Abstract
The synapsin III gene, SYN3, which belongs to the family of synaptic vesicle-associated proteins, has been implicated in the modulation of neurotransmitter release and in synaptogenesis, suggesting a potential role in several neuropsychiatric diseases. The human SYN3 gene is located on chromosome 22q12–13, a candidate region implicated in previous linkage studies of schizophrenia. However, association studies of SYN3 and schizophrenia have produced inconsistent results. In this study, four SYN3 SNPs (rs133945(−631c>g), rs133946(−196g>a), rs9862 and rs1056484) were tested in three sets of totally 3759 samples that comprise 655 affected subjects and 626 controls in the Irish Case-Control Study of Schizophrenia (ICCSS), 1350 samples incorporating 273 pedigrees in the Irish Study of High Density Schizophrenia Families (ISHDSF), and 564 unrelated schizophrenia patients and 564 healthy individuals in a Chinese case-control sample. The expression levels of SYN3 in schizophrenic patients and unaffected controls were compared using postmortem brain cDNAs provided by the Stanley Medical Research Institute (SMRI). There was no significant association in either the Irish or Chinese case-control samples, nor in the combined samples. Consistent with this finding, we did not find any significant difference in allele or haplotype frequencies when we used the pedigree disequilibrium test to analyze the Irish family sample. In the expression studies, no significant difference (p=0.507) was observed between patients and controls. Both the association studies and expression studies didn’t support a major role for SYN3 in the susceptibility of schizophrenia in Irish and Chinese populations.
Keywords: synapsin III, schizophrenia, association, Irish, gene expression, Chinese
2 INTRODUCTION
Schizophrenia is a complex neuropsychiatric disorder, and both genetics and neurodevelopmental abnormalities are considered to contribute to its etiology [7;14]. The synapsin III (SYN3) gene, which belongs to the family of synaptic vesicle-associated proteins, has been implicated in the modulation of neurotransmitter release and in synaptogenesis, thus suggesting a potential role in several neuropsychiatric diseases [4;10;10]. Recently, Kao and colleagues reported that SYN3 plays a role in early neural progenitor cell development, which implies the regulation of adult neurogenesis, with possible relevance to neuropsychiatric disease[9]. The human SYN3 gene is located on chromosome 22q12–13, a candidate region implicated in linkage studies of schizophrenia[6;23]. Based on function and location, SYN3 seems to be a candidate gene to schizophrenia.
There are several association studies of SYN3 and schizophrenia in different populations, including those of Asian, European and African ancestry, but most of the results are negative, and positive findings are not consistent across studies [8;12;13;17–20;22]. For example, Porton and colleagues found that a rare, missense polymorphism, S470N, appeared more frequently in European individuals with schizophrenia (5%) than in controls (0.9%) (p=0.0048)[19]. Lachman and colleagues[13] on the other hand, found that none of their control subjects of European descent had the S470N polymorphism and that only two out of 131 subjects in their European-American schizophrenia sample had one copy of the allele; the numbers were too small for them to do a valid statistical analysis. However, they did report an increase in the frequency of the SNP 469G/A AA genotype in African-American patients compared to controls (p=0.04), while the frequency of the A allele is extremely low in schizophrenic and healthy Europeans. The small sample sizes of these studies and the very low frequencies of S470N and 496G/A (minor allele frequency, MAF<0.005) make type I error more likely.
Vawter and colleagues reported that, compared to controls, there were reductions in synapsin IIa and IIIa proteins in the hippocampus of schizophrenia patients (p=0.034) [24]. Furthermore, Porton and colleagues found that synapsin III protein levels were reduced in patients in the prefrontal cortex, an area believed to be the major locus of dysfunction in schizophrenia[20;24].
In this study, we tested for an association between four SYN3 SNPs and schizophrenia in more than 3750 samples, including three Irish sets of 655 cases and 626 controls, an Irish family sample of 273 pedigrees totally about 1350 subjects, and 564 Chinese unrelated patients with schizophrenia and 564 Chinese controls. We also measured the mRNA levels in postmortem brain from schizophrenia patients and unaffected controls to further assess the relationship between SYN3 and schizophrenia.
3 MATERIALS AND METHODS
3.1 The ICCSS sample
The Irish Case-Control Study of Schizophrenia (ICCSS) sample was collected in Northern Ireland, the United Kingdom and the Republic of Ireland. The affected subjects were selected from in-patient and out-patient psychiatric facilities. Subjects were eligible for inclusion if they had a diagnosis of schizophrenia or schizoaffective disorder by DSM-III-R criteria. Controls, selected from several sources, including blood donation centers, were included if they denied a lifetime history of schizophrenia. Both cases and controls were included only if they reported all four grandparents as being born in Ireland or the United Kingdom. In this study, we used 655 (436 males and 219 females) affected subjects and 626 (354 males, 269 females) controls.
3.2 The ISHDSF sample
The Irish Study of High Density Schizophrenia Families (ISHDSF) sample was collected in the same geographic regions as the ICCSS sample. The sample contained 273 pedigrees and about 1350 subjects had DNA sample for genotyping. Historically, the ISHDSF sample had multiple disease definitions reflecting the probable genetic relationship of the syndromes to classic schizophrenia. In this study, we used the narrow definition, which includes schizophrenia, poor-outcome schizoaffective disorder and simple schizophrenia according to DSM-III criteria[11], so we could compare the results with that of the ICCSS sample directly. Using this definition, 522 subjects were classified as affected.
3.3 The Chinese sample
We tested 564 (male 339, female 225) unrelated patients with schizophrenia and 564 (male 291, female 273) control individuals. All subjects were of Han Chinese origin. A clinical interview was administered by two independent senior psychiatrists to all subjects including cases and controls according to the criteria of the DSM-IV. All patients were recruited from in-patient and out-patient of the Shanghai Mental Health Center, East China. The healthy controls were drawn from the general population of East China. None had a history of psychotic disorders. Participants were fully informed of, and gave written consent for, the genetic analysis, which was reviewed and approved by the Shanghai Ethics Committee of Human Genetic Resources.
3.4 Marker selection and genotyping
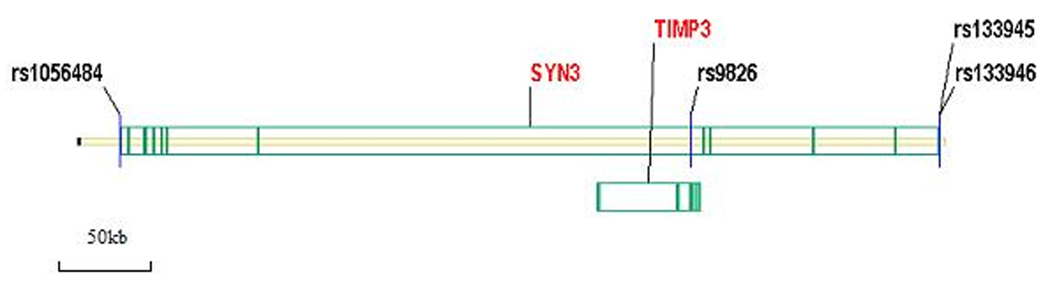
A total of four SNPs were tested in this study. Following previous studies, we chose rs133946 (−636 C/G) and rs133945 (−196 G/A), whose frequencies are higher than 10% in populations of European ancestry. They are located in the 5’ promoter region of SYN3. SNP rs9862 is located in intron 5 of SYN3. A synonymous polymorphism is located in exon 2 of the TIMP3 gene. TIMP3 is in the SYN3 gene region but it is in the reverse orientation. TIMP3 gene (Tissue Inhibitor of Metalloproteinase 3) belongs to the TIMP gene family, which encodes the proteins that are inhibitors of the matrix metalloproteinases, a group of peptidases involved in degradation of the extracellular matrix (ECM). Mutations in this gene have been associated with the autosomal dominant disorder Sorsby's fundus dystrophy. SNP rs1056484 is located in the 3’ untranslated region of SYN3. The distribution of the SNPs in the gene is shown in Figure 1. All genotyping was conducted with the TaqMan method[15]. The genotyping assays were developed by Applied BioSystems Corporation (Foster City, CA). Genotypes were scored using an Excel template developed in our lab. All typed SNPs were checked for Mendelian consistency and Hardy-Weinberg Equilibrium (HWE)[25].
Figure 1.
Genomic structure and location of genotyped SNPs in SYN3. SYN3 spans over 49.4 kb and includes 12 exons (green bars). Four markers were genotyped in this study. TIMP3 locates in SYN3.
3.5 Expression studies
The expression studies were carried out with postmortem brain cDNAs from the Stanley Medical Research Institute (SMRI) (http://www.stanleyresearch.org/). The mRNA isolation and reverse transcription to cDNA were carried out by SMRI researchers. The SMRI panel consisted of 104 subjects: thirty-five individuals diagnosed with schizophrenia (26 males and 9 females; mean ±SD age, 42.6 ±8.5 years; postmortem interval (PMI), 31.4± 15.5 h; brain pH, 6.5±0.2), thirty-four individuals with bipolar disorder (16 males and 18 females; mean±SD age, 45.4±10.6 years; PMI, 37.9±18.6 h; brain pH, 6.4 0.3), and thirty-five unaffected controls (26 males and 9 females; mean±SD age, 44.2±7.6 years; PMI, 29.4±12.9 h; brain pH, 6.6±0.3). Diagnoses were made according to DSM-IV criteria. There were no significant demographic differences between the schizophrenia, bipolar disorder and control subjects. All schizophrenic patients were medicated with antipsychotics.
Quantitative PCRs were conducted with a TaqMan expression probe (Hs01022312_m1) for the SYN3 gene, and human TATA box binding protein (TBP) gene was used as internal reference. Each sample was amplified in triplicate, and for each reaction, 0.25 ng of cDNA were used in a 15 µL PCR reaction also containing the FAM-labeled SYN3 probe and VIC-labeled TBP probe. PCR was conducted with the Roche Lightcycler 480(Switzerland). PCR cycling parameters were 95°C for 2 minutes, followed by 55 cycles of 92°C for 15 seconds and 60°C for 1 minute. The expression level of each reaction was determined by the CT value (calculated by the iCycler software, version 3.1). The results from three repeat assays were averaged to produce a single mean CT value for each individual. The relative expression level between the SYN3 and TBP for each individual was calculated by the 2−ΔC T method, where ΔCT = CT SYN3 - CT TBP[16].
3.6 Statistical analyses
For the case-control samples, the COCAPHASE module of the UNPHASED program (version 2.4)[3] was used to analyze both single marker and multi-marker haplotype associations. The PDTPHASE module was used to analyze the ISHDSF sample. In this analysis, both vertical and horizontal transmissions were included. We used the HAPLOVIEW program [1] to estimate pairwise LD and to illustrate haplotype blocks. The p-values reported were based on weighting all families equally (the ave option in the program). For both the case-control and family samples, haplotypes with frequencies less than 1% were aggregated.
For the SMRI samples, potentially confounding variables were tested for association with SYN3 expression in all individuals using bivariate correlation analysis with the SPSS software (version 10 for Windows). For the schizophrenia and healthy control samples, we found that only brain pH was marginally correlated with SYN3 expression level (R=0.238, p=0.052), therefore brain pH was used as a covariate in subsequent analyses. ANCOVAs were used to compare the expression levels between controls and schizophrenia patients. A power calculation was performed with a web-based statistical program, Genetic Power Calculator[21]. Power was estimated under multiplicative model of inheritance, assuming the disease prevalence to be 1% and the population susceptibility allele frequencies to be the values observed in control samples.
4 RESULTS
4.1 Association analyses of the ICCSS sample
Marker information and allele frequencies are presented in Table 1. For all markers typed, no deviations from HWE were observed. There is no allele and genotype frequency difference for any of the four markers between cases and controls (Table 1). For rs133945( −196 g/a), the A allele is the minor allele in schizophrenia, with a frequency of 0.477, while in controls the A allele is the more common allele with a frequency of 0.506. However, the difference in genotype and allele distribution did not reach statistic significance (p=0.285, 0.197 respectively). We examined the LD and haplotype structure with the HAPLOVIEW program, found the D’ of rs133495 and rs133496 to be 0.99, so we did a haplotype analysis with rs133945 and rs133946. Only the global p-value indicated marginal positive significance (p=0.0234) (data not shown).
Table 1.
Marker characteristics and single marker associations (p-values) in the ICCSS sample.
| Marker | Function | HWE | Genotype(freq.) | p value | Allele (freq.) | p value | Odds ratio [95%CI] |
||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| rs133946 | 5'-promoter | 0.29 | GG | GC | CC | G | C | ||||
| Case | 178(0.276) | 320(0.495) | 148(0.229) | 0.319 | 676(0.523) | 616(0.477) | 0.170 | 1.115 | |||
| Control | 160(0.257) | 297(0.477) | 165(0.265) | 617(0.496) | 627(0.504) | (0.954 –1.303) | |||||
| rs133945 | 5'-promoter | 0.52 | GG | GA | AA | G | A | ||||
| Case | 178(0.273) | 325(0.499) | 148(0.227) | 0.293 | 681(0.523) | 621(0.477) | 0.136 | 1.125 | |||
| Control | 159(0.251) | 308(0.486) | 167(0.263) | 626(0.494) | 642(0.506) | (0.963–1.313) | |||||
| rs9862 | SYN3 intron; TIMP1 exon |
0.56 | CC | CT | TT | C | T | ||||
| Case | 170(0.267) | 314(0.492) | 154(0.241) | 0.558 | 654(0.513) | 622(0.487) | 0.901 | 0.993 | |||
| Control | 161(0.254) | 331(0.522) | 142(0.224) | 651(0.515) | 615(0.485) | (0.851–1.160) | |||||
| rs1056484 | 3'-UTR | 0.59 | CC | CT | TT | C | T | ||||
| Case | 238(0.371) | 306(0.477) | 97(0.151) | 0.717 | 782(0.610) | 500(0.390) | 0.572 | 1.047 | |||
| Control | 231(0.366) | 294(0.466) | 106(0.168) | 756(0.599) | 506(0.401) | (0.893–1.227) | |||||
4.2 Association analyses of the ISHDSF sample
The genotype frequencies of all four markers studied were in HWE. Using the pedigree disequilibrium test (PDT) to examine the associations, we found none of the markers reached 5% nominal significance. Similar to our case-control sample, only rs133945 and rs133946 were grouped in a single LD block. We conducted haplotype analyses; however, no significant associations were observed. (Data not shown).
4.3 Association analyses of the Chinese case-control sample
The genotype frequencies of all four markers were in the same direction as ICCSS, with nodeviation from HWE. No marker reached 5% nominal significance. The LD group was the same as in the Irish samples. No significant association of haplotypes was found (Data not shown).
4.4 Expression studies of the SYN3 gene in the brain
We used quantitative PCR to determine the expression levels of SYN3. First, variables potentially affecting postmortem mRNA levels were evaluated separately from the expression of the SYN3 gene by bivariate correlation analysis. SYN3 was not significantly correlated with PMI, freezer storage time, age, percent of protein extracted or gender (data not shown). Brain pH had marginal effect on the expression of the SYN3 gene (R=0.238, p=0.052). Based on this finding, we used brain pH as a covariate in subsequent analyses. No significance difference (p=0.507) in SYN3 expression levels was observed between patients(mean ± standard error: 11.002 ± 1.65, n=34) and controls (10.660 ± 1.289,n=31).
5 DISCUSSION
Functional studies and its location imply that SYN3 may play a role in schizophrenia neurodevelopmental etiology. In this study, we were unable to confirm an association between SYN3 and schizophrenia in the Irish and Chinese populations. We tested four SNPs in 3759 individuals from three sets of samples that comprise the Irish Case-Control Study of Schizophrenia (ICCSS), the Irish Study of High-Density Schizophrenia Families (ISHDSF), and a Chinese case-control sample.
In our Irish sample, we found the A allele of rs133945 to be the minor allele among schizophrenia patients, with a frequency of 0.477, which is very similar to what Lachman and colleagues reported for their European sample (minor allele frequency of 0.48) Among controls in our study, the A allele was the more common allele with a frequency of 0.506. However, the difference in genotype and allele distribution did not reach statistical significance (p=0.285, 0.197 respectively). The frequencies reported in the Lachman's study were different from the studies of Asian populations which contain a frequency A allele of rs133945 of 0.31 in a Chinese schizophrenic population[22] and of 0.28 in a Japanese one [17]. However, neither did these two Asian studies show any positive association. We also tested the 1128 Chinese case control samples. The frequency (31.9%) is close to Tsai and colleague's result[22]. Our Chinese samples' results indicated that no association of these four markers with schizophrenia exists.
We combined the Irish and Chinese case-control samples for a total of 2409 individuals, and still found no significant association SYN3 SNPs and schizophrenia (Table 4). The result from our Irish family samples also showed no association between any of these four markers and schizophrenia. The haplotype analysis of the Irish case-control sample indicated a positive result (p=0.023) only before correction for multiple testing. The haplotype analyses of the Chinese case-control and the Irish family samples both produced negative results. Power analyses showed that the power was more than 80% when genotype relative risk (GRR) was set at 1.3–1.5 under a multiplicative model of inheritance.
Table 4.
Association analyses of the Chinese and Irish case-control Samples, totaling 2409 samples.
| Marker | Sample | Allele (freq.) | p value | Odds ratio [95%CI] |
|
|---|---|---|---|---|---|
| rs133946 | G | C | |||
| Case | 1372(0.593) | 942(0.407) | 0.065 | 1.119 | |
| Control | 1192(0.565) | 916(0.435) | [0.993–1.261] | ||
| rs133945 | G | A | |||
| Case | 1403(0.600) | 935(0.400) | 0.113 | 1.099 | |
| Control | 1317(0.577) | 965(0.423) | [0.978–1.236] | ||
| rs9862 | C | T | |||
| Case | 1304(0.561) | 1022(0.439) | 0. 52 | 1.039 | |
| Control | 1187(0.551) | 967(0.449) | [0.924–1.17] | ||
| rs1056484 | C | T | |||
| Case | 1733(0.746) | 591(0.254) | 0.688 | 1.028 | |
| Control | 1635(0.740) | 573(0.260) | [0.899– 1.174] | ||
We compared the expression level of SYN3 between the postmortem brain cDNAs from schizophrenic patients and unaffected controls provided by the SMRI using quantitative real-time PCR. Vawter[24] and Porton [20]used western immunoblotting method and found that protein SYN3 levels were significantly decreased in the hippocampus or dorsolateral prefrontal cortex (DLPFC) of individuals with schizophrenia compared to controls. However, we did not find a significant mRNA expression difference between cases and controls.
In summary, data from our family and case-control association studies and expression study suggest that SYN3 might not play a major role in schizophrenia.
Table 2.
Association analyses in the ISHDSF
| Marker | Allele | Trio-T+NT | AffSib+UnafSib | Z | p |
|---|---|---|---|---|---|
| rs133946 | C | 77+660 | 86+652 | 1.109 | 0.268 |
| G | 87+684 | 78+692 | |||
| rs133945 | A | 75+666 | 78+654 | 0.882 | 0.378 |
| G | 79+670 | 76+682 | |||
| rs9862 | C | 68+655 | 63+643 | −0.451 | 0.652 |
| T | 94+695 | 99+707 | |||
| rs1056484 | C | 63+539 | 59+577 | 1.095 | 0.274 |
| T | 85+773 | 89+773 |
Table 3.
Association analyses of the Chinese case-control samples.
| Marker | Sample | HWE P |
Genotype(freq.) | P | Allele(freq.) | P | Odds ratio [95%CI] |
|||
|---|---|---|---|---|---|---|---|---|---|---|
| GG | GC | CC | G | C | ||||||
| Case | 0.07 | 228(0.446) | 240(0.470) | 43(0.084) | 0.574 | 696(0.681) | 326(0.319) | 0.474 | 1.073 | |
| rs133946 | Control | 0.47 | 188(0.435) | 199(0.461) | 45(0.104) | 575(0.666) | 289(0.334) | [0.885~1.302] | ||
| GG | GA | AA | G | A | ||||||
| Case | 0.46 | 248(0.479) | 226(0.436) | 44(0.085) | 0.437 | 722(0.697) | 314(0.303) | 0.450 | 1.075 | |
| rs133945 | Control | 0.47 | 239(0.471) | 213(0.420) | 55(0.108) | 691(0.681) | 323(0.319) | [0.891~1.296] | ||
| CC | CT | TT | C | T | ||||||
| Case | 0.55 | 198(0.377) | 254(0.484) | 73(0.139) | 0.335 | 650(0.619) | 400(0.381) | 0.487 | 1.067 | |
| rs9862 | Control | 0.22 | 168(0.378) | 200(0.450) | 76(0.171) | 536(0.604) | 352(0.396) | [0.888~1.282 | ||
| CC | CT | TT | C | T | ||||||
| Case | 0.28 | 432(0.829) | 87(0.167) | 2(0.004) | 0.213 | 951(0.913) | 91(0.087) | 0.173 | 0.797 | |
| rs1056484 | Control | 0.1 | 406(0.858) | 67(0.142) | 0(0.000) | 879(0.929) | 67(0.071) | [0.573~1.106] | ||
ACKNOWLEDGEMENTS
This study is supported by an Independent Investigator Award and research grant 07R-1770 to XC from the NARSAD and the Stanley Medical Research Institute, and by grant RO1MH41953 to KSK from National Institute of Mental Health. We thank the patients and their families for participating in this study. The Northern Ireland Blood Transfusion Service assisted with collection of control sample. We thank Kay Grennan from the University of Chicago for reviewing English of the paper.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- 1.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 2.Chin LS, Li L, Ferreira A, Kosik KS, Greengard P. Impairment of Axonal Development and of Synaptogenesis in Hippocampal-Neurons of Synapsin I-Deficient Mice. Proceedings of the National Academy of Sciences of the United States of America. 1995;92:9230–9234. doi: 10.1073/pnas.92.20.9230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dudbridge F. Pedigree disequilibrium tests for multilocus haplotypes. Genet. Epidemiol. 2003;25:115–121. doi: 10.1002/gepi.10252. [DOI] [PubMed] [Google Scholar]
- 4.Feng J, Chi P, et al. Regulation of neurotransmitter release by synapsin III. Journal of Neuroscience. 2002;22:4372–4380. doi: 10.1523/JNEUROSCI.22-11-04372.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ferreira A, Kao HT, Feng J, Rapoport M, Greengard P. Synapsin III: Developmental expression, subcellular localization, and role in axon formation. Journal of Neuroscience. 2000;20:3736–3744. doi: 10.1523/JNEUROSCI.20-10-03736.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gill M, et al. A combined analysis of D22S278 marker alleles in affected sib-pairs: Support for a susceptibility locus for schizophrenia at chromosome 22q12. American Journal of Medical Genetics. 1996;67:40–45. doi: 10.1002/(SICI)1096-8628(19960216)67:1<40::AID-AJMG6>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 7.Harrison PJ. Schizophrenia susceptibility genes and neurodevelopment. Biological Psychiatry. 2007;61:1119–1120. doi: 10.1016/j.biopsych.2007.02.021. [DOI] [PubMed] [Google Scholar]
- 8.Imai K, Harada S, Kawanishi Y, Tachikawa H, Okubo T, Suzuki T. Polymorphisms in the promoter and coding regions of the synapsin III gene. A lack of association with schizophrenia. Neuropsychobiology. 2001;43:237–241. doi: 10.1159/000054896. [DOI] [PubMed] [Google Scholar]
- 9.Kao HT, Li P, Chao HM, Janoschka S, Pham K, Feng J, Mcewen BS, Greengard P, Pieribone VA, Porton B. Early involvement of synapsin III in neural progenitor cell development in the adult hippocampus. Journal of Comparative Neurology. 2008;507:1860–1870. doi: 10.1002/cne.21643. [DOI] [PubMed] [Google Scholar]
- 10.Kao HT, Porton B, Czernik AJ, Feng J, Yiu G, Haring M, Benfenati F, Greengard P. A third member of the synapsin gene family. Proc. Natl. Acad. Sci. U. S. A. 1998;95:4667–4672. doi: 10.1073/pnas.95.8.4667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kendler KS, Myers JM, O'neill FA, Martin R, Murphy B, MacLean CJ, Walsh D, Straub RE. Clinical features of schizophrenia and linkage to chromosomes 5q, 6p, 8p, and 10p in the Irish Study of High-Density Schizophrenia Families. Am. J. Psychiatry. 2000;157:402–408. doi: 10.1176/appi.ajp.157.3.402. [DOI] [PubMed] [Google Scholar]
- 12.Lachman HM, Stopkova P, Papolos DF, Pedrosa E, Margolis B, Aghalar MR, Saito T. Analysis of synapsin III-196 promoter mutation in schizophrenia and bipolar disorder. Neuropsychobiology. 2006;53:57–62. doi: 10.1159/000091720. [DOI] [PubMed] [Google Scholar]
- 13.Lachman HM, Stopkova P, Rafael MA, Saito T. Association of schizophrenia in African Americans to polymorphism in synapsin III gene. Psychiatr. Genet. 2005;15:127–132. doi: 10.1097/00041444-200506000-00009. [DOI] [PubMed] [Google Scholar]
- 14.Lewis DA, Levitt P. Schizophrenia as a disorder of neurodevelopment. Annual Review of Neuroscience. 2002;25:409–432. doi: 10.1146/annurev.neuro.25.112701.142754. [DOI] [PubMed] [Google Scholar]
- 15.Livak KJ. Allelic discrimination using fluorogenic probes and the 5' nuclease assay. Genet. Anal. 1999;14:143–149. doi: 10.1016/s1050-3862(98)00019-9. [DOI] [PubMed] [Google Scholar]
- 16.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(T)(-Delta Delta C) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 17.Ohmori O, Shinkai T, Hori H, Kojima H, Nakamura J. Synapsin III gene polymorphisms and schizophrenia. Neurosci. Lett. 2000;279:125–127. doi: 10.1016/s0304-3940(99)00970-2. [DOI] [PubMed] [Google Scholar]
- 18.Ohtsuki T, Ichiki R, Toru M, Arinami T. Mutational analysis of the synapsin III gene on chromosome 22q12-q13 in schizophrenia. Psychiatry Res. 2000;94:1–7. doi: 10.1016/s0165-1781(00)00123-2. [DOI] [PubMed] [Google Scholar]
- 19.Porton B, Ferreira A, DeLisi LE, Kao HT. A rare polymorphism affects a mitogen-activated protein kinase site in synapsin III: possible relationship to schizophrenia. Biol. Psychiatry. 2004;55:118–125. doi: 10.1016/j.biopsych.2003.07.002. [DOI] [PubMed] [Google Scholar]
- 20.Porton B, Wetsel WC. Reduction of synapsin III in the prefrontal cortex of individuals with schizophrenia. Schizophr. Res. 2007;94:366–370. doi: 10.1016/j.schres.2007.04.016. [DOI] [PubMed] [Google Scholar]
- 21.Purcell S, Cherny SS, Sham PC. Genetic Power Calculator: design of linkage and association genetic mapping studies of complex traits. Bioinformatics. 2003;19:149–150. doi: 10.1093/bioinformatics/19.1.149. [DOI] [PubMed] [Google Scholar]
- 22.Tsai MT, Hung CC, Tsai CY, Liu MY, Su YC, Chen YH, Hsiao KJ, Chen CH. Mutation analysis of synapsin III gene in schizophrenia. Am. J. Med. Genet. 2002;114:79–83. doi: 10.1002/ajmg.10116. [DOI] [PubMed] [Google Scholar]
- 23.Vallada H, et al. A transmission disequilibrium and linkage analysis of D22S278 marker alleles in 574 families: further support for a susceptibility locus for schizophrenia at 22q12. Schizophrenia Research. 1998;32:115–121. doi: 10.1016/s0920-9964(98)00048-6. [DOI] [PubMed] [Google Scholar]
- 24.Vawter MP, Thatcher L, Usen N, Hyde TM, Kleinman JE, Freed WJ. Reduction of synapsin in the hippocampus of patients with bipolar disorder and schizophrenia. Molecular Psychiatry. 2002;7:571–578. doi: 10.1038/sj.mp.4001158. [DOI] [PubMed] [Google Scholar]
- 25.Wigginton JE, Cutler DJ, Abecasis GR. A note on exact tests of Hardy-Weinberg equilibrium. Am. J. Hum. Genet. 2005;76:887–893. doi: 10.1086/429864. [DOI] [PMC free article] [PubMed] [Google Scholar]