Abstract
The SRS2 (Suppressor of RAD Six screen mutant 2) gene encodes an ATP-dependent DNA helicase that regulates homologous recombination in Saccharomyces cerevisiae. Mutations in SRS2 result in a hyper-recombination phenotype, sensitivity to DNA damaging agents and synthetic lethality with mutations that affect DNA metabolism. Several of these phenotypes can be suppressed by inactivating genes of the RAD52 epistasis group that promote homologous recombination, implicating inappropriate recombination as the underlying cause of the mutant phenotype. Consistent with the genetic data, purified Srs2 strongly inhibits Rad51-mediated recombination reactions by disrupting the Rad51-ssDNA presynaptic filament. Srs2 interacts with Rad51 in the yeast two-hybrid assay and also in vitro. To investigate the functional relevance of the Srs2-Rad51 complex, we have generated srs2 truncation mutants that retain full ATPase and helicase activities, but differ in their ability to interact with Rad51. Importantly, the srs2 mutant proteins attenuated for Rad51 interaction are much less capable of Rad51 presynaptic filament disruption. An internal deletion in Srs2 likewise diminishes Rad51 interaction and anti-recombinase activity. We also present evidence that deleting the Srs2 C-terminus engenders a hyper-recombination phenotype. These results highlight the importance of Rad51 interaction in the anti-recombinase function of Srs2, and provide evidence that this Srs2 function can be uncoupled from its helicase activity.
INTRODUCTION
Homologous recombination (HR) is a major pathway for the elimination of DNA double-strand breaks (DSBs) induced by genotoxic agents or that arise from endogenous damage and replication fork demise. As such, HR is critical for maintaining genome integrity and important for cancer avoidance in humans (1–3). Paradoxically, intermediates generated by the HR machinery can trigger prolonged arrest of the cell cycle (4), and the aberrant resolution of these intermediates can lead to gross chromosomal rearrangements, such as translocations (5,6). Moreover, the HR machinery can interfere with other DNA repair pathways, such as RAD6/RAD18-mediated post-replicative repair (PRR) (4,7). For these reasons, cells possess multiple mechanisms to prevent untimely and deleterious HR events. One such mechanism is mediated by the SRS2 (Suppressor of RAD Six screen mutant 2) gene in Saccharomyces cerevisiae.
A mutant variant of SRS2 was first recognized as a suppressor of the DNA damage sensitivity of rad6 or rad18 mutants (8). Suppression of the rad6 or rad18 DNA repair defect by the srs2 mutation requires that HR be functional (9), suggesting that SRS2 negatively regulates HR. By attenuating HR, it is thought that SRS2 helps ensure the channeling of DNA lesions encountered by the DNA replication machinery into the Rad6/Rad18-mediated PRR pathway. Yeast strains harboring certain SRS2 mutations also exhibit a hyper-recombination phenotype, failure to recover from DNA damage checkpoint-mediated G2/M arrest, and synthetic lethality with a variety of other mutations, such as sgs1, that affect DNA metabolism (10). The synthetic lethality of these double mutants often can be overcome by eliminating HR (11,12), implicating inappropriate HR events as the cause of lethality.
As a member of the SF1 family of nucleic acid unwinding enzymes (13), Srs2 possesses ssDNA-dependent ATPase and DNA helicase activities (14,15). In concordance with the genetic data, Srs2 strongly inhibits the formation of DNA joints in in vitro recombination reactions that are mediated by the Rad51 recombinase (16,17). Extensive biochemical and electron microscopic analyses have shown that Srs2 accomplishes this feat by disrupting the Rad51 presynaptic filament, comprised of a right-handed Rad51 helical polymer assembled on ssDNA (16,17), that catalyzes recombination reactions (18). The Rad51 presynaptic filament dissociative function and HR attenuating role of Srs2 require its ATPase activity (19). Srs2 interacts with the sumoylated form of the proliferating cell nuclear antigen (PCNA) at DNA replication forks (20,21). This serves to target Srs2 to DNA replication forks to prevent Rad51 presynaptic filament assembly, thereby avoiding unwanted recombination during DNA replication (20,21). Interestingly, when PCNA is not able to be sumoylated, Srs2 recruitment to S phase replication fork foci is decreased, while its recruitment to recombination foci is unaffected (22). This reveals that recruitment of Srs2 to replication forks or sites of recombination are independent processes.
Interestingly, even though Srs2 suppresses spontaneous recombination events, it facilitates DSB repair by the synthesis-dependent single-strand annealing (SDSA) pathway of HR (23–25). In addition to its DSB repair function, Srs2 has been shown to have a role in DNA damage checkpoint activation during S phase (26) and to act with DNA polymerase δ to suppress DNA triplet repeat expansion, possibly by unwinding DNA stem-loop structures formed by the repeats (27,28).
By yeast two hybrid and biochemical analyses, Srs2 was found to physically interact with Rad51 (16). However, the significance of the Rad51-Srs2 complex with regards to the anti-recombinase function of Srs2 is not clear. Here, we finely map the Rad51 interaction domain in Srs2 and generate mutant srs2 forms that retain ATP hydrolysis and helicase activities, but differ in their ability to interact with Rad51. We show that the Rad51 interaction-deficient mutant srs2 proteins are attenuated for the Rad51 presynaptic filament dissociative function. Our results thus reveal a requirement for complex formation with Rad51 in the anti-recombinase function of Srs2.
MATERIALS AND METHODS
Yeast two-hybrid assay
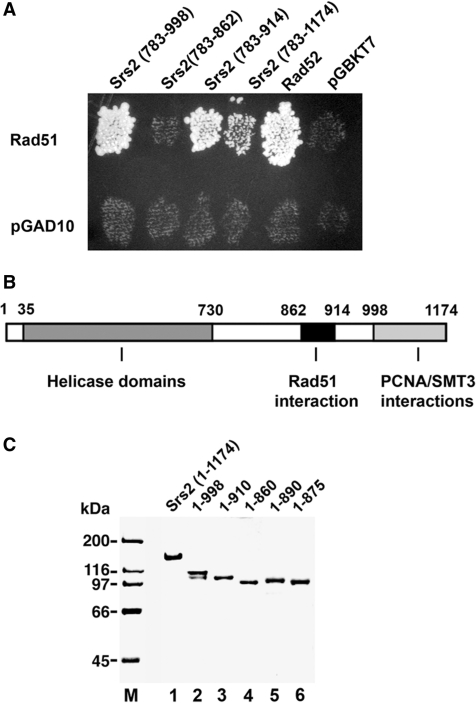
RAD51 was fused to the GAL4 transcription activation domain in the vector pGAD10, and a C-terminal SRS2 fragment encompassing residues 783–1174 harboring the Rad51 interaction domain (16) was fused to the GAL4 DNA-binding domain in pGBKT7. The pGAD10-RAD51 and pGBKT7-srs2 plasmids were introduced into haploid yeast strains PJ69-4a (MATa trp1-901 leu2-3,112 ura3-52 his3-200 gal4Δ gal80Δ LYS2::GAL1-HIS3 GAL2-ADE2 met2::GAL7-lacZ) and PJ69-4α (MATα trp1-901 leu2-3,112 ura3-52 his3-200 gal4Δ gal80Δ LYS2::GAL1-HIS3 GAL2-ADE2 met2::GAL7-lacZ), respectively (29,30). Diploid strains obtained by mating plasmid-bearing PJ69-4a and PJ69-4α haploids were grown on synthetic medium lacking tryptophan and leucine (30). To select for two-hybrid interactions, which would result in the activation of the ADE2 or HIS3 reporter genes, diploid cells were replica-plated on synthetic medium lacking tryptophan, leucine and adenine, or on synthetic medium lacking tryptophan, leucine and histidine, respectively. Both plating gave the same results. Only the plating on the tryptophan, leucine and adenine dropout medium is shown in Figure 1A. Additional pGBKT7-based plasmids containing Srs2 residues 783–998, 783–914 and 783–862 were also tested for interaction with Rad51.
Figure 1.
Two-hybrid interactions of Rad51 with Srs2. (A) Fragments of Srs2 were tested for two-hybrid interaction with Rad51 by plating on medium lacking leucine, tryptophan and adenine. The empty vector (pGBKT7) and plasmid harboring RAD52 were included as negative and positive controls, respectively. (B) A schematic of the helicase and protein-protein interaction domains in Srs2. (C) Purified Srs2, srs2 1–998, srs2 1–910, srs2 1–860, srs2 1–890 and srs2 1–875, 1 µg each, were analyzed by SDS-PAGE and stained with Coomassie Blue.
Plasmid construction
The pET11c-(His)9-SRS2 plasmid was generated by site-directed mutagenesis (Stratagene) of pET11c-SRS2, resulting in the addition of an N-terminal 9-histidine tag to the open reading frame of Srs2 (21). The srs2 variants were generated by site-directed mutagenesis (Stratagene) using the pET11c-(His)9-SRS2 plasmid as template. The srs2 783–998 and srs2 783–1174 cloned in pGBKT7 were described elsewhere (16). The other two constructs, srs2 783–862 and srs2 783–914 in pGBKT7 were generated by site-directed mutagenic insertion of a stop codon in srs2 783–998/pGBKT7.
Protein purification
The purification of the (His)9-tagged Srs2 and mutant srs2 proteins from Escherichia coli cells (50 g of cell paste, from 10 L of culture) was conducted as previously described (15), except that an affinity step on nickel NTA agarose was included to exploit the (His)9 tag. Peak fractions from the SP Sepharose column were combined and incubated with 2 ml of nickel-NTA agarose (Qiagen) for 2 h at 4°C with gentle mixing. The beads were washed with 50 ml of K buffer (20 mM K2HPO4, pH 7.4, 10% glycerol, 0.5 mM DTT and 0.5 mM EDTA) containing 300 mM KCl and 10 mM imidazole. Srs2 or mutant was eluted with 12 ml (8 fractions of 1.5 ml each) of 300 mM imidazole in K buffer containing 300 mM KCl. The peak fractions were diluted with 2.5 volumes of T buffer (20 mM Tris–HCl, pH 7.5, 10% glycerol, 0.5 mM DTT and 0.5 mM EDTA), before being fractionated in a 1 ml Mono S column, as described (16). The nearly homogeneous Srs2 and mutant protein preparations were concentrated to ∼5 mg/ml and stored in small aliquots at −80°C.
Rad51 and RPA were over-expressed in yeast and purified to near homogeneity, as described (31). Rad54 and PCNA were expressed in E. coli and purified to near homogeneity, as described (32,33).
DNA substrates
The DNA helicase substrate was constructed using the unlabeled H3 oligonucleotide and 32P-labeled H1 oligonucleotide, as previously described (15). The ϕX174 (+) strand DNA was purchased from New England Biolabs. The D1 oligonucleotide used in the D-loop assay and the 150-mer E oligonucleotide used in electron microscopy have been described (16,34).
Affinity pulldown assays
Affi-gel 15 beads containing Rad51 (Affi-Rad51; 5 mg/ml) and bovine serum albumin (Affi-BSA; 12 mg/ml) were prepared as described previously (35). (His)9-tagged Srs2 or the indicated mutant variant (5 µg) was incubated with 5 µl of Affi-Rad51 or Affi-BSA in 30 µl of K buffer (20 mM K2HPO4, 10% glycerol, 0.5 mM EDTA, 0.01% Igepal and 1 mM DTT) containing 150 mM KCl for 1 hr on ice, with gentle mixing. The beads were washed twice with 150 µl of the same buffer before being treated with 30 µl of 2% SDS to elute the bound protein. The supernatant that contained unbound Srs2 or mutant, the second wash, and the SDS eluate, 10 µl of each, were analyzed by 7.5% SDS-PAGE and staining with Coomassie Blue. For the pull-down with srs2 Δ875–902 and Rad51, purified (His)9-tagged Srs2 or srs2 Δ875–902 (5 µg) was mixed with purified Rad51 (5 µg) in buffer K containing 150 mM KCl for 1 hr on ice, before adding 10 µl Ni-NTA agarose beads (Qiagen) and continuing the incubation for 30 min on ice with gentle mixing. The beads were washed twice with 150 µl of the same buffer before being treated with 30 µl of 2% SDS to elute the bound protein. The supernatant that contained unbound proteins, the second wash and the SDS eluate, 10 µl each, were analyzed by 7.5% SDS-PAGE and staining with Coomassie Blue. For examining Srs2-PCNA complex formation, nickel pull-downs were conducted as described above for srs2 Δ875–902 and Rad51, however purified PCNA (5 µg) was used in place of Rad51.
ATPase assay
Purified Srs2 or truncated srs2 (35 nM) was incubated with 1 mM (γ-32P) ATP and ϕX174 (+) strand DNA (25 µM nucleotides) in 10 µl of buffer A (30 mM Tris–HCl, pH 7.5, 2.5 mM MgCl2, 1 mM DTT, 150 mM KCl and 100 µg/ml BSA) for 7.5 min at 37°C. The level of ATP hydrolysis was determined by thin layer chromatography with phosphorimaging analysis in a Personal Molecular Imager FX (Bio-Rad), as described (35).
DNA helicase assay
Purified Srs2 or mutant srs2 (40 nM) was incubated at 30°C for 5 min with the helicase substrate (300 nM nucleotides) in 10 µl of buffer H (30 mM Tris–HCl, pH 7.5, 2.5 mM MgCl2, 1 mM DTT, 100 mM KCl, 2 mM ATP and 100 µg/ml BSA). The reaction mixtures were stopped, deproteinized and resolved in a 10% non-denaturing polyacrylamide gel run in TAE buffer (40 mM Tris–HCl, pH 7.4, 0.5 mM EDTA) at 4°C. The gel was dried onto Whatman DE81 paper and subjected to phosphorimaging analysis.
D-loop reaction
Reactions were carried out in Buffer D (35 mM Tris–HCl, pH 7.5, 2 mM ATP, 2.5 mM MgCl2, 1 mM DTT, including an ATP-regenerating system consisting of 20 mM creatine phosphate and 20 µg/ml creatine kinase) and 50, 100 or 150 mM KCl in a final volume of 12.5 µl, as described previously (16). The 32P-labeled D1 oligonucleotide (3 µM nucleotides) was incubated with Rad51 (1 µM) for 5 min at 37°C to assemble the Rad51-ssDNA nucleoprotein filaments, followed by the incorporation of RPA (200 nM) and a 4 min incubation. Then, Rad54 (150 nM) was added, and following a 3 min incubation at 23°C, the reaction was initiated by adding pBluescript replicative form I DNA (50 µM base pairs), followed by a 6 min incubation at 30°C. The reaction was stopped by treatment with SDS (0.5%) and proteinase K (0.5 mg/ml) at 37°C for 5 min and resolved in a 1% agarose gel in TAE buffer. The gel was dried and subjected to phosphorimaging analysis. The percentage D-loop refers to the quantity of the D1 oligonucleotide substrate that has been converted into D-loop. When present, Srs2 or mutant srs2 (7.5 or 15 nM, as indicated) was added to the reaction with RPA.
DNA strand exchange reaction
Reactions were conducted at 37°C in Buffer R (35 mM Tris–HCl, pH 7.4, 2 mM ATP, 2.5 mM MgCl2, 50 mM KCl, 1 mM DTT and an ATP-regenerating system) in a final volume of 12.5 µl, as described previously (36). Rad51 (10 µM) was incubated with ϕX174 circular (+) strand (30 µM nucleotides) for 4 min, followed by a 6 min incubation with RPA (2 µM). The reaction was completed by adding spermidine hydrochloride (4 mM) and linear ϕX dsDNA (30 µM nucleotides). After an 80 min incubation, the reaction mixtures were deproteinized and resolved in 0.9% agarose gels, followed by ethidium bromide staining of the DNA species. When present, Srs2 or srs2 mutant (30–60 nM, as indicated) was added to the reaction with RPA.
Topoisomerase I-linked assay
Reactions were carried out in 10 µl of Buffer R at 37°C. Rad51 (4 µM) was incubated for 5 min with ϕX174 circular (+) strand DNA (20 µM nucleotides). Srs2 or mutant srs2 (60 or 80 nM, as indicated) and RPA (1 µM) were added, followed by a 4 min incubation. Topologically relaxed ϕX174 DNA (12.5 µM nucleotides) and 2.5 U calf thymus topoisomerase I (Invitrogen) were then incorporated to complete the reaction. After 8 min of incubation, the reaction mixtures were deproteinized and then subject to electrophoresis in 0.9% agarose gels. The DNA species were stained with ethidium bromide.
Electron microscopy
Reaction mixtures were assembled as described for the DNA strand exchange reaction except that the 150-mer E oligonucleotide (34) (7.2 µM nucleotides), Rad51 (2.4 µM), RPA (0.5 µM), Srs2 or mutant srs2 (100 nM) were used. After a 3 min incubation, the reaction mixtures were diluted 10 times with buffer R lacking the ATP regenerating system, and a 3 µl aliquot was applied to 400-mesh grids coated with carbon film and which had been glow-discharged in air. The grids were stained with 2% uranyl acetate for 30 sec and rinsed with water before being examined with a Tecnai 12 Biotwin electron microscope (FEI company) equipped with a tungsten filament at 100 keV. Digital images were captured with a Morada (Olympus Soft Imaging Solutions) charge-coupled device camera at a nominal magnification of 87 000X.
Yeast strains
All yeast strains were in the W303 background (leu2-3, 112 his3-11,15 ade2-1 ura3-1 trp1-1 can1-100 RAD5). All constructs were verified by DNA sequencing. The strains were: HKY2070-4 (srs2 1–860), HKY2082-4 (srs2 Δ875–902), HKY590-1D (srs2Δ), HKY579-10A (SRS2), and F28-1A (rad18Δ).
Methyl methane sulfonate sensitivity
Methyl methane sulfonate (MMS) sensitivity was determined using freshly made plates containing 0.005% MMS. Overnight cultures of strains were serially diluted and 4 µl aliquots of each dilution were applied onto YPD and YPD + MMS plates. Growth was assessed after 2 and 3 days at 30°C. The rad18Δ strain used has been described (37).
Determination of recombination rates
Recombination rates were calculated by the median method of Lea and Coulson (38) using the leu2-ecoRI::URA3::leu2-bstEII recombination system as described previously (39). Three different strains derived from the same parental strains were analyzed for each genotype studied. For each strain, nine independent colonies were analyzed for each fluctuation test.
RESULTS
Mapping the Srs2-Rad51 interaction in the yeast two-hybrid assay
In previously published work, we and others demonstrated an interaction between Rad51 and the carboxy-terminal region of Srs2 in the yeast two-hybrid assay, such that Srs2 fragments encompassing residues 783–1174 or residues 783–998 are capable of Rad51 association (16,40) (Figure 1A). Based on these two-hybrid data, additional constructs harboring two other portions of the Srs2 C-terminus were created and tested for Rad51 interaction. As shown in Figure 1A, while the 783–914 fragment showed robust interaction with Rad51, no signal could be detected with the 783–862 fragment (Figure 1A). Based on these results, we suspected the Rad51 interaction domain in Srs2 to lie between amino acid residues 862 and 914.
C-terminally truncated variants of Srs2 deficient for Rad51 interaction
To verify the yeast two-hybrid data in vitro and for additional biochemical analyses, we constructed various C-terminally truncated Srs2 variants—1–998, 1–910, 1–890, 1–875 and 1–860—by introducing stop codons within the pET11c-(His)9-SRS2 plasmid. The tagged full length Srs2 and the truncated variants were expressed in E. coli by the use of the IPTG-inducible T7 promoter. The chromatographic procedure for purification of the (His)9-tagged proteins was modified from the original protocol (16) to include a nickel affinity step. By this method, we were able to purify full length Srs2 and the truncated srs2 variants to near homogeneity (Figure 1C).
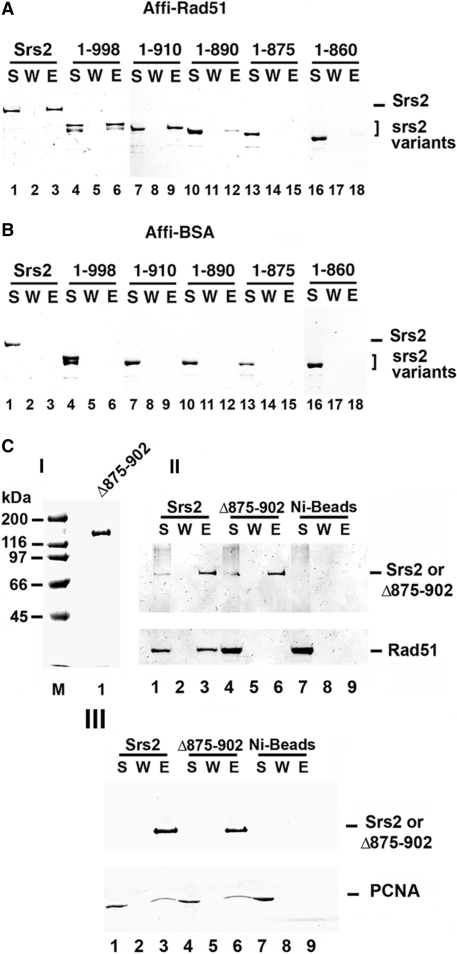
To test for interaction with Rad51, purified Srs2 and the five srs2 truncation mutant proteins were each mixed with Affi-gel beads that contained covalently conjugated Rad51 protein (Affi-Rad51) or bovine serum albumin (Affi-BSA). The beads were washed with buffer before being treated with SDS to elute the bound Srs2 or srs2 mutant protein, followed by SDS-PAGE analysis. Consistent with results from the yeast two-hybrid analysis, full length Srs2, srs2 1–998, and srs2 1–910 were retained on the Affi-Rad51 beads but not on the Affi-BSA beads (Figure 2A and B). Importantly, and consistent with the yeast two-hybrid results, srs2 1–860 did not bind the Affi-Rad51 beads (Figure 2A), indicating that it is defective for Rad51 interaction. Moreover, while srs2 1–890 showed only a slightly attenuated affinity for Affi-Rad51, little or no srs2 1–875 was retained on the Affi-Rad51 beads (Figure 2A). These results indicated that the region between amino acid residues 875 and 910 in Srs2 is likely critical for Rad51 interaction.
Figure 2.
Mapping of Rad51 interaction by in vitro pull-down assay. Purified Srs2, srs2 1–998, srs2 1–910, srs2 1–890, srs2 1–875 and srs2 1–860 were mixed with Affi-Rad51 beads (A) or Affi-BSA beads (B) The supernatant that contained unbound protein (S), wash (W) and the SDS eluate (E) were resolved by SDS-PAGE and stained with Coomassie Blue. (C) In panel I, Purified srs2 Δ875–902, 1 µg, analyzed by SDS-PAGE and stained with Coomassie Blue. In panel II, Srs2 and srs2 Δ875–902 were combined with purified Rad51 and mixed with nickel NTA agarose beads. The supernatant that contained unbound protein (S), wash (W) and the SDS eluate (E) were resolved by SDS-PAGE and stained with Coomassie Blue (lanes 1–6). As control, Rad51 alone was incubated with nickel NTA agarose beads (lanes 7–9). In panel III, Srs2 and srs2 Δ875–902 were examined for PCNA interaction (lanes 1–9) following the same procedure in panel II.
Based on the above domain mapping results, we created a form of Srs2 that lacks amino acid residues 875–902, but is otherwise full length. The srs2 Δ875–902 mutant was expressed and purified to near homogeneity following the strategy used for other Srs2 forms (Figure 2C, panel I) and examined for interaction with Rad51. The internal deletion mutant exhibited some non-specific binding to Affi-BSA beads, thus the pulldown analysis was conducted using nickel NTA affinity beads to isolate the protein complex via the (His)9-tag on Srs2. Full-length Srs2 was able to bind Rad51 as expected, but little or no Rad51 associated with srs2 Δ875–902 (Figure 2C, panel II). This provides evidence that the region between residues 875–902 of Srs2 is needed for Rad51 interaction. Additionally, we tested the ability of the internal deletion mutant to bind PCNA. The PCNA interaction domain is known to lie in the far C-terminus of Srs2 (20,41), and consistent with this, we found that srs2 Δ875–902 is just as adept as Srs2 in PCNA interaction (Figure 2C, panel III).
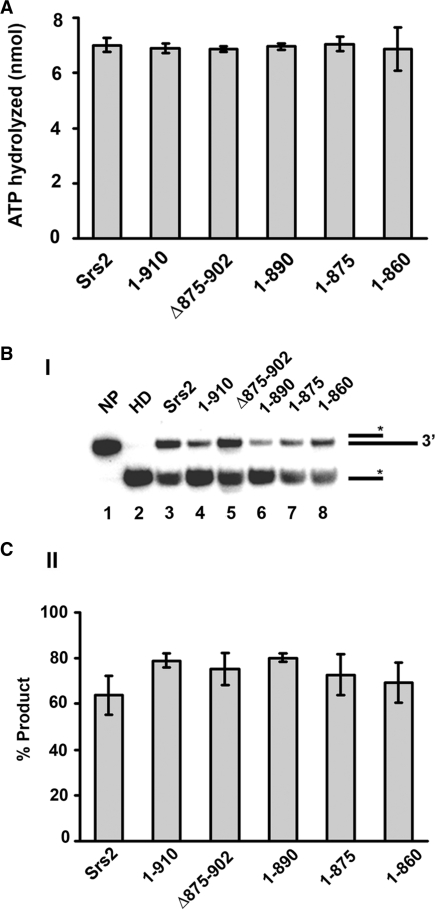
C-terminal truncations do not affect ATPase and helicase activities
The deduced Rad51 interaction domain in Srs2 resides distally to the catalytic domains that are concerned with ATP hydrolysis and helicase activity (Figure 1B), so we expected even the shortest of our srs2 truncation variants, i.e. srs2 1–860, to retain the ability to hydrolyze ATP and unwind DNA. To verify this expectation, the purified srs2 mutant proteins were first tested for their ability to hydrolyze ATP. We showed previously that ATP hydrolysis by Srs2 is ssDNA-dependent (15), and, as summarized in Figure 3A, all of the srs2 truncation variants show a level of ssDNA-dependent ATPase activity comparable to that of the wild-type protein. We next examined the various srs2 truncation mutant proteins for DNA helicase activity using a 32P-labeled substrate that contained a 40 bp duplex region with a 40-nucleotide 3′-ssDNA overhang [Figure 3B and (15)]. As shown in Figure 3B, the srs2 mutants were as adept as the full length Srs2 protein in unwinding the DNA substrate. Based on the above results, we can conclude that all of the truncated forms of srs2 that we have generated possess ATP hydrolysis and DNA helicase activities comparable to those of the wild-type counterpart.
Figure 3.
Proficiency of the srs2 mutants in ATP hydrolysis and DNA unwinding. (A) Graphical representation of the ssDNA-dependent ATPase activity of Srs2 and srs2 mutants. (B) The DNA helicase activity of Srs2 and srs2 mutants was assayed using a 3′-tailed duplex substrate (panel I) and the results quantified (panel II). NP, no protein control; HD, heat-denatured substrate. In all cases, error bars represent the standard deviations derived from three independent experiments.
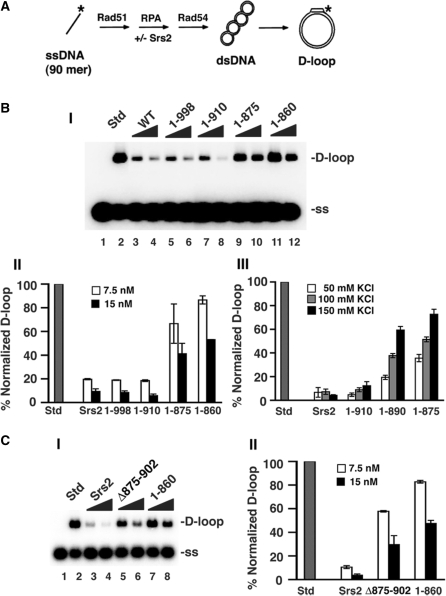
Interaction of Srs2 with Rad51 is critical for its anti-recombinase function
We (16) and others (17) demonstrated previously that Srs2 inhibits the recombinase activity of Rad51 (42). We used the D-loop assay to test the anti-recombinase activity of our srs2 truncation mutants (Figure 4A). As we reported previously, the addition of a small amount of Srs2 (7.5–15 nM) to the D-loop reaction containing Rad51, Rad54 and the heterotrimeric ssDNA-binding factor RPA led to pronounced inhibition of the reaction (Figure 4B). The two srs2 truncation mutants, srs2 1–998 and srs2 1–910, that are able to interact with Rad51 also inhibited the D-loop reaction to a comparable degree as Srs2 (Figure 4B). However, the srs2 1–875 and srs2 1–860 mutants, which are deficient in Rad51 interaction, exerted only a slight inhibitory effect on the D-loop reaction (Figure 4B). The slight inhibition observed with increasing amounts of srs2 1–875 or srs2 1–860 could be due to a residual interaction with Rad51. Likewise, the srs2 Δ875–902 mutant that is also deficient in Rad51 interaction is again significantly impaired for the ability to suppress the D-loop reaction (Figure 4C). Interestingly, the srs2 1–890 truncation mutation that is partially attenuated for Rad51 interaction showed an intermediate level of inhibition of the D-loop reaction, with the defect being much more obvious at higher concentrations of KCl (Figure 4B, panel III), which very likely weakened the residual interaction between Rad51 and this srs2 mutant. Altogether, these results revealed interaction with Rad51 as important for the anti-recombinase activity of Srs2.
Figure 4.
Relevance of the Srs2-Rad51 complex in the attenuation of the D-loop reaction. (A) D-loop reaction scheme. (B) In panel I, the radiolabeled D1 oligonucleotide was incubated with Rad51, Rad54, RPA (lanes 2–12) and with or without Srs2, srs2 1–998, srs2 1–910, srs2 1–875 or srs2 1–860 (7.5 or 15 nM) and then pBluescript form I DNA was incorporated. The reaction (lane 2) without Srs2 or mutant srs2 is designated as standard (Std) and corresponded to conversion of 43% of the input D1 oligonucleotide into the D-loop product. Lane 1 contained the DNA substrates but no protein. The KCl concentration was 50 mM KCl in these reactions and the results were quantified and plotted in panel II. In panel III, the results from D-loop reactions carried out at 50, 100, and 150 mM KCl with 15 nM of Srs2 or srs2 mutant were quantified and plotted. The D-loop product in the standard (Std) was 58%, 61% and 66% at 50, 100 and 150 mM KCl, respectively. (C) Panel I shows D-loop reactions with 7.5 and 15 nM Srs2, srs2 Δ875–902 or srs2 1–860 at 50 mM KCl. The D-loop in the standard (Std) was 54%. The results were quantified and plotted in panel II. In all cases, error bars represent the standard deviations derived from three independent experiments.
Examination of Rad51-interaction defective srs2 mutants in the DNA strand exchange reaction
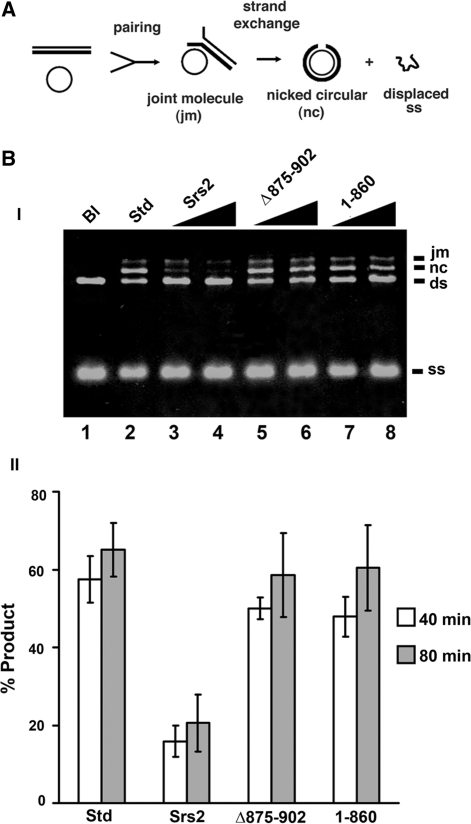
We applied a DNA strand exchange system that uses plasmid length DNA substrates to seek independent verification that interaction with Rad51 is critical for the anti-recombinase attribute of Srs2. In this system, Rad51-made DNA joint molecules are processed by DNA strand exchange to yield nicked circular duplex as the final reaction product (16,18) (Figure 5A). As expected (16), the addition of a catalytic quantity of Srs2 (30 and 60 nM) strongly suppressed the DNA strand exchange reaction (Figure 5B). In contrast, little or no attenuation of DNA strand exchange efficiency was seen when srs2 1–860 or srs2 Δ875–902 was tested (Figure 5B). The above results support our conclusion that the efficacy of Srs2’s anti-recombinase activity is contingent upon Rad51 interaction.
Figure 5.
Anti-recombinase activity examined by DNA strand exchange. (A) The DNA strand exchange scheme. (B) In panel I, circular ϕX174 (+) strand DNA was incubated with Rad51, RPA, and in the absence or presence of Srs2, srs2 Δ875–902 or srs2 1–860 (30 and 60 nM), and then spermidine and linear ϕX dsDNA were added and the reaction incubated for either 40 or 80 min. A reaction without Srs2 or mutant srs2, designated as standard (Std; lane 2) served as a control. The DNA substrates were also incubated without any protein (Bl; lane 1). Only the results from the 80 min time-point are shown. In panel II, the sum of joint molecules and nicked circular duplex from the 40 min and 80 min time-points of reactions that contained 30 nM Srs2 or srs2 mutant was plotted. In all cases, error bars represent the standard deviations derived from the results of three independent experiments.
Relevance of protein–protein interaction in Rad51 presynaptic filament disruption
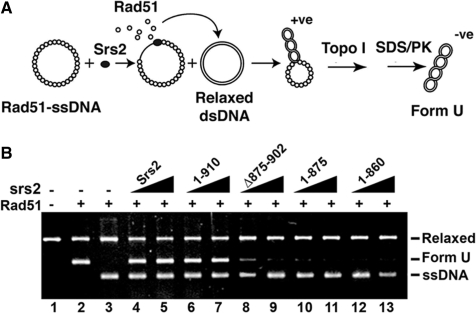
The above results obtained using C-terminally truncated srs2 mutant proteins helped establish that interaction with Rad51 is critical for Srs2’s ability to attenuate Rad51-mediated HR homologous DNA pairing and strand exchange. We employed a biochemical assay to provide evidence that the impairment of anti-recombinase activity in the truncated srs2 mutants stems from an inability to disrupt the Rad51 presynaptic filament. In this biochemical test, pre-assembled presynaptic filaments are incubated with Srs2 or one of the srs2 mutants together with RPA, and then topologically relaxed duplex DNA is added to trap the Rad51 molecules freed from ssDNA as a result of anti-recombinase function. Since Rad51 binding induces lengthening of the DNA trap, the level of anti-recombination function can be conveniently monitored as a DNA linking number change upon treatment of the duplex with topoisomerase I [Figure 6A, (16)]. The product of this reaction, an underwound DNA species called form U (Figure 6B, lane 2), is resolved from other DNA species by agarose gel electrophoresis and then revealed by ethidium bromide staining.
Figure 6.
Rad51 presynaptic filament disruption as measured by a topoisomerase-linked assay. (A) Reaction scheme for detecting Rad51 presynaptic filament disruption. (B) Rad51 presynaptic filaments were incubated without or with Srs2, srs2 1–910, srs2 Δ875–902, srs2 1–875 or srs2 1–860 (60 and 80 nM) before the addition of relaxed DNA and topoisomerase I (lanes 3–13). Lane 2 contains Form U DNA made by incubating topologically relaxed DNA with Rad51 and topoisomerase I. Lane 1 contains topologically relaxed DNA.
As expected, the addition of Srs2 to the presynaptic filaments led to the generation of form U, indicative of the release of Rad51 from the ssDNA [Figure 6B, (15)]. Abundant Form U DNA was also seen when the Rad51 interaction proficient srs2 1–910 mutant, which we showed earlier to possess the wild-type level of anti-recombinase activity (e.g. Figure 4B), was incubated with the presynaptic filaments (Figure 6B). Importantly, the Rad51-interaction deficient srs2 1–860, srs2 1–875 and srs2 Δ875–902 mutants, which we found to be impaired for anti-recombinase activity, were incapable of producing a significant amount of Form U DNA (Figure 6B). These results thus provide direct evidence that dissociation of the Rad51 presynaptic filament by Srs2 is reliant upon the interaction between Srs2 and Rad51.
Analysis by electron microscopy
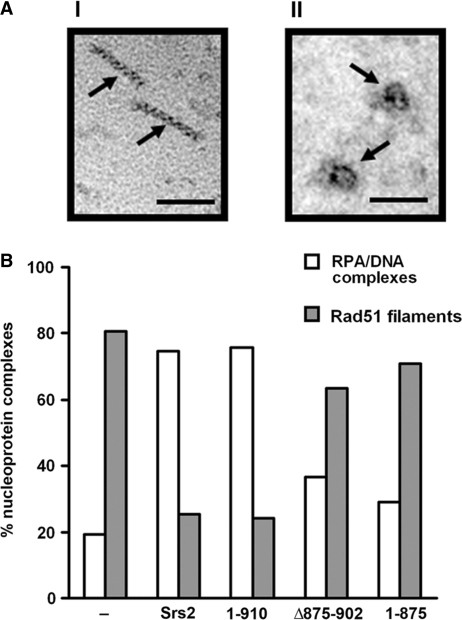
We also employed electron microscopy to examine and quantify the disruption of Rad51 presynaptic filaments by Srs2 and several of the srs2 truncation mutants. For this purpose, we assembled Rad51 presynaptic filaments on a 150-mer oligonucleotide (43) and then incubated these filaments with Srs2 or one of several srs2 mutants in conjunction with RPA. The reaction mixtures were diluted with buffer, applied onto carbon grids, and then examined by electron microscopy to quantify the levels of Rad51 presynaptic filament and RPA–ssDNA complex in each case. As summarized in Figure 7, Rad51 presynaptic filaments accounted for ∼80% of the nucleoprotein complexes in Srs2’s absence, whereas these filaments represented only a little over 20% of the nucleoprotein complexes upon the addition of Srs2. As we had anticipated, srs2 1–910 was just as adept as the full-length protein in Rad51 presynaptic filament dissociation, but, importantly, srs2 1–875 or srs2 Δ875–902 was much less effective in this regard (Figure 7). Overall, the results from the EM analysis provided independent support for Rad51 interaction as a key determinant in the efficacy of Srs2 as an anti-recombinase.
Figure 7.
Quantification of Rad51 presynaptic filament disruption by electron microscopy. (A) Representative Rad51 nucleoprotein filaments (I) or RPA-ssDNA complexes (II) assembled on 150-mer ssDNA. The scale bar represents 50 nm. (B) Distribution of Rad51 presynaptic filaments and RPA-ssDNA complexes observed following incubation without or with 100 nM Srs2, srs2 1–910, srs2 Δ875–902 or srs2 1–875.
Phenotypic analysis of srs2 mutants
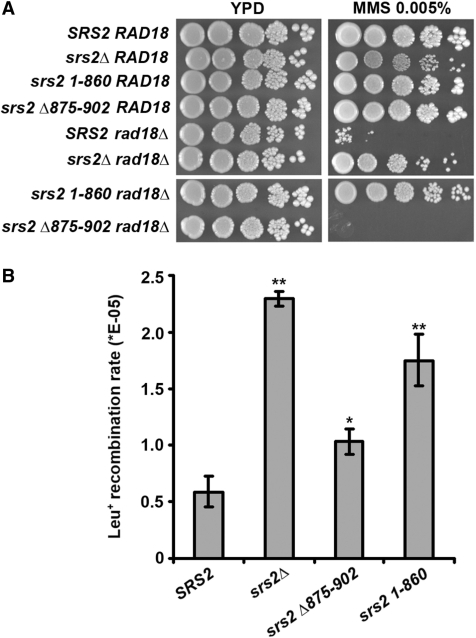
We wished to examine the genetic behavior of some of our srs2 mutants. Yeast strains were created which lacked Srs2 (srs2Δ), or that lacked amino acid residues 861–1174 (the 1–860 allele) or 875–902 (the Δ875–902 allele) of Srs2. As expected (8), the deletion of SRS2 suppressed the DNA damage sensitivity of the rad18Δ mutation (Figure 8A). DNA damage sensitivity of rad18Δ mutants can be suppressed by nonfunctional srs2 mutations in a RAD51-dependent manner (13). Suppression of the rad18Δ MMS sensitivity was seen with the srs2 1–860 but not the srs2 Δ875–902 allele (Figure 8A).
Figure 8.
Phenotypic analysis of srs2 1-860 and srs2 Δ875-902. (A) The ability of the srs2 1–860 and srs2 Δ875–902 alleles to suppress the MMS sensitivity of rad18Δ cells was examined. Platings on both YPD and YPD containing 0.005% MMS are shown. (B) Rates of recombination between the leu2-RI and leu2-bsteII alleles were determined and the results graphed. The rates shown represent the mean from three independent fluctuation tests performed for each strain. Error bars are the standard deviations derived from this mean of three independent rate determinations. Asterisks denote significant results, as determined by t-tests: **P < 0.005; *P < 0.05. Specifically, the rates for SRS2 and srs2Δ875–902 are significantly different (P = 0.00004), the rates for SRS2 and srs2 Δ875–902 are marginally different (P = 0.0117), and the rates for SRS2 and srs2 1–860 are significantly different (P = 0.0016).
We next examined the srs2 mutant strains for a hyper-recombination phenotype. Consistent with published results (10), we observed a 4-fold increase in the rate of gene conversion events in srs2Δ cells (Figure 8B). A similarly elevated gene conversion rate, one that was three-fold that of wild-type (Figure 8B), was found for the srs2 1–860 mutant. In contrast, the srs2 Δ875–902 gave a gene conversion rate that is only slightly higher than the wild-type control (Figure 8B). These findings fit with our observations that the srs2 1–860 mutant protein is quite defective in Rad51 interaction and in the inhibition of Rad51-mediated reactions, while the srs2 Δ875–902 mutant protein retains some ability to interact with Rad51 and possesses residual anti-recombinase activity.
DISCUSSION
Despite the important contributions that HR makes to the elimination of deleterious DNA lesions and the re-establishment of injured DNA replication forks, it must be tightly regulated so as to prevent untimely events that interfere with other DNA and replication fork repair processes, or to avoid the generation of toxic nucleoprotein intermediates that trigger prolonged cell cycle arrest (6,7,11,44). Moreover, crossover HR events are disfavored in mitotic cells, as they can give rise to undesirable chromosome rearrangements, such as translocations. Genetic and other companion analyses have found several pathways able to attenuate undesirable HR events or promote the usage of the conservative SDSA pathway that does not entail the generation of crossovers. Importantly, all of these HR regulatory pathways are dependent on a DNA helicase. Specifically, the human BLM helicase, and likely its S. cerevisiae counterpart Sgs1, functions with Topo IIIα (Top 3 in yeast), a type 1A topoisomerase, and other associated factors to resolve key HR DNA intermediates, such as the double Holliday structure, to non-crossover recombinants in a process termed double Holliday junction dissolution (45). Interestingly, the Srs2 helicase suppresses spurious HR events and also prevents crossover formation via a unique ability to disrupt the Rad51 presynaptic filament (16,17). Genetic and biochemical evidence has been presented to implicate the mammalian RECQ5 helicase in HR regulation via the same general mechanism as Srs2 (43). More recently, a separate means of HR regulation has been documented for the S. cerevisiae Mph1 helicase and its orthologues human FANCM and Schizosaccharomyces pombe Fml1; these orthologous helicases have been suggested to promote the use of the SDSA pathway of HR by disrupting the D-loop intermediate made by the Rad51 recombinase protein (34,46). Finally, the human RTEL1 helicase has also been shown to disrupt Rad51-made D-loops, but this novel attribute of RETL1 is thought to exert a general suppressive effect on HR frequency rather than fulfilling a regulatory role. Importantly, mutations in all the above helicases compromise genome instability and in some cases are associated with the tumor phenotype (43,47,48).
Several of the aforementioned helicases, including Srs2, RECQ5 and BLM/Sgs1 are known to physically interact with Rad51 (16,43,49), although the functional significance of these protein-protein interactions has not been addressed. In this study, we have strived to address the functional relevance of the Srs2-Rad51 complex at the biochemical level. Specifically, by a combination of yeast two-hybrid analysis and biochemical mapping, we have narrowed the Rad51 interaction domain to a short region within the Srs2 C-terminus, and have taken advantage of this information to generate several truncation mutants of Srs2 that retain wild-type levels of ATPase and helicase activities but are specifically compromised for Rad51 interaction. Importantly, with both biochemical means and electron microscopy, we have demonstrated a diminished ability of these srs2 mutants to attenuate Rad51-mediated HR reactions and to dismantle the Rad51 presynaptic filament, in concordance with the degree of Rad51 interaction deficiency. We note that, by employing a kinetic assay involving the use of a fluorescently labeled DNA substrate, Antony et al. (50) also showed recently that the srs2 1–860 mutant is deficient in anti-recombinase activity. Thus, in aggregate, our findings and those of Antony et al. (50) reveal that the efficacy of Srs2 as an anti-recombinase is dependent on its physical interaction with Rad51. Consistent with our biochemical results, we found that the srs2 1–860 mutant displays a hyper-recombination phenotype. Since the srs2 1–860 mutant protein retains wild-type levels of ATPase and helicase activities, our results thus provide evidence that the Srs2 helicase domain alone is insufficient for anti-recombinase function in cells. We have shown that the srs2 Δ875–902 mutant protein is less impaired for Rad51 interaction or for the ability to inhibit Rad51-mediated reactions in vitro. The phenotype of srs2 Δ875–902 cells is accordingly mild or near wild-type.
Like Srs2, the human RECQ5 helicase associated with Rad51 physically, inhibits the Rad51-mediated D-loop reaction, and catalyzes the removal of Rad51 from ssDNA in a manner that is dependent on its ATPase activity (43). Based on these functional similarities between Srs2 and RECQ5, it seems reasonable to suggest that Rad51 interaction is also a determinant of the anti-recombinase activity of RECQ5. The results of Antony et al. have suggested that Srs2 enhances ATP hydrolysis by Rad51 within the presynaptic filament, thereby hastening Rad51 dissociation from DNA (50). It will be interesting to test whether RECQ5 also acts by a similar mechanism.
We note that Srs2 becomes phosphorylated on multiple sites in response to intra-S phase DNA damage (26), and several of these phosphorylation sites are located within the C-terminus of the protein, including two such sites in the region spanning residues 875–902 (51) that we know are important for complex formation with Rad51 (this work). It will be particularly interesting to ascertain how C-terminal phosphorylation of Srs2 in response to DNA damage regulates its anti-recombinase function via an influence on complex formation with Rad51.
FUNDING
Research grants (GM074739, ES07061, R01GM57814, GM053738 and R01CA146940) from the US National Institutes of Health, by a Wellcome International Senior Research Fellowship (WT076476, GACR 301/09/317, and GACR 203/09/H046), a National Science Foundation Graduate Research Fellowship and a Department of Defense National Defense Science and Engineering Graduate Fellowship. Funding for open access charge: US National Institutes of Health grant R01CA146940.
Conflict of interest statement. None declared.
REFERENCES
- 1.Richards RI. Fragile and unstable chromosomes in cancer: causes and consequences. Trends Genet. 2001;17:339–345. doi: 10.1016/s0168-9525(01)02303-4. [DOI] [PubMed] [Google Scholar]
- 2.Popescu NC. Genetic alterations in cancer as a result of breakage at fragile sites. Cancer Lett. 2003;192:1–17. doi: 10.1016/s0304-3835(02)00596-7. [DOI] [PubMed] [Google Scholar]
- 3.Kennedy RD, D'A;ndrea AD. The Fanconi Anemia/BRCA pathway: new faces in the crowd. Genes Dev. 2005;19:2925–2940. doi: 10.1101/gad.1370505. [DOI] [PubMed] [Google Scholar]
- 4.Smirnova M, Klein HL. Role of the error-free damage bypass postreplication repair pathway in the maintenance of genomic stability. Mutat. Res. 2003;532:117–135. doi: 10.1016/j.mrfmmm.2003.08.026. [DOI] [PubMed] [Google Scholar]
- 5.Hickson ID. RecQ helicases: caretakers of the genome. Nat. Rev. Cancer. 2003;3:169–178. doi: 10.1038/nrc1012. [DOI] [PubMed] [Google Scholar]
- 6.Sung P, Klein H. Mechanism of homologous recombination: mediators and helicases take on regulatory functions. Nat. Rev. Mol. Cell Biol. 2006;7:739–750. doi: 10.1038/nrm2008. [DOI] [PubMed] [Google Scholar]
- 7.Fabre F, Chan A, Heyer WD, Gangloff S. Alternate pathways involving Sgs1/Top3, Mus81/Mms4, and Srs2 prevent formation of toxic recombination intermediates from single-stranded gaps created by DNA replication. Proc. Natl Acad. Sci. USA. 2002;99:16887–16892. doi: 10.1073/pnas.252652399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lawrence CW, Christensen RB. Metabolic suppressors of trimethoprim and ultraviolet light sensitivities of Saccharomyces cerevisiae rad6 mutants. J. Bacteriol. 1979;139:866–876. doi: 10.1128/jb.139.3.866-876.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schiestl RH, Prakash S, Prakash L. The SRS2 suppressor of rad6 mutations of Saccharomyces cerevisiae acts by channeling DNA lesions into the RAD52 DNA repair pathway. Genetics. 1990;124:817–831. doi: 10.1093/genetics/124.4.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Krogh BO, Symington LS. Recombination proteins in yeast. Annu. Rev. Genet. 2004;38:233–271. doi: 10.1146/annurev.genet.38.072902.091500. [DOI] [PubMed] [Google Scholar]
- 11.Gangloff S, Soustelle C, Fabre F. Homologous recombination is responsible for cell death in the absence of the Sgs1 and Srs2 helicases. Nat. Genet. 2000;25:192–194. doi: 10.1038/76055. [DOI] [PubMed] [Google Scholar]
- 12.Klein HL. Spontaneous chromosome loss in Saccharomyces cerevisiae is suppressed by DNA damage checkpoint functions. Genetics. 2001;159:1501–1509. doi: 10.1093/genetics/159.4.1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aboussekhra A, Chanet R, Adjiri A, Fabre F. Semidominant suppressors of Srs2 helicase mutations of Saccharomyces cerevisiae map in the RAD51 gene, whose sequence predicts a protein with similarities to procaryotic RecA proteins. Mol. Cell Biol. 1992;12:3224–3234. doi: 10.1128/mcb.12.7.3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rong L, Klein HL. Purification and characterization of the SRS2 DNA helicase of the yeast Saccharomyces cerevisiae. J. Biol. Chem. 1993;268:1252–1259. [PubMed] [Google Scholar]
- 15.Van Komen S, Reddy MS, Krejci L, Klein H, Sung P. ATPase and DNA helicase activities of the Saccharomyces cerevisiae anti-recombinase Srs2. J. Biol. Chem. 2003;278:44331–44337. doi: 10.1074/jbc.M307256200. [DOI] [PubMed] [Google Scholar]
- 16.Krejci L, Van Komen S, Li Y, Villemain J, Reddy MS, Klein H, Ellenberger T, Sung P. DNA helicase Srs2 disrupts the Rad51 presynaptic filament. Nature. 2003;423:305–309. doi: 10.1038/nature01577. [DOI] [PubMed] [Google Scholar]
- 17.Veaute X, Jeusset J, Soustelle C, Kowalczykowski SC, Le Cam E, Fabre F. The Srs2 helicase prevents recombination by disrupting Rad51 nucleoprotein filaments. Nature. 2003;423:309–312. doi: 10.1038/nature01585. [DOI] [PubMed] [Google Scholar]
- 18.Sung P. Catalysis of ATP-dependent homologous DNA pairing and strand exchange by yeast RAD51 protein. Science. 1994;265:1241–1243. doi: 10.1126/science.8066464. [DOI] [PubMed] [Google Scholar]
- 19.Krejci L, Macris M, Li Y, Van Komen S, Villemain J, Ellenberger T, Klein H, Sung P. Role of ATP hydrolysis in the antirecombinase function of Saccharomyces cerevisiae Srs2 protein. J. Biol. Chem. 2004;279:23193–23199. doi: 10.1074/jbc.M402586200. [DOI] [PubMed] [Google Scholar]
- 20.Pfander B, Moldovan GL, Sacher M, Hoege C, Jentsch S. SUMO-modified PCNA recruits Srs2 to prevent recombination during S phase. Nature. 2005;436:428–433. doi: 10.1038/nature03665. [DOI] [PubMed] [Google Scholar]
- 21.Papouli E, Chen S, Davies AA, Huttner D, Krejci L, Sung P, Ulrich HD. Crosstalk between SUMO and ubiquitin on PCNA is mediated by recruitment of the helicase Srs2p. Mol. Cell. 2005;19:123–133. doi: 10.1016/j.molcel.2005.06.001. [DOI] [PubMed] [Google Scholar]
- 22.Burgess RC, Lisby M, Altmannova V, Krejci L, Sung P, Rothstein R. Localization of recombination proteins and Srs2 reveals anti-recombinase function in vivo. J. Cell Biol. 2009;185:969–981. doi: 10.1083/jcb.200810055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ira G, Malkova A, Liberi G, Foiani M, Haber JE. Srs2 and Sgs1-Top3 suppress crossovers during double-strand break repair in yeast. Cell. 2003;115:401–411. doi: 10.1016/s0092-8674(03)00886-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aylon Y, Liefshitz B, Bitan-Banin G, Kupiec M. Molecular dissection of mitotic recombination in the yeast Saccharomyces cerevisiae. Mol. Cell Biol. 2003;23:1403–1417. doi: 10.1128/MCB.23.4.1403-1417.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ira G, Haber JE. Characterization of RAD51-independent break-induced replication that acts preferentially with short homologous sequences. Mol. Cell Biol. 2002;22:6384–6392. doi: 10.1128/MCB.22.18.6384-6392.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liberi G, Chiolo I, Pellicioli A, Lopes M, Plevani P, Muzi-Falconi M, Foiani M. Srs2 DNA helicase is involved in checkpoint response and its regulation requires a functional Mec1-dependent pathway and Cdk1 activity. EMBO J. 2000;19:5027–5038. doi: 10.1093/emboj/19.18.5027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang M, Elledge SJ. The FHA domain, a phosphoamino acid binding domain involved in the DNA damage response pathway. Cold Spring Harb. Symp. Quant. Biol. 2000;65:413–421. doi: 10.1101/sqb.2000.65.413. [DOI] [PubMed] [Google Scholar]
- 28.Bhattacharyya S, Lahue RS. Saccharomyces cerevisiae Srs2 DNA helicase selectively blocks expansions of trinucleotide repeats. Mol. Cell Biol. 2004;24:7324–7330. doi: 10.1128/MCB.24.17.7324-7330.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.James P, Halladay J, Craig EA. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krejci L, Damborsky J, Thomsen B, Duno M, Bendixen C. Molecular dissection of interactions between Rad51 and members of the recombination-repair group. Mol. Cell Biol. 2001;21:966–976. doi: 10.1128/MCB.21.3.966-976.2001. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 31.Sung P. Yeast Rad55 and Rad57 proteins form a heterodimer that functions with replication protein A to promote DNA strand exchange by Rad51 recombinase. Genes Dev. 1997;11:1111–1121. doi: 10.1101/gad.11.9.1111. [DOI] [PubMed] [Google Scholar]
- 32.Raschle M, Van Komen S, Chi P, Ellenberger T, Sung P. Multiple interactions with the Rad51 recombinase govern the homologous recombination function of Rad54. J. Biol. Chem. 2004;279:51973–51980. doi: 10.1074/jbc.M410101200. [DOI] [PubMed] [Google Scholar]
- 33.Ayyagari R, Impellizzeri KJ, Yoder BL, Gary SL, Burgers PM. A mutational analysis of the yeast proliferating cell nuclear antigen indicates distinct roles in DNA replication and DNA repair. Mol. Cell Biol. 1995;15:4420–4429. doi: 10.1128/mcb.15.8.4420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Prakash R, Satory D, Dray E, Papusha A, Scheller J, Kramer W, Krejci L, Klein H, Haber JE, Sung P, et al. Yeast Mph1 helicase dissociates Rad51-made D-loops: implications for crossover control in mitotic recombination. Genes Dev. 2009;23:67–79. doi: 10.1101/gad.1737809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Petukhova G, Stratton S, Sung P. Catalysis of homologous DNA pairing by yeast Rad51 and Rad54 proteins. Nature. 1998;393:91–94. doi: 10.1038/30037. [DOI] [PubMed] [Google Scholar]
- 36.Sung P, Robberson DL. DNA strand exchange mediated by a RAD51-ssDNA nucleoprotein filament with polarity opposite to that of RecA. Cell. 1995;82:453–461. doi: 10.1016/0092-8674(95)90434-4. [DOI] [PubMed] [Google Scholar]
- 37.Palladino F, Klein HL. Analysis of mitotic and meiotic defects in Saccharomyces cerevisiae SRS2 DNA helicase mutants. Genetics. 1992;132:23–37. doi: 10.1093/genetics/132.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lea DE, Coulson CA. The distribution of the numbers of mutants in bacterial populations. J. Genet. 1949;49:264–285. doi: 10.1007/BF02986080. [DOI] [PubMed] [Google Scholar]
- 39.Aguilera A, Klein HL. Genetic control of intrachromosomal recombination in Saccharomyces cerevisiae. I. Isolation and genetic characterization of hyper-recombination mutations. Genetics. 1988;119:779–790. doi: 10.1093/genetics/119.4.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jentsch S, McGrath JP, Varshavsky A. The yeast DNA repair gene RAD6 encodes a ubiquitin-conjugating enzyme. Nature. 1987;329:131–134. doi: 10.1038/329131a0. [DOI] [PubMed] [Google Scholar]
- 41.Le Breton C, Dupaigne P, Robert T, Le Cam E, Gangloff S, Fabre F, Veaute X. Srs2 removes deadly recombination intermediates independently of its interaction with SUMO-modified PCNA. Nucleic Acids Res. 2008;36:4964–4974. doi: 10.1093/nar/gkn441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sung P, Krejci L, Van Komen S, Sehorn MG. Rad51 recombinase and recombination mediators. J. Biol. Chem. 2003;278:42729–42732. doi: 10.1074/jbc.R300027200. [DOI] [PubMed] [Google Scholar]
- 43.Hu Y, Raynard S, Sehorn MG, Lu X, Bussen W, Zheng L, Stark JM, Barnes EL, Chi P, Janscak P, et al. RECQL5/Recql5 helicase regulates homologous recombination and suppresses tumor formation via disruption of Rad51 presynaptic filaments. Genes Dev. 2007;21:3073–3084. doi: 10.1101/gad.1609107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Magner DB, Blankschien MD, Lee JA, Pennington JM, Lupski JR, Rosenberg SM. RecQ promotes toxic recombination in cells lacking recombination intermediate-removal proteins. Mol. Cell. 2007;26:273–286. doi: 10.1016/j.molcel.2007.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bussen W, Raynard S, Busygina V, Singh AK, Sung P. Holliday junction processing activity of the BLM-Topo IIIalpha-BLAP75 complex. J. Biol. Chem. 2007;282:31484–31492. doi: 10.1074/jbc.M706116200. [DOI] [PubMed] [Google Scholar]
- 46.Sun W, Nandi S, Osman F, Ahn JS, Jakovleska J, Lorenz A, Whitby MC. The FANCM ortholog Fml1 promotes recombination at stalled replication forks and limits crossing over during DNA double-strand break repair. Mol. Cell. 2008;32:118–128. doi: 10.1016/j.molcel.2008.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ellis NA, Groden J, Ye TZ, Straughen J, Lennon DJ, Ciocci S, Proytcheva M, German J. The Bloom's; syndrome gene product is homologous to RecQ helicases. Cell. 1995;83:655–666. doi: 10.1016/0092-8674(95)90105-1. [DOI] [PubMed] [Google Scholar]
- 48.Barber LJ, Youds JL, Ward JD, McIlwraith MJ, O'N;eil NJ, Petalcorin MI, Martin JS, Collis SJ, Cantor SB, Auclair M, et al. RTEL1 maintains genomic stability by suppressing homologous recombination. Cell. 2008;135:261–271. doi: 10.1016/j.cell.2008.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu L, Davies SL, Levitt NC, Hickson ID. Potential role for the BLM helicase in recombinational repair via a conserved interaction with RAD51. J. Biol. Chem. 2001;276:19375–19381. doi: 10.1074/jbc.M009471200. [DOI] [PubMed] [Google Scholar]
- 50.Antony E, Tomko EJ, Xiao Q, Krejci L, Lohman TM, Ellenberger T. Srs2 disassembles Rad51 filaments by a protein-protein interaction triggering ATP turnover and dissociation of Rad51 from DNA. Mol. Cell. 2009;35:105–115. doi: 10.1016/j.molcel.2009.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chiolo I, Carotenuto W, Maffioletti G, Petrini JH, Foiani M, Liberi G. Srs2 and Sgs1 DNA helicases associate with Mre11 in different subcomplexes following checkpoint activation and CDK1-mediated Srs2 phosphorylation. Mol. Cell Biol. 2005;25:5738–5751. doi: 10.1128/MCB.25.13.5738-5751.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]