Abstract
FLOWERING LOCUS T (FT) plays a key role as a mobile floral induction signal that initiates the floral transition. Therefore, precise control of FT expression is critical for the reproductive success of flowering plants. Coexistence of bivalent histone H3 lysine 27 trimethylation (H3K27me3) and H3K4me3 marks at the FT locus and the role of H3K27me3 as a strong FT repression mechanism in Arabidopsis have been reported. However, the role of an active mark, H3K4me3, in FT regulation has not been addressed, nor have the components affecting this mark been identified. Mutations in Arabidopsis thaliana Jumonji4 (AtJmj4) and EARLY FLOWERING6 (ELF6), two Arabidopsis genes encoding Jumonji (Jmj) family proteins, caused FT-dependent, additive early flowering correlated with increased expression of FT mRNA and increased H3K4me3 levels within FT chromatin. Purified recombinant AtJmj4 protein possesses specific demethylase activity for mono-, di-, and trimethylated H3K4. Tagged AtJmj4 and ELF6 proteins associate directly with the FT transcription initiation region, a region where the H3K4me3 levels were increased most significantly in the mutants. Thus, our study demonstrates the roles of AtJmj4 and ELF6 as H3K4 demethylases directly repressing FT chromatin and preventing precocious flowering in Arabidopsis.
Introduction
Flowering, a critical developmental transition in plants, is controlled by both environmental cues and internal developmental signals. Photoperiod exerts profound effects on flowering in numerous plant species including Arabidopsis. Generation of the photoperiodic floral induction signal in the leaves is mediated by light and circadian-clock signaling and relayed through the photoperiod pathway. GIGANTEA (GI) [1], [2] and CONSTANS (CO) [3] act as upstream activators of FLOWERING LOCUS T (FT) [4], [5] in the photoperiod pathway. On the other hand, FT expression is repressed by FLOWERING LOCUS C (FLC) [1], [2], and this repression is mediated possibly by a protein complex between FLC and SHORT VEGETATIVE PHASE [6]. Thus, FT acts not only as a component in the photoperiod pathway but also as a floral integrator that combines the perception of inductive photoperiods and the FLC-mediated floral repression signal. FT protein, as a graft-transmissible signal, is translocated from the vascular tissue of leaves to the shoot apex [7], where it interacts with FD and stimulates the floral transition [8], [9].
Recent studies have shown that FT expression is affected by histone modifications. The FT locus was shown to be enriched with trimethylated histone H3 lysine 27 (H3K27me3) [10], [11], and loss of putative Polycomb Repressive Complex 2 (PRC2) components results in decreased H3K27me3 within FT chromatin, which in turn increases FT expression [12]. Furthermore, lack of LIKE-HETEROCHROMATIN PROTEIN1 (LHP1), which can bind to H3K27me3 and silence chromatin [10], [11], also causes increased FT expression [13], [14]. Therefore, FT transcription is repressed by H3K27me3 and its effector protein (LHP1).
Methylation at histone residues can contribute to mitotically stable epigenetic changes in gene expression. In contrast it has recently been demonstrated that at least two classes of enzymes are capable of removing methyl groups from either histone lysine or arginine (R) residues and potentially reversing epigenetic changes in gene expression. Human Lysine-Specific Demethylase1 (LSD1), a nuclear amine oxidase, specifically demethylates mono- and dimethylated but not trimethylated H3K4 [15]. After the discovery of LSD1, a human Jmj C domain-containing protein, JHDM1A, was first shown to be able to remove methyl groups from H3K36 [16]. Soon after the identification of JHDM1A, a number of JmjC domain-containing proteins have been demonstrated to be H3K4, H3K9, H3K27, H3K36, H3R2, and H4R3 demethylases [17]–[19]. Unlike LSD1, JmjC domain-containing proteins are capable of demethylating all of the mono-, di- and trimethylated lysines of histones [20]. Thus, JmjC family proteins are considered as the major histone demethylases in eukaryotic cells.
Arabidopsis has twenty-one genes encoding JmjC family proteins (Arabidopsis thaliana Jumonji (AtJmj) 1∼21) [21]. To date, three of these genes have been functionally characterized. EARLY FLOWERING6 (ELF6; AtJmj1) and RELATIVE OF EARLY FLOWERING6 (REF6; AtJmj2) were shown to be involved in photoperiodic flowering and FLC regulation, respectively [22]. INCREASED EXPRESSION OF BONSAI METHYLATION 1 (IBM1; AtJmj15), represses genic cytosine methylation, possibly through demethylation of H3K9me [23]. In this report, we show that ELF6 and another Arabidopsis JmjC family protein (AtJmj4) directly repress FT expression via demethylation of H3K4me. Thus, our study demonstrates the presence of an H3K4me demethylation-mediated mechanism in addition to the previously characterized H3K27 methylation-mediated mechanism in the chromatin repression of a key flowering time regulator, FT.
Results
Mutations in AtJmj4 Cause Early Flowering
To address the biological roles of Arabidopsis JmjC domain-containing proteins, we obtained T-DNA insertion lines of the corresponding genes from the SALK T-DNA collection and evaluated their phenotypes. Two independent homozygous T-DNA insertion mutants of Arabidopsis thaliana Jumonji4 (AtJmj4 or At4g20400) [21] showed an early flowering phenotype both in long days (LD; 16 h light/8 h dark) and short days (SD; 8 h light/16 h dark; Figures 1A–1C ). The early flowering phenotype was not due to an accelerated leaf initiation rate (Figure S1) but resulted from a more rapid developmental transition of the shoot apical meristem (SAM) from the vegetative to the reproductive phase as characterized by a lower number of rosette and cauline leaves at the onset of flowering ( Figure 1C ). No other visible phenotypic traits were apparent in atjmj4 mutants. Plants heterozygous for the T-DNA insertions displayed a wild-type (wt) flowering time (data not shown), indicating that atjmj4-1 and atjmj4-2 are fully recessive mutations with respect to flowering time. Because both alleles displayed similar early flowering behaviors, atjmj4-1 was selected for the genetic and molecular analyses we report herein.
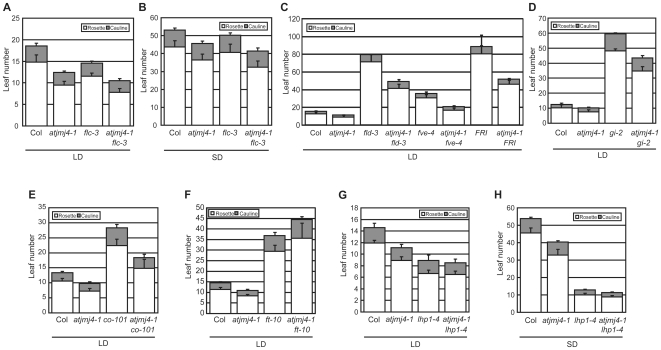
Figure 1. Early flowering of atjmj4 mutants.
A) Domain organization of AtJmj4. Domains were predicted by SMART (http://smart.embl-heidelberg.de/). Lines indicate interdomain regions. T-DNA insertion sites on the genomic sequence of AtJmj4 in atjmj4-1 and atjmj4-2 are marked on the corresponding positions of their translated protein products. B) Early flowering phenotype of atjmj4-1 mutant plants grown in either LD or SD. C) Flowering time of atjmj4 mutants. Wt Col and atjmj4 mutant plants were grown in either LD or SD and their flowering times were determined as the number of primary rosette and cauline leaves formed at bolting. At least 12 individuals were scored for each genotype. Error bars represent sd.
Early Flowering of atjmj4 Is Due to Increased Expression of FT
Because of the early-flowering phenotype of loss of AtJmj4, we evaluated whether it might have a role in the expression of key flowering time regulators, such as GI, CO, FT, FLC, and SUPPRESSOR OF OVEREXPRESSION OF CO1 (SOC1), a floral integrator [24], [25] ( Figure 2 ). In LD, the mRNA levels of GI and CO were not affected by the atjmj4-1 mutation. However, the mRNA levels of the floral integrators FT and SOC1 were up-regulated in atjmj4-1 and that of FLC was slightly reduced ( Figure 2A ). To further explore these observations, we monitored the expression of these genes in SD ( Figure 2B ). FT mRNA levels were consistently higher in the atjmj4-1 mutants in SD, but the expression of SOC1 mRNA was not affected significantly. Because FT induction by CO requires the stabilization of CO protein by light, which occurs only in LD in Arabidopsis, the up-regulation of FT by the atjmj4-1 mutation in SD might be caused by de-repression instead of induction. The mRNA level of FLC was slightly reduced in the atjmj4-1 mutants in SD compared to wt as was the case in LD.
Figure 2. Increased expression of FT in atjmj4 mutants.
A and B) Expression of flowering genes in atjmj4-1 mutants. Col and atjmj4-1 plants were grown in LD (A) for 10 days (d) or in SD (B) for 15 d, harvested every 4 hours (h) at indicated zeitgeber (ZT; h after light-on) for one d, and used for RT-PCR analyses. Ubiquitin (UBQ) was included as an expression control. Identical results were obtained from two independent experiments, and one of them is shown. White and black bars represent light and dark periods, respectively. C and D) Temporal expression of flowering genes in atjmj4-1 mutants. Col and atjmj4-1 plants were grown for up to 12 days after germination (DAG) in LD (C) or 18 DAG in SD (D). Plants were harvested during the growth period at ZT14 (LD) or ZT8 (SD) of designated DAG and used for RT-PCR analyses because FT mRNA expression peaked at the ZT in each photoperiod. E) Histochemical GUS staining of transgenic plants harboring FT::GUS fusion construct in Col or atjmj4-1 plants. Plants were grown for 16 d either in LD or SD before GUS staining.
We then compared the mRNA levels of FLC, FT, and SOC1 in wt versus atjmj4-1 at different developmental stages both in LD and SD ( Figures 2C and 2D ). Notably, FT mRNA levels were higher in atjmj4-1 than in wt throughout the developmental stages tested. Moreover, the FT promoter showed increased activity both in LD and SD in the atjmj4-1 homozygous mutants when the construct containing the FT promoter fused with β-glucuronidase (FT::GUS) [14] was compared to expression levels in wt Col ( Figure 2E ). FT::GUS expression which was detected in the marginal minor veins of wt leaves was also observed in the central minor veins of atjmj4-1 mutant leaves in LD. In SD, FT::GUS expression was robust in the major veins of atjmj4-1 mutant leaves, although its expression was weak in the same wt leaf tissue. Thus, the atjmj4 mutation leads to increased expression of FT mRNA through enhanced activity of FT promoter. However, the cell-type specific expression pattern of FT in leaf veins was not affected by the atjmj4 mutation.
Because the atjmj4 mutation also caused a slight reduction in FLC mRNA levels ( Figures 2A–2D ), we tested whether the decrease in FLC expression was a cause for the increased expression of FT in the mutants. For this, we first compared FT mRNA levels between an flc null mutant (flc-3) [26] and atjmj4-1 (Figure S2). Although the expression levels of GI, CO, and SOC1 mRNAs were similar in the two genotypes, FT mRNA level was clearly higher in atjmj4-1. Furthermore, when we compared the flowering times between atjmj4-1 single and atjmj4-1 flc-3 double mutants both in LD and SD, the double mutants flowered earlier than the single mutants in both photoperiodic conditions ( Figures 3A and 3B ). Because vernalization can reduce FLC expression and VERNALIZATION INSENSITIVE3 (VIN3) is essential for the process [27]–[29], we tested a possibility of constitutive vernalization response in atjmj4 using atjmj4 single and atjmj4 vin3 double mutants. atjmj4 showed a normal response to vernalization, whereas atjmj4 vin3 did not (Figure S3). Further, atjmj4 vin3 flowered earlier than vin3 without or with vernalization (Figure S3). Therefore, data above altogether indicate that the atjmj4 mutation causes an early flowering independently of FLC expression.
Figure 3. Genetic interaction between Atjmj4 and other flowering time genes.
A and B) Flowering time of atjmj4-1 flc-3 double mutants. C) Genetic interaction between AtJmj4 and FLC regulators. D to F) Genetic interaction between AtJmj4 and photoperiod-pathway genes. G and H) Genetic interaction between AtJmj4 and LHP1. Flowering times were determined in LD (A and C to G) or SD (B and H). At least 12 individuals were scored for each genotype (A to H). Error bars represent sd (A to H).
That AtJmj4 acted downstream of FLC was reinforced by our studies on genetic interactions between atjmj4 and several autonomous-pathway mutants [30] ( Figure 3C ). Double mutants between atjmj4 and flowering locus d (fld) [31] or fve [32] exhibited flowering times that were intermediate relative to each single mutant. When a functional FRIGIDA (FRI) [33], [34] allele was introduced into atjmj4, atjmj4-1 FRI also flowered at intermediate time between atjmj4-1 and FRI. These results indicate that AtJmj4 controls flowering mainly through an FLC-independent pathway.
To test interactions between AtJmj4 and genes acting in the photoperiod pathway the following double mutants were analyzed: atjmj4-1 gi-2, atjmj4-1 co-101, and atjmj4-1 ft-10. The early-flowering phenotype of atjmj4-1 was attenuated by the LD-specific late flowering phenotypes of gi-2 and co-101 ( Figures 3D and 3E ). However, the early flowering of atjmj4-1 was fully suppressed by the ft-10 mutation ( Figure 3F ). Therefore, AtJmj4 might affect FT expression independently of GI and CO. Consistent with this hypothesis, FT mRNA level in atjmj4-1 gi-2 or atjmj4-1 co-101 double mutants was higher compared to that in gi-2 or co-101 single mutants, respectively (Figure S4). LHP1 directly represses FT chromatin [10], [11] such that FT is strongly de-repressed in lhp1 mutants [13]. Consistent with the repressive roles of AtJmj4 and LHP1 in FT expression, atjmj4-1 lhp1-4 double mutants flowered at similar times with the severe early flowering mutant lhp1-4 in both LD and SD ( Figures 3G and 3H ).
The data above indicated that AtJmj4 acts as an FT repressor, and thus it was of interest to test if AtJmj4 affects FT expression indirectly through controlling the expression of FT regulators. To address this, we compared the mRNA levels of known FT regulators, namely TARGET OF EAT1 (TOE1), TOE2, TOE3 [35], [36], SCHLAFMÜTZE (SMZ), SCHNARCHZAPFEN (SNZ) [35], [37], CRYPTOCHROME-INTERACTING BASIC-HELIX-LOOP-HELIX1 (CIB1) [38], TEMPRANILLO1 (TEM1), TEM2 [39], SHORT VEGETATIVE PHASE (SVP) [6], [40], [41], AGAMOUS-like15 (AGL15), and AGL18 [42], between wt and atjmj4 mutants, but none showed detectable differences (Figure S5).
AtJm4 Is a Nuclear Protein Preferentially Expressed in Vascular Tissues and Shoot/Root Apices
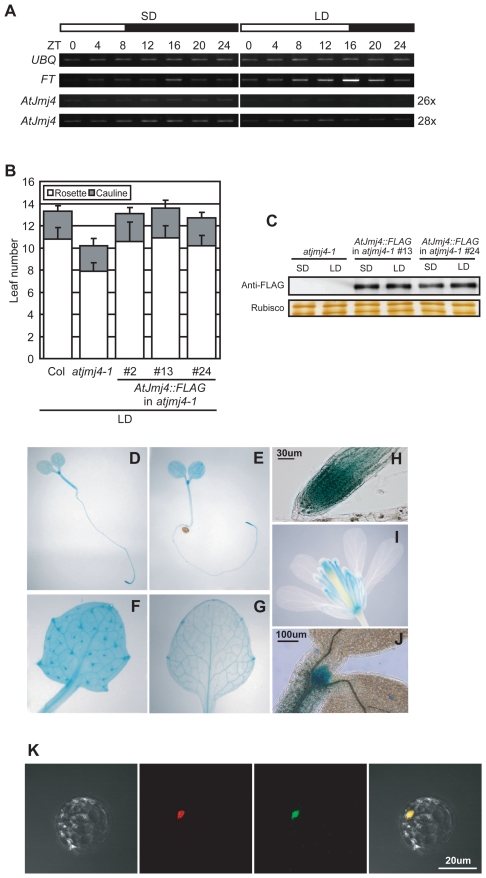
When we compared the mRNA levels of AtJmj4 between SD- and LD-grown seedlings at a similar developmental stage, we could observe higher expression of AtJmj4 mRNA in SD-grown seedlings, although the FT mRNA levels were clearly higher in LD-grown seedlings ( Figure 4A ). This observation was consistent with the higher AtJmj4 promoter activity in SD than in LD as studied by using an AtJmj4 promoter::GUS fusion construct [21]. This might indicate a preferential repressive role of AtJmj4 in FT expression in SD, and thus we further evaluated whether the expression level of AtJmj4 protein was also higher in SD than in LD. For this, we made an AtJmj4::FLAG fusion construct that contained an AtJmj4 promoter fragment, 3 copies of the FLAG tag, and the AtJmj4 cDNA with the entire coding sequence. The construct fully rescued the early-flowering phenotype of atjmj4-1 when introduced into the mutants ( Figure 4B ). When we measured the expression levels of the AtJmj4::FLAG fusion protein in transgenic plants at the same developmental stage with the one used to study the expression of AtJmj4 mRNA ( Figure 4A ), however, there was no difference in the expression level between SD- and LD-grown seedlings ( Figure 4C ). Thus, AtJmj4 expression might be further controlled at posttranscriptional level(s), although its promoter activity per se is affected by day-length, and AtJmj4 protein exerts its repressive role for FT in both LD and SD.
Figure 4. AtJmj4 expression.
A) mRNA expression of AtJmj4 in SD and LD. Wt Col plants were grown in SD for 12 d or in LD for 8 d and used for RT-PCR analyses. UBQ was used as an expression control. Number of PCR cycles used for AtJmj4 is indicated on the right. B) Genomic complementation of atjmj4-1. Three independent transgenic lines of atjmj4-1 containing AtJmj4::FLAG (see text for details) were grown in LD and their flowering times were determined as the number of rosette and cauline leaves formed at bolting. At least 12 individuals were scored for each genotype. Error bars represent sd. C) Expression of the AtJmj4::FLAG fusion protein in SD and LD. Plants of atjmj4-1 and two of the complementation lines shown in (B) were grown for 12 d in SD or 8 d in LD, harvested at ZT12, and used for Western blot analyses. Upper panel: Western blot with anti-FLAG antibody. Lower panel: Silver stained gel image of rubisco subunits. D to J) Histochemical GUS staining of transgenic Col plants harboring AtJmj4::GUS. Plants grown in LD (D and I) or SD (E, F, G, H, and J) were used for GUS staining. (D) In 4 d-old seedling. (E) In 6-d old seedling. (F) In trichomes. (G) In leaf. (H) In root tip. (I) In floral organs. (J) In shoot apex. K) Subcellular localization of AtJmj4 in Arabidopsis mesophyll protoplast. From left to right; bright-field image, LHP1::RFP fusion protein, AtJmj4::GFP fusion protein, merged image of the left three images.
Because AtJmj4 protein levels are constant at the whole plant level despite day-length dependent mRNA levels, we studied the spatial expression pattern of AtJmj4 protein using a construct harboring the entire genomic region of AtJmj4 including 1.5 kb promoter in frame with GUS (AtJmj4::GUS). In seedlings, AtJmj4::GUS expression pattern was similar in SD and LD ( Figures 4D and 4E ). The GUS activity was detected in most organs, but strong activity was observed in the shoot apex ( Figure 4J ), primary root tip ( Figure 4H ), trichomes of young leaves ( Figure 4F ), and leaf vascular tissues ( Figure 4G ). In floral organs, strong GUS activity was detected in anther filaments and styles ( Figure 4I ). Importantly, the AtJmj4::GUS expression domain showed an overlap with the FT expression domain [14] in leaves.
The subcellular localization of AtJmj4 protein was evaluated by a protoplast transfection assay using a fusion protein between AtJmj4 and green fluorescence protein (GFP; AtJmj4::GFP) expressed from the Cauliflower Mosaic Virus 35S (CaMV35) promoter. A fusion protein between LHP1 and red fluorescence protein (RFP; LHP1::RFP), which was known to be localized into the nucleus [43], was co-expressed with AtJmj4::GFP. Both RFP and GFP signals were detected only in the nucleus ( Figure 4K ). This result is in agreement with the possible role of AtJmj4 as a chromatin and/or transcriptional regulator.
AtJmj4 and ELF6 Play Redundant Roles in FT Repression as H3K4-Specific Demethylases
In our previous study, we reported that ELF6 (At5g04240), a gene encoding an Arabidopsis Jmj-domain protein, acts as a repressor in the photoperiodic flowering pathway [22]. Therefore, it was of interest to study the relationship between AtJmj4 and ELF6 in the regulation of photoperiodic flowering. For this, an elf6-4 atjmj4-1 double mutant was generated and assayed for flowering time. The double-mutant plants flowered earlier than either single mutants as well as the wt Col plants both in LD and SD ( Figures 5A–5C ). Because both ELF6 and AtJmj4 have repressive roles in the photoperiod pathway, we then evaluated the mRNA levels of genes acting in the photoperiod pathway, namely GI, CO and FT, using RNAs isolated from SD-grown plants. mRNA levels of GI and CO were similar among wt, the elf6-4 and atjmj4-1 single mutants, and the elf6-4 atjmj4 double mutants at ZT4 and ZT11 ( Figure 5D ). However, FT mRNA level was increased in the elf6-4 and atjmj4-1 single mutants by at least 3 fold compared to that in wt, and this increase was more significant in the elf6-4 atjmj4-1 double mutants ( Figures 5D and 5E ). Therefore, the data for FT mRNA expression as well as the flowering time ( Figures 5A–5C ) indicate that ELF6 and AtJmj4 have redundant repressive roles in photoperiodic flowering through negatively regulating FT expression.
Figure 5. Additive effect of elf6 and atjmj4 mutations on FT-dependent early flowering.
A) Early flowering phenotype of elf6-4 atjmj4-1 double mutant. All plants were grown in SD for 63 d before taken picture. B and C) Flowering time of wt Col, elf6-4, atjmj4-1, and elf6-4 atjmj4-1 double mutants in LD and SD as determined by number of leaves formed at bolting. At least 15 individuals were scored for each genotype. Error bars represent sd. D) Expression of flowering genes in elf6-4 atjmj4 double mutants. Plants of each genotype were grown in SD for 57 d and harvested at ZT4 or ZT11 for RT-PCR analyses. Actin1 was included as an expression control. Identical results were obtained from two independent experiments and one of them is shown. E) qPCR analysis of FT expression. The same RNAs used in (D) were evaluated. The wt Col levels were set to 1 after normalization by Actin1 for qPCR analysis. Error bars represent sd.
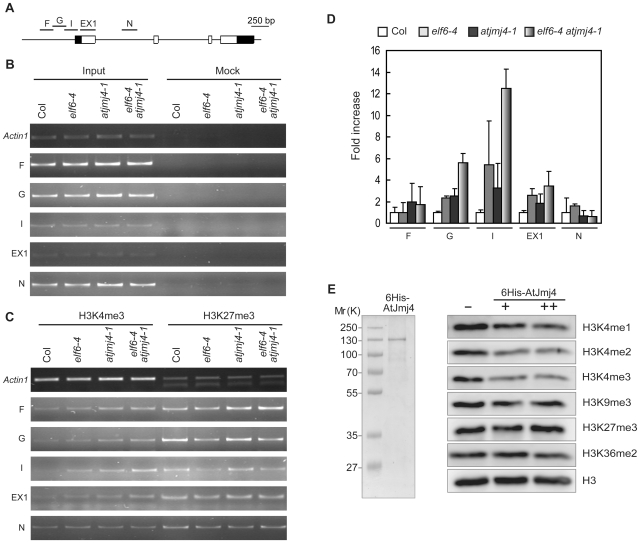
Our unpublished phylogenetic analysis of the JmjC domains of ELF6, AtJmj4, and human Jmj proteins showed that the JmjC domains of ELF6 and AtJmj4 are clustered along with the JmjC domains of human JARID1 family which is known to specifically demethylate H3K4me3 and H3K4me2 [17], [19]. Hence, we evaluated whether the level of H3K4me within FT chromatin is affected by elf6 and atjmj4 mutations using chromatin immunoprecipitation (ChIP) assays. Sets of primers covering different regions of FT locus were used ( Figure 6A ). H3K4me3 levels were increased in G, I, and EX1 regions of FT locus by the elf6-4 and atjmj4-1 mutations, and the increase was more significant when both the mutations were combined ( Figures 6B–6D ). However, H3K4me3 levels in regions F and N were not affected significantly by these mutations. The level of another histone methylation, H3K27me3, which was reported to be enriched within FT chromatin [10], [11], was slightly reduced by the elf6-4 but not by the atjmj4-1 mutation in some of the FT regions tested ( Figure 6C ). These results indicate that ELF6 and AtJmj4 repress FT expression by negatively affecting the methylation of H3K4 but not H3K27 within FT chromatin.
Figure 6. Increased trimethylation of H3K4 at FT locus by elf6 and atjmj4 mutations.
A) Schematic of FT locus showing regions (F, G, I, EX1, and N) amplified by the primers used for ChIP analysis. The front and the rear black boxes indicate 5′ and 3′ UTRs, respectively. White boxes indicate exons, while lines indicate introns and intergenic regions. B and C) ChIP assay of FT chromatin with antibody against H3K4me3 or H3K27me3. Plants of each genotype were grown in SD for 57 d and harvested for ChIP assay. ‘Input’ indicates chromatins before immunoprecipitation. ‘Mock’ refers to control samples lacking antibody. Actin1 was used as an internal control. D) qPCR analysis of the ChIP assay for H3K4me3 described in (B and C). The wt Col levels were set to 1 after normalization by input. Error bars represent sd. E) Coomassie-blue stained 6His-AtJmj4 protein purified from sf9 cells (left), and in vitro histone demethylation activity assay using the purified protein (right). Assays were performed without (−) or with either two (+) or four (++) µg of purified 6His-AtJmj4 protein. Mr (K), molecular mass in kilo-daltons.
To test if ELF6 and AtJmj4 are active histone demethylases, we tried to express the full-length ELF6 and AtJmj4 proteins in several expression systems. Although we could not express the full-length ELF6 in any systems employed, we could express the full-length AtJmj4 in insect sf9 cells as an amino-terminal 6 histidine-tagged protein (6His-AtJmj4) which had a molecular mass of 130 kilo-daltons. 6His-AtJmj4 was purified to near homogeneity ( Figure 6E ) and subjected to in vitro histone demethylase activity assay. 6His-AtJmj4-mediated histone demethylase activity was analyzed by decreased signals in western blots with antibodies specific to methylated histone H3 residues ( Figure 6E ). Incubation of the recombinant 6His-AtJmj4 protein with histone substrates in the demethylase assays resulted in reduced levels of H3K4me1, H3K4me2, H3K4me3, but not of H3K9me3, H3K36me2, and H3K27me3 ( Figure 6E ). The levels of H3K4me3 and H3K4me2 were decreased more than the level of H3K4me1. These results indicate that AtJmj4 is an intrinsic H3K4-specific demethylase which has higher activity for H3K4me3 and H3K4me2 than H3K4me1.
The results in Figure 5 and Figure 6 suggested that AtJmj4 and ELF6 might directly target FT chromatin and repress the transcription activity of FT by reducing the methylation level of H3K4. To test if FT chromatin is directly targeted by ELF6 and AtJmj4, we preformed ChIP assays using transgenic plants expressing functional ELF6::GUS [22] and AtJmj4::FLAG ( Figures 4B and 4C ) as demonstrated by the complementation of the elf6-4 and atjmj4-1 mutant phenotypes, respectively. PCR was then carried out using primers amplifying various regions of FT locus ( Figure 7A ). ELF6::GUS showed binding to broad regions of the FT locus around the transcription start site with strongest binding to region I ( Figures 7B ). However, ELF6::GUS did not show binding to region N which is a part of the first intron of FT and the transcription initiation region (−117 to +71 from the transcription start site) of CO. AtJmj4::FLAG showed a similar binding pattern with ELF6::GUS to FT chromatin ( Figures 7C ). It showed strong bindings to regions I to EX1 like ELF6::GUS, but its binding to regions F and G was not detected unlike ELF6::GUS. AtJmj4::FLAG binding to region N and the transcription initiation region of CO was not detected as for the case of ELF6::GUS. In summary, both ELF6::GUS and AtJmj4::FLAG can associate directly and specifically with the transcription initiation region of FT locus where the H3K4me3 levels showed the largest increase in elf6, atjmj4, and elf6 atjmj4 mutants ( Figure 6D ). Thus, ELF6 and AtJmj4 proteins directly target FT chromatin and regulate flowering time via demethylation of H3K4me.
Figure 7. Direct association of ELF6 and AtJmj4 with FT chromatin.
A) FT regions tested for ChIP assay. Schematic is as described in Figure 6A except for the region Ia, which was added in assays in (C). B) ELF6 binding to FT chromatin. LD grown 16 d-old wt Col and ELF6::GUS–containing transgenic elf6-4 plants [22] were harvested and used for ChIP assay using GUS-specific antibody. Amount of immunoprecipitated chromatin was measured by qPCR (B and C). Actin1 and CO were used as internal controls, and the level of Actin1 in each sample was set to 1 for normalization (B and C). Error bars represent se of three independent biological replicates (B and C). C) AtJmj4 binding to FT chromatin. LD grown 16 d-old wt Col and AtJmj4::FLAG–containing atjmj4-1 plants were harvested and used for ChIP assay using FLAG-specific antibody.
Discussion
Recent studies have shown that the expression of some key flowering genes, such as FLC and FT, are regulated through chromatin modifications and have also identified many of the factors involved in the chromatin modification processes [44], [45]. In this study, we show that AtJmj4 and ELF6 play a role in the repression of FT transcription by removing methyl groups from H3K4 at the FT locus.
FT chromatin contains bivalent marks; the active mark (H3K4me3) and the repressive mark (H3K27me3) exist simultaneously [12]. Of these, only H3K27me3 has been studied. It plays a critical role in preventing precocious floral transition by establishing and maintaining repressive FT chromatin as default state. The PRC2-like complex comprised of CURLY LEAF, SWINGER, EMBRYONIC FLOWER2, and FERTILIZATION INDEPENDENT ENDOSPERM, is required for H3K27me and the repression of FT [12].
Unlike H3K27me3, H3K4me3 has been known to be positively associated with transcriptional activity [46]. H3K4me3 can be recognized by the TFIID complex via the PHD finger of TAF3, which in turn recruits RNA polymerase II, leading to transcription activation [47]. In this study, we demonstrate that AtJmj4 is involved in FT repression as an H3K4-specific demethylase directly targeting FT locus with the following data: 1) The loss of AtJmj4 function increased FT expression via enhanced FT promoter activity ( Figure 2 ); 2) The loss of AtJmj4 function increased H3K4me3 level in the transcription initiation region of FT ( Figures 6C and 6D ); 3) The purified AtJmj4 protein can specifically demethylate H3K4me1, H3K4me2, and H3K4me3 in vitro ( Figure 6E ); 4) The AtJmj4::FLAG binds to the transcription initiation region of FT. In addition, up-regulation of FT expression could also be responsible, at least in part, for the increased H3K4me3 level in atjmj4 at the FT locus because chromatin modification often involves positive feedback loops [44].
It is notable that the increase of H3K4me3 at FT caused by the loss AtJmj4 is modest. This indicated that there might be other histone demethylases having redundant roles with AtJmj4 in the demethylation of H3K4 at the FT locus. Indeed, we found that ELF6 is another histone demethylase with similar activity towards FT based on the following data: 1) The loss of ELF6 function increased FT expression and the H3K4me3 level in the transcription initiation region of FT, and these increases were more significant when AtJmj4 and ELF6 functions were lost together ( Figures 5D, 5E , 6C, and 6D ); 2) ELF6::GUS binds to the promoter and transcription initiation region of FT. Previously we reported that both AtJmj4 and ELF6 belong to the same group (Group I) of Arabidopsis Jmj family proteins [21]. Our unpublished phylogenetic analysis indicates that eight Arabidopsis Jmj family proteins belonging to this group have JmjC domains clustered together with the JmjC domains of human JARID1 family that are H3K4 demethylases [17], [19]. Therefore, not only AtJmj4 and ELF6 but also other members of the Arabidopsis Group I Jmj family proteins have a potential to be H3K4 demethylases acting at FT locus. Their genetic and biochemical roles will be addressed in the future studies.
According to recent studies, the antagonistic histone marks, H3K27me3 and H3K4me3, are coordinately regulated by protein complexes containing both histone methyltransferases and histone demethylases [48], [49]. Pasini et al [49] reported that the RBP2 H3K4 demethylase is recruited by the PRC2 to repress the expression of target genes in mouse embryonic stem cells, and the loss of RBP2 increases expression of target genes. At this moment, it is not clear if a similar interaction between AtJmj4/ELF6 and the Arabidopsis PRC2 components occurs in FT repression. However, the report that the level of H3K4me3 within FT chromatin is increased in the absence of CURLY LEAF activity [12] suggests such scenario is plausible. In this study, we did not observe a significant reduction of H3K27me3 level within FT chromatin in elf6 atjmj4 double mutants. Thus, H3K4 demethylases might be recruited by PRC2, but the PRC2 recruitment might not be affected by H3K4 demethylases.
The coexistence of bivalent H3K27me3 and H3K4me3 marks at the same locus has been proposed to poise genes for activation upon appropriate developmental cues [50], [51]. Thus, the existence of bivalent chromatin marks within FT chromatin might be a strategy for plants to achieve reproductive success by a precise regulation of FT expression and flowering time. It is possible that enriched H3K27me3 favors constitutive FT repression, while a proper level of H3K4me3 provides appropriate accessibility for transcription factors controlled temporally such that FT expression can be regulated by changing developmental or environmental cues. Interestingly, the region I of FT locus, in which both AtJmj4::FLAG and ELF6::GUS showed strongest binding ( Figures 7A–7C ) and the largest increase of H3K4me3 by the elf6 and atjmj4 mutations was observed ( Figures 6A, 6C, and 6D ), contains binding sites for FT transcriptional regulators, namely TEM1/TEM2 [39] and a CO-containing protein complex [52]. Thus, it would be of interest in the future to test if the binding of these FT transcriptional regulators is altered by the activity of AtJmj4 and ELF6.
Materials and Methods
Plant Materials and Growth
atjmj4 T-DNA insertion lines in the Col background were obtained from the SALK collection (http://signal.salk.edu/; atjmj4-1, SALK_135712; atjmj4-2, SALK_136058). The following mutants are in the Col background and were described previously: elf6-4 [22], vin3-5 [53], flc-3 [29], gi-2 [1], co-101 [14], ft-10 [54], lhp1-4 [55], fld-3 [31], fve-4 [32], FRI [33]. All plants were grown under 100 µE m−2 s−1 cool white fluorescent light at 22°C.
T-DNA Flanking Sequence Analysis
The T-DNA borders of atjmj4-1 and atjmj4-2 alleles were defined by sequencing PCR products obtained using a T-DNA border primer (SALKLB1; Table S1) and gene-specific primers. For atjmj4-1, AtJmj4-1-R and AtJmj4-1-F primer pair (Table S1) was used to detect wt allele while AtJmj4-1-R and SALKLB1 primer pair was used to detect atjmj4-1 allele. For atjmj4-2, AtJmj4-2-F and AtJmj4-2-R primer pair (Table S1) was used to detect wt allele while AtJmj4-2-F and SALKLB1 primer pair was used to detect atjmj4-2 allele.
RT-PCR and qPCR Analyses
Total RNA was isolated from seedlings using TRI Reagent (Molecular Research Center, INC.) according to the manufacturer's instructions. RT was performed with M-MuLV Reverse Transcriptase (Fermentas) according to the manufacturer's instructions using 3 µg of total RNA. PCR was performed on first strand DNA with i-Taq DNA polymerase (iNtRON Biotechnology). Primers used for RT-PCR and qPCR analyses are listed in Table S2. qPCR was performed in 96-well blocks with an Applied Biosystems 7300 real-time PCR system using the SYBR Green I master mix (Bio-Rad) in a volume of 20 µl. The reactions were performed in triplicate for each run. The comparative ΔΔCT method was used to evaluate the relative quantities of each amplified product in the samples. The threshold cycle (Ct) was automatically determined for each reaction by the system set with default parameters.
GUS and GFP Assays
The FTpro::GUS [54] and the ELF6::GUS [22] were described previously. For the construction of the AtJmj4::GUS translational fusion construct, a 6-kb genomic DNA fragment of AtJmj4 containing 1.5-kb 5′ upstream region and the entire coding region was generated by PCR amplification using AtJmj4GUS-F and AtJmj4GUS-R as primers (Table S3). After restriction digestion with SalI-SmaI, the PCR product was ligated into pPZP211-GUS [56] at SalI-SmaI sites. The final construct was introduced into wt Col plants by the floral dip method [57] through Agrobacterium tumefaciens strain C58C1, and transformants were selected on MS media supplemented with 1% sucrose and 50 µg ml−1 kanamycin. Histochemical GUS staining was performed as described [58].
AtJmj4 cDNA obtained from Col RNA through RT-PCR using AtJmj4OE-F and AtJmj4OE-R as primers (Table S3) was used for the construction of the CaMV35Spro::AtJmj4::GFP. The cDNA was cloned into the SalI site of p35SsGFP/pGEM which has the open reading frame of sGFP [59] behind the Cauliflower Mosaic Virus 35S promoter (CaMV35Spro). Mesophyll protoplasts were isolated from rosette leaves of Col plants grown for 4 weeks in LD as described [60]. The LHP1::RFP [43] was included as a nuclear protein control. Protoplasts were co-transformed with the CaMV35Spro::AtJmj4::GFP and the LHP1::RFP constructs, each with 10 µg of plasmid DNA prepared with Nucleo Bond Xtra Midi Kit (Macherey-Nagel). After 16 h incubation at 22°C in dark, protoplasts were observed with LSM 510 confocal microscope (Zeiss). The GFP and RFP fusion proteins were excited at 488 nm and 543 nm, respectively. The autofluorescence of chlorophylls, GFP, and RFP were analyzed with LP650, BP500-530IR, and BP565-615IR filters, respectively. The merged image was obtained using the LSM Image Browser (Zeiss).
AtJmj4::FLAG
The AtJmj4::FLAG construct is consisted of a 0.8 kb 5′ upstream region of AtJmj4 (AtJmj4pro), the sequence for 3xFLAG tags, and the full coding sequence of AtJmj4 cDNA. AtJmj4 cDNA was obtained from Col RNA through RT-PCR using AtJmj4OE-F and AtJmj4OE-R1 as primers (Table S3), digested with SalI, and cloned into the SalI site of a construct containing 3xFLAG behind the CaMV35Spro in pPZP211 vector. The CaMV35Spro was replaced with the AtJmj4pro obtained from Col genomic DNA through PCR using Atjmj4FLAG-F and Atjmj4FLAG-R as primers (Table S3) at PstI site. Then the AtJmj4pro::3xFLAG::AtJmj4 cDNA was PCR-amplified using AtJmj4FLAG-F1 and Atjmj4FLAG-R1 as primers (Table S3). After restriction digestion with NheI, the PCR fragment was ligated into the SmaI-XbaI sites of the binary vector pPZP221B [61]. The final construct was introduced into atjmj4-1 mutants by the floral dip method through Agrobacterium tumefaciens strain C58C1, and transformants were selected on MS media supplemented with 1% sucrose and 25 µg ml−1 glufosinate ammonium.
Protein samples were extracted using 2xloading buffer from wt Col and transgenic plants harboring the AtJmj4::FLAG construct, and their concentrations were determined by Protein Assay (Bio-Rad). 3.75 µg of protein samples were size-fractionated on a 7% SDS-PAGE gel, transferred to Pure Nitrocellulose (GE Water & Process Technologies), and blocked with 10% skim milk power in TTBS (0.1% tween 20, 20 mM Tris-HCl pH7.4, 150 mM NaCl). AtJmj4::FLAG protein was detected using Anti-FLAG M2-Peroxidase (HRP) antibody (Sigma), ECL Western Blotting Detection Kit (GE Healthcare), and JP/LAS-3000 Luminescent Image Analyzer (Fujifilm).
AtJmj4 Protein Expression and Purification
For the expression of AtJmj4 protein in insect cells, the full length AtJmj4 coding region was PCR amplified from a cDNA clone using JMJ4_pENTR_For and JMJ4_pENTR_Rev as primers (Table S3) and ligated into the Klenow-filled EcoRI site of pFastBac HT A vector (Invitrogen). The resulting AtJmj4::pFastBac HT A construct with amino-terminal 6xHis tag was used to transform DH10Bac E. coli competent cells (Invitrogen), and the recombinant baculovirus DNA was selected and used for the infection of sf9 cells following the Bac-to-Bac system instructions (Invitrogen). Cells positive for the recombinant-protein expression as tested by western blot with anti-His antibody (Santa Cruz) was used to infect cells to produce 6His-AtJmj4 baculovirus stocks. Viral stocks were stored at 4°C. For protein expression, 2.5 ml of viral stock was used to infect approximately 2×106 adherent sf9 cells in 400 ml of sf-900 II SFM serum free medium (Gibco) and cultured at 27°C for 48 h. Then, cells were harvested and washed with PBS and frozen at −80°C until further purification. Frozen cells were thawed on ice and resuspended with 20 ml equilibration buffer (80 mM Na2HP04, 20 mM NaH2PO4, 300 mM NaCl, 10 mM imidazole, 100 µM PMSF, 10% glycerol). Cells were disrupted by sonication, and the lysate was clarified by centrifugation at 10,000× g for 20 min at 4°C. The supernatant was applied into a Ni-NTA-Agarose (Qiagen) chromatography column and washed with 10× column volume of equilibration buffer. Protein was eluted from the column with 3× bed volume of elution buffer (80 mM Na2HP04, 20 mM NaH2PO4, 300 mM NaCl, 250 mM imidazole, 100 µM PMSF, 10% glycerol). Purified recombinant 6His-AtJmj4 protein was dialyzed against dialysis buffer (40 mM HEPES-KOH pH 7.9, 50 mM KCl, 10% glycerol, 1 mM DTT, 0.2 mM PMSF) overnight at 4°C. Next-day, dialyzed 6His-AtJmj4 proteins was quantified and stored at −20°C.
Histone Demethylase Assay
In vitro histone demethylation assay was performed as previously described [62] with minor modifications. Briefly, two or four µg of purified 6His-AtJmj4 protein was incubated with 4 µg of calf thymus histones type II-A (Sigma) in the DeMTase reaction buffer 1 (20 mM Tris-HCl pH 7.3, 150 mM NaCl, 50 mM (NH4)2Fe(SO4)2·6(H2O), 1 mM α-ketoglutarate, 2 mM ascorbic acid) for 5 h at 37°C. Histone modifications were detected by western blot analysis with antibodies as follows: anti-H3K4me1 (Upstate 07-436), anti-H3K4me2 (Upstate 07-030), anti-H3K4me3, (Abcam ab8580), anti-H3K9me3 (Upstate 07-442), anti-H3K36me2 (Upstate 07-369), anti-H3K27me3 (Upstate 07-449), and anti-H3 (Abcam ab1791-100).
ChIP Assay
ChIP was performed as described by Han et al. [63] using 55- to 60-d-old plants grown in SD. Briefly, leaves were vacuum infiltrated with 1% formaldehyde for cross-linking and ground in liquid nitrogen after quenching the cross-linking process. Chromatin was isolated and sonicated into ∼0.5 to 1 kb fragments. Specific antibody against GUS (Invitrogen A5790), FLAG (Sigma A8592-0.2MG), H3K4me3 (Upstate 07-473), or H3K27me3 (Upstate 07-449) was added to the chromatin solution, which had been precleared with salmon sperm DNA/Protein A agarose beads (Upstate 16-157). After subsequent incubation with salmon sperm DNA/Protein A agarose beads, immunocomplexes were precipitated and eluted from the beads. Cross-links were reversed, and residual proteins in the immunocomplexes were removed by incubation with proteinase K, followed by phenol/chloroform extraction. DNA was recovered by ethanol precipitation. The amount of immunoprecipitated FT, CO, and Actin1 chromatins was determined by PCR with primer pairs in Table S4.
Supporting Information
Leaf initiation rate of atjmj4-1 mutants: Wt Col (black circles) and atjmj4-1 mutant plants (white squares) were grown in SD and their leaf numbers were scored every week from four weeks after planting. At least 10 individuals were scored for each genotype. Error bars represent sd.
(0.13 MB TIF)
FLC-independent function of AtJmj4: Expression of flowering genes in flc-3 and atjmj4-1 mutant plants grown in SD for 12 d as determined by RT-PCR analysis. UBQ was used as an expression control.
(0.25 MB TIF)
Vernalization response of atjmj4 mutants: Plants of each genotype were treated with vernalization for 40 d as described previously [56]. Flowering time was scored as leaf number for plants either without (V0) or after (V40) vernalization treatment. At least 12 individuals were scored for each genotype. Error bars represent sd.
(0.36 MB TIF)
CO- and GI-independent increase of FT expression in atjmj4: Plants of each genotype were grown in LD for 14 d and harvested at ZT8 for RT-PCR analyses. UBQ was used as an expression control.
(0.22 MB TIF)
Expression of FT regulators in atjmj4: A and B) Temporal expression of FT regulators in atjmj4-1. Col and atjmj4-1 plants were grown in SD (A) or in LD (B) until indicated DAG and harvested at ZT14 (LD) or ZT8 (SD) for RT-PCR analyses. UBQ was used as an expression control.
(0.60 MB TIF)
Oligonucleotides used for T-DNA flanking sequence analysis
(0.03 MB DOC)
Oligonucleotides used for RT-PCR analysis
(0.05 MB DOC)
Oligonucleotides used for constructs
(0.03 MB DOC)
Oligonucleotides used for ChIP assay
(0.04 MB DOC)
Acknowledgments
We would like to thank the Salk Institute Genome Analysis Laboratory for providing knockout pools containing atjmj4 alleles. We are grateful to Ji-Yeon Hong for technical assistance in mutant isolation.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by the Global Research Laboratory Program of the Ministry of Education, Science, and Technology, by the Plant Diversity Research Center, and by the Bio-Green21 Program of the Rural Development Administration. B.N. was supported by a grant from the Ministry of Education, Science, and Technology to the Environmental Biotechnology National Core Research Center (R15-2003-012-01001-0) and by grants from the Korea Research Foundation (KRF-2007-313-C00703 and KRF-2008-314-C00359). J.-H.J and H.-R.S. were supported by the BK21 Program. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Park DH, Somers DE, Kim YS, Choy YH, Lim HK, et al. Control of circadian rhythms and photoperiodic flowering by the Arabidopsis GIGANTEA gene. Science. 1999;285:1579–1582. doi: 10.1126/science.285.5433.1579. [DOI] [PubMed] [Google Scholar]
- 2.Fowler S, Lee K, Onouchi H, Samach A, Richardson K, et al. GIGANTEA: a circadian clock-controlled gene that regulates photoperiodic flowering in Arabidopsis and encodes a protein with several possible membrane-spanning domains. EMBO J. 1999;18:4679–4688. doi: 10.1093/emboj/18.17.4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Putterill J, Robson F, Lee K, Simon R, Coupland G. The CONSTANS gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell. 1995;80:847–857. doi: 10.1016/0092-8674(95)90288-0. [DOI] [PubMed] [Google Scholar]
- 4.Kobayashi Y, Kaya H, Goto K, Iwabuchi M, Araki T. A pair of related genes with antagonistic roles in mediating flowering signals. Science. 1999;286:1960–1962. doi: 10.1126/science.286.5446.1960. [DOI] [PubMed] [Google Scholar]
- 5.Kardailsky I, Shukla VK, Ahn JH, Dagenais N, Christensen SK, et al. Activation tagging of the floral inducer FT. Science. 1999;286:1962–1965. doi: 10.1126/science.286.5446.1962. [DOI] [PubMed] [Google Scholar]
- 6.Li D, Liu C, Shen L, Wu Y, Chen H, et al. A repressor complex governs the integration of flowering signals in Arabidopsis. Dev Cell. 2008;15:110–120. doi: 10.1016/j.devcel.2008.05.002. [DOI] [PubMed] [Google Scholar]
- 7.Corbesier L, Vincent C, Jang S, Fornara F, Fan Q, et al. FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science. 2007;316:1030–1033. doi: 10.1126/science.1141752. [DOI] [PubMed] [Google Scholar]
- 8.Abe M, Kobayashi Y, Yamamoto S, Daimon Y, Yamaguchi A, et al. FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science. 2005;309:1052–1056. doi: 10.1126/science.1115983. [DOI] [PubMed] [Google Scholar]
- 9.Wigge PA, Kim MC, Jaeger KE, Busch W, Schmid M, et al. Integration of spatial and temporal information during floral induction in Arabidopsis. Science. 2005;309:1056–1059. doi: 10.1126/science.1114358. [DOI] [PubMed] [Google Scholar]
- 10.Turck F, Roudier F, Farrona S, Martin-Magniette ML, Guillaume E, et al. Arabidopsis TFL2/LHP1 specifically associates with genes marked by trimethylation of histone H3 lysine 27. PLoS Genet. 2007;3:e86. doi: 10.1371/journal.pgen.0030086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang X, Germann S, Blus BJ, Khorasanizadeh S, Gaudin V, et al. The Arabidopsis LHP1 protein colocalizes with histone H3 Lys27 trimethylation. Nat Struct Mol Biol. 2007;14:869–871. doi: 10.1038/nsmb1283. [DOI] [PubMed] [Google Scholar]
- 12.Jiang D, Wang Y, He Y. Repression of FLOWERING LOCUS C and FLOWERING LOCUS T by the Arabidopsis Polycomb repressive complex 2 components. PLoS ONE. 2008;3:e3404. doi: 10.1371/journal.pone.0003404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kotake T, Takada S, Nakahigashi K, Ohto M, Goto K. Arabidopsis TERMINAL FLOWER 2 gene encodes a heterochromatin protein 1 homolog and represses both FLOWERING LOCUS T to regulate flowering time and several floral homeotic genes. Plant Cell Physiol. 2003;44:555–564. doi: 10.1093/pcp/pcg091. [DOI] [PubMed] [Google Scholar]
- 14.Takada S, Goto K. Terminal flower2, an Arabidopsis homolog of heterochromatin protein1, counteracts the activation of flowering locus T by constans in the vascular tissues of leaves to regulate flowering time. Plant Cell. 2003;15:2856–2865. doi: 10.1105/tpc.016345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, et al. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 16.Tsukada Y, Fang J, Erdjument-Bromage H, Warren ME, Borchers CH, et al. Histone demethylation by a family of JmjC domain-containing proteins. Nature. 2006;439:811–816. doi: 10.1038/nature04433. [DOI] [PubMed] [Google Scholar]
- 17.Christensen J, Agger K, Cloos PA, Pasini D, Rose S, et al. RBP2 belongs to a family of demethylases, specific for tri-and dimethylated lysine 4 on histone 3. Cell. 2007;128:1063–1076. doi: 10.1016/j.cell.2007.02.003. [DOI] [PubMed] [Google Scholar]
- 18.Chang B, Chen Y, Zhao Y, Bruick RK. JMJD6 is a histone arginine demethylase. Science. 2007;318:444–447. doi: 10.1126/science.1145801. [DOI] [PubMed] [Google Scholar]
- 19.Klose RJ, Zhang Y. Regulation of histone methylation by demethylimination and demethylation. Nat Rev Mol Cell Biol. 2007;8:307–318. doi: 10.1038/nrm2143. [DOI] [PubMed] [Google Scholar]
- 20.Klose RJ, Yamane K, Bae Y, Zhang D, Erdjument-Bromage H, et al. The transcriptional repressor JHDM3A demethylates trimethyl histone H3 lysine 9 and lysine 36. Nature. 2006;442:312–316. doi: 10.1038/nature04853. [DOI] [PubMed] [Google Scholar]
- 21.Hong EH, Jeong YM, Ryu JY, Amasino RM, Noh B, et al. Temporal and spatial expression patterns of nine Arabidopsis genes encoding Jumonji C-domain proteins. Mol Cells. 2009;27:481–490. doi: 10.1007/s10059-009-0054-7. [DOI] [PubMed] [Google Scholar]
- 22.Noh B, Lee SH, Kim HJ, Yi G, Shin EA, et al. Divergent roles of a pair of homologous jumonji/zinc-finger-class transcription factor proteins in the regulation of Arabidopsis flowering time. Plant Cell. 2004;16:2601–2613. doi: 10.1105/tpc.104.025353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saze H, Shiraishi A, Miura A, Kakutani T. Control of genic DNA methylation by a jmjC domain-containing protein in Arabidopsis thaliana. Science. 2008;319:462–465. doi: 10.1126/science.1150987. [DOI] [PubMed] [Google Scholar]
- 24.Samach A, Onouchi H, Gold SE, Ditta GS, Schwarz-Sommer Z, et al. Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis. Science. 2000;288:1613–1616. doi: 10.1126/science.288.5471.1613. [DOI] [PubMed] [Google Scholar]
- 25.Lee H, Suh SS, Park E, Cho E, Ahn JH, et al. The AGAMOUS-LIKE 20 MADS domain protein integrates floral inductive pathways in Arabidopsis. Genes Dev. 2000;14:2366–2376. doi: 10.1101/gad.813600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Michaels SD, Amasino RM. Loss of FLOWERING LOCUS C activity eliminates the late-flowering phenotype of FRIGIDA and autonomous pathway mutations but not responsiveness to vernalization. Plant Cell. 2001;13:935–941. doi: 10.1105/tpc.13.4.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sung S, Amasino RM. Vernalization in Arabidopsis thaliana is mediated by the PHD finger protein VIN3. Nature. 2004;427:159–164. doi: 10.1038/nature02195. [DOI] [PubMed] [Google Scholar]
- 28.Sheldon CC, Burn JE, Perez PP, Metzger J, Edwards JA, et al. The FLF MADS box gene: a repressor of flowering in Arabidopsis regulated by vernalization and methylation. Plant Cell. 1999;11:445–458. doi: 10.1105/tpc.11.3.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Michaels SD, Amasino RM. FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell. 1999;11:949–956. doi: 10.1105/tpc.11.5.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Koornneef M, Hanhart CJ, van der Veen JH. A genetic and physiological analysis of late flowering mutants in Arabidopsis thaliana. Mol Gen Genet. 1991;229:57–66. doi: 10.1007/BF00264213. [DOI] [PubMed] [Google Scholar]
- 31.He Y, Michaels SD, Amasino RM. Regulation of flowering time by histone acetylation in Arabidopsis. Science. 2003;302:1751–1754. doi: 10.1126/science.1091109. [DOI] [PubMed] [Google Scholar]
- 32.Ausin I, Alonso-Blanco C, Jarillo JA, Ruiz-Garcia L, Martinez-Zapater JM. Regulation of flowering time by FVE, a retinoblastoma-associated protein. Nat Genet. 2004;36:162–166. doi: 10.1038/ng1295. [DOI] [PubMed] [Google Scholar]
- 33.Lee I, Michaels SD, Amasino RM. The late-flowering phenotype of FRIGIDA and mutations in LUMINIDEPENDENS is suppressed in the Landsberg erecta strain of Arabidopsis. Plant J. 1994;6:903–909. [Google Scholar]
- 34.Koornneef M, Vrles HB, Hanhart C, Soppe W, Peeters T. The phenotype of some late-flowering mutants is enhanced by a locus on chromosome 5 that is not effective in the Landsberg erecta wild-type. Plant J. 1994;6:911–919. [Google Scholar]
- 35.Jung JH, Seo YH, Seo PJ, Reyes JL, Yun J, et al. The GIGANTEA-regulated microRNA172 mediates photoperiodic flowering independent of CONSTANS in Arabidopsis. Plant Cell. 2007;19:2736–2748. doi: 10.1105/tpc.107.054528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Aukerman MJ, Sakai H. Regulation of flowering time and floral organ identity by a MicroRNA and its APETALA2-like target genes. Plant Cell. 2003;15:2730–2741. doi: 10.1105/tpc.016238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schmid M, Uhlenhaut NH, Godard F, Demar M, Bressan R, et al. Dissection of floral induction pathways using global expression analysis. Development. 2003;130:6001–6012. doi: 10.1242/dev.00842. [DOI] [PubMed] [Google Scholar]
- 38.Liu H, Yu X, Li K, Klejnot J, Yang H, et al. Photoexcited CRY2 interacts with CIB1 to regulate transcription and floral initiation in Arabidopsis. Science. 2008;322:1535–1539. doi: 10.1126/science.1163927. [DOI] [PubMed] [Google Scholar]
- 39.Castillejo C, Pelaz S. The balance between CONSTANS and TEMPRANILLO activities determines FT expression to trigger flowering. Curr Biol. 2008;18:1338–1343. doi: 10.1016/j.cub.2008.07.075. [DOI] [PubMed] [Google Scholar]
- 40.Lee JH, Park SH, Lee JS, Ahn JH. A conserved role of SHORT VEGETATIVE PHASE (SVP) in controlling flowering time of Brassica plants. Biochim Biophys Acta. 2007;1769:455–461. doi: 10.1016/j.bbaexp.2007.05.001. [DOI] [PubMed] [Google Scholar]
- 41.Hartmann U, Hohmann S, Nettesheim K, Wisman E, Saedler H, et al. Molecular cloning of SVP: a negative regulator of the floral transition in Arabidopsis. Plant J. 2000;21:351–360. doi: 10.1046/j.1365-313x.2000.00682.x. [DOI] [PubMed] [Google Scholar]
- 42.Adamczyk BJ, Lehti-Shiu MD, Fernandez DE. The MADS domain factors AGL15 and AGL18 act redundantly as repressors of the floral transition in Arabidopsis. Plant J. 2007;50:1007–1019. doi: 10.1111/j.1365-313X.2007.03105.x. [DOI] [PubMed] [Google Scholar]
- 43.Choi K, Kim S, Kim SY, Kim M, Hyun Y, et al. SUPPRESSOR OF FRIGIDA3 encodes a nuclear ACTIN-RELATED PROTEIN6 required for floral repression in Arabidopsis. Plant Cell. 2005;17:2647–2660. doi: 10.1105/tpc.105.035485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Farrona S, Coupland G, Turck F. The impact of chromatin regulation on the floral transition. Semin Cell Dev Biol. 2008;19:560–573. doi: 10.1016/j.semcdb.2008.07.015. [DOI] [PubMed] [Google Scholar]
- 45.Schatlowski N, Creasey K, Goodrich J, Schubert D. Keeping plants in shape: polycomb-group genes and histone methylation. Semin Cell Dev Biol. 2008;19:547–553. doi: 10.1016/j.semcdb.2008.07.019. [DOI] [PubMed] [Google Scholar]
- 46.Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 47.Vermeulen M, Mulder KW, Denissov S, Pijnappel WW, van Schaik FM, et al. Selective anchoring of TFIID to nucleosomes by trimethylation of histone H3 lysine 4. Cell. 2007;131:58–69. doi: 10.1016/j.cell.2007.08.016. [DOI] [PubMed] [Google Scholar]
- 48.Lee MG, Norman J, Shilatifard A, Shiekhattar R. Physical and functional association of a trimethyl H3K4 demethylase and Ring6a/MBLR, a polycomb-like protein. Cell. 2007;128:877–887. doi: 10.1016/j.cell.2007.02.004. [DOI] [PubMed] [Google Scholar]
- 49.Pasini D, Hansen KH, Christensen J, Agger K, Cloos PA, et al. Coordinated regulation of transcriptional repression by the RBP2 H3K4 demethylase and Polycomb-Repressive Complex 2. Genes Dev. 2008;22:1345–1355. doi: 10.1101/gad.470008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 51.Azuara V, Perry P, Sauer S, Spivakov M, Jorgensen HF, et al. Chromatin signatures of pluripotent cell lines. Nat Cell Biol. 2006;8:532–538. doi: 10.1038/ncb1403. [DOI] [PubMed] [Google Scholar]
- 52.Wenkel S, Turck F, Singer K, Gissot L, Le Gourrierec J, et al. CONSTANS and the CCAAT box binding complex share a functionally important domain and interact to regulate flowering of Arabidopsis. Plant Cell. 2006;18:2971–2984. doi: 10.1105/tpc.106.043299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mylne JS, Barrett L, Tessadori F, Mesnage S, Johnson L, et al. LHP1, the Arabidopsis homologue of HETEROCHROMATIN PROTEIN1, is required for epigenetic silencing of FLC. Proc Natl Acad Sci U S A. 2006;103:5012–5017. doi: 10.1073/pnas.0507427103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yoo SK, Chung KS, Kim J, Lee JH, Hong SM, et al. CONSTANS activates SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1 through FLOWERING LOCUS T to promote flowering in Arabidopsis. Plant Physiol. 2005;139:770–778. doi: 10.1104/pp.105.066928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Larsson AS, Landberg K, Meeks-Wagner DR. The TERMINAL FLOWER2 (TFL2) gene controls the reproductive transition and meristem identity in Arabidopsis thaliana. Genetics. 1998;149:597–605. doi: 10.1093/genetics/149.2.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Noh YS, Amasino RM. PIE1, an ISWI family gene, is required for FLC activation and floral repression in Arabidopsis. Plant Cell. 2003;15:1671–1682. doi: 10.1105/tpc.012161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 58.Schomburg FM, Patton DA, Meinke DW, Amasino RM. FPA, a gene involved in floral induction in Arabidopsis, encodes a protein containing RNA-recognition motifs. Plant Cell. 2001;13:1427–1436. doi: 10.1105/tpc.13.6.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Niwa Y, Hirano T, Yoshimoto K, Shimizu M, Kobayashi H. Non-invasive quantitative detection and applications of non-toxic, S65T-type green fluorescent protein in living plants. Plant J. 1999;18:455–463. doi: 10.1046/j.1365-313x.1999.00464.x. [DOI] [PubMed] [Google Scholar]
- 60.Yoo SD, Cho YH, Sheen J. Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc. 2007;2:1565–1572. doi: 10.1038/nprot.2007.199. [DOI] [PubMed] [Google Scholar]
- 61.Kang BH, Busse JS, Dickey C, Rancour DM, Bednarek SY. The arabidopsis cell plate-associated dynamin-like protein, ADL1Ap, is required for multiple stages of plant growth and development. Plant Physiol. 2001;126:47–68. doi: 10.1104/pp.126.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Whetstine JR, Nottke A, Lan F, Huarte M, Smolikov S, et al. Reversal of Histone Lysine Trimethylation by the JMJD2 Family of Histone Demethylases. Cell. 2006;125:467–481. doi: 10.1016/j.cell.2006.03.028. [DOI] [PubMed] [Google Scholar]
- 63.Han SK, Song JD, Noh YS, Noh B. Role of plant CBP/p300-like genes in the regulation of flowering time. Plant J. 2007;49:103–114. doi: 10.1111/j.1365-313X.2006.02939.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Leaf initiation rate of atjmj4-1 mutants: Wt Col (black circles) and atjmj4-1 mutant plants (white squares) were grown in SD and their leaf numbers were scored every week from four weeks after planting. At least 10 individuals were scored for each genotype. Error bars represent sd.
(0.13 MB TIF)
FLC-independent function of AtJmj4: Expression of flowering genes in flc-3 and atjmj4-1 mutant plants grown in SD for 12 d as determined by RT-PCR analysis. UBQ was used as an expression control.
(0.25 MB TIF)
Vernalization response of atjmj4 mutants: Plants of each genotype were treated with vernalization for 40 d as described previously [56]. Flowering time was scored as leaf number for plants either without (V0) or after (V40) vernalization treatment. At least 12 individuals were scored for each genotype. Error bars represent sd.
(0.36 MB TIF)
CO- and GI-independent increase of FT expression in atjmj4: Plants of each genotype were grown in LD for 14 d and harvested at ZT8 for RT-PCR analyses. UBQ was used as an expression control.
(0.22 MB TIF)
Expression of FT regulators in atjmj4: A and B) Temporal expression of FT regulators in atjmj4-1. Col and atjmj4-1 plants were grown in SD (A) or in LD (B) until indicated DAG and harvested at ZT14 (LD) or ZT8 (SD) for RT-PCR analyses. UBQ was used as an expression control.
(0.60 MB TIF)
Oligonucleotides used for T-DNA flanking sequence analysis
(0.03 MB DOC)
Oligonucleotides used for RT-PCR analysis
(0.05 MB DOC)
Oligonucleotides used for constructs
(0.03 MB DOC)
Oligonucleotides used for ChIP assay
(0.04 MB DOC)