Abstract
Impairments in folate-mediated 1-carbon metabolism are associated with several common diseases and developmental anomalies including intestinal cancers, vascular disease, cognitive decline, and neural tube defects. The etiology of folate-associated pathologies involves interactions among multiple genetic risk alleles and environmental factors, although the causal mechanisms that define the role of folate and other B-vitamins in these complex disorders remain to be established. Folate and other B-vitamins fundamentally differ from other nutrients that interact with the genome in determining health and disease outcomes in that their interaction is reciprocal. Common gene variants influence the activity of folate-dependent enzymes and anabolic pathways; folate-mediated 1-carbon metabolism is essential for the high-fidelity synthesis of DNA and activated methyl groups that are required for DNA methylation and regulation of chromatin structure. This review focuses on the regulation of folate-mediated 1-carbon metabolism and its role in maintaining genome integrity and on strategies for establishing the metabolic pathways and mechanisms that underlie folate-associated pathologies.
Folate-mediated 1-carbon metabolism
Folate is a B-vitamin that is present in cells as a family of enzyme cofactors that carry and chemically activate 1-carbons at the oxidation level of formate, formaldehyde, and methanol (1). Folate-activated 1-carbons are required for the de novo synthesis of purines and thymidylate and for the remethylation of homocysteine to methionine. Methionine is an essential amino acid that is used for protein synthesis or can be adenosylated to S-adenosylmethionine (known as AdoMet or SAM), which is required for polyamine synthesis and for numerous AdoMet-dependent methylation reactions including the methylation of proteins (including histones), cytosine bases on DNA, neurotransmitters, phospholipids, and numerous small molecules (2,3).
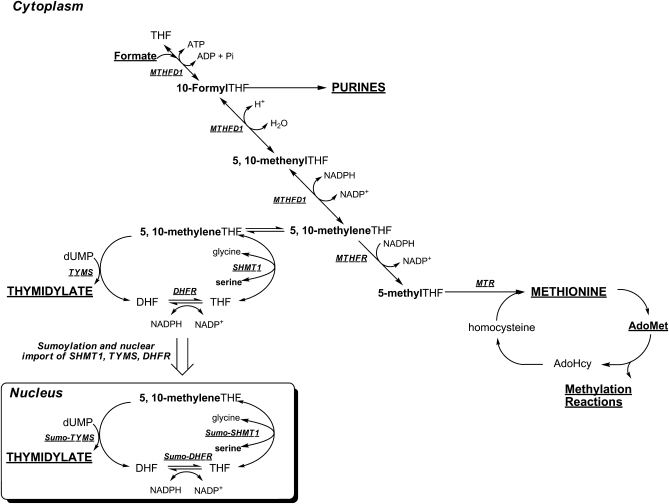
Folate-dependent metabolic pathways are compartmentalized in the cell, and many of these pathways are associated with multienzyme complexes (1). Approximately 40% of total cellular folate is located in mitochondria and is sequestered in that compartment (4). A primary function of 1-carbon metabolism in mitochondria is to generate 1-carbon units in the form of formate for cytoplasmic 1-carbon metabolism (Fig. 1) (5). The amino acids serine, glycine, dimethylglycine, and sarcosine are catabolized in mitochondria to produce formaldehyde, which is condensed with tetrahydrofolate (THF) to generate methylene-THF. The activated formaldehyde of methylene-THF is oxidized, forming 10-formyl-THF, which serves to formylate MET-tRNA for mitochondrial protein synthesis, or alternatively, 10-formyl-THF is hydrolyzed to THF and formate, which enters the cytoplasm (Fig. 1).
FIGURE 1 .
Folate-mediated 1-carbon metabolism. Folate-activated 1-carbons are utilized in the synthesis of purines and thymidylate (dTMP) and in the methylation of homocysteine to methionine. Methionine can be converted to a methyl donor through its adenosylation to S-adenosylmethionine (AdoMet). Mitochondrial-derived formate traverses to the cytoplasm, where it is incorporated into the folate-activated 1-carbon pool. Nuclear folate metabolism occurs through the SUMO-dependent import of the de novo thymidylate synthase pathway from the cytoplasm to the nucleus (8,27). SHMT isozymes are present in each of these 3 compartments and function to generate single carbons from the hydroxymethyl group of serine in the form of 5,10-methylene-THF. The 1-carbon unit is labeled in bold. MTHFD1, methylenetetrahydrofolate dehydrogenase; MTR, methionine synthase; MTHFR, methylenetetrahydrofolate reductase; SHMT1, cytoplasmic serine hydroxymethyltransferase; TYMS, thymidylate synthase; DHFR, dihydrofolate reductase.
In the cytoplasm, folate-activated 1-carbon units function in an interdependent anabolic network comprised of 3 biosynthetic pathways: de novo purine biosynthesis, which requires 10-formyl-THF for the C2 and C8 carbons of the purine ring; de novo thymidylate biosynthesis, which requires methylene-THF for the reductive methylation of deoxyuridylate (dUMP) to form thymidylate (dTMP); and the remethylation of homocysteine to methionine, which requires 5-methyl-THF. Formate enters the folate-activated 1-carbon pool through its ATP-dependent conversion to 10-formyl-THF catalyzed by 10-formyl-THF synthetase, which is encoded by MTHFD1 (Fig. 1). 10-Formyl-THF-dependent purine biosynthesis occurs only when purine nucleotides are not available from synthesis through the purine nucleotide salvage pathway. Recently, it was demonstrated that the enzymes that constitute the de novo purine biosynthetic pathway are present in a complex termed a “purinosome,” which assembles only when exogenous purines are not available (6). 10-Formyl-THF is also a source of other folate 1-carbon derivatives. The formyl moiety of 10-formyl-THF can be reduced to methylene-THF by methenyltetrahydrofolate dehydrogense (also encoded by MTHFD1) in a NADPH-dependent reaction. Alternatively, methylene-THF (and glycine) can also be generated in the cytoplasm from serine and THF in a reaction catalyzed by serine hydroxymethyltransferase, encoded by SHMT1. Methylene-THF is a cofactor for the methylation of dUMP to dTMP in a reaction catalyzed by thymidylate synthase, encoded by TYMS. In this reaction, methylene-THF serves as both a 1-carbon donor (in the form of formaldehyde) and the THF cofactor serves as a donor of 2 electrons, thereby generating dihydrofolate (DHF). Dihydrofolate reductase, encoded by DHFR, catalyzes the NADPH-dependent conversion of DHF to THF and thereby regenerates the functional THF cofactor. Alternatively, the 1-carbon moiety of methylene-THF can be further reduced to 5-methyl-THF in a NADPH-dependent reaction catalyzed by methylenetetrahydrofolate reductase, encoded by MTHFR. 5-Methyl-THF serves as 1-carbon donor for the vitamin B-12-dependent conversion of homocysteine to methionine in a reaction catalyzed by methionine synthase, which is encoded by MTR.
As observed for de novo purine biosynthesis, there is also evidence that de novo thymidylate biosynthesis occurs through a multienzyme complex (7). Furthermore, the enzymes involved in this pathway may function in both the cytoplasm and nucleus (Fig. 1). Evidence for nuclear nucleotide biosynthesis through an enzyme complex termed a “replitase” was first proposed by Prem veer Reddy and Pardee (7), although an intact metabolic pathway was never identified. About 10% of cellular folates are present in the nucleus (4), and recently, folate-mediated 1-carbon metabolism was demonstrated to occur in the nucleus (8,9). During S-phase of the cell cycle, the enzymes that constitute the de novo dTMP pathway, TYMS, SHMT1, SHMT2, and DHFR, undergo posttranslational modification by the small ubiquitin-like modifier (SUMO) and nuclear import (8). Isolated, intact mouse liver nuclei can convert serine, NADPH, and dUMP to dTMP, whereas nuclei disrupted by sonication cannot convert dUMP to dTMP under the same experimental conditions, suggesting that the presence of a metabolic complex in the nucleus is essential for nuclear dTMP synthesis (9). This finding indicates that dTMP synthesis occurs in the nucleus during DNA synthesis and raises the possibility that folate-dependent dTMP synthesis may not occur in the cytoplasm.
Impaired 1-carbon metabolism and human pathology
Disruptions in folate metabolism are linked to several common human pathologies including developmental anomalies and gastrointestinal cancers (10,11). Neural tube closure defects (NTD), which include the birth defects anencephaly and spina bifida, arise from the failure of neurulation during early human embryonic development. NTD are among the most common human birth defects and have a heterogeneous and multifactorial etiology with interacting genetic and environmental risk factors. Clinical trials and folic acid fortification initiatives indicate that up to 70% of NTD can be prevented by maternal folic acid supplementation, and human gene variants in the folate-mediated 1-carbon network have been identified as risk factors (10,12). However, the metabolic pathways and associated mechanisms underlying the association between folate-mediated 1-carbon metabolism and NTD pathogenesis are still unknown.
Gastrointestinal cancers are a leading cause of cancer deaths globally, accounting for ∼20% of all cancer incidences (13). In the United States, colorectal cancer is the second leading cause of cancer-related mortality in men and the third leading cause in women (14). Several studies support an association of folate intake with colon cancer risk. Low circulating folate concentrations increase risk of colon cancer (15), and an interaction between folate status and gene variants of 5,10-methylenetetrahydrofolate reductase (MTHFR), an enzyme required for homocysteine remethylation, have been reported. Although the C677T variant of the MTHFR gene is associated with reduced risk of developing colorectal cancer compared with carriers of the more common allele (16), this protective effect is diminished by folate deficiency (17). Low MTHFR activity reduces DNA methylation (18) but may enhance de novo thymidylate biosynthesis (19).
Impaired folate metabolism and genome integrity
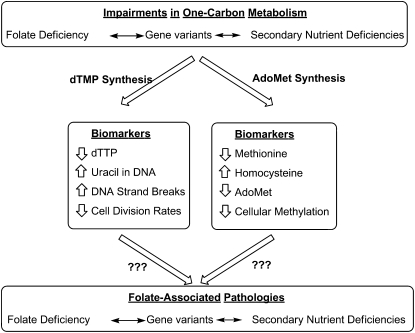
Current experimental and epidemiological evidence indicates that neural tube defects and colon cancer arise from deleterious gene-nutrient interactions (Fig. 2). However, causal mechanisms have yet to be established, including the identification of the folate-dependent pathway(s) that are directly involved in pathogenesis. There are discrete biomarkers of impaired folate metabolism that are sensitive readouts of metabolic efficiency. Decreased rates of thymidylate synthesis result in dUMP accumulation and increased rates of uracil nucleotide misincorporation into nuclear DNA (20). Likewise, insufficient rates of homocysteine remethylation result in elevated plasma homocysteine, decreases in AdoMet, and elevations in S-adensoylhomocysteine (AdoHcy) leading to a decreased cellular AdoMet/AdoHcy ratio, which may be an indicator of cellular methylation capacity (3), and decreased levels of methylcytosine in nuclear DNA (18). Despite the availability of these sensitive biomarkers, progress in deciphering causal metabolic pathways in folate-associated pathways has been limited because of the interconnectedness of the network, such that perturbations in 1 biosynthetic pathway influence the entire 1-carbon network. Furthermore, there are no established sensitive biomarkers for folate-dependent purine nucleotide biosynthesis, which is a major gap in our understanding of the relationships among impairments in 1-carbon metabolism, genome integrity, and folate-associated pathologies.
FIGURE 2 .
Biomarkers of impaired folate-mediated 1-carbon metabolism.
Impairments in the folate-dependent 1-carbon network can arise from a primary folate deficiency, secondary B-vitamin nutrient deficiencies, and genetic variations that influence cellular folate accumulation and/or utilization. Many studies have shown that folate cofactors are limiting in the cell and that the concentration of folate-dependent enzymes and folate-binding proteins exceeds the concentration of folate cofactors, which is estimated to be in the range of 25–35 μmol/L (21,22). Given that folate-dependent enzymes and folate-binding proteins exhibit binding constants (Kd values) in the nanomolar range, all cellular folate cofactors are expected to be protein bound, and folate-dependent anabolic pathways must compete for a limiting pool of folate cofactors (23). Therefore, all folate anabolic pathways are anticipated to be sensitive to primary folate deficiency. Furthermore, genetic variation that alters the partitioning of folate cofactors through any folate-dependent pathway influences the entire 1-carbon network. For example, the common 677 C→T human variant of MTHFR results in decreased MTHFR specific activity, elevated homocysteine, and depressed levels of nuclear methylcytosine but potentially enhances rates of de novo thymidylate biosynthesis (19). Last, secondary nutrient deficiencies can also impair folate-dependent pathways. Vitamin B-12 deficiency diminishes MTR activity and methionine synthesis but also impairs nucleotide biosynthesis through the accumulation of cellular folate cofactors such as 5-methyl-THF. This accumulation of 5-methyl-THF, referred to as a “methyl trap,” results because the MTHFR reaction is essentially irreversible in vivo, and MTR is the only enzyme that can regenerate THF from 5-methyl-THF. Therefore, it is often not possible to establish which biomarkers are “causal” in folate-associated pathologies and which biomarkers are bystanders.
Mouse models of impaired folate metabolism to elucidate mechanisms of folate-associated pathologies
Genetic mouse models offer an opportunity to establish the causal pathways in the folate-dependent 1-carbon network that underlie the associations between impaired folate metabolism and human pathologies that involve gene-nutrient interactions. Targeted manipulation of genes that encode key folate-dependent enzymes that regulate the partitioning of folate-activated 1-carbons among the 3 key biosynthetic pathways allows for precise manipulation of a single biosynthetic pathway in the absence of severe perturbations to the entire network (24). SHMT1 and MTHFD1 represent 2 key enzymes that are potential targets to elucidate the causal metabolic pathways associated with impairments in the 1-carbon network. SHMT1 and the 10-formyl-THF synthetase activity of MTHFD1 comprise the primary entry point of 1-carbon units into the network, and these 2 enzymes may compete for a limiting pool of THF (25). The 1-carbon units generated by MTHFD1 are preferentially partitioned to homocysteine remethylation and purine biosynthesis, whereas SHMT1-derived folate-activated 1-carbons, in the form of methylene-THF, are preferentially directed to thymidylate biosynthesis (23,26). Mice lacking SHMT1 expression exhibit impaired de novo thymidylate biosynthesis and several-fold increases in uracil content in nuclear DNA but exhibit normal levels of AdoMet (26). Disruption of a single MTHFD1 allele results in lower hepatic AdoMet levels, which is consistent with formate serving as a source of 1-carbons for AdoMet synthesis and cellular methylation reactions. These mice also exhibit decreased levels of uracil in nuclear DNA, indicating enhanced de novo thymidylate synthesis and confirming other studies that indicate that SHMT1 and MTHFD1 compete for a limiting pool of unsubstituted THF cofactors (Fig. 1). Therefore, by selectively repressing the entry of folate-activated 1-carbons by varying MTHFD1 and SHMT1 enzyme levels, thymidylate biosynthesis or homocysteine remethylation can be selectively impaired. Ongoing studies will determine whether MTHFD1- and SHMT1-deficient animals are sensitized to folate-associated pathologies including NTD and intestinal cancers and thereby establish the causal pathways for folate-associated pathologies. Better understanding of the causal pathways that underpin folate-associated pathways may lead to more targeted and effective dietary interventions for certain at-risk populations by providing the end product of 1-carbon metabolism (e.g., thymidylate or methionine) rather than the cofactor required for its synthesis.
Other articles in the supplement include references (28–31).
Acknowledgments
The sole author had responsibility for all parts of the manuscript.
Presented as part of the symposium entitled “Nutrients and Epigenetic Regulation of Gene Expression” at the Experimental Biology 2009 meeting, April 20, 2009, in New Orleans, LA. This symposium was sponsored by the American Society for Nutrition (ASN) and had no outside support declared. The Guest Editor for this symposium publication was Kevin Schalinske. Guest Editor disclosure: no relationships to disclose.
This work was supported by Public Health Service grant DK58144 to P.J.S.
Abbreviations used: DHF, dihydrofolate; dTMP, thymidylate; dUMP, deoxyuridylate; MTHFR, methylenetetrahydrofolate reductase; NTD, neural tube closure defect; SUMO, small ubiquitin-like modifier; THF, tetrahydrofolate.
References
- 1.Fox JT, Stover PJ. Folate-mediated one-carbon metabolism. Vitam Horm. 2008;79:1–44. [DOI] [PubMed] [Google Scholar]
- 2.Stover PJ. Physiology of folate and vitamin B12 in health and disease. Nutr Rev. 2004;62:S3–12; discussion S3. [DOI] [PubMed] [Google Scholar]
- 3.Finkelstein JD. Homocysteine: a history in progress. Nutr Rev. 2000;58:193–204. [DOI] [PubMed] [Google Scholar]
- 4.Shin YS, Chan C, Vidal AJ, Brody T, Stokstad EL. Subcellular localization of gamma-glutamyl carboxypeptidase and of folates. Biochim Biophys Acta. 1976;444:794–801. [DOI] [PubMed] [Google Scholar]
- 5.Appling DR. Compartmentation of folate-mediated one-carbon metabolism in eukaryotes. FASEB J. 1991;5:2645–51. [DOI] [PubMed] [Google Scholar]
- 6.An S, Kumar R, Sheets ED, Benkovic SJ. Reversible compartmentalization of de novo purine biosynthetic complexes in living cells. Science. 2008;320:103–6. [DOI] [PubMed] [Google Scholar]
- 7.Prem veer Reddy G, Pardee AB. Multienzyme complex for metabolic channeling in mammalian DNA replication. Proc Natl Acad Sci USA. 1980;77:3312–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Woeller CF, Anderson DD, Szebenyi DM, Stover PJ. Evidence for small ubiquitin-like modifier-dependent nuclear import of the thymidylate biosynthesis pathway. J Biol Chem. 2007;282:17623–31. [DOI] [PubMed] [Google Scholar]
- 9.Anderson DD, Stover PJ. SHMT1 and SHMT2 are functionally redundant in nuclear de novo thymidylate biosynthesis. PLoS One. 2009;4:e5839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beaudin AE, Stover PJ. Insights into metabolic mechanisms underlying folate-responsive neural tube defects: A minireview. Birth Defects Res A Clin Mol Teratol. 2009;85:274–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.MacFarlane AJ, Stover PJ. Convergence of genetic, nutritional and inflammatory factors in gastrointestinal cancers. Nutr Rev. 2007;65:S157–66. [DOI] [PubMed] [Google Scholar]
- 12.Ray JG. Efficacy of Canadian folic acid food fortification. Food Nutr Bull. 2008;29:S225–30. [DOI] [PubMed] [Google Scholar]
- 13.Murray CJ, Lopez AD. Alternative projections of mortality and disability by cause 1990–2020: Global Burden of Disease Study. Lancet. 1997;349:1498–504. [DOI] [PubMed] [Google Scholar]
- 14.Jemal A, Siegel R, Ward E, Murray T, Xu J, Smigal C, Thun MJ. Cancer statistics, 2006. CA Cancer J Clin. 2006;56:106–30. [DOI] [PubMed] [Google Scholar]
- 15.Eichholzer M, Luthy J, Moser U, Fowler B. Folate and the risk of colorectal, breast and cervix cancer: the epidemiological evidence. Swiss Med Wkly. 2001;131:539–49. [DOI] [PubMed] [Google Scholar]
- 16.Chen J, Giovannucci E, Kelsey K, Rimm EB, Stampfer MJ, Colditz GA, Spiegelman D, Willett WC, Hunter DJ. A methylenetetrahydrofolate reductase polymorphism and the risk of colorectal cancer. Cancer Res. 1996;56:4862–4. [PubMed] [Google Scholar]
- 17.Ma J, Stampfer MJ, Giovannucci E, Artigas C, Hunter DJ, Fuchs C, Willett WC, Selhub J, Hennekens CH, Rozen R. Methylenetetrahydrofolate reductase polymorphism, dietary interactions, and risk of colorectal cancer. Cancer Res. 1997;57:1098–102. [PubMed] [Google Scholar]
- 18.Friso S, Choi SW, Girelli D, Mason JB, Dolnikowski GG, Bagley PJ, Olivieri O, Jacques PF, Rosenberg IH, et al. A common mutation in the 5,10-methylenetetrahydrofolate reductase gene affects genomic DNA methylation through an interaction with folate status. Proc Natl Acad Sci USA. 2002;99:5606–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Quinlivan EP, Davis SR, Shelnutt KP, Henderson GN, Ghandour H, Shane B, Selhub J, Bailey LB, Stacpoole PW, Gregory JF 3rd. Methylenetetrahydrofolate reductase 677C→T polymorphism and folate status affect one-carbon incorporation into human DNA deoxynucleosides. J Nutr. 2005;135:389–96. [DOI] [PubMed] [Google Scholar]
- 20.Blount BC, Mack MM, Wehr CM, MacGregor JT, Hiatt RA, Wang G, Wickramasinghe SN, Everson RB, Ames BN. Folate deficiency causes uracil misincorporation into human DNA and chromosome breakage: implications for cancer and neuronal damage. Proc Natl Acad Sci USA. 1997;94:3290–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Suh JR, Herbig AK, Stover PJ. New perspectives on folate catabolism. Annu Rev Nutr. 2001;21:255–82. [DOI] [PubMed] [Google Scholar]
- 22.Strong WB, Tendler SJ, Seither RL, Goldman ID, Schirch V. Purification and properties of serine hydroxymethyltransferase and C1-tetrahydrofolate synthase from L1210 cells. J Biol Chem. 1990;265:12149–55. [PubMed] [Google Scholar]
- 23.Herbig K, Chiang EP, Lee LR, Hills J, Shane B, Stover PJ. Cytoplasmic serine hydroxymethyltransferase mediates competition between folate-dependent deoxyribonucleotide and S-adenosylmethionine biosyntheses. J Biol Chem. 2002;277:38381–9. [DOI] [PubMed] [Google Scholar]
- 24.Stover PJ, MacFarlane AJ. Mouse models to elucidate mechanisms of folate-related cancer pathologies. Nutr Rev. 2008;66: Suppl 1:S54–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.MacFarlane AJ, Perry CA, Girnary HH, Gao D, Allen RH, Stabler SP, Shane B, Stover PJ. Mthfd1 is an essential gene in mice and alters biomarkers of impaired one-carbon metabolism. J Biol Chem. 2009;284:1533–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.MacFarlane AJ, Liu X, Perry CA, Flodby P, Allen RH, Stabler SP, Stover PJ. Cytoplasmic serine hydroxymethyltransferase regulates the metabolic partitioning of methylenetetrahydrofolate but is not essential in mice. J Biol Chem. 2008;283:25846–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Anderson DD, Woeller CF, Stover PJ. Small ubiquitin-like modifier-1 (SUMO-1) modification of thymidylate synthase and dihydrofolate reductase. Clin Chem Lab Med. 2007;45:1760–3. [DOI] [PubMed] [Google Scholar]
- 28.Ho E, Zempleni J. Overview to symposium “Nutrients and Epigenetic Regulation of Gene Expression.”. J Nutr. 2009;139:2387–8. [DOI] [PubMed] [Google Scholar]
- 29.Zempleni J, Chew YC, Bao B, Pestinger V, Wijeratne SSK. Repression of transposable elements by histone biotinylation. J Nutr. 2009;139:2389–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ho E, Clarke JD, Dashwood RH. Dietary sulforaphane, a histone deacetylase inhibitor for cancer prevention. J Nutr. 2009;139:2393–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kirkland JB. Niacin status impacts chromatin structure. J Nutr. 2009;139:2397–401. [DOI] [PubMed] [Google Scholar]