Abstract
APOA5 -1131T > C and S19W single nucleotide polymorphisms (SNP) have been consistently associated with plasma lipid concentration and metabolic syndrome (MetS), alone and in modulation by dietary factors. Puerto Ricans have a high prevalence of metabolic conditions and high minor allele frequency for these SNP, suggesting a possible role in disease for this population. We aimed to determine the association of APOA5 -1131T > C and S19W with plasma lipids and markers of MetS, alone and in interaction with total fat intake, as a percent of total energy intake, in Puerto Ricans. Anthropometric and demographic data, FFQ, and blood samples were collected at baseline from participants in the Boston Puerto Rican Health Study (n = 802, 45–75 y). APOA5 S19W was associated with plasma HDL cholesterol (HDL-C) (P = 0.044); minor allele carriers had lower HDL-C [1.12 ± 0.03 (mean ± SE)] than those with the common variant (1.18 ± 0.01 mmol/L), even after adjustment for plasma triglycerides (TG) (P = 0.012). Neither polymorphism was associated with TG or other lipids. Interaction of the -1131T > C SNP with total fat energy intake was observed for plasma TG (P = 0.032) and total cholesterol (P = 0.034). APOA5 S19W interacted with total fat intake in association with systolic (P = 0.002) and diastolic (P = 0.007) blood pressure. Neither SNP was associated with MetS in the overall analysis or after stratifying by total energy intake as fat. In conclusion, Puerto Ricans present a distinctive lipid profile in association with APOA5 polymorphisms. Dietary fat intake seems to modulate these associations. The results contribute to the understanding of health disparities in this population.
Introduction
Apolipoprotein A-V is a protein produced in the liver that enhances the catabolism of triglyceride (TG)7 rich lipoprotein and that is positively associated with the plasma TG concentration (1,2). Two haplotype-tagging single nucleotide polymorphisms (SNP) mapping to the APOA5 gene, -1131T > C and c.56C > G (S19W), consistently predict high plasma TG, cholesterol, HDL cholesterol (HDL-C), LDL cholesterol (LDL-C), and VLDL cholesterol (VLDL-C) (1,3).
Several studies have explored the association of these 2 polymorphisms with metabolic syndrome (MetS), as this condition comprises lipid, cardiovascular, and anthropometric markers that have been previously linked to APOA5 function. A recent candidate gene study in elderly Japanese identified the -1131T > C SNP as a strong candidate for association with MetS (4). Other groups have shown that this SNP, as well as the S19W variant, are associated with this condition in several Caucasian ethnic groups (5–8).
Variations in lipid profiles and other metabolic markers may result not only from genetic variants but also in response to diet, including fat intake (9). Several possible mechanisms for an effect modifying the action of dietary fat on APOA5 phenotypic effects have been proposed (10), including a differential gene regulation by thyroid hormones or PPAR (11,12). Various studies found interactions of the APOA5 SNP with dietary fat intake in relation to obesity (10,13), plasma lipids (14), and the capacity to clear chylomicron-TG or hydrolyze TG (15), suggesting that APOA5 variants may play a role in the individual sensitivity of circulating lipids diet (16).
Tai and Ordovas (1) noted that the association between the plasma TG concentration and APOA5 variants seems consistent across studies, but that the strength of the association varies among studies. Interactions with environmental factors may provide a key to the observed differences in genetic risk (1). Differences by ethnic group in minor allele frequency (MAF) in disease-associated genetic loci could also play a role in the effect of genetic contribution to an outcome (17). Puerto Ricans have significantly higher MAF than non-Hispanic whites for the APOA5 -1131T > C and S19W SNP (18). Puerto Ricans living in the United States have a high prevalence of diabetes (19,20), hypertension (21), large body mass (22,23), and MetS (24). In contrast, Puerto Rican women have lower plasma total cholesterol, LDL-C, HDL-C, and apolipoprotein B than non-Hispanic whites (25). The observed differences in MAF suggest that Puerto Ricans may have a distinctive association between risk alleles and disease.
Studies of genetic effects and gene-diet associations on metabolic conditions in Puerto Ricans are very limited. The aim of this study was to determine the association of 2 genetic variants of the APOA5 gene (-1131T > C and S19W) with plasma lipid concentrations and markers of MetS (waist circumference, fasting glucose, blood pressure, and plasma TG and HDL-C), alone and in combination with dietary fat intake in Puerto Rican older adults.
Methods
Study population.
The sample consisted of 1020 unrelated participants, at the time of analysis, of the Boston Puerto Rican Health Study, a longitudinal study on stress, nutrition, health, and aging (26). Eligible participants had to be able to answer interview questions in either English or Spanish, be of Puerto Rican descent, be between the ages of 45 and 75 y, and be living in the Boston, MA area at the time of the study. Ethnicity was self-reported. The Institutional Review Board for Human Research at Tufts Medical Center approved the protocol of the study. All participants signed consent forms. Baseline data obtained during 2004 to 2006 were used for this analysis.
Measurements.
All data were obtained during a home-based interview by trained staff. A comprehensive questionnaire was used to obtain demographic information, use of medications, levels of physical activity, alcohol use, and smoking habits. Physical activity was measured with a modified version of the Paffenbarger questionnaire of the Harvard Alumni Activity Survey. A physical activity score was calculated as the sum of hours spent on typical 24-h activities (heavy, moderate, light, or sedentary activity as well as sleeping) multiplied by weighing factors that parallel the rate of oxygen consumption associated with each category of physical activity. A physical activity score of <30 was considered indicative of a sedentary lifestyle. Questions about past and current drinking and smoking behaviors were used to determine smoking (never, <100 cigarettes in entire life; past, previously smoked but not currently; and current smoker) and drinking status (never, <12 alcoholic beverages in entire life; past, previous drinking but not currently; and current drinker).
Blood pressure was taken by trained interviewers using an electronic sphignomanometer TM (Dinamap Model 8260, Critikon) after short rests at 3 time points, in duplicate, during the home visit: once near the beginning, during the middle, and near the end of the interview. The mean of the second and 3rd readings was used as the blood pressure variable. Anthropometric measures including height, weight, and waist circumference were taken in duplicate, following the techniques used in NHANES III (27). The mean of the 2 readings was used for this analysis. BMI was calculated as body weight (kg)/height (m)2. Blood samples for biochemical analyses were drawn by a certified phlebotomist on the day following the home interview. Participants were asked to fast 12 h prior to the blood draw. Blood samples were immediately cooled to 4°C and the plasma separated within 4 h in a refrigerated centrifuge (3421 × g; 15 min). The RBC were washed twice with cold saline. Plasma aliquots were saved in 1-mL cryogenic, screw-cap tubes and stored at −70°C for later batch processing for HDL-C. Serum glucose was measured using an enzymatic kinetic reaction on the Olympus AU400e with Olympus Glucose reagents (OSCR6121) (Olympus America). Plasma total cholesterol, TG, and HDL-C were analyzed using EDTA plasma with the enzymatic endpoint reaction on the Olympus AU400e with Olympus cholesterol reagents (OSR6116), Olympus TG reagents (OSR6133), and HDL-C reagents (OSR6195), respectively. VLDL-C was calculated as TG/5 and LDL-C as cholesterol – (VLDL-C + HDL-C).
Dietary assessment.
Dietary intake was assessed with a FFQ specially designed for this population (28). FFQ were scanned using the OPSCAN program and linked with a portion size entry program. Data were then linked with the Minnesota Nutrient Data System (version 5.0_35) for nutrient analyses.
Genotyping.
Genotyping has been previously described in detail elsewhere (18). Briefly, genomic DNA was purified using the QIAamp DNA Blood Mini kits (Qiagen) from buffy coats of nucleated cells obtained from blood samples. Genotyping for SNP APOA5 S19W (c.56C > G, rs3135506) and APOA5 -1131T > C (rs662799) was carried using TaqMan SNP genotyping assays (Applied Biosystems). Quality control estimated the genotyping error rate as <1%.
Population admixture.
Unequal levels of ancestral proportions among individuals in a population may result in misleading associations in genetic and epidemiological studies (29). Thus, we have controlled for population admixture in this study. Calculation for population admixture in this sample has been described previously (30). Briefly, individual ancestry was calculated based on the genotypes of 100 ancestry informative markers using 2 programs: STRUCTURE 2.2 and IAE3CI with reference to 3 ancestral populations: West African, European, and Native American. Ancestry is expressed as the percent that each ancestral population contributes to total composition. For this study, the variables for African and European ancestry were included as covariates in the models to adjust for population admixture, as they were the 2 major ancestral components in this Puerto Rican sample. Those 2 components were more likely to be confounders in models for association with chronic conditions such as diabetes, CVD, and hypertension, than adjusting for Native-American ancestry (30).
Statistical analysis.
At the time of analysis, there were 924 participants with available genotyping data. Of those, 892 participants had complete data on dietary intake and MetS. Participants were excluded from analysis if they had energy intake considered implausible [between 2512 and 20,097 kJ (600 and 4800 kcal)], or BMI ≤18. A total of 802 participants were included in the analysis. Initial exploratory analysis was conducted to check for normality of distribution of continuous variables. Glucose, TG, and VLDL-C concentrations were log transformed to normalize the distribution of the data. Pearson's chi-square statistic was used to test Hardy-Weinberg Equilibrium for each SNP and to test the differences in percentages. We used t tests to compare unadjusted means. Pairwise linkage disequilibria (LD) between the 2 SNP at the APOA5 locus, as well as others in the APOA1/C3/A4/A5 cluster, were estimated with the coefficient R using PowerMarker software (31).
The relationship between each APOA5 genotype with plasma lipids and markers of MetS was evaluated using analysis of covariance and the adjusted means were estimated after controlling for potential confounders, including age, sex, alcohol intake (never, past, current), smoking (never, past, current), medications (diabetes, hypertension, or lipid-lowering drugs), BMI (or physical activity for waist circumference outcome), and population admixture. Additional adjustment for plasma TG concentrations was performed. Interaction terms with sex were explored separately to determine the homogeneity of the sample. The interactions between total dietary fat intake and polymorphisms were also tested in multivariate interaction models after adjusting for the same covariates and total energy intake. Macronutrient intake was expressed as percentage of total energy intake. Dietary fat intake was included in the analyses as a categorical variable, defined as 2 groups divided by the median intake of the population.
A dichotomous outcome for MetS was defined following the 2001 Expert Panel on Detection, Adult Treatment Panel Guidelines, modified to reflect recommendations by the American Diabetes Association (32). Participants were classified as having MetS if they had ≥3 of the following conditions: waist circumference ≥102 cm in men or ≥88 cm in women; fasting glucose ≥5.6 mmol/L; elevated blood pressure [systolic blood pressure (SBP) ≥130 or diastolic blood pressure (DBP) ≥85 mm Hg); high TG (≥1.7 mmol/L); low HDL-C (<1.0 mmol/L in men or <1.3 mmol/L in women). We tested the association of the presence of each APOA5 gene variant with MetS with logistic regression models, fitted to estimate odds ratio (OR) and 95% CI, controlling for age, sex, alcohol intake, smoking, population admixture, and medication use. Further adjustment for physical activity did not alter the models. The significance of the polymorphisms and MetS, as modulated by total fat energy intake, was also tested by analyzing the effect for each category of total fat energy intake separately.
Carriers of the minor allele for each polymorphism were grouped and compared against common allele homozygotes due to the low frequency of the minor homozygous alleles. We used several different statistical models to test the consistency of results and to adjust for potential confounders, including stepwise logistic regression and adjustment for other macronutrients (carbohydrate, fiber, and alcohol intake) in secondary analysis. Standard regression diagnostic procedures (residual distribution and multicollinearity tests) were used to ensure the appropriateness of these models. Statistical analyses were performed using SPSS version 15.0 software. All reported probability tests were 2-sided. Tests with P < 0.05 were considered significant. Values in the text are mean ± SE or geometric mean (95% CI) for anti-log transformed measures for glucose, TG, and VLDL-C; logistic regression results are expressed as OR (95% CI).
Results
The pairwise LD coefficient R between the APOA5 -1131T > C and S19W SNP was 0.016 in the Puerto Rican sample, suggesting that the markers are independent (Supplemental Table 1). Both SNP were in Hardy-Weinberg Equilibrium.
Baseline characteristics are presented by the -1131T > C and S19W gene variant (Table 1). None of the variables significantly differed by genotype, except for the percent of women by S19W polymorphism. There were significantly more women carrying the minor allele for APOA5 S19W. However, the results for both SNP were comparable by sex and there were no significant interactions between SNP and sex (data not shown), so men and women were combined to ensure adequate statistical power.
TABLE 1.
Baseline anthropometric and biological characteristics for participants of the Boston Puerto Rican Health Study1
|
APOA5-1131T > C |
APOA5 S19W |
||||
|---|---|---|---|---|---|
| TT | TC+CC | CC | CG+GG | All participants | |
| n | 618 | 184 | 639 | 163 | 802 |
| Female, n (%) | 458 (74.1) | 129 (70.1) | 452 (70.7) | 135 (82.8)* | 587 (73.2) |
| Age, y | 57.9 ± 7.2 | 57.5 ± 7.1 | 57.9 ± 7.2 | 57.6 ± 7.2 | 57.8 ± 7.2 |
| BMI, kg/m2 | 32.2 ± 6.7 | 32.1 ± 7.0 | 32.0 ± 6.8 | 33.0 ± 6.6 | 32.2 ± 6.8 |
| Waist circumference, cm | 101.7 ± 15.1 | 101.8 ± 14.6 | 101.4 ± 15.0 | 103.2 ± 15.0 | 101.8 ± 15.0 |
| Physical activity score | 31.4 ± 4.5 | 32.0 ± 4.5 | 31.5 ± 4.5 | 31.8 ± 4.4 | 31.5 ± 4.5 |
| SBP, mm Hg | 136.2 ± 18.2 | 135.3 ± 19.7 | 136.1 ± 18.5 | 135.6 ± 19.0 | 136.0 ± 18.6 |
| DBP, mm Hg | 81.2 ± 10.6 | 80.7 ± 10.2 | 81.0 ± 10.5 | 81.5 ± 10.6 | 81.1 ± 10.5 |
| Biochemistry | |||||
| Serum glucose, mmol/L | 6.78 ± 2.69 | 6.69 ± 3.25 | 6.73 ± 2.88 | 6.87 ± 2.64 | 6.76 ± 2.83 |
| Plasma TG, mmol/L | 1.80 ± 1.23 | 1.95 ± 1.82 | 1.87 ± 1.49 | 1.71 ± 0.90 | 1.84 ± 1.39 |
| Plasma HDL-C, mmol/L | 1.16 ± 0.33 | 1.15 ± 0.33 | 1.17 ± 0.34 | 1.14 ± 0.29 | 1.16 ± 0.33 |
| Plasma LDL-C, mmol/L | 2.80 ± 0.84 | 2.73 ± 0.95 | 2.77 ± 0.91 | 2.81 ± 0.89 | 2.78 ± 0.90 |
| Plasma VLDL-C, mmol/L | 0.75 ± 0.38 | 0.79 ± 0.38 | 0.77 ± 0.39 | 0.74 ± 0.35 | 0.76 ± 0.38 |
| Plasma total cholesterol, mmol/L | 4.77 ± 1.09 | 4.70 ± 1.13 | 4.77 ± 1.11 | 4.70 ± 1.02 | 4.75 ± 1.10 |
| Energy intake, kJ/d | 9039 ± 3603 | 8938 ± 3859 | 8959 ± 3640 | 9239 ± 3744 | 9016 ± 3661 |
| Total fat, % energy | 31.1 ± 5.1 | 30.5 ± 5.6 | 30.9 ± 5.3 | 31.1 ± 5.1 | 52.0 ± 7.6 |
| Carbohydrate, % energy | 51.9 ± 7.3 | 52.3 ± 8.5 | 51.9 ± 7.7 | 52.4 ± 7.2 | 17.3 ± 3.5 |
| Protein, % energy | 17.2 ± 3.4 | 17.5 ± 3.6 | 17.3 ± 3.5 | 17.1 ± 7.2 | 31.0 ± 5.2 |
| European ancestry,2% | 56.9 ± 15.2 | 52.3 ± 8.5 | 57.3 ± 15.4 | 56.4 ± 14.6 | 27.4 ± 15.7 |
| African ancestry,2% | 27.7 ± 15.7 | 26.5 ± 15.8 | 27.2 ± 15.9 | 28.4 ± 14.9 | 57.1 ± 15.3 |
| Native American ancestry,2% | 15.4 ± 6.6 | 15.5 ± 6.7 | 15.5 ± 6.7 | 15.1 ± 6.1 | 15.4 ± 6.6 |
| Current smoker, % | 23.0 | 22.4 | 22.6 | 23.8 | 22.8 |
| Current drinker, % | 40.3 | 37.0 | 40.6 | 35.2 | 39.5 |
| Diabetes medication use, % | 32.6 | 36.6 | 34.4 | 30.2 | 33.5 |
| Hypertension medication use, % | 56.3 | 55.7 | 56.5 | 54.9 | 56.2 |
| Lipid-lowering medication use, % | 41.4 | 38.3 | 40.2 | 42.6 | 40.7 |
| MetS3 |
67.3 |
64.3 |
66.5 |
67.1 |
66.6 |
Values are mean ± SD or percent; n = 802. *Different from participants with CC genotype for APOA5 S19W, P < 0.05.
Expressed as the percent that each ancestral population contributes to total composition.
Three or more of the following conditions must be present: waist circumference ≥102 cm in men or ≥88 cm in women; fasting glucose ≥ 5.6 mmol/L; elevated blood pressure (SBP ≥ 130 or DBP ≥ 85 mm Hg); high TG (≥1.7 mmol/L); low HDL-C (<1.0 mmol/L in men or <1.3 mmol/L in women).
Independent associations between APOA5 -1131T > C and S19W gene variants and plasma lipids and markers of MetS were adjusted for potential confounders (Table 2). S19W SNP and HDL-C were associated (P = 0.044), with participants carrying the minor allele having a lower plasma HDL-C concentration (1.12 ± 0.03) than those with the common variant (1.18 ± 0.01 mmol/L). Because TG and HDL-C metabolism are highly interconnected, we further adjusted for the plasma TG concentration in the model for HDL-C. The S19W SNP remained associated with HDL-C even after adjusting for TG (P = 0.012). There were no other markers significantly related to either SNP.
TABLE 2.
Association of APOA5 gene variants with markers of MetS in participants of the Boston Puerto Rican Health Study1
|
APOA5-1131T > C |
APOA5 S19W |
|||||
|---|---|---|---|---|---|---|
| TT | TC+CC | P-value | CC | CG+GG | P-value | |
| Waist circumference, cm | 101.7 ± 0.6 | 101.9 ± 1.1 | 0.863 | 101.3 ± 0.6 | 103.7 ± 1.2 | 0.064 |
| SBP, mm Hg | 136.1 ± 0.7 | 135.5 ± 1.4 | 0.721 | 135.9 ± 0.7 | 136.1 ± 1.5 | 0.929 |
| DBP, mm Hg | 81.9 ± 0.4 | 80.8 ± 0.8 | 0.702 | 81.0 ± 0.4 | 81.6 ± 0.8 | 0.554 |
| Serum glucose, mmol/L | 6.42 (6.29–6.55) | 6.23 (5.98–6.48) | 0.164 | 6.35 (6.23–6.48) | 6.55 (6.23–6.81) | 0.289 |
| Plasma TG, mmol/L | 1.56 (1.50–1.63) | 1.63 (1.50–1.74) | 0.387 | 1.58 (1.52–1.64) | 1.53 (1.41–1.66) | 0.485 |
| Plasma HDL-C, mmol/L | 1.17 ± 0.01 | 1.16 ± 0.02 | 0.859 | 1.18 ± 0.01 | 1.12 ± 0.03 | 0.044 |
| Plasma HDL-C (adjusted for TG), mmol/L | 1.17 ± 0.01 | 1.17 ± 0.02 | 0.847 | 1.18 ± 0.01 | 1.11 ± 0.02 | 0.012 |
| Plasma total cholesterol, mmol/L | 4.77 ± 0.04 | 4.74 ± 0.08 | 0.735 | 4.79 ± 0.04 | 4.67 ± 0.09 | 0.194 |
| Plasma LDL-C, mmol/L | 2.80 ± 0.04 | 2.74 ± 0.07 | 0.489 | 2.78 ± 0.04 | 2.79 ± 0.07 | 0.898 |
| Plasma VLDL-C, mmol/L |
0.67 (0.65–0.70) |
0.71 (0.66–0.76) |
0.195 |
0.69 (0.66–0.71) |
0.67 (0.63–0.72) |
0.721 |
Values are the estimated mean ± SE, or geometric mean (95% CI) for anti-log transformed measures for glucose, TG, and VLDL-C, n = 802. P-value shown for APOA5 polymorphism as main predictor, with adjustment for age, sex, smoking, alcohol intake, medication use (lipid-lowering, diabetes, and hypertension), population admixture, and BMI (or physical activity for waist circumference outcome).
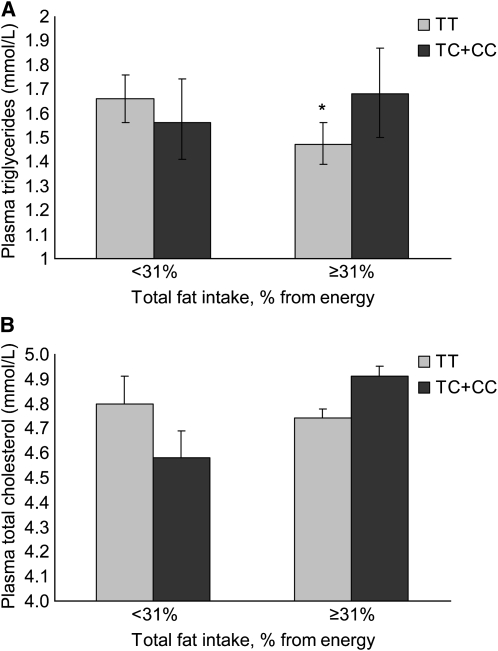
We then examined whether total dietary fat energy intake modulates the association between these gene variants and the same markers, with additional adjustment for total energy intake (Table 3). Total fat intake was categorized using the median fat intake as a percent of total energy intake (31%), which also corresponded to the mean of the population. There were 2 interactions between the -1131T > C polymorphism and total fat energy intake: for plasma TG (P = 0.032) and plasma total cholesterol (P = 0.034) (Fig. 1). Participants with the 1131C allele had higher plasma TG concentrations when they consumed ≥31% of total energy as fat (P = 0.038), whereas there was no difference by genotype at the lower total fat energy intake category (P = 0.334). Carriers of the 1131T > C minor allele tended to have lower plasma total cholesterol in the low fat energy intake group (P = 0.085). There was no difference by variant, with total fat intake ≥ 31% of energy (P = 0.201). Secondary adjustment of other macronutrient intakes, including carbohydrate, fiber, and alcohol, did not alter the models (data not shown).
TABLE 3.
Interaction between APOA5 gene variants and total fat intake, and markers of MetS in participants of the Boston Puerto Rican Health Study1
|
APOA5-1131T > C |
APOA5 S19W |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Total fat | TT | TC+CC | SNP | SNP × fat interaction | CC | CG+GG | SNP | SNP × fat interaction | |
| % energy | P-value | P-value | |||||||
| Waist circumference, cm | <31 | 100.5 ± 0.9 | 101.5 ± 1.5 | 0.788 | 0.600 | 100.0 ± 0.8 | 103.7 ± 1.7 | 0.075 | 0.286 |
| ≥31 | 102.8 ± 0.8 | 102.5 ± 1.6 | 102.5 ± 0.8 | 103.4 ± 1.6 | |||||
| Serum glucose, mmol/L | <31 | 6.42 (6.23–6.61) | 6.29 (5.98–6.68) | 0.168 | 0.468 | 6.35 (6.16–6.55) | 6.61 (6.16–7.02) | 0.315 | 0.639 |
| ≥31 | 6.42 (6.23–6.61) | 6.10 (5.75–6.48) | 6.35 (6.16–6.55) | 6.42 (6.04–6.81) | |||||
| SBP, mm Hg | <31 | 135.5 ± 1.1 | 136.5 ± 1.9 | 0.720 | 0.293 | 134.7 ± 1.0 | 140.1 ± 2.2 | 0.802 | 0.002 |
| ≥31 | 136.6 ± 1.0 | 134.4 ± 2.0 | 137.1 ± 1.0 | 132.5 ± 2.0 | |||||
| DBP, mm Hg | <31 | 80.0 ± 0.6 | 80.5 ± 1.1 | 0.772 | 0.423 | 79.5 ± 0.6 | 82.7 ± 1.2 | 0.496 | 0.007 |
| ≥31 | 82.2 ± 0.6 | 81.3 ± 1.1 | 82.4 ± 0.6 | 80.5 ± 1.1 | |||||
| Plasma TG, mmol/L | <31 | 1.66 (1.56–1.76) | 1.56 (1.41–1.74) | 0.432 | 0.032 | 1.66 (1.56–1.74) | 1.61 (1.43–1.82) | 0.521 | 0.926 |
| ≥31 | 1.47 (1.39–1.56) | 1.68 (1.50–1.87) | 1.52 (1.44–1.61) | 1.47 (1.32–1.64) | |||||
| Plasma HDL-C, mmol/L | <31 | 1.14 ± 0.02 | 1.15 ± 0.03 | 0.928 | 0.618 | 1.15 ± 0.02 | 1.11 ± 0.04 | 0.043 | 0.444 |
| ≥31 | 1.20 ± 0.02 | 1.18 ± 0.03 | 1.21 ± 0.02 | 1.13 ± 0.03 | |||||
| Plasma HDL-C adjusted for TG, mmol/L | <31 | 1.15 ± 0.02 | 1.15 ± 0.03 | 0.803 | 0.684 | 1.16 ± 0.02 | 1.12 ± 0.03 | 0.012 | 0.379 |
| ≥31 | 1.18 ± 0.02 | 1.19 ± 0.03 | 1.20 ± 0.02 | 1.11 ± 0.03 | |||||
| Plasma total cholesterol, mmol/L | <31 | 4.80 ± 0.06 | 4.58 ± 0.11 | 0.756 | 0.034 | 4.76 ± 0.06 | 4.70 ± 0.13 | 0.196 | 0.484 |
| ≥31 | 4.74 ± 0.06 | 4.91 ± 0.04 | 4.82 ± 0.06 | 4.63 ± 0.12 | |||||
| Plasma LDL-C, mmol/L | <31 | 2.77 ± 0.05 | 2.64 ± 0.09 | 0.542 | 0.293 | 2.72 ± 0.05 | 2.79 ± 0.10 | 0.864 | 0.416 |
| ≥31 | 2.82 ± 0.05 | 2.85 ± 0.09 | 2.84 ± 0.05 | 2.79 ± 0.10 | |||||
| Plasma VLDL-C, mmol/L | <31 | 0.71 (0.67–0.74) | 0.70 (0.64–0.77) | 0.225 | 0.216 | 0.70 (0.67–0.74) | 0.71 (0.63–0.79) | 0.758 | 0.618 |
| ≥31 |
0.65 (0.62–0.68) |
0.72 (0.65–0.79) |
0.67 (0.63–0.70) |
0.65 (0.58–0.72) |
|||||
Values are the estimated mean ± SE, or geometric mean (95% CI) for anti-log transformed measures for fasting glucose, TG, and VLDL-C, n = 802. P-values shown for APOA5 polymorphism as main predictor and in interaction term with category of total fat as % from energy (below or above population median intake), after adjustment for age, sex, smoking, alcohol intake, medication use (lipid-lowering, diabetes, and hypertension), population admixture, BMI (or physical activity for waist circumference outcome), and total energy intake.
FIGURE 1 .
Interaction between total fat intake as percent of energy intake (below or above the population median) and APOA5 -1131T > C polymorphism for plasma TG (A) and total cholesterol concentrations (B) in Puerto Rican older adults. Values are geometric mean and 95% CI for the antilog of TG and mean and SE for total cholesterol; adjusted for age, sex, smoking, alcohol intake, medication use (lipid-lowering, diabetes, and hypertension), population admixture, BMI and total energy intake, n = 802. P for interaction was 0.032 (A) and 0.034 (B). *Different from TC+CC within the total fat energy intake category, P < 0.05.
The APOA5 S19W polymorphism interacted with total fat energy intake in association with SBP (P = 0.002) and DBP (P = 0.007) (Fig. 2). Carriers of the G minor allele had higher SBP (P = 0.023) with low total fat intake and lower SBP (P = 0.042) with high total fat intake compared with participants with the common S19 genotype. Similarly, carriers of the minor allele had higher DBP when they consumed <31% of total energy as fat (P = 0.020). The difference at the higher dietary fat intake category was not significant (P = 0.141).
FIGURE 2 .
Interaction between total fat intake category (below or above the population median) and APOA5 S19W polymorphism for SBP (A) and DBP (B) in Puerto Ricans older adults. Values are mean and SE; adjusted for age, sex, smoking, alcohol intake, medication use (lipid-lowering, diabetes, and hypertension), population admixture, BMI, and total energy intake, n = 802. P for interaction was 0.002 (A) and 0.007 (B). *Different from CG+GG within the total fat energy intake category, P < 0.05.
Logistic regression models tested whether the APOA5 polymorphisms were associated with the odds of MetS in this population (Table 4). Neither the -1131T > C nor the S19W minor alleles had an association with MetS when all participants were analyzed [OR (95% CI) = 0.87 (0.59–1.29) and 0.94 (0.62–1.41), respectively]. Further stratification by total fat intake category was not significantly associated with MetS for the -1131C minor allele or the S19W minor allele, suggesting that there was no modulation by this dietary factor on APOA5 SNP and MetS in this Puerto Rican group.
TABLE 4.
OR (95% CI) for MetS in association with APOA5 polymorphisms, overall and by category of total fat intake12
|
APOA5 -1131T > C |
SNP, P-value |
APOA5 S19W |
SNP, P-value | |||
|---|---|---|---|---|---|---|
| TT | TC+CC | CC | CG+GG | |||
| Overall | 1.00 | 0.87 (0.59–1.29) | 0.490 | 1.00 | 0.94 (0.62–1.41) | 0.749 |
| <31% fat energy3 | 1.00 | 0.95 (0.54–1.66) | 0.847 | 1.00 | 1.00 (0.54–1.84) | 0.999 |
| ≥31% fat energy |
1.00 |
0.84 (0.49–1.47) |
0.546 |
1.00 |
0.91 (0.52–1.60) |
0.746 |
n = 802, adjusted for age, sex, smoking, alcohol intake, population admixture, and medication use (lipid-lowering, diabetes, and hypertension).
Participants were classified as having MetS if they had ≥3 of the following conditions: waist circumference ≥102 cm in men or ≥88 cm in women; fasting glucose ≥5.6 mmol/L; elevated blood pressure (SBP ≥ 130 or DBP ≥ 85 mm Hg); high TG (≥1.7 mmol/L); low HDL-C (<1.0 mmol/L in men or <1.3 mmol/L in women).
Category of total fat intake defined as below or above the population median.
Discussion
We have shown that APOA5 S19W was significantly associated with plasma HDL-C but not with other lipid concentrations, whereas the -1131T > C SNP was not associated with any plasma lipid or metabolic marker in a cohort of Puerto Rican older adults living in the US. We found significant interactions between total fat intake as percent of total energy intake and the -1131T > C SNP associated with plasma TG and total cholesterol, and between fat intake and the S19W SNP, associated with SBP and DBP. MetS was not associated with either polymorphism. Puerto Rican carriers of the APOA5 -1131C variant may benefit from a low-fat diet more than carriers of the common allele, resulting in a protective lipid profile. Alternatively, the benefit of a low-fat diet in maintaining low blood pressure was observed for carriers of the S19W common allele.
Our results agree with previous observations showing that carriers of the APOA5 S19W minor allele had lower plasma HDL-C (6,33). The association in our sample remained significant after adjusting for plasma TG, suggesting that the effect of the SNP on the HDL-C concentration is not through an association with TG in Puerto Ricans. This Puerto Rican cohort has a higher MAF for the S19W SNP than non-Hispanic whites (18), which may translate into a less favorable HDL-C concentration in Puerto Ricans and may potentially explain some of the observed disparities in HDL-C–related metabolic conditions in this population, such as diabetes and MetS.
Several studies on the association with other MetS markers or plasma lipids showed independent associations of APOA5 SNP with plasma total cholesterol, LDL-C, VLDL-C, and fasting glucose (6,8,34–36) but not with waist circumference (5,36). Reports on blood pressure are contradictory (5,36). We did not observe any associations of the individual APOA5 SNP with any of these measures, including plasma TG concentration, as has been often reported in other populations. In fact, studies showed different associations of the gene variants with TG according to ethnic group (33). Whereas European white individuals with diabetes and the -1131C variant had higher plasma TG concentrations than major homozygotes, the same was not observed for Indian Asians or Afro-Caribbeans with diabetes. On the other hand, Indian Asian, but not European white or Afro-Caribbean, diabetes patients with the minor allele of S19W had higher TG than carriers of the common genotype. Another study showed that both APOA5 SNP were significantly associated with plasma TG concentrations in whites in the United Kingdom, but only the -1131T > C, and not the S19W, was associated with plasma TG concentrations in Pune Indians (34). In our Puerto Rican cohort, neither SNP was associated with TG. This suggests that in Puerto Ricans, the 2 APOA5 SNP may not play a direct role in the modulation of plasma TG or other plasma lipids.
One possible way by which APOA5 influences lipid and metabolic outcomes may be through interactions with dietary fat intake. In U.S. Caucasians, the -1131T > C SNP interacts with dietary fat in modulation of BMI (10), with individuals homozygous for the major allele having increased BMI with higher total fat and monounsaturated fatty acid intakes and with PUFA intake in modulation of the plasma TG concentration, lipoprotein particle size, and remnant lipoprotein concentration (14). The minor allele of the -1131T > C SNP imparted a more disadvantageous lipid profile at high PUFA intake. In our cohort, carriers of the -1131T > C variant allele had the less favorable lipid profile at higher fat intake. Although the nature of our study cannot determine the specific effect-modifying mechanism of dietary fat intake, Aberle et al. (13) proposed that the C allele may impair ribosomal translation efficiency, which could reduce the lipoprotein lipase-mediated TG uptake into adipocytes. Their finding of a larger reduction in BMI in individuals with the -1131C allele who consume a short-term, fat-restricted diet agrees with our observations, suggesting that such a mechanism may be operating here.
To the best of our knowledge, there are no previous reports of an interaction between an APOA5 variant and total dietary fat intake in relation to SBP and DBP. In one study, the S19W SNP was independently associated with both SBP and DBP in a dominant but not in an additive model (5). Participants with the minor allele had significantly higher blood pressure than those homozygous for the common allele. Those authors suggested that a direct effect of APOA5 on blood pressure is unlikely and attributed the association to evidence that chronic hypertriglyceridemia leads to endothelial dysfunction, which is associated with an impaired response to vasodilator stimulation and decreased nitric oxide availability. Those last 2 factors contribute to increased blood pressure. We did not observe an independent association between the S19W SNP and the plasma TG concentration, suggesting that the role of S19W on blood pressure could be more direct; a potential mechanism should be explored in detail.
The 2 SNP analyzed were not significantly associated with MetS, either alone or by total dietary fat energy intake category in this Puerto Rican population. Dallongeville et al. (5) showed that the association of the APOA5 S19W variant with MetS was via an association with TG. Neither SNP was significantly associated with plasma TG, partly explaining the lack of association of the SNP with MetS in our cohort. The high prevalence of MetS in Puerto Ricans may likely be due to other genetic and environmental factors that do not involve APOA5.
In addition to the significant gene-diet interactions that we report here, it is possible that interactions between the APOA5 SNP and other dietary factors could play a role in the modulation of metabolic outcomes. Individuals carrying the -1131C and 19W alleles have been shown to have defective regulation of plasma nonesterified fatty acid concentrations after a glucose load (37), suggesting a possible effect of carbohydrate intake. In our Puerto Rican cohort, among participants with higher complex carbohydrate intake, those with the perilipin SNP minor allele were less likely to be obese than those without the minor allele (38). In our analysis, we controlled for carbohydrate, fiber, and alcohol intakes, as well as total energy intake; thus, the observed interactions likely are not mediated by other macronutrient intakes. Future studies on APOA5 polymorphisms are needed to elucidate the role of other nutritional and environmental factors modifying the outcomes.
Because APOA5 is part of the APOA1/C3/A4/A5 gene cluster, it is possible that the observed associations for APOA5 SNP could be operating through its linkage with other SNP in the cluster, particularly with APOC3 (3,39). Yet, the pairwise LD coefficient R suggested that the APOA5 SNP act independently. This partly agrees with other observations (40), but it is possible that other SNP in the cluster not tested for LD in our study could mediate the observations we have reported. Also, other studies show associations between other APOA5 SNP with markers of MetS that we did not test here (36,41,42).
Our results may partly explain the contrasting profile of cardio-metabolic risk factors reported in Puerto Ricans, namely protective total cholesterol concentrations but high prevalence of hypertension. The common alleles for both APOA5 SNP, carried by close to 80% of this Puerto Rican cohort, present the most detrimental blood pressure profile at the higher dietary fat intake; yet at that same level of consumption, their lipid profile is more cardio-protective. As Puerto Ricans living in the US assimilate more into a Westernized lifestyle characterized by high fat intake, those interactions may be driving the observed pervasiveness of hypertension but fairly favorable plasma cholesterol concentrations that are reported for this group. The cross-sectional nature of our data limited our ability to test for changes in plasma lipids and MetS markers or to establish directionality. Longitudinal studies are needed to clarify the role of genes and dietary changes on the occurrence of chronic disease and their risk factors. Future studies could focus on the difference between traditional dietary habits of Puerto Ricans and those adopted when moving to the mainland US. Moreover, the ability of individuals with a certain genotype to modify their lipid profiles by changing their diets should be tested in intervention studies.
In conclusion, Puerto Ricans have a distinctive profile of association of APOA5 gene variants with metabolic markers relative to other populations; the SNP are not associated with plasma TG concentrations, a common observation in other groups. Dietary fat intake seems to modulate these associations, with Puerto Ricans who carry the APOA5 variants having lower plasma TG and cholesterol concentrations but higher blood pressure, but only with high-fat energy intake. The growing number of Puerto Ricans living in the mainland US makes them a priority population for studying genetic and dietary contributions to disease to help develop interventions that may reduce observed disparities in chronic disease.
Supplementary Material
Acknowledgments
J.M. and J.M.O. designed the research; J.M. performed statistical analysis and wrote the paper; J.M., K.L.T., and J.M.O. provided the data; J.M., S.D., K.L.T., and J.M.O. interpreted the results; J.M. and J.M.O. had primary responsibility for final content. All authors read and approved the final manuscript.
Supported by the NIH, National Institute on Aging, grant nos. P01AG023394 and P01AG023394-S1 and by the USDA, Agricultural Research Service agreement no. 58-1950-7-707.
Author disclosures: J. Mattei, S. Demissie, K. L. Tucker, and J. M. Ordovas, no conflicts of interest.
Supplemental Table 1 is available with the online posting of this paper at jn.nutrition.org.
Abbreviations used: DBP, diastolic blood pressure; HDL-C, HDL cholesterol; LD, linkage disequilibria; LDL cholesterol; LDL-C, MAF, minor allele frequency; MetS, metabolic syndrome; OR, odds ratio; SBP, systolic blood pressure; SNP, single nucleotide polymorphism; TG, triglyceride; VLDL-C, VLDL cholesterol.
References
- 1.Tai ES, Ordovas JM. Clinical significance of apolipoprotein A5. Curr Opin Lipidol. 2008;19:349–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kluger M, Heeren J, Merkel M. Apoprotein A-V: an important regulator of triglyceride metabolism. J Inherit Metab Dis. 2008;31:281–8. [DOI] [PubMed] [Google Scholar]
- 3.Lai CQ, Parnell LD, Ordovas JM. The APOA1/C3/A4/A5 gene cluster, lipid metabolism and cardiovascular disease risk. Curr Opin Lipidol. 2005;16:153–66. [DOI] [PubMed] [Google Scholar]
- 4.Yamada Y, Kato K, Hibino T, Yokoi K, Matsuo H, Segawa T, Watanabe S, Ichihara S, Yoshida H, et al. Prediction of genetic risk for metabolic syndrome. Atherosclerosis. 2007;191:298–304. [DOI] [PubMed] [Google Scholar]
- 5.Dallongeville J, Cottel D, Wagner A, Ducimetiere P, Ruidavets JB, Arveiler D, Bingham A, Ferrieres J, Amouyel P, et al. The APOA5 Trp19 allele is associated with metabolic syndrome via its association with plasma triglycerides. BMC Med Genet. 2008;9:84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grallert H, Sedlmeier EM, Huth C, Kolz M, Heid IM, Meisinger C, Herder C, Strassburger K, Gehringer A, et al. APOA5 variants and metabolic syndrome in Caucasians. J Lipid Res. 2007;48:2614–21. [DOI] [PubMed] [Google Scholar]
- 7.Maasz A, Kisfali P, Horvatovich K, Mohas M, Marko L, Csongei V, Farago B, Jaromi L, Magyari L, et al. Apolipoprotein A5 T-1131C variant confers risk for metabolic syndrome. Pathol Oncol Res. 2007;13:243–7. [DOI] [PubMed] [Google Scholar]
- 8.Niculescu LS, Fruchart-Najib J, Fruchart JC, Sima A. Apolipoprotein A-V gene polymorphisms in subjects with metabolic syndrome. Clin Chem Lab Med. 2007;45:1133–9. [DOI] [PubMed] [Google Scholar]
- 9.Corella D, Ordovas JM. Single nucleotide polymorphisms that influence lipid metabolism: interaction with dietary factors. Annu Rev Nutr. 2005;25:341–90. [DOI] [PubMed] [Google Scholar]
- 10.Corella D, Lai CQ, Demissie S, Cupples LA, Manning AK, Tucker KL, Ordovas JM. APOA5 gene variation modulates the effects of dietary fat intake on body mass index and obesity risk in the Framingham Heart Study. J Mol Med. 2007;85:119–28. [DOI] [PubMed] [Google Scholar]
- 11.Prieur X, Coste H, Rodriguez JC. The human apolipoprotein AV gene is regulated by peroxisome proliferator-activated receptor-alpha and contains a novel farnesoid X-activated receptor response element. J Biol Chem. 2003;278:25468–80. [DOI] [PubMed] [Google Scholar]
- 12.Prieur X, Huby T, Coste H, Schaap FG, Chapman MJ, Rodriguez JC. Thyroid hormone regulates the hypotriglyceridemic gene APOA5. J Biol Chem. 2005;280:27533–43. [DOI] [PubMed] [Google Scholar]
- 13.Aberle J, Evans D, Beil FU, Seedorf U. A polymorphism in the apolipoprotein A5 gene is associated with weight loss after short-term diet. Clin Genet. 2005;68:152–4. [DOI] [PubMed] [Google Scholar]
- 14.Lai CQ, Corella D, Demissie S, Cupples LA, Adiconis X, Zhu Y, Parnell LD, Tucker KL, Ordovas JM. Dietary intake of n-6 fatty acids modulates effect of apolipoprotein A5 gene on plasma fasting triglycerides, remnant lipoprotein concentrations, and lipoprotein particle size: the Framingham Heart Study. Circulation. 2006;113:2062–70. [DOI] [PubMed] [Google Scholar]
- 15.Kim JY, Kim OY, Koh SJ, Jang Y, Yun SS, Ordovas JM, Lee JH. Comparison of low-fat meal and high-fat meal on postprandial lipemic response in non-obese men according to the -1131T>C polymorphism of the apolipoprotein A5 (APOA5) gene (randomized cross-over design). J Am Coll Nutr. 2006;25:340–7. [DOI] [PubMed] [Google Scholar]
- 16.Hubacek JA, Bohuslavova R, Skodova Z, Pitha J, Bobkova D, Poledne R. Polymorphisms in the APOA1/C3/A4/A5 gene cluster and cholesterol responsiveness to dietary change. Clin Chem Lab Med. 2007;45:316–20. [DOI] [PubMed] [Google Scholar]
- 17.Lan Q, Shen M, Garcia-Rossi D, Chanock S, Zheng T, Berndt SI, Puri V, Li G, He X, et al. Genotype frequency and F ST analysis of polymorphisms in immunoregulatory genes in Chinese and Caucasian populations. Immunogenetics. 2007;59:839–52. [DOI] [PubMed] [Google Scholar]
- 18.Mattei J, Parnell L, Lai CQ, Garcia-Bailo B, Adiconis X, Shen J, Arnett D, Demissie S, Tucker K, et al. Disparities in allele frequencies and population differentiation for 101 disease-associated single nucleotide polymorphisms between Puerto Ricans and non-Hispanic whites. BMC Genet. 2009;10:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cleghorn GD, Nguyen M, Roberts B, Duran G, Tellez T, Alecon M. Practice-based interventions to improve health care for Latinos with diabetes. Ethn Dis. 2004;14:S117–21. [PubMed] [Google Scholar]
- 20.Tucker KL, Bermudez OI, Castaneda C. Type 2 diabetes is prevalent and poorly controlled among Hispanic elders of Caribbean origin. Am J Public Health. 2000;90:1288–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin H, Bermudez OI, Falcon LM, Tucker KL. Hypertension among Hispanic elders of a Caribbean origin in Massachusetts. Ethn Dis. 2002;12:499–507. [PubMed] [Google Scholar]
- 22.Denney JT, Krueger PM, Rogers RG, Boardman JD. Race/ethnic and sex differentials in body mass among US adults. Ethn Dis. 2004;14:389–98. [PubMed] [Google Scholar]
- 23.Bermudez OI, Tucker KL. Total and central obesity among elderly Hispanics and the association with Type 2 diabetes. Obes Res. 2001;9:443–51. [DOI] [PubMed] [Google Scholar]
- 24.Gomez M, Ramirez M, Disdier O. Prevalence of the metabolic syndrome among a determined Puerto Rican population. P R Health Sci J. 2006;25:111–6. [PubMed] [Google Scholar]
- 25.Bermudez OI, Velez-Carrasco W, Schaefer EJ, Tucker KL. Dietary and plasma lipid, lipoprotein, and apolipoprotein profiles among elderly Hispanics and non-Hispanics and their association with diabetes. Am J Clin Nutr. 2002;76:1214–21. [DOI] [PubMed] [Google Scholar]
- 26.Tucker KL. Stress and nutrition in relation to excess development of chronic disease in Puerto Rican adults living in the Northeastern USA. J Med Invest. 2005;52 Suppl:252–8. [DOI] [PubMed] [Google Scholar]
- 27.Chumlea WC, Guo SS, Wholihan K, Cockram D, Kuczmarski RJ, Johnson CL. Stature prediction equations for elderly non-Hispanic white, non-Hispanic black, and Mexican-American persons developed from NHANES III data. J Am Diet Assoc. 1998;98:137–42. [DOI] [PubMed] [Google Scholar]
- 28.Tucker KL, Bianchi LA, Maras J, Bermudez OI. Adaptation of a food frequency questionnaire to assess diets of Puerto Rican and non-Hispanic adults. Am J Epidemiol. 1998;148:507–18. [DOI] [PubMed] [Google Scholar]
- 29.Enoch MA, Shen PH, Xu K, Hodgkinson C, Goldman D. Using ancestry-informative markers to define populations and detect population stratification. J Psychopharmacol. 2006;20:19–26. [DOI] [PubMed] [Google Scholar]
- 30.Lai CQ, Tucker KL, Choudhry S, Parnell LD, Mattei J, Garcia-Bailo B, Beckman K, Burchard EG, Ordovas JM. Population admixture associated with disease prevalence in the Boston Puerto Rican health study. Hum Genet. 2009;125:199–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu K, Muse SV. PowerMarker: an integrated analysis environment for genetic marker analysis. Bioinformatics. 2005;21:2128–9. [DOI] [PubMed] [Google Scholar]
- 32.Grundy SM, Cleeman JI, Daniels SR, Donato KA, Eckel RH, Franklin BA, Gordon DJ, Krauss RM, Savage PJ, et al. Diagnosis and management of the metabolic syndrome: an American Heart Association/National Heart, Lung, and Blood Institute Scientific Statement. Circulation. 2005;112:2735–52. [DOI] [PubMed] [Google Scholar]
- 33.Dorfmeister, B, Cooper, JA, Stephens, JW, Ireland, H, Hurel, SJ, Humphries, SE, Talmud, PJ. The effect of APOA5 and APOC3 variants on lipid parameters in European Whites, Indian Asians and Afro-Caribbeans with type 2 diabetes. Biochim Biophys Acta. 2007;1772:355–63. [DOI] [PubMed] [Google Scholar]
- 34.Chandak GR, Ward KJ, Yajnik CS, Pandit AN, Bavdekar A, Joglekar CV, Fall CH, Mohankrishna P, Wilkin TJ, et al. Triglyceride associated polymorphisms of the APOA5 gene have very different allele frequencies in Pune, India compared to Europeans. BMC Med Genet. 2006;7:76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jang Y, Kim JY, Kim OY, Lee JE, Cho H, Ordovas JM, Lee JH. The -1131T→C polymorphism in the apolipoprotein A5 gene is associated with postprandial hypertriacylglycerolemia; elevated small, dense LDL concentrations; and oxidative stress in nonobese Korean men. Am J Clin Nutr. 2004;80:832–40. [DOI] [PubMed] [Google Scholar]
- 36.Martin S, Nicaud V, Humphries SE, Talmud PJ. Contribution of APOA5 gene variants to plasma triglyceride determination and to the response to both fat and glucose tolerance challenges. Biochim Biophys Acta. 2003;1637:217–25. [DOI] [PubMed] [Google Scholar]
- 37.Kovar J, Adamkova V. Lipoprotein lipase activity determined in vivo is lower in carriers of apolipoprotein A-V gene variants 19W and -1131C. Physiol Res. 2008;57:555–61. [DOI] [PubMed] [Google Scholar]
- 38.Smith CE, Tucker KL, Yiannakouris N, Garcia-Bailo B, Mattei J, Lai CQ, Parnell LD, Ordovas JM. Perilipin polymorphism interacts with dietary carbohydrates to modulate anthropometric traits in hispanics of Caribbean origin. J Nutr. 2008;138:1852–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Talmud PJ, Hawe E, Martin S, Olivier M, Miller GJ, Rubin EM, Pennacchio LA, Humphries SE. Relative contribution of variation within the APOC3/A4/A5 gene cluster in determining plasma triglycerides. Hum Mol Genet. 2002;11:3039–46. [DOI] [PubMed] [Google Scholar]
- 40.Olivier M, Wang X, Cole R, Gau B, Kim J, Rubin EM, Pennacchio LA. Haplotype analysis of the apolipoprotein gene cluster on human chromosome 11. Genomics. 2004;83:912–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yamada Y, Ichihara S, Kato K, Yoshida T, Yokoi K, Matsuo H, Watanabe S, Metoki N, Yoshida H, et al. Genetic risk for metabolic syndrome: examination of candidate gene polymorphisms related to lipid metabolism in Japanese people. J Med Genet. 2008;45:22–8. [DOI] [PubMed] [Google Scholar]
- 42.Kisfali P, Mohas M, Maasz A, Hadarits F, Marko L, Horvatovich K, Oroszlan T, Bagosi Z, Bujtor Z, et al. Apolipoprotein A5 IVS3+476A allelic variant associates with increased trigliceride levels and confers risk for development of metabolic syndrome in Hungarians. Circ J. 2008;72:40–3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.