Abstract
Poxviruses employ many strategies to evade and neutralize the host immune response. In this study, we have identified two vaccinia virus ORFs, termed A46R and A52R, that share amino acid sequence similarity with the Toll/IL-1 receptor (TIR) domain, a motif that defines the IL-1/Toll-like receptor (TLR) superfamily of receptors, which have a key role in innate immunity and inflammation. When expressed in mammalian cells, the protein products of both ORFs were shown to interfere specifically with IL-1 signal transduction. A46R partially inhibited IL-1-mediated activation of the transcription factor NFκB, and A52R potently blocked both IL-1- and TLR4-mediated NFκB activation. MyD88 is a TIR domain-containing adapter molecule known to have a central role in both IL-1 and TLR4 signaling. A52R mimicked the dominant-negative effect of a truncated version of MyD88 on IL-1, TLR4, and IL-18 signaling but had no effect on MyD88-independent signaling pathways. Therefore, A46R and A52R are likely to represent a mechanism used by vaccinia virus of suppressing TIR domain-dependent intracellular signaling.
Poxviruses are family of complex DNA viruses that includes variola virus, the causative agent of smallpox, and the antigenically related virus used to eradicate this disease, vaccinia virus (VV; ref. 1). Orthopoxviruses such as VV display unique strategies for the evasion of host immune responses, such as the ability to produce secreted decoy receptors for cytokines such as IL-1, tumor necrosis factor (TNF), CC chemokines, IFN-α/β, and IFN-γ (2, 3). The study of the mechanism of immune evasion by poxviruses has provided insights into the physiological role of immune regulatory molecules such as IL-1 (4) and has identified previously uncharacterized proteins and potential strategies for therapeutic intervention in immune responses and inflammatory diseases.
The IL-1 receptor/Toll-like receptor (TLR) superfamily comprises an expanding group of molecules that participate in host responses to injury and infection. The family is defined by the presence of an intracellular Toll/IL-1 receptor (TIR) domain that appears in proteins in insects, plants, and mammals that have the related function of translating the detection of injury and infection into the induction of immune response genes (5). The family splits broadly into two subgroups, based on extracellular sequence similarity to the type I IL-1 receptor (IL-1RI), the signaling receptor for IL-1 (6), or the Drosophila receptor Toll, which controls the potent antifungal response in adult flies (7). Other mammalian receptors in the family involved in immune function include the IL-18 receptor and IL-18 receptor accessory protein (AcPL), which are involved in Th1 cell activation (8). Another family member, T1/ST2, has been proposed to have a role in directing Th2 function (9), although this role remains controversial (10, 11).
Recently, mammalian TLRs have been identified (12). Two in particular, TLR2 and TLR4, have been studied and are now implicated in innate immunity, in that they have been shown to be required for responses to bacterial products (13, 14). Most recently, TLR4 has been shown to mediate the host response to lipopolysaccharide and hence Gram-negative bacteria (15–17). A wider role for TLR4 in inflammation is also suggested given that its expression and signaling is increased in the injured myocardium in the absence of any infection (18).
Both IL-1RI and TLR4 trigger the activation of the transcription factor NFκB through signaling pathways that use similar intermediates (5, 19, 20). Binding of IL-1 to IL-1RI induces the recruitment of the IL-1 receptor accessory protein (IL-1RAcP; refs. 21 and 22), whereas TLR4 does not seem to need a signaling transmembrane accessory protein (19). MyD88, which also has a TIR domain, has been shown recently to have an essential role in both IL-1 and lipopolysaccharide/TLR4 signaling (23, 24). MyD88 had been implicated previously as an adaptor molecule that associates with both IL-1 receptor complexes and TLR4 via homotypic interactions mediated by its TIR domain (19, 25, 26). MyD88 can subsequently recruit the IL-1 receptor-associated kinase (IRAK) and IRAK2 through a death domain interaction (19, 26, 27), which then leads to TNF-receptor-associated factor 6 activation (28). TNF-receptor-associated factor 6, possibly by activating both NFκB-inducing kinase and mitogen-activated protein kinase/ERK kinase kinase-1 (19, 29–31), bridges both the IL-1RI and TLR4 pathway to the IκB kinase complex, which is responsible for NFκB activation; recently, however, the role of NFκB-inducing kinase in proinflammatory signaling to NFκB has been disputed (see Science's Signal Transduction Knowledge Environment at www.stke.org/cgi/content/full/OC_sigtrans;1999/5/re1).
Given the importance of IL-1RI and TLRs in the host response to infection, we addressed whether additional poxvirus mechanisms would exist to target IL-1 and TLR intracellular signaling pathways. Herein, we describe the identification and initial characterization of A46R and A52R as potential viral antagonists of IL-1 and TLR signaling. Both A46R and A52R have putative TIR domains and are shown to inhibit NFκB activation by IL-1RI in the case of A46R or that driven by IL-1RI, TLR4, and IL-18 in the case of A52R. This study of these proteins represents, to our knowledge, the first demonstration of a specific viral inhibitory effect on intracellular IL-1R/TLR signaling.
Materials and Methods
DNA Expression and Reporter Vectors.
IL-1R1 and IL-1RAcP expression vectors were gifts from W. Falk (University of Regensburg, Regensburg, Germany). Full-length MyD88, the truncated ΔMyD88 (amino acids 152–296) lacking the death domain, full-length TLR4, and the mutant ΔTLR4 (amino acids 1–666) lacking the TIR domain were provided by M. Muzio (Mario Negri Institute, Milan, Italy; refs. 19 and 26). IRAK and pRK5 were from Tularik (South San Francisco). The AcPL expression vector was a gift from C. Dinarello (University of Colorado Health Sciences Center, Denver). IL-1R1 and IL-1RAcP expression vectors used in the HeLa experiments have been described (21, 32), and MyD88 used in these cells was a gift from F. Volpe (Glaxo Wellcome).
The NFκB-luciferase reporter construct (NFκB-luc) containing five κB elements was a gift from R. Hofmeister (University of Regensburg). The IL-8 promoter reporter plasmid was constructed by subcloning the 5′ noncoding region of the human IL-8 gene, including part of the first exon into pGL3-basic vector (Promega). The lactogenesis hormone response element (LHRE)-luc reporter gene containing the STAT5-binding element LHRE fused to luciferase has been described (33).
Cloning of A46R and A52R.
The VV ORFs A46R and A52R, termed SalF9R and SalF15R, respectively, in Western Reserve (WR) strain (34), were cloned by PCR amplification from WR DNA with primers incorporating restriction sites for EcoRI upstream and HindIII downstream of the ORFs. The primers used for SalF9R were 5′-CGTGAATTCCGAGAATGGCGTTTGA (sense) and 5′-CGGAAGCTTTTATACATCCGTTTCCCT (antisense) and for SalF15R were 5′-CGTGAATTCGTGATCACCATGGAC (sense) and 5′-CGCAAGCTTCTATGACATTTCCAC (antisense). The restriction sites and start and stop codons are underlined. The resulting EcoRI–HindIII fragments were ligated into the multiple cloning site of the mammalian expression vector pRK5. For immunoblot analysis, epitope-tagged A46R and A52R expression vectors were constructed, employing the same strategy, except that the 8-amino acid Flag coding sequence was inserted into the antisense primer 5′ of the stop codon.
Immunoblotting.
Human embryonic 293 cells (1.3 × 106) were seeded in 100-mm dishes and transfected 24 h later with plasmids encoding Flag-tagged A46R or A52R with FuGENE 6 (Roche Molecular Biochemicals). The total amount of DNA (12 μg) was kept constant by supplementation with pRK5. At 24 or 48 h after transfection, cells were lysed in 100 μl of SDS sample buffer [62.5 mM Tris⋅HCl (pH 6.8)/2% (wt/vol) SDS/10% (wt/vol) glycerol/50 mM DTT/0.1% (wt/vol) bromophenol blue] and then sonicated. Lysates were then resolved by SDS/PAGE, transferred to poly(vinylidene difluoride) membranes, and probed with anti-Flag mAb according to the manufacturer's instructions.
Reporter Assays.
Human embryonic 293 cells (4 × 104 per well) were seeded into 24-well plates and transfected 24 h later with 40 ng of NFκB-luc, 60 ng of β-galactosidase, and the indicated amount of expression vectors with FuGENE 6. The total amount of DNA (700 ng) was kept constant by supplementation with pRK5. At the indicated times, cells were harvested in passive lysis buffer (Promega), and the relative stimulation of NFκB activity was calculated by normalizing luciferase activity with β-galactosidase activity.
HeLa cells (1.5 × 104 per well) were seeded into 96-well tissue culture plates 24 h before transfection. Transfections were performed with SuperFect (Qiagen, West Sussex, U.K.); a total amount of 1 μg of DNA was used, consisting of 0.5 μg of reporter construct, the stated amount of the expression vector, and the appropriate amount of pcDNA3.1 (Invitrogen) to keep the total amount of DNA constant. Samples were analyzed by dual luciferase assay (Promega) 24–30 h after transfection.
In all cases, data shown are from one of two to four independent experiments with similar qualitative results. Data from experiments performed in triplicate are expressed as means ± SD.
Results and Discussion
We were interested in finding previously unidentified members of the IL-1R/TLR family, and using profilesearch (Genetics Computer Group, Madison, WI), we identified a VV ORF, A46R, that was related to the family at a statistically significant level. A blast search with A46R then identified a further VV ORF as the highest scoring sequence producing significant alignment, termed A52R. The names A46R and A52R are based on the standard VV nomenclature of the Copenhagen strain (35). A46R and A52R were cloned from the laboratory VV strain WR, where they were previously called SalF9R and SalF15R, respectively (34), into the mammalian expression vector pRK5 (see Materials and Methods). Fig. 1A shows the predicted amino acid sequences of A46R and A52R from WR, together with the region of sequence similarity detected by blast. The region of the putative TIR domain in each protein determined in the alignment shown in Fig. 1B is also indicated. The predicted A46R WR protein differs from the Copenhagen version in that the former predicts a protein of 240 amino acids, whereas the latter predicts a protein with a C terminus truncated by 26 amino acids because of a missing base at position 152,701 in the Copenhagen genome (35) that creates a stop codon in frame upstream of the WR stop codon. In both strains, the predicted A52R protein has 190 amino acids, the one difference being an F-to-S substitution in Copenhagen at amino acid position 57 (35).
Figure 1.
Identification of A46R and A52R as potential members of the IL-1 receptor/TLR family. (A) Predicted amino acid sequence of A46R and A52R. The region of sequence similarity detected in a blast search with A46R is boxed. Identical amino acids are indicated by lines, and conservative substitutions are indicated by dots. The regions aligned with the family TIR domain in B are underlined. (B) Sequence comparison of the TIR domain of IL-1 receptor/TLR family members with A52R and A46R. For clarity, only those family members referred to in this article are shown. IL-1Rrp is the IL-18 receptor. Three conserved regions thought to be important in signaling are indicated by boxes.
Fig. 1B shows an alignment of the putative TIR domains of A46R and A52R with the TIR domain from some known IL-1R/TLR family members, abridged for clarity from a larger alignment; 33 family members were aligned by using pileup (Genetics Computer Group), and a consensus sequence was generated by using pretty (Genetics Computer Group). The consensus sequence is based on the criterion of a residue appearing in 15 of 33 of the family members. The TIR domain is a hallmark of the family and is likely to mediate homotypic interactions with other TIR domain-containing proteins (25, 27). With reference to the alignment, although there are other regions of similarity that appear in the consensus sequence, the three boxed regions shown, termed boxes 1, 2, and 3, are particularly important. Box B1 is a signature sequence of the family, and boxes 2 and 3 contain amino acids shown to be important in signaling, based on mutational analysis of mainly IL-1R1 (36). Recently, it has also been shown that a single P-to-H mutation in box 2 of TLR4 renders mice insensitive to lipopolysaccharide (15, 16). A46R and A52R display sequence similarity with boxes 1 and 3 in particular, but the alignment predicts an absence of box 2. Box B1 is particularly strong in A46R, the sequence DTFISY being as closely related to the box 1 consensus as other proven family members. A52R has two putative box 1 sequences, DKFTVT, which aligns with the other box 1 regions from the family, and also ADNFIDY, just upstream of its box 3, whose position may be spatially conserved at the tertiary structure level. Box B3 is more marked in A52R compared with A46R, with VWRN fitting the broader box 3 residue consensus of hydrophobic-W-basic-basic.
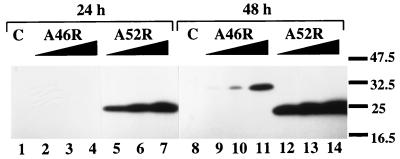
Given the sequence similarity of the viral ORFs with the TIR domain, the similarity between A46R and A52R, and the fact that the region in the VV genome from which these ORFs are expressed is rich in immunomodulatory genes (3, 34), we hypothesized that these ORFs might represent a previously unidentified VV strategy directed against IL-1/TLR signaling. To confirm that the ORFs were capable of expressing stable proteins in mammalian cells, an expression plasmid encoding either C-terminal Flag-tagged A46R or A52R was introduced into human 293 cells in 100-mm culture dishes. Fig. 2 shows that subsequent immunoblotting with anti-Flag antibody 24 h and 48 h after transfection revealed bands of the predicted molecular masses of 28.5 and 23.6 kDa for A46R-Flag and A52R-Flag, respectively. Compared with A52R, expression of A46R was not detectable at 24 h, but at 48 h, expression of both ORFs was measurable and dose-dependent, with 3 μg of A52R-Flag DNA leading to greater protein expression than 12 μg of A46R-Flag DNA (Fig. 2, compare lane 12 to lane 11). This result suggested that A52R accumulated intracellularly to a much greater degree than A46R.
Figure 2.
Ectopic expression of epitope-tagged A46R and A52R in mammalian cells. 293 cells were transfected with equal amounts of DNA (12 μg) comprising empty vector (lanes 1 and 8); 3, 6, or 12 μg of A46R-Flag (lanes 2–4 and 9–11); or 3, 6, or 12 μg A52R-Flag (lanes 5–7 and 12–14). Proteins were then detected by immunoblotting either 24 h (lanes 1–7) or 48 h (lanes 8–14) later by using an anti-Flag antibody. The relevant molecular mass markers (in kDa) are shown on the right.
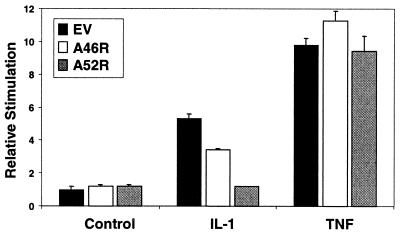
To test whether A46R and A52R were capable of antagonizing IL-1 signaling, we examined their ability to inhibit IL-1-induced NFκB activation in a reporter gene assay. Fig. 3 shows that incubation of 293 cells with 100 ng/ml IL-1α for 6 h led to a 5-fold stimulation of NFκB activation. Cells were transfected with either A46R or A52R for 48 h, because, at this time, expression of both proteins was measurable (Fig. 2). The concentrations of DNA used were within the range tested in Fig. 2 but scaled down for 24-well plate assays. A46R had a marginal inhibitory effect on IL-1-stimulated NFκB activation (Fig. 3) but did not affect basal levels of reporter gene expression. A52R had a more profound effect, abolishing the effect of IL-1, even though four times less DNA was used (compared with A46R) to compensate for its stronger expression profile. TNF activates a signal transduction pathway distinct from IL-1 and does not require MyD88. Neither A46R nor A52R had an inhibitory effect on TNF-mediated NFκB activation. We tested a range of plasmid doses for both A46R and A52R. The effect of A52R on IL-1 was dose-dependent, with the dose of plasmid shown having the optimal effect, and A46R did not show further inhibition than that shown in Fig. 3 (not shown).
Figure 3.
Effect of A46R and A52R on IL-1 signaling. 293 cells were transfected with 600 ng of empty vector (EV; black bars), 600 ng of vector encoding A46R (white bars), or 150 ng of vector encoding A52R (gray bars) for 48 h. At 6 h before harvesting, cells were stimulated with 100 ng/ml IL-1α or TNFα. NFκB reporter gene activity was then measured.
Thus A52R, when expressed in mammalian cells, is capable of interfering with host immune signaling, possibly by interfering with homotypic TIR domain interactions. Inhibition by A52R was more potent than that by A46R. Although similar at the amino acid level (Fig. 1), fold-prediction analysis programs suggest important differences in secondary structure between A46R and A52R (not shown). In addition, A52R is expressed more strongly. These factors may explain the difference in potency between the two expressed ORFs. Differences in structure between A46R and A52R may be important in targeting the proteins to different TIR domain-containing signaling molecules. Because of its greater potency, we next focused on the effect of A52R on IL-1RI signaling.
To investigate further the inhibition of signaling by A52R, we compared the inhibitory effects of A52R with those of ΔMyD88, which contains only the C-terminal TIR domain of MyD88 and acts as a dominant negative of IL-1-mediated NFκB activation (26). These experiments were performed 24 h after transfection, because expression of both A52R (Fig. 2) and ΔMyD88 (not shown) was strong at this time. Fig. 4A shows that both A52R and ΔMyD88 inhibited IL-1-induced NFκB activation with comparable potency. Activation of NFκB by TNF was insensitive to ΔMyD88, whereas A52R had little effect (Fig. 4A). Next, we examined the effect of both inhibitors on activation of NFκB by overexpression of IL-1 signal intermediates. Fig. 4B shows that overexpression of IL-1RI together with IL-1RAcP or of IL-1RAcP, MyD88, or IRAK alone was sufficient to drive NFκB activation. A52R was effective in inhibiting IL-1RI/IL-1RAcP-, IL1RAcP-, and MyD88-mediated NFκB activation but had no effect on IRAK-mediated activation (Fig. 4B Upper). These results are consistent with a TIR-dependent mechanism of antagonism, because IRAK acts downstream of MyD88 and IL-1RI and IL-1RAcP act upstream of it (25–27). ΔMyD88 potently antagonized IL-1RI/IL-1RAcP- or IL-1RAcP-induced NFκB but showed only marginal inhibition of wild-type MyD88 and no inhibitory effect on IRAK, consistent with a mechanism of action of preventing MyD88 binding to receptor complexes (26, 27). Where inhibition by A52R and ΔMyD88 was observed, these effects were confirmed to be dose-dependent, optimal doses being shown.
Figure 4.
A52R and ΔMyD88 inhibit IL-1 signaling to NFκB. (A) Both A52R and ΔMyD88 inhibit the IL-1 but not the TNF pathway to NFκB. 293 cells were transfected with 300 ng of empty vector (EV; black bars), A52R (white bars), or ΔMyD88 (gray bars) for 24 h. At 6 h before harvesting, cells were stimulated with 100 ng/ml IL-1α or TNFα. NFκB reporter gene activity was then measured. (B) 293 cells were transfected with vectors encoding IL-1 signaling intermediates (150 ng each of IL-1RI and IL-1RAcP, 300 ng of IL-1RAcP or MyD88, or 150 ng of IRAK) together with 300 ng (or 450 ng for IRAK) of a vector encoding either A52R (Upper) or ΔMyD88 (Lower) for 24 h. NFκB reporter gene activity was then measured. (C) A52R inhibits IL-1-induced IL-8 promoter activation but not GH-induced lactogenesis hormone response element activation. (Left) HeLa cells were transfected with 10 ng of IL-1R1 and IL-1RAcP or MyD88 in the presence of 100 ng of empty vector (black bars) or vector encoding A52R (white bars). After 24–30 h, IL-8 promoter activity was measured by a reporter gene assay. (Right) HeLa cells were transfected with 100 ng of empty vector (black bar) or vector encoding A52R (white bar) 18 h before stimulation with 1 ng/ml growth hormone (GH) for 6 h. Lactogenesis hormone response element activity was measured by a reporter gene assay. Data are expressed as stimulation by GH over control in the absence or presence of A52R.
It was interesting that A52R could inhibit activation of NFκB induced by MyD88 overexpression, but ΔMyD88 could not, suggesting that A52R acts at the level of MyD88 whereas ΔMyD88 might act upstream of it. MyD88 self-associates in vitro, and activation of NFκB by ectopic expression of MyD88 probably requires MyD88 dimerization (25). Thus, ΔMyD88, which contains only the TIR domain, may be unable to affect these interactions, which involve the death domain of MyD88. Alternatively, A52R might be effective in either disrupting dimerization or sequestering MyD88. Further experiments will have to be carried out to test this hypothesis. A46R failed to inhibit MyD88-driven NFκB activation but had some inhibitory effect on IL-1RAcP (not shown), consistent with the data from Fig. 3. Thus, it may have been acting on the IL-1 pathway at a point upstream of MyD88, for example, by interfering with the formation of the IL-1RI/IL-1RAcP/MyD88 complex.
Further evidence for the ability of A52R to inhibit TIR-dependent and not other signaling pathways was obtained from reporter gene studies in HeLa cells. We tested the ability of A52R to inhibit an IL-8 promoter-dependent reporter gene. Induction of IL-8 is a downstream consequence of activation of both the IL-1 and TLR4 pathways (13, 37). Fig. 4C shows that the induction of this promoter by ectopic expression of either IL-1RI and IL-1RAcP or MyD88 was potently inhibited by coexpression of A52R, whereas the relative induction of a reporter gene linked to the STAT5-dependent lactogenesis hormone response element (33) by the recombinant human GH genotropin, which is not TIR-dependent, was the same in the presence and absence of A52R.
We next addressed whether other TIR-dependent pathways to NFκB would also be inhibited by A52R. Given the emerging importance of TLR4 in host responses to injury and infection (15–18) and the role of MyD88 in this pathway (19, 20, 24), we compared the ability of A52R and ΔMyD88 to impair TLR4 signaling. Fig. 5A shows that ectopic expression of TLR4 led to activation of NFκB as has been shown (19). This activation was dose-dependent (not shown) and was not observed on overexpression of ΔTLR4 (not shown), which lacks most of the cytoplasmic portion of the receptor including the TIR domain. Both A52R and ΔMyD88 potently blocked TLR4-mediated NFκB activation (Fig. 5A), optimal doses being shown. Another TIR-dependent signaling pathway, IL-18 (8), was also antagonized by A52R. Fig. 5B shows that, when 293 cells were transfected with AcPL to sensitize them to IL-18, subsequent stimulation with IL-18 was sensitive to both A52R and ΔMyD88, although the effect of A52R was not as marked as that observed against IL-1R1 or TLR-4. A52R may therefore also act to block the antiviral cytokine IL-18 (38).
Figure 5.
Inhibition of TLR4 and IL-18 signaling by A52R and ΔMyD88. (A) 293 cells were cotransfected with 150 ng of a vector encoding TLR4 together with 450 ng of empty vector (EV; black bars), A52R (white bars), or ΔMyD88 (gray bars) for 24 h. NFκB reporter gene activity was then measured. (B) 293 cells were cotransfected with 300 ng of a vector encoding AcPL together with 300 ng of empty vector (black bars), A52R (white bars), or ΔMyD88 (gray bars) for 48 h. At 6 h before harvesting, cells were stimulated with 20 ng/ml IL-18. NFκB reporter gene activity was then measured.
Hence, A52R is capable of antagonizing MyD88-dependent signaling by IL-1, TLR4, and IL-18 but has little effect on MyD88-independent pathways triggered by TNF and GH. Further, A52R effectively mimicked the effect of ΔMyD88, a known dominant-negative inhibitor of IL-1/TLR signaling, that structurally would be comparable to A52R in that it consists largely of a TIR domain (26). In fact, A52R was capable of inhibiting NFκB activation induced by MyD88 overexpression, whereas ΔMyD88 had only a slight effect. The particular intracellular targets of A46R and A52R together with their effects on other TIR-dependent signaling pathways will have to be investigated further. To our knowledge, our study of A46R and A52R represents the first demonstration of a specific viral inhibitory effect on intracellular IL-1/TLR signaling. The advantage of A52R in particular for VV would be that it could act to block multiple stimuli (IL-1, IL-18, and activators of TLRs) that use receptors with TIR domains. Therefore, A52R could have a broader effect on host defense. In contrast, viral cytokine decoy receptors such as the soluble TNF receptor would be specific for a single pathway.
In conclusion, A46R and A52R are likely to be useful tools in further defining the importance of TIR-domain-dependent signaling pathways in the host response to injury and infection, particularly with regard to viral pathogenesis.
Acknowledgments
We are grateful to Prof. W. Falk, Dr. M. Muzio, Prof. C. Dinarello, Dr. F. Volpe, Dr. R. Hofmeister, and Tularik for providing various plasmids.
Abbreviations
- VV
vaccinia virus
- TLR
Toll-like receptor
- TIR
Toll/IL-1 receptor
- TNF
tumor necrosis factor
- IL-1RI
type I IL-1 receptor
- IL-1RAcP
IL-1 receptor accessory protein
- IRAK
IL-1 receptor-associated kinase
- AcPL
IL-18 receptor accessory protein
- WR
Western Reserve
- GH
growth hormone
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.160027697.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.160027697
References
- 1.Moss B. In: Virology. Fields B N, Knipe D M, Howley P M, Chanock R M, Melnick J, Monath T P, Roizman B, Straus S E, editors. Vol. 2. New York: Lippincott–Raven; 1996. pp. 2637–2671. [Google Scholar]
- 2.Alcamí A, Symons J A, Khanna A, Smith G L. Semin Virol. 1998;5:419–427. [Google Scholar]
- 3.Smith G L, Symons J A, Alcamí A. Semin Virol. 1998;8:409–418. [Google Scholar]
- 4.Alcamí A, Smith G L. Proc Natl Acad Sci USA. 1996;93:11029–11034. doi: 10.1073/pnas.93.20.11029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.O'Neill L A J, Greene C. J Leukocyte Biol. 1998;63:650–657. [PubMed] [Google Scholar]
- 6.Stylianou E, O'Neill L A J, Rawlinson L, Edbrooke M R, Woo P, Saklatvala J. J Biol Chem. 1992;267:15836–15841. [PubMed] [Google Scholar]
- 7.Lemaitre B, Nicolas E, Michaut L, Reichhart J M, Hoffman J A. Cell. 1996;86:973–983. doi: 10.1016/s0092-8674(00)80172-5. [DOI] [PubMed] [Google Scholar]
- 8.Hoshino K, Tsutsui H, Kawai T, Takeda K, Nakanishi K, Takeda Y, Akira S. J Immunol. 1999;162:5041–5044. [PubMed] [Google Scholar]
- 9.Coyle A J, Lloyd C, Tian J, Nguyen T, Erikkson C, Wang L, Ottoson P, Persson P, Delaney T, Lehar S, et al. J Exp Med. 1999;190:895–902. doi: 10.1084/jem.190.7.895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hoshino K, Kashiwamura S, Kuribayashi K, Kodama T, Tsujimura T, Nakanishi K, Matsuyama K, Akira S. J Exp Med. 1999;190:1541–1547. doi: 10.1084/jem.190.10.1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Townsend M J, Fallon P G, Matthews D J, Jolin H E, McKenzie A N J. J Exp Med. 2000;191:1069–1075. doi: 10.1084/jem.191.6.1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rock F L, Hardiman G, Timans J C, Kastelein R A, Bazan F. Proc Natl Acad Sci USA. 1998;95:588–593. doi: 10.1073/pnas.95.2.588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Medzhitov R, Preston-Hurlburt P, Janeway C A. Nature (London) 1997;388:394–397. doi: 10.1038/41131. [DOI] [PubMed] [Google Scholar]
- 14.Yang R B, Mark M R, Gray A, Huang A, Xie M H, Zhang M, Goddard A, Wood W I, Gurney A L, Godowski P J. Nature (London) 1998;395:284–288. doi: 10.1038/26239. [DOI] [PubMed] [Google Scholar]
- 15.Poltorak A, He X, Smirnova I, Lie M Y, Huffel C V, Du X, Birdwell D, Alejos E, Silva M, Galanos C, et al. Science. 1998;282:2085–2088. doi: 10.1126/science.282.5396.2085. [DOI] [PubMed] [Google Scholar]
- 16.Qureshi S T, Lariviere L, Leveque G, Clermont S, Moore K J, Gros P, Malo D. J Exp Med. 1999;189:615–625. doi: 10.1084/jem.189.4.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoshino K, Takeuchi O, Kawai T, Sanjo H, Ogawa T, Takeda Y, Takeda K, Akira S. J Immunol. 1999;162:3749–3752. [PubMed] [Google Scholar]
- 18.Frantz S, Kobzik L, Kim Y D, Fukazawa R, Medzhitov R, Lee R T, Kelly R A. J Clin Invest. 1999;104:271–280. doi: 10.1172/JCI6709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Muzio M, Natoli G, Saccani S, Levrero M, Mantovani A. J Exp Med. 1998;187:2097–2101. doi: 10.1084/jem.187.12.2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang F X, Kirschning C J, Mancinelli R, Xu X-P, Jin Y, Faure E, Mantovani A, Rothe M, Muzio M, Arditi M. J Biol Chem. 1999;274:7611–7614. doi: 10.1074/jbc.274.12.7611. [DOI] [PubMed] [Google Scholar]
- 21.Greenfeder S A, Nunes P, Kwee L, Labow M, Chizzonite P A, Ju G. J Biol Chem. 1995;270:13757–13765. doi: 10.1074/jbc.270.23.13757. [DOI] [PubMed] [Google Scholar]
- 22.Wesche H, Korherr C, Kracht M, Falk W, Resch K, Martin M U. J Biol Chem. 1997;272:7727–7731. doi: 10.1074/jbc.272.12.7727. [DOI] [PubMed] [Google Scholar]
- 23.Adachi O, Kawai T, Takeda K, Matsumoto M, Tsutsui H, Sakagami M, Nakanishi K, Akira S. Immunity. 1998;9:143–150. doi: 10.1016/s1074-7613(00)80596-8. [DOI] [PubMed] [Google Scholar]
- 24.Kawai T, Adachi O, Ogawa T, Takeda K, Akira S. Immunity. 1999;11:115–122. doi: 10.1016/s1074-7613(00)80086-2. [DOI] [PubMed] [Google Scholar]
- 25.Burns K, Martinon F, Esslinger C, Pahl H, Schneider P, Bodmer J-L, Di Marco F, French L, Tschopp J. J Biol Chem. 1998;273:12203–12209. doi: 10.1074/jbc.273.20.12203. [DOI] [PubMed] [Google Scholar]
- 26.Muzio M, Ni J, Feng P, Dixit V M. Science. 1997;278:1612–1615. [Google Scholar]
- 27.Wesche H, Henzel W, Shillinglaw W, Li S, Cao Z. Immunity. 1997;7:837–847. doi: 10.1016/s1074-7613(00)80402-1. [DOI] [PubMed] [Google Scholar]
- 28.Cao Z, Henzel W, Gao X. Science. 1996;271:1128–1131. doi: 10.1126/science.271.5252.1128. [DOI] [PubMed] [Google Scholar]
- 29.Malinin N L, Boldin M P, Kovalenko A V, Wallach D. Nature (London) 1997;385:540–544. doi: 10.1038/385540a0. [DOI] [PubMed] [Google Scholar]
- 30.Baud V, Liu Z-G, Bennett B, Suzuki N, Xia Y, Karin M. Genes Dev. 1999;13:1297–1308. doi: 10.1101/gad.13.10.1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kopp E, Medzhitov R, Carothers J, Xiao C, Douglas I, Janeway C A, Ghosh S. Genes Dev. 1999;13:2059–2071. doi: 10.1101/gad.13.16.2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mitcham J L, Parnet P, Bonnert T P, Garka K E, Gerhart M J, Slack J L, Gayle M A, Dower S K, Sims J E. J Biol Chem. 1996;271:5777–5783. doi: 10.1074/jbc.271.10.5777. [DOI] [PubMed] [Google Scholar]
- 33.Maamra M, Finidori J, Von Laue S, Simon S, Justice S, Webster J, Dower S, Ross R. J Biol Chem. 1999;274:14791–14798. doi: 10.1074/jbc.274.21.14791. [DOI] [PubMed] [Google Scholar]
- 34.Smith G L, Chan Y S, Howard S T. J Gen Virol. 1991;72:1349–1376. doi: 10.1099/0022-1317-72-6-1349. [DOI] [PubMed] [Google Scholar]
- 35.Goebel S J, Johnson G P, Perkus M E, Davis S W, Winslow J P, Paoletti E. Virology. 1990;179:247–266. doi: 10.1016/0042-6822(90)90294-2. [DOI] [PubMed] [Google Scholar]
- 36.Slack J L, Schooley K, Bonnert T P, Mitcham J L, Qwarnstrom E E, Sims J E, Dower S K. J Biol Chem. 2000;275:4670–4678. doi: 10.1074/jbc.275.7.4670. [DOI] [PubMed] [Google Scholar]
- 37.Dinarello C A. Blood. 1996;87:2095–2147. [PubMed] [Google Scholar]
- 38.Novick D, Kim S H, Fantuzzi G, Reznikov L L, Dinarello C A, Rubinstein M. Immunity. 1999;10:127–136. doi: 10.1016/s1074-7613(00)80013-8. [DOI] [PubMed] [Google Scholar]