Tumorigenesis is a multistep process controlled by a number of proteins involved in diverse pathways. Traditionally, proteins are either considered as oncogenes, which promote tumorigenesis or as tumor suppressors, which prevent tumorigenesis. However, recent studies revealed quite a few proteins that could function as oncogene as well as tumor suppressor. A new member of such proteins is p30 DBC (deleted in breast cancer 1, also called DBC1). p30 DBC is one of the proteins involved in tumorigenesis that does not clearly adhere to either descriptions. Several studies show that p30 DBC is involved in cell proliferation, apoptosis and histone modification, all processes important for regulating tumorigenesis. However, there are other conflicting results regarding how p30 DBC contributes to tumorigenesis. The most interesting aspect of this is that p30 DBC is a strong inhibitor of SIRT1 protein deacetylase, whose exact role in tumorigenesis is currently under debate. This review summarizes the current understandings on p30 DBC functions, with a focus on the proposed roles of p30 DBC in tumorigenesis.
p30 DBC Expression
The p30 DBC (KIAA1967) gene was originally found to be homozygously deleted in human chromosome 8p21 in breast cancer.1 Therefore, it has been named Deleted in Breast Cancer 1 (DBC1). The symbol of this gene is identical to another gene DBCCR1 also called DBC1, which stands for deleted in bladder cancer 1. To avoid further confusion, we will use the nomenclature p30 DBC in this review.
In the same study that identified p30 DBC to be deleted in breast cancer specimens, Hamaguchi et al.1 also found that p30 DBC mRNA determined by RT-PCR was downregulated in some lung and colon cancer cell lines. However, in contrast to these findings, several microarray studies found that p30 DBC mRNA is upregulated in breast cancers.2,3 In a different study, no significant overexpression of p30 DBC was found in more than 100 prostate cancer specimens.4 Therefore the p30 DBC gene expression appears to be either overexpressed or downregulated, which may reflect the different etiologies of the cancer specimens used in these studies. Another complicating factor is the lack of data regarding the presence or absence of mutations in the p30 DBC gene in these studies. Thus, based exclusively on observed expression levels it is currently unclear whether the role of p30 DBC should be described as a tumor suppressor or promoter. It is critical, therefore, to understand the cellular functions of p30 DBC, which might provide clues as to how p30 DBC affects tumorigenesis.
p30 DBC in SIRT1 Regulation
Although p30 DBC was identified in 2002, its cellular functions are just emerging. We recently identified p30 DBC as a SIRT1-interacting protein through the affinity purification of SIRT1-associated protein complexes. SIRT1, a member of the sirtuin family of proteins, is considered to be the mammalian ortholog of yeast sir2. SIRT1 has protein deacetylase activity and is involved in diverse cellular pathways, such as metabolism and cellular stress response.5 p30 DBC directly interacts with the catalytic domain of SIRT1 and inhibits the deacetylase activity of SIRT1.6,7 In doing so, p30 DBC promotes the acetylation of p53 and FOXO3 following cellular stress in several cancer cell lines and inhibits SIRT1-dependent cell survival through p53 or FOXO pathways. These findings imply that p30 DBC attenuates the survival of cancer cells following genotoxic stress through its ability to inhibit SIRT1 activity.
Our group also performed the affinity purification to identify p30 DBC-associated proteins. Again, SIRT1 was identified as the major p30 DBC binding protein. Recently, p30 DBC has also been demonstrated to inhibit the activity of SUV39H1 methyltransferase and regulate heterochromatin formation via its inhibitory effect toward both SIRT1 and SUV39H1.8 In addition, p30 DBC contains the Nudix hydrolase (MutT) domains, which is predicted to bind nucleoside diphosphate sugars and nicotinamide adenine dinucleotide (NAD), a co-substrate for SIRT1 enzyme.9 These results suggest that a major cellular function of p30 DBC is to regulate SIRT1.
This raises the question: does p30 DBC affect tumorigenesis through its association with SIRT1? As described, SIRT1 possesses protein deacetylating activity and deacetylates diverse substrates including p53, FOXO, NFκB, Ku70, Rb as well as histones.10 With regard to cancer, it is still controversial whether SIRT1 acts as a tumor promoter or tumor suppressor.11,12 Up until several years ago, SIRT1 was considered to be a tumor promoter, primarily due to the fact that the expression of SIRT1 protein was shown to increase in cancer tissues, cancer cell lines, as well as in mouse tumors.13–18 Secondly, SIRT1 inhibitors or depletion of SIRT1 by siRNA induce the death of cancer cells and sensitizes cells to the anti-cancer drugs.19–23 Thirdly, SIRT1 mediates the silencing of tumor suppressor genes without affecting hypermethylation of their promoter regions.24 Finally, SIRT1 deacetylates p53 and FOXO and inhibits p53 and/or FOXO-dependent transcription or apoptosis following genotoxic stress.6,7,25 All these data suggest that SIRT1 acts by promoting tumorigenesis.
However, these tumor-promoting activities of SIRT1 were challenged by a number of other studies. Notably, overexpression of SIRT1 in APC mutated transgenic mice is able to inhibit the formation of colon cancer by deacetylating and inactivating the constitutively activated oncogenic β-catenin in these mice.26 In addition, HCT116 colon cancer cells with SIRT1 knock-down induces tumor formation in xenograft model.27 In addition, SIRT1−/− MEFs (mouse embryonic fibroblasts) show chromosomal instability due to impaired DNA repair and histone modification,28,29 both hallmarks of the genetic instability that precedes the formation of many types of cancers. Furthermore, SIRT1 deficiency led to a significant increase of tumorigenesis in p53+/− mouse, while SIRT1 expression was found to be reduced in breast tumor samples when compared to normal breast tissue.28 SIRT1 is also able to deacetylate RelA/p65 subunit of NFκB and inhibit its transactivation activity, and thus augment apoptosis in response to tumor necrosis factor-alpha (TNFα).17 Similarly, SIRT1 deacetylates both androgen receptor (AR) and histone in the promoter of prostate specific antigen (PSA), and inhibits the dihydrotestosterone (DHT)-dependent growth of LNCaP prostate cancer cells, which is mediated by AR signaling.30,31 Finally, SIRT1 deacetylates c-Myc, destabilizes c-Myc, and thereby inhibits transformation activity of c-Myc.32 This series of studies indicate that SIRT1 is a tumor suppressor.
The data above would suggest that SIRT1 functions in both tumor promotion and tumor suppression. Based on the fact that p30 DBC is a major SIRT1-binding protein and negatively regulates SIRT1 activity, it is entirely possible that p30 DBC may also participate in tumorigenesis via its role in modulating SIRT1 activity. While it remains to be determined as whether all of SIRT1 functions are regulated by p30 DBC in all cellular contexts, it is likely that p30 DBC counteracts SIRT1 functions and acts as either tumor promoter or tumor suppressor in specific tumor environment.
Other Cellular Functions of p30 DBC
In addition to regulating SIRT1, p30 DBC may have other cellular functions. In response to apoptosis-inducing signals, such as exposure to TNFα, etoposide or staurosporine, p30 DBC is cleaved into C-terminal p120 and p66 fragments in a caspase-dependent manner.33 The C-terminal fragment then relocalizes from nucleus to cytosol and mitochondria, and sensitizes cells to apoptotic stimuli. These findings suggest that p30 DBC promotes apoptosis through a positive feedback mechanism, which might suppress tumorigenesis by facilitating cell death in response to cellular stresses.
Recently, p30 DBC was found to act as a transcriptional coactivator of retinoic acid receptor α (RARα).34 The induction of RARα target genes such as Sox9 and HoxA1 gene in response to retinoic acid requires p30 DBC in MCF-7 breast cancer cells. This transcriptional activity of p30 DBC is not affected by SIRT1 inhibitor nicotinamide, suggesting that at least this transcriptional regulation function of p30 DBC is independent of SIRT1. Since retinoids inhibit cell growth by inducing tumor suppressor genes through RAR in some breast cancers, it appears that p30 DBC enhances a RA-mediated inhibition of cell growth in breast cancer cells and thus functions as a tumor suppressor.
While the above findings imply that p30 DBC may inhibit tumor growth or survival, a recent study demonstrates that through an interaction with estrogen receptor (ERα), p30 DBC acts as a survival factor in breast cancer cells. The first 150 amino acids of p30 DBC has been shown to interact with ERα through its hormone-binding domain in an estrogen-independent manner.35 It is thought that this interaction may enhance the stability of unliganded ERα, with no effect on the mRNA level of ERα. However, the mechanism by which p30 DBC modulates ERα protein stability remains unclear. Depletion of p30 DBC by siRNA enhances the death of MCF-7 breast cancer cells in the absence of estrogen, suggesting a positive role of p30 DBC in cell proliferation. In addition, tamoxifen is able to disrupt the interaction between p30 DBC and ERα, which may lead to the destabilization of ERα. These findings imply that the anti-cancer activity of tamoxifen would be more effective in breast cancers with low expression of p30 DBC.
In addition to regulating ERα activity, p30 DBC could also act as an androgen receptor (AR) coactivator.4 The ligand binding domain (LBD) of AR interacts with the N-terminus of p30 DBC (residues 1~265) in the presence of AR ligand. This interaction enhances AR—DNA binding and facilitates AR’s transcriptional activity. Knocking-down of p30 DBC decreases the induction of AR target genes including prostate specific antigen (PSA) in LNCaP prostate cancer cells. It is not clear whether p30 DBC activates AR through its inhibitory effect toward SIRT1, since SIRT1 could suppress AR signaling.30,31 Nevertheless, this suggests that p30 DBC may act as a tumor promoter and that its ablation may delay the progression of androgen hormone-mediated prostate cancer.
p30 DBC is also present in several other protein complexes, although the physiological functions of p30 DBC in these complexes have not been studied. For example, a large-scale purification of c-Myc complex identified p30 DBC as an associated protein through Myc-box II36 which raises the possibility that p30 DBC may be involved in various functions of proto-oncogene c-Myc. We recently found that SIRT1 directly interacts with and inhibits c-Myc.32 It is possible that p30 DBC, SIRT1 and c-Myc form a complex in cells. Another large-scale functional proteomics study revealed that p30 DBC interacts with IKKβ (IκB kinase β subunit) following the treatment of TNFα.37 This interaction could be one of the mechanisms by which p30 DBC regulates the NFκB pathway.
All of the above-mentioned studies suggest that p30 DBC is likely to regulate the survival or death of cancer cells, which might contribute to tumorigenic process. However, exactly how p30 DBC affects tumorigenesis might be tissue specific, with its promoting tumor formation in some cases while suppressing tumor growth or survival in others. The major caveat of the current studies is the use of cancer cell lines, which have distinct genetic background that make it difficult to draw any consistent conclusions. The physiological functions of p30 DBC in vivo remain to be determined.
Conclusions
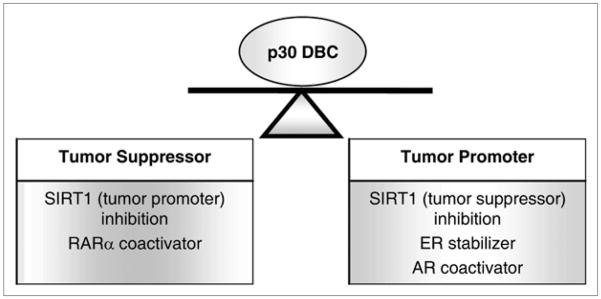
p30 DBC has been implicated in cancer cell proliferation and death in SIRT1-dependent or independent manner (Fig. 1). The role of p30 DBC in tumorigenesis suggests that it may be a potential therapeutic target. The development of compounds that can modulate the interaction between p30 DBC and its binding proteins could be a therapeutic approach to control tumorigenesis. However, a more thorough understanding of p30 DBC biology is urgently required, since p30 DBC could function as either a tumor suppressor or a tumor promoter in a cell type-specific manner. The further study of in vivo physiological function of p30 DBC and how it affects tumorigenesis in tissue-specific manner will not only provide novel insights into this interesting protein, but also allow us to design the best strategies to use any potential p30 DBC modulators for clinical applications.
Figure 1.
The possible mechanisms of p30 DBC in regulating tumorigenesis.
Acknowledgments
This work was supported by the Korea Science and Engineering Foundation (KOSEF) grant funded by the Korea government (MEST) (No. 2009-0059054 and 2009-0063279, J.E.K.) and by grant from the National Institution of Health (1 R01 CA129344, to Z.L.).
Abbreviations
- p30 DBC
DBC1, deleted in breast cancer 1
- SIRT1
sirtuin (silent mating type information regulation 2 homolog) 1
References
- 1.Hamaguchi M, Meth JL, von Klitzing C, Wei W, Esposito D, Rodgers L, et al. DBC2, a candidate for a tumor suppressor gene involved in breast cancer. Proc Natl Acad Sci USA. 2002;99:13647–52. doi: 10.1073/pnas.212516099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Radvanyi L, Singh-Sandhu D, Gallichan S, Lovitt C, Pedyczak A, Mallo G, et al. The gene associated with trichorhinophalangeal syndrome in humans is overexpressed in breast cancer. Proc Natl Acad Sci USA. 2005;102:11005–10. doi: 10.1073/pnas.0500904102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Richardson AL, Wang ZC, De Nicolo A, Lu X, Brown M, Miron A, et al. X chromosomal abnormalities in basal-like human breast cancer. Cancer Cell. 2006;9:121–32. doi: 10.1016/j.ccr.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 4.Fu J, Jiang J, Li J, Wang S, Shi G, Feng Q, et al. The deleted in breast cancer 1 (DBC-1): A novel AR coactivator that promotes AR DNA binding activity. J Biol Chem. 2009;284:6832–40. doi: 10.1074/jbc.M808988200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dali-Youcef N, Lagouge M, Froelich S, Koehl C, Schoonjans K, Auwerx J. Sirtuins: the ‘magnificent seven’, function, metabolism and longevity. Ann Med. 2007;39:335–45. doi: 10.1080/07853890701408194. [DOI] [PubMed] [Google Scholar]
- 6.Kim JE, Chen J, Lou Z. DBC1 is a negative regulator of SIRT1. Nature. 2008;451:583–6. doi: 10.1038/nature06500. [DOI] [PubMed] [Google Scholar]
- 7.Zhao W, Kruse JP, Tang Y, Jung SY, Qin J, Gu W. Negative regulation of the deacetylase SIRT1 by DBC1. Nature. 2008;451:587–90. doi: 10.1038/nature06515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li Z, Chen L, Kabra N, Wang C, Fang J, Chen J. Inhibition of SUV39H1 methyltransferase activity by DBC1. J Biol Chem. 2009;284:10361–6. doi: 10.1074/jbc.M900956200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Anantharaman V, Aravind L. Analysis of DBC1 and its homologs suggests a potential mechanism for regulation of sirtuin domain deacetylases by NAD metabolites. Cell Cycle. 2008;7:1467–72. doi: 10.4161/cc.7.10.5883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kwon HS, Ott M. The ups and downs of SIRT1. Trends Biochem Sci. 2008;33:517–25. doi: 10.1016/j.tibs.2008.08.001. [DOI] [PubMed] [Google Scholar]
- 11.Deng CX. SIRT1, is it a tumor promoter or tumor suppressor? Int J Biol Sci. 2009;5:147–52. doi: 10.7150/ijbs.5.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deppert W. SIRT1 protein levels in cancer: tuning SIRT1 to the needs of a cancer cell. Cell Cycle. 2008;7:2947–8. doi: 10.4161/cc.7.19.7010. [DOI] [PubMed] [Google Scholar]
- 13.Chen WY, Wang DH, Yen RC, Luo J, Gu W, Baylin SB. Tumor suppressor HIC1 directly regulates SIRT1 to modulate p53-dependent DNA-damage responses. Cell. 2005;123:437–48. doi: 10.1016/j.cell.2005.08.011. [DOI] [PubMed] [Google Scholar]
- 14.Ford J, Ahmed S, Allison S, Jiang M, Milner J. JNK2-dependent regulation of SIRT1 protein stability. Cell Cycle. 2008;7:3091–7. doi: 10.4161/cc.7.19.6799. [DOI] [PubMed] [Google Scholar]
- 15.Jung-Hynes B, Nihal M, Zhong W, Ahmad N. Role of Sirtuin Histone Deacetylase SIRT1 in Prostate Cancer: A TARGET FOR PROSTATE CANCER MANAGEMENT VIA ITS INHIBITION? J Biol Chem. 2009;284:3823–32. doi: 10.1074/jbc.M807869200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuzmichev A, Margueron R, Vaquero A, Preissner TS, Scher M, Kirmizis A, et al. Composition and histone substrates of polycomb repressive group complexes change during cellular differentiation. Proc Natl Acad Sci USA. 2005;102:1859–64. doi: 10.1073/pnas.0409875102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yeung F, Hoberg JE, Ramsey CS, Keller MD, Jones DR, Frye RA, Mayo MW. Modulation of NFkappaB-dependent transcription and cell survival by the SIRT1 deacetylase. EMBO J. 2004;23:2369–80. doi: 10.1038/sj.emboj.7600244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huffman DM, Grizzle WE, Bamman MM, Kim JS, Eltoum IA, Elgavish A, Nagy TR. SIRT1 is significantly elevated in mouse and human prostate cancer. Cancer Res. 2007;67:6612–8. doi: 10.1158/0008-5472.CAN-07-0085. [DOI] [PubMed] [Google Scholar]
- 19.Ford J, Jiang M, Milner J. Cancer-specific functions of SIRT1 enable human epithelial cancer cell growth and survival. Cancer Res. 2005;65:10457–63. doi: 10.1158/0008-5472.CAN-05-1923. [DOI] [PubMed] [Google Scholar]
- 20.Kojima K, Ohhashi R, Fujita Y, Hamada N, Akao Y, Nozawa Y, et al. A role for SIRT1 in cell growth and chemoresistance in prostate cancer PC3 and DU145 cells. Biochem Biophys Res Commun. 2008;373:423–8. doi: 10.1016/j.bbrc.2008.06.045. [DOI] [PubMed] [Google Scholar]
- 21.Heltweg B, Gatbonton T, Schuler AD, Posakony J, Li H, Goehle S, et al. Antitumor activity of a small-molecule inhibitor of human silent information regulator 2 enzymes. Cancer Res. 2006;66:4368–77. doi: 10.1158/0008-5472.CAN-05-3617. [DOI] [PubMed] [Google Scholar]
- 22.Ota H, Tokunaga E, Chang K, Hikasa M, Iijima K, Eto M, et al. Sirt1 inhibitor, Sirtinol, induces senescence-like growth arrest with attenuated Ras-MAPK signaling in human cancer cells. Oncogene. 2006;25:176–85. doi: 10.1038/sj.onc.1209049. [DOI] [PubMed] [Google Scholar]
- 23.Liang XJ, Finkel T, Shen DW, Yin JJ, Aszalos A, Gottesman MM. SIRT1 contributes in part to cisplatin resistance in cancer cells by altering mitochondrial metabolism. Mol Cancer Res. 2008;6:1499–506. doi: 10.1158/1541-7786.MCR-07-2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pruitt K, Zinn RL, Ohm JE, McGarvey KM, Kang SH, Watkins DN, et al. Inhibition of SIRT1 reactivates silenced cancer genes without loss of promoter DNA hypermethylation. PLoS Genet. 2006;2:40. doi: 10.1371/journal.pgen.0020040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brunet A, Sweeney LB, Sturgill JF, Chua KF, Greer PL, Lin Y, et al. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science. 2004;303:2011–5. doi: 10.1126/science.1094637. [DOI] [PubMed] [Google Scholar]
- 26.Firestein R, Blander G, Michan S, Oberdoerffer P, Ogino S, Campbell J, et al. The SIRT1 deacetylase suppresses intestinal tumorigenesis and colon cancer growth. PLoS ONE. 2008;3:2020. doi: 10.1371/journal.pone.0002020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kabra N, Li Z, Chen L, Li B, Zhang X, Wang C, et al. SirT1 is an inhibitor of proliferation and tumor formation in colon cancer. J Biol Chem. 2009;284:18210–7. doi: 10.1074/jbc.M109.000034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang RH, Sengupta K, Li C, Kim HS, Cao L, Xiao C, et al. Impaired DNA damage response, genome instability and tumorigenesis in SIRT1 mutant mice. Cancer Cell. 2008;14:312–23. doi: 10.1016/j.ccr.2008.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yuan J, Pu M, Zhang Z, Lou Z. Histone H3-K56 acetylation is important for genomic stability in mammals. Cell Cycle. 2009;8:1747–53. doi: 10.4161/cc.8.11.8620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dai Y, Ngo D, Forman LW, Qin DC, Jacob J, Faller DV. Sirtuin 1 is required for antagonist-induced transcriptional repression of androgen-responsive genes by the androgen receptor. Mol Endocrinol. 2007;21:1807–21. doi: 10.1210/me.2006-0467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fu M, Liu M, Sauve AA, Jiao X, Zhang X, Wu X, et al. Hormonal control of androgen receptor function through SIRT1. Mol Cell Biol. 2006;26:8122–35. doi: 10.1128/MCB.00289-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yuan J, Minter-Dykhouse K, Lou Z. A c-Myc-SIRT1 feedback loop regulates cell growth and transformation. J Cell Biol. 2009;185:203–11. doi: 10.1083/jcb.200809167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sundararajan R, Chen G, Mukherjee C, White E. Caspase-dependent processing activates the proapoptotic activity of deleted in breast cancer-1 during tumor necrosis factor-alpha-mediated death signaling. Oncogene. 2005;24:4908–20. doi: 10.1038/sj.onc.1208681. [DOI] [PubMed] [Google Scholar]
- 34.Garapaty S, Xu CF, Trojer P, Mahajan MA, Neubert TA, Samuels HH. Identification and characterization of a novel nuclear protein complex Involved In nuclear hormone receptor-mediated gene regulation. J Biol Chem. 2009;284:7542–52. doi: 10.1074/jbc.M805872200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Trauernicht AM, Kim SJ, Kim NH, Boyer TG. Modulation of estrogen receptor alpha protein level and survival function by DBC-1. Mol Endocrinol. 2007;21:1526–36. doi: 10.1210/me.2007-0064. [DOI] [PubMed] [Google Scholar]
- 36.Koch HB, Zhang R, Verdoodt B, Bailey A, Zhang CD, Yates JR, 3rd, et al. Large-scale identification of c-MYC-associated proteins using a combined TAP/MudPIT approach. Cell Cycle. 2007;6:205–17. doi: 10.4161/cc.6.2.3742. [DOI] [PubMed] [Google Scholar]
- 37.Bouwmeester T, Bauch A, Ruffner H, Angrand PO, Bergamini G, Croughton K, et al. A physical and functional map of the human TNFalpha/NFkappaB signal transduction pathway. Nat Cell Biol. 2004;6:97–105. doi: 10.1038/ncb1086. [DOI] [PubMed] [Google Scholar]