutamyl-tRNA synthetase. Predominant and distinct roles of the mnm5 and s2 modifications of U34. Eur. J. Biochem. 1999;266:1128–1135. doi: 10.1046/j.1432-1327.1999.00965.x. [DOI] [PubMed] [Google Scholar]
- 21.Sekine S, Nureki O, Sakamoto K, Niimi T, Tateno M, Go M, Kohno T, Brisson A, Lapointe J, Yokoyama S. Major identity determinants in the “augmented D helix” of tRNA(Glu) from Escherichia coli. J. Mol. Biol. 1996;256:685–700. doi: 10.1006/jmbi.1996.0118. [DOI] [PubMed] [Google Scholar]
- 22.Hayase Y, Jahn M, Rogers MJ, Sylvers LA, Koizumi M, Inoue H, Ohtsuka E, Söll D. Recognition of bases in Escherichia coli tRNA(Gln) by glutaminyl-tRNA synthetase: a complete identity set. EMBO J. 1992;11:4159–4165. doi: 10.1002/j.1460-2075.1992.tb05509.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jahn M, Rogers MJ, Söll D. Anticodon and acceptor stem nucleotides in tRNA(Gln) are major recognition elements for E. coli glutaminyl-tRNA synthetase. Nature. 1991;352:258–260. doi: 10.1038/352258a0. [DOI] [PubMed] [Google Scholar]
- 24.Sekine S, Nureki O, Shimada A, Vassylyev DG, Yokoyama S. Structural basis for anticodon recognition by discriminating glutamyl-tRNA synthetase. Nat. Struct. Biol. 2001;8:203–206. doi: 10.1038/84927. [DOI] [PubMed] [Google Scholar]
- 25.Oshikane H, Sheppard K, Fukai S, Nakamura Y, Ishitani R, Numata T, Sherrer RL, Feng L, Schmitt E, Panvert M, et al. Structural basis of RNA-dependent recruitment of glutamine to the genetic code. Science. 2006;312:1950–1954. doi: 10.1126/science.1128470. [DOI] [PubMed] [Google Scholar]
- 26.Charron C, Roy H, Blaise M, Giege R, Kern D. Non-discriminating and discriminating aspartyl-tRNA synthetases differ in the anticodon-binding domain. EMBO J. 2003;22:1632–1643. doi: 10.1093/emboj/cdg148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Giege R, Sissler M, Florentz C. Universal rules and idiosyncratic features in tRNA identity. Nucleic Acids Res. 1998;26:5017–5035. doi: 10.1093/nar/26.22.5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Crothers DM, Seno T, Söll G. Is there a discriminator site in transfer RNA? Proc. Natl Acad. Sci. USA. 1972;69:3063–3067. doi: 10.1073/pnas.69.10.3063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nakamura A, Yao M, Chimnaronk S, Sakai N, Tanaka I. Ammonia channel couples glutaminase with transamidase reactions in GatCAB. Science. 2006;312:1954–1958. doi: 10.1126/science.1127156. [DOI] [PubMed] [Google Scholar]
- 30.Bailly M, Giannouli S, Blaise M, Stathopoulos C, Kern D, Becker HD. A single tRNA base pair mediates bacterial tRNA-dependent biosynthesis of asparagine. Nucleic Acids Res. 2006;34:6083–6094. doi: 10.1093/nar/gkl622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Namgoong S, Sheppard K, Sherrer RL, Söll D. Co-evolution of the archaeal tRNA-dependent amidotransferase GatCAB with tRNA(Asn) FEBS Lett. 2007;581:309–314. doi: 10.1016/j.febslet.2006.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Francklyn C, Schimmel P. Aminoacylation of RNA minihelices with alanine. Nature. 1989;337:478–481. doi: 10.1038/337478a0. [DOI] [PubMed] [Google Scholar]
- 33.Ribas de Pouplana L, Schimmel P. Aminoacyl-tRNA synthetases: potential markers of genetic code development. Trends Biochem Sci. 2001;26:591–596. doi: 10.1016/s0968-0004(01)01932-6. [DOI] [PubMed] [Google Scholar]
- 34.Schimmel P, Ribas De Pouplana L. Footprints of aminoacyl-tRNA synthetases are everywhere. Trends Biochem Sci. 2000;25:207–209. doi: 10.1016/s0968-0004(00)01553-x. [DOI] [PubMed] [Google Scholar]
- 35.Ribas de Pouplana L, Schimmel P. Two classes of tRNA synthetases suggested by sterically compatible dockings on tRNA acceptor stem. Cell. 2001;104:191–193. doi: 10.1016/s0092-8674(01)00204-5. [DOI] [PubMed] [Google Scholar]
- 36.Siatecka M, Rozek M, Barciszewski J, Mirande M. Modular evolution of the Glx-tRNA synthetase family—rooting of the evolutionary tree between the bacteria and archaea/eukarya branches. Eur. J. Biochem. 1998;256:80–87. doi: 10.1046/j.1432-1327.1998.2560080.x. [DOI] [PubMed] [Google Scholar]
- 37.Dubois DY, Blais SP, Huot JL, Lapointe J. A C-truncated glutamyl-tRNA synthetase specific for tRNA(Glu) is stimulated by its free complementary distal domain: mechanistic and evolutionary implications. Biochemistry. 2009;48:6012–6021. doi: 10.1021/bi801690f. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.