Abstract
The lipoxygenase family of lipidperoxidizing, nonheme iron dioxygenases form products that are precursors for diverse physiological processes in both plants and animals. In soybean (Glycine max), five vegetative isoforms, VLX-A, VLX-B, VLX-C, VLX-D, VLX-E, and four seed isoforms LOX-1, LOX-2, LOX-3a, LOX-3b have been identified. In this study, we determined the crystal structures of the substrate-free forms of two major vegetative isoforms, with distinct enzymatic characteristics, VLX-B and VLX-D. Their structures are similar to the two seed isoforms, LOX-1 and LOX-3, having two domains with similar secondary structural elements: a β-barrel N-terminal domain containing highly flexible loops and an α-helix-rich C-terminal catalytic domain. Detailed comparison of the structures of these two vegetative isoforms with the structures of LOX-1 and LOX-3 reveals important differences that help explain distinct aspects of the activity and positional specificity of these enzymes. In particular, the shape of the three branches of the internal subcavity, corresponding to substrate-binding and O2 access, differs among the isoforms in a manner that reflects the differences in positional specificities.
Keywords: crystal, structure, soybean, lipoxygenase
INTRODUCTION
Lipoxygenases (LOXs) are nonheme, iron-containing dioxygenases that catalyze the regio- and stereo-specific insertion of molecular oxygen into polyunsaturated lipids containing 1z-4z-pentadiene systems to yield conjugated hydroperoxide products.1 These enzymes are ubiquitous in both plants and animals, participating in diverse physiological processes2,3 and recently have been discovered in fungal4–6 and bacterial species.7,8
In plants, LOXs function in multiple aspects of plant growth and development including lipid metabolism,9–11 nitrogen storage,12–14 plant defense responses,15–17 stress responses,18 organ development,19 mechanical wounding,20 and plant–microbe interactions.15,21 Plant LOXs are characterized based on their positional specificity against the substrates, linoleic (18:2) and linolenic (18:3) acids. There are two types of LOXs in plants, 9-LOX and 13-LOX, which oxygenate at the C-9 or C-13 position of the fatty acid, respectively. In addition to this regiospecificity, most lipoxygenases control the stereochemistry of hydroperoxide products.22,23
The most detailed characterizations of plant LOXs have been carried out on the LOX isoforms from soybean (Glycine max cv. Wye) seed, LOX-1, LOX-2, LOX-3a, and LOX-3b.24 In addition, five LOX isoforms were detected in mature soybean leaves and were termed the vegetative lipoxygenases (VLX-A, -B, -C, -D, -E).13 VLX-A, VLX-B, and VLX-C are localized in the cytosol of paravenial mesophyll (PVM) cells and VLX-D is localized in the vacuole of the PVM as well as in the bundle sheath cells.25 So far the location of VLX-E is not clear. VLX-C was also shown to be present in the vacuole in epidermal cells at low levels.25 These data indicate that VLX-A, VLX-B, and VLX-C are likely involved in the hydroperoxidation of lipids in the PVM.25 All five vegetative isoforms are active against the two predominant free fatty acids in plants, linoleic and linolenic acids. In addition, VLX-C and VLX-D are active against triglycerides.25,26 VLX-A, VLX-B, and VLX-E have highest activity against linoleic acid (18:2) and VLX-C and VLX-D have highest activity against linolenic acid (18:3).25,26 The hydroperoxide products produced by VLX activity are predominantly 13(S)-hydroperoxy octadecadienoic acid (HPOD), and 13(S)-hydroperoxy octadecatrienoic acid (HPOT), which characterizes them as 13(S) LOXs.26 VLX-A and VLX-B also produce 9-HPOD at a ratio of approximately 60% 13-HPOD and 40% 9-HPOD from 18:2.26 All these 9-LOX products are a racemic mixture of R and S stereoisomers.26
Unusual features of catalysis by LOX-1 have been the subject of detailed study.27 A large kinetic isotope effect, when deuterium-labeled substrate is used, points to hydrogen tunneling in the first step of the reaction cycle. Also, there is a lag phase associated with LOX catalysis that appears to result from an activation process preceding initiation of the reaction. The catalytic iron atom in resting lipoxygenase is ferrous Fe2+. Upon activation of the enzyme by one equivalent of the hydroperoxy product, the iron is converted to the ferric Fe3+ oxidation state.1,27,28 The initial and rate limiting step involves concerted proton abstraction and a one electron oxidation of the fatty acid substrate producing a radical that is delocalized across the pentadiene system.27 Next, the fatty acid radical is trapped by oxygen molecule resulting in a peroxy radical intermediate that is then reduced and protonated by the ferrous center and an associated water molecule, respectively.27 The enzyme is then primed for another round of catalysis.
Persistent questions about the structural bases for lipoxygenase catalysis remain. The circumstances under which lipoxygenases associate with membranes peripherally are not universal. No structure containing the fatty acid substrate in the large internal cavities of the enzyme has been obtained. Although substrate entry and product exit sites have been suggested, definitive proof of them is also lacking. The crystal structures of several LOXs have been determined, including soybean seed LOX-1 and LOX-3, an 8-R-LOX from coral, and a rabbit reticulocyte 15-LOX.29–33 The family of well-characterized lipoxygenase isoforms in soybean presents a unique opportunity to correlate isolated structural changes with known differences in subcellular location, substrate specificity, and product variation in the isoforms and to extend the insights to the entire lipoxygenase family. Here, we report the crystal structures of two lipoxygenases, VLX-B (Protein ID: AAB67732) and VLX-D (CAA39604), which represent the soybean vegetative lipoxygenases. Our discussion focuses on several structure–function relationships of soybean lipoxygenases, such as the contribution of the N-terminal β-barrel to calcium binding and the relation of cavity shape to differences in reactivity. We considered it instructive to conduct a detailed comparison of these vegetative lipoxygenases with two seed lipoxygenases, LOX-1 and LOX-3, particularly with respect to their putative binding site geometries and their unique regio- and stereospecificity.
METHODS
VLX Expression and Purification
Previously, the VLX isoforms were cloned into the pSBETa vector.26 The DNA sequences of plasmids harboring VLX-B (AAB67732) and VLX-D (CAA39604) were verified to be identical to the corresponding gene sequences, U50075.1 and U50075.1 respectively, and then were used to individually transform BL21(DE3)pLysS Escherichia coli cells (Stratagene, La Jolla, CA), and the cells were grown at 37°C with shaking (250 rpm) in M9 medium (42 mM Na2HPO4, 22 mM KH2PO4, 19 mM NH4Cl, 8.5 mM NaCl, 1 mM MgSO4, 0.2% (w/v) glucose, 50 μM Fe(NH4)2(SO4)2) supplemented with chloroamphenicol (20 μg ml–1) and kanamycin (30 μg ml–1). Overexpression of VLX-B and VLX-D was induced by addition of isopropyl β-d-thiogalactopyranoside (IPTG) to a final concentration of 0.4 mM at midlog phase (A600 = 0.4–0.5). After agitation (250 rpm) for 24 h at 20°C, the cells were harvested individually by centrifugation (4000g for 10 min). The cell pellets were resuspended individually in lysis buffer (50 mM Tris pH 7.4, 500 mM NaCl, 10% glycerol and 0.1% Tween-20), lysed by sonication (5 × 15 s, model 450 sonifier1, Branson Ultrasonics), and the lysates were cleared by centrifugation (10,000g for 30 min). Supernatant was then fractionated by (NH4)2SO4. The resulting 30–70% fraction was recovered by centrifugation (20,000g for 5 min). The pellet was resuspended in a minimal volume of Buffer A (20 mM Tris pH 8.0, 25 mM NaCl) and dialyzed overnight at 4°C against the same buffer to remove the remaining (NH4)2SO4. This desalted protein was loaded onto a MonoQ™ GL10/100 anion exchange column (Amersham Biosciences/BioCad 700E preparative HPLC, Applied Biosystems) equilibrated in Buffer A at a flow rate of 4 mL min–1, and the proteins were eluted with a linear NaCl gradient. The catalytically active VLX-B and VLX-D fractions were eluted in a 0.07–0.12M NaCl gradient (over 10 min) and a 0.09–0.14M NaCl gradient (over 10 min), respectively. Fractions containing VLXs were pooled and dialyzed into Buffer B (10 mM Na phosphate, pH 6.8.) and passed over a CHT-II hydroxyapatite column (BioRad), which was pre-equilibrated in Buffer B, at a flow rate of 10.0 mL min–1. Each column was eluted with a linear gradient of Na phosphate (from 10 to 500mM). VLX-B and VLX-D were individually eluted at 15–65 mM and 10–50 mM Na phosphate (over 5 min), respectively. The VLX-B and VLX-D fractions from the CHT-II step were then individually dialyzed into Buffer C (20 mM Tris pH 8.0, 5 mM Na acetate) and loaded onto a MonoQ™ GL10/100 anion exchange column (Amersham Biosciences), pre-equilibrated in Buffer C at a flow rate of 4 mL min–1. The active VLX-B and VLX-D were eluted with a 0.01–0.1M Na acetate gradient (over 10 min) and 0.15–0.2M Na acetate gradient (over 15 min), respectively. Confirmation of the presence and purity of the VLX-B and VLX-D were made by SDS-PAGE. The yields for VLX-B and VLX-D were approximately 0.5 and 1.25 mg/L, respectively.
Crystallization of VLX-B and VLX-D
For crystallization of VLXs, a solution of purified VLX-B (9 mg mL–1) or VLX-D (6 mg mL–1)in 10 mM MOPS (pH 7.5) containing 25 mM NaCl was prepared. Crystallization trials were performed using the hanging drop vapor diffusion method at two temperatures (277 and 293 K). Optimized VLX-B and VLX-D crystals were obtained at 293 K by mixing the above protein solution (1.5 μL) with an equal volume of reservoir solution containing 12% (w/v) PEG 4000, 0.1M MES (pH 6.5 for VLX-B), and 20–22% (w/v) PEG 3350, 0.25M potassium sulfate (pH 6.6 for VLX-D), respectively. Crystals usually appeared after 3 days, and larger crystals with dimensions of ~0.2 × 0.3 × 0.7 mm were obtained after 2 weeks. The VLX-B crystal belongs to the orthorhombic space group, P212121 (a = 99.99, b 105.31, c = 105.92 Å), with one molecule in the asymmetric unit. On the other hand, the VLX-D crystal belongs to the monoclinic group, P21 (a = 92.76, b = 115.10, c = 120.22 Å, β = 112.34°) with two molecules in the asymmetric unit. The solvent contents of VLX-B and VLX-D are approximately 53% and 51% by volume, respectively. Enzyme recovered from dissolved crystals had normal catalytic activity. For the cryodiffraction experiments, the crystals were soaked for 10 min in cryoprotectants (25% glycerol in each reservoir solution) through three solutions with intermediate concentrations of these reagents. After incubation in the final solution, the crystal was mounted in a nylon loop and placed in a nitrogen gas stream at 100 K to achieve flash freezing. The VLX-B (2.4 Å resolution) and VLX-D (2.4 Å resolution) diffraction data were collected from the Berkeley Advanced Light Source (ALS, beam line 8.2.1) and Rigaku Saturn 92/MicroMax-007 diffractometer, respectively, at a temperature of 100 K. Data were indexed, integrated, and scaled with Crystal-Clear 1.3.6 (Rigaku MSC).
Structural solution and refinement
The structure of VLX-B was solved by the molecular replacement method using coordinates of the soybean lipoxygenase L-1 (1YGE) and the software package AMoRe.34 The VLX-B coordinates were used for VLX-D phase determination. Amino acid substitutions, insertions, and deletions were performed by using the graphics program O.35 The rigid body refinements of the initial position for VLX-B and VLX-D were carried out by using 15.0–3.5-Å resolution data and gave an R-value of 42.0% and 41.3%, respectively. After several cycles of positional and temperature factor refinements using the program X-PLOR36 and a series of simulated annealing omit maps, most residues were fitted against the electron density, except for seven residues that are located in N-terminus and one loop area. The noncrystallographic symmetry (NCS) restraints were not used during refinement of VLX-D, and two molecules in the asymmetric unit were refined independently. Both coordinates have been deposited in the Protein Data Bank (VLX-B: 2IUJ, VLX-D: 2IUK).
Modeling of linoleic acid
The coordinates of one of a cluster of linoleic acid molecules previously docked in LOX-137 were chosen to begin modeling the substrate into the structures of VLX-B and VLX-D. In short, the previous docking used the LOX-1 coordinates 1YGE31 with all waters removed except the one bound to iron. Docking was achieved using Autodock 3.03/Autogrid.38 In the cluster of six low-energy docked molecules with similar orientation, the average distance from C-11 to the oxygen of the water bound to iron is 3.9 ± 0.5 Å and the corresponding C-10 distance is 3.9 ± 0.7 Å.
For VLX-B and VLX-D, after removing all the water molecules in both structures and docking one of these linoleic acid molecules, the quick energy minimization using X-PLOR36 with potential function parameters of CHARMM1939 was applied; especially, the close contact between carboxyl group of linoleic acid and the side chain of residue 567 (leucine) in VLX-D was revealed after this process.
Dynamic light scattering
The radius and molecular weight of VLX-B and VLX-D were estimated using a DynaPro-Titan (Wyatt Technology) instrument at 22°C. Purified VLX-B or VLX-D (2 mg mL–1) in a freshly prepared PBS buffer were filtered through polyvinylidene difluoride filter (0.2 μm, Millipore). Scattering data were acquired through accumulation (5 times) of 10 scans with 10 s/scan, with the laser intensity set to a range of 50–60% (30–36 mW). The corresponding molecular weight and radius were calculated using the software package ‘DYNAMICS V6’ supplied with the instrument.
RESULTS
Structure Determinations
Two recombinant soybean vegetative lipoxygenases, VLX-B, VLX-D were crystallized and their corresponding structures were determined at 2.4-Å resolution. VLX-B was solved by molecular replacement using coordinates of soybean LOX-1 (1YGE). In turn, the structure of VLX-D was determined using the coordinates of the deduced VLX-B structure. The final R factors (Table I) for the VLX-B and VLX-D were 20.0% (Rfree = 23.9% for the random 5% data) and 20.3% (Rfree 23.5% for the random 5% data), respectively. The number of reflections above 2σ level were 41,906 (89.5% completeness) between 15.0- and 2.4-Å resolution for VLX-B and 87,392 (96.1% completeness) between 15.0- and 2.4-Å resolution for VLX-D. The root mean square deviations (RMSD) from ideal geometry of the final coordinates corresponding to VLX-B and VLX-D were 0.014, and 0.015 Å for bonds and 3.3° and 3.5° for angles, respectively.
TABLE I.
Crystallographic Data for VLX-B and VLX-D
| VLX-B | VLX-D | |
|---|---|---|
| Data | ||
| Beam line | ALS 8.2.1 | WSU (MM007) |
| Wavelength (Å) | 1.07812 | 1.5418 |
| Resolution (Å) | 50–2.4 | 50–2.4 |
| Space group | P212121 | P21 |
| Unit cell dimensions a, b, c (Å) β (deg) | 99.99, 105.31, 105.92 | 92.76, 115.10, 120.22 112.34 |
| Asymmetric unit | 1 molecule | 2 molecule |
| Total observations | 994,342 | 312,353 |
| Unique reflections | 480,752 | 111,693 |
| Completeness (%) | 99.8 (52.5) | 99.5 (67.6) |
| Rsym (%)ab | 8.4 (19.2) | 7.7 (18.3) |
| Refinement | ||
| Resolution (Å) | 15– 2.4 | 15–2.4 |
| Number of reflections | 41,906 | 87,392 |
| Rcryst (%)c | 20.0 (28.2) | 20.3 (29.9) |
| Rfree (%)d | 23.9 (31.5) | 23.5 (33.1) |
| rmsde bonds (Å) | 0.014 | 0.015 |
| rmsd angles (°) | 3.3 | 3.5 |
| Number of atoms | ||
| Protein & ion | 6,693 | 13,302 |
| Water | 134 | 229 |
Numbers in parentheses refer to the highest shell.
Rsym= Σ|Ih – <Ih>|/ΣIh, where <Ih> is the average intensity over symmetry equivalent reflections.
Rcryst = Σ|Fobs – Fcalc|/ΣFobs, where summation is over the data used for refinement.
Rfree was calculated as for Rcryst using 5% of the data that was excluded from refinement.
Root mean square deviations.
The residues 32–44 in VLX-B and 32–53 in VLX-D are disordered, and the corresponding electron density was not visible from the early stage of refinement.
There is one molecule of VLX-B in asymmetric unit, but that of VLX-D is composed of two molecules, which are virtually superimposable on each other with a root mean square deviation (RMSD) of 0.71 Å between the Cα atoms of two molecules in asymmetric unit. VLX-B and VLX-D were studied by light scattering to determine their tendencies to form oligomers. Dynamic light scattering experiments clearly indicated the monomeric nature of both VLX-B and VLX-D molecules in solution at 2 mg ml–1 (see Fig. 1).
Fig. 1.
Molecular mass determination of VLX-B and VLX-D and Dynamic light scattering data for VLX-B (solid line) and VLX-D (dotted line) (~2 mg/ml each). The calculated molecular radius and molecular weight are 4.14 nm and 95 kDa for VLX-B and 4.25 nm and 96 kDa for VLX-D, respectively.
Overall Structure
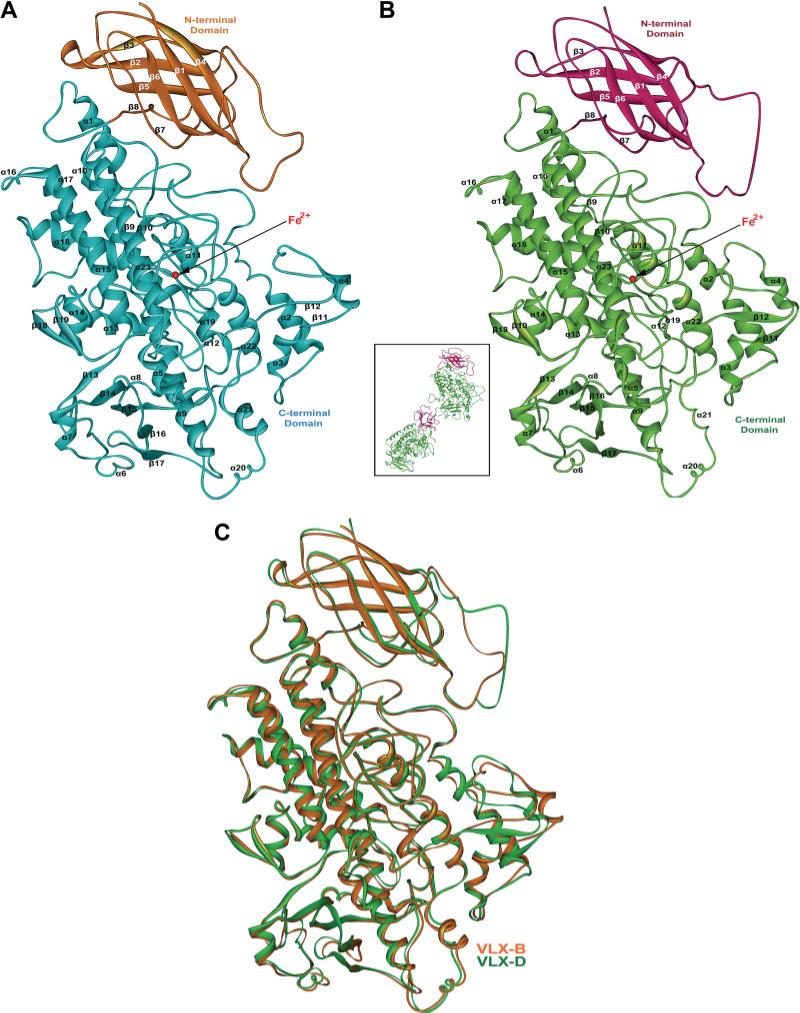
Consistent with structures of other lipoxygenases, both VLX-B and VLX-D are composed of two distinct domains: an N-terminal domain, residues 1–160 in VLX-B (1–172 in VLX-D), and a C-terminal globular domain, residues 161–853 (173–864 in VLX-D) [Fig. 2 and Fig. 3(A,B)]. As expected from the high level of sequence similarity, the overall folds of the two structures show no major conformational differences in terms of their backbone structures [Fig. 3 (A,B)]. The Cα carbons of the VLX-B and VLX-D are superimposable with a RMSD of 0.70 Å excluding one region of disorder, residues 32–53 [Fig. 3(C)].
Fig. 2.
Amino acid sequence comparisons of VLX-B, VLX-D, LOX-1, LOX-3, Oryctolagus cuniculus 15-LOX (1LOX) and Plexaura homomalla 8R-LOX (1ZQ4). Secondary structural elements of VLX-B are highlighted in colored bars on top of the corresponding sequence. The N-terminal domain and C-terminal domain are separated by a blue vertical line. A vertical red dotted line is marked at every 20 amino acids, and every 100th amino acids are indicated by their corresponding residue numbers.
Fig. 3.
Crystal structure of VLX-B (A) and VLX-D (B). The asymmetric unit in the lattice of VLX-D is composed of two molecules (inset of (B)). The N-terminal and C-terminal domains are colored in orange and light blue for VLX-B and violet and light green for VLX-D, respectively. Secondary structural elements have been numbered sequentially as α1–α23 and β1–β19 for the α-helices and β-strands, respectively. The Fe2+ ion is indicated by the red sphere. (C) Superimposed views of VLX-B (orange) and VLX-D (green).
The N-terminal domain of VLX-B and VLX-D forms a small eight-stranded β-barrel composed of two four-stranded antiparallel sheets. The larger α-helical catalytic C-domain is composed of 23 helices and two antiparallel β-sheets and contains the active site iron (see Fig. 3). In both domains, several regions of structural heterogeneity are apparent between two soybean lipoxygenases. This heterogeneity involves one loop connecting strands β1 and β2 (residues 27–54 in VLX-B) in the N-terminal domain, and three loops connecting α7 and β13 (residues 382–407 in VLX-B), α4 and β12 (residues 321–336 in VLX-B), and β16 and β17 (residues 467–476 in VLX-B) in C-terminal domain. In addition, two structures show a difference in one peripheral region composed of three secondary elements of α4, β11, and β12 (residues 307–340).
The nonheme iron cofactor of VLX-B and VLX-D is octahedrally coordinated. VLX-B and VLX-D were crystallized in their resting Fe2+ states and the determined Fe2+–ligand bond lengths are summarized in Table II. No water molecule ligated to Fe2+ was found in the Fo-Fc map of either VLX-B or VLX-D at any stage of the refinement.
TABLE II.
Coordination Geometry of the Active Site Fe2+
| Distances (Å) and Angle (deg) |
||
|---|---|---|
| Defining atomsa | VLX-B | VLX-Db |
| Fe2+- H513 (525) NE2 | 2.39 | 2.23 |
| Fe2+- H518 (530) NE2 | 2.29 | 2.30 |
| Fe2+- H704 (716) NE2 | 2.37 | 2.34 |
| Fe2+- N708 (720) OD1 | 3.07 | 3.05 |
| Fe2+- I853 (864) OXT | 2.28 | 2.50 |
| H513 (525) NE2-Fe2+-H518 (530) NE2 | 87 | 84 |
| H513 (525) NE2-Fe2+-H704 (716) NE2 | 96 | 99 |
| H518 (530) NE2-Fe2+-H704 (716) NE2 | 91 | 96 |
| I853 (864) OXT-Fe2+-N708 (720) OD1 | 113 | 87 |
| H513 (525) NE2-Fe2+-N708 (720) OD1 | 85 | 77 |
| H518 (530) NE2-Fe2+-I853 (864) OXT | 75 | 102 |
| H704 (716) NE2-Fe2+-N708 (720) OD1 | 96 | 95 |
| H704 (716) NE2-Fe2+-I853 (864) OXT | 89 | 126 |
| H513 (525) NE2-Fe2+-I853 (864) OXT | 161 | 133 |
| H518 (530) NE2-Fe2+-N708 (720) OD1 | 169 | 159 |
VLX-B numbering. VLX-D atom numbers are in parentheses.
VLX-D values are average of two molecules in an asymmetric unit.
DISCUSSION
The size of the N-terminal domain of the soybean lipoxygenases is in general larger than that of mammalian lipoxygenases (115–125 amino acids). In this regard, the size of the N-terminal domain of VLX-D is the largest among the four soybean lipoxygenases with determined structures, VLX-B, VLX-D, LOX-1 and LOX-3. For the sake of clarity, throughout this discussion, the residue numbering system of VLX-B is used and the corresponding numbers in VLX-D and LOX-1 are given in superscripts. Except for several loop regions, the nomenclature of the secondary structural elements of the two VLXs can be conveniently described by comparison with LOX-1 nomenclature because of the similarity between the vegetative form and seed form lipoxygenases.
Putative Ca2+ Binding Sites
The N-terminal domain of VLX-B and VLX-D folds into an eight-stranded β-barrel (or β-sandwich); a structure conserved among the seed lipoxygenases as well. This motif is classified as a PLAT (Polycystin-1, Lipoxygenase, Alpha Toxin) domain.40 PLAT domains were suggested to associate directly with membranes, as in the process of Ca2+-mediated membrane binding of alpha toxin and plant lipoxygenases,41 or to mediate interaction with a membrane protein, as in the interaction of the mammalian 5-lipoxygenase β-barrel with the membrane protein FLAP. A surface lysine, K114K126,K100, in β5 of PLAT domains that are conserved in all soybean isoforms and mammalian 15-lipoxygenase, may be important for association with negatively charged membrane or lipoprotein-sequestered substrates as noticed previously.40
As shown in Figure 2, a sequence alignment of the two vegetative lipoxygenases with the sequences of the two seed lipoxygenases, LOX-1 and LOX-3, revealed one major region of sequence heterogeneity in the loop area connecting the first two β-strands of the N-terminal PLAT domain. This loop region (residue 27–54 in VLX-B, 27–64 in VLX-D) also displays the largest conformational differences among the four structures as reflected by the high temperature factors and/or disorder in all isoforms. In this N-terminal domain, another region of high flexibility was noticed in a shorter loop connecting β3 and β4 strands (residue 83–91 in VLX-B, 94–103 in VLX-D), although it is not as severe as the loop connecting β1 and β2. In coral 8R-lipoxygenase, these two flexible loops connecting β1 to β2, and β3 to β4, are located at one edge of the molecule and are stabilized by three calcium ions, apparently for membrane localization and activation.33 The crystals of both VLX-B and VLX-D were obtained in buffers that contain no calcium ion. In addition, the proposed calcium coordinating residues in coral lipoxygenase, such as aspartic acid, glutamic acid, and asparagines, are not conserved in either VLX-B or VLX-D. Consequently, both loops are poorly aligned with those of coral 8R-lipoxygenase.
Calcium is known to promote activity and membrane association of both mammalian and plant lipoxygenases probably by forming salt bridges between the acidic residues and negatively charged lipids in the membrane.42–46 The putative calcium-binding site that was also recognized in LOX1 corresponds to D24, E120, E193 in VLX-B and D24, E132, D205 in VLX-D. Noticeably, in all four soybean lipoxygenases (LOX-1, LOX-3 VLX-B, VLX-D), D24D24,E21 is flanked by hydrophobic resides. The N-side residue is identically leucine in four isoforms and other plant lipoxygenases. The significance of these flanking hydrophobic residues has been suggested to further reinforce membrane binding by being inserted into the membrane.45 In VLX-B, the positions of E120E132,E106 and E193D205,E179 superimpose well with those of LOX-1, but D24D24,E21 is not superimposed, but instead is relatively distant from those two glutamic acids, which is also observed in LOX-3.45 In VLX-D, only E132 is superimposable with the corresponding residue of LOX-1, and the other two aspartic acid residues are too distant for Ca2+ binding. But considering the extreme flexibility of the loop in which D24D24,E21 residues, coordination of Ca2+ can still happen by a concomitant ionic attraction and positional movement among the participating residues.
Catalytic Domain: Iron Coordination and Second-Coordination Sphere
Like LOX-1 and LOX-3, the nonheme iron cofactor of VLX-B and VLX-D, is coordinated by the side chains of the highly conserved residues H513H525,H499, H518H530,H504, H704H716,H690, N708N720,N694, and the C-terminal carboxylate group of I853I864,I839 (Table II). These histidines and aspargine belong to two central α-helices of the catalytic domain, α9 and α18. The side chains of these five residues superimpose well except for the imidazole ring of H513H525,H499, the high mobility of which was previously noticed.47 Also, there has been debate about an iron coordination role for N708N720,N694 in seed lipoxygenases due to the relatively longer bond distance (~3.0 Å); however, upon activation to the Fe3+ state, conformational changes might occur that place the side chain of this residue within the proper bonding distance,31 as observed in the case of LOX-3 (1IK3).32
In the high-resolution (1.4 Å) crystal structure of LOX-1 (1YGE), one water molecule binds Fe2+ in the equatorial plane of the coordination complex and is further stabilized by hydrogen bonding with the oxygen atom of the C-terminal carboxylate group.31 A density corresponding to a water molecule coordinated to Fe2+ in the VLX structures could not be seen.
Around this iron coordination sphere in both VLX-B and VLX-D is a second-coordination sphere composed of a hydrogen bond network that was previously examined in the crystal structures of LOX-1, LOX-3, and rabbit 15-lipoxygenase.48 The second coordination sphere is formed by the side chains of two completely conserved residues, Q509Q521,Q495 and Q711Q723,Q697, two iron coordinating residues, H513H525,H499 and N708N720,N694, and the main chain carbonyl oxygen of residue L768L780,L754. The functional significance of this hydrogen bond pattern may be for the proper positioning of the substrate (or productive conformation of substrate)30,48,49 and will be discussed later.
Catalytic Domain: Substrate-Entry Site
There are several differences in the substrate-entrance site among the four soybean lipoxygenases in terms of polarity and bulkiness of the constituting residues and local structures. For example, in LOX-1, a network of strong hydrogen bonds among the sidechains of residues H248-E256-N534,32 which correspond to H261H271,H248, A269T279,E256, and D548D560,N534, was noticed. The replacement of glutamate in LOX-1 by shorter side chains alanine and threonine in VLX-B and VLX-D and consequent destruction of that hydrogen bond net significantly open up the corresponding entry point in VLX-B, VLX-D, and LOX-3 compared with that observed in LOX-1. In addition, LOX-1 lacks several bulky aromatic residues residing around this entry point, F250V261,V237, F267F777,A254, and Y270Y280,I257, which could help properly register the C-13 position of the phosphoester forms of fatty acid in LOX-1. The disordered loop of sequence heterogeneity (residues 27–54 in VLX-B, 27–64 in VLX-D) noticed in N-terminal PLAT domain is located near the outside entry port; thus the speculated membrane interaction with those flexible loops could also control substrate entry.
Minor et al. suggested one possible entry of linoleic acid into LOX-1 through movement of the side chains of T259 and L541, in α2 and α11, respectively.31 Likewise, in both LOX-3 and VLX-B, an access port for substrate entry into the substrate-binding channel can easily be generated by simple manipulation of the side chains of the corresponding residues, the different conformations of which possibly reflect their intrinsic flexibility.
Noticeably, the α2 of VLX-B, VLX-D, and LOX-3 are well superimposable, but that of LOX-1 is slightly shifted. The composition of residues around this entry site immediately over the modeled linolate also shows similarity among VLX-B, VLX-D, and LOX-3. That is, two residues at the putative entrance, F267F277,A254 and Y270Y280,I257, are conserved as phenylalanine and tyrosine among VLX-B, VLX-D, and LOX-3, but they are amino acids of smaller side chains, alanine and isoleucine, in LOX-1. Regarding another residue constituting the entry port, A269T279,E256, the conformation of E256 in LOX-1 is locked in the open position through a hydrogen bond network, establishing a similarly sized port equivalent to that made of alanine in VLX-B. In VLX-D and LOX-3, it is threonine that somewhat restricts this putative entry site.
Uniquely in VLX-D, there is one more phenylalanine, F25, that is at the beginning part of the disordered loop connecting β1 and β2, that forms a hydrophobic cluster with two other aromatic amino acids, F267F277,A254 and Y270Y280,I257 at the cavity entrance. As a result of these multiple differences, a simple manipulation of the two above-mentioned residues cannot expose the entry port of VLX-D, and this might be related to its unique enzymatic capability for the triglyceride form of fatty acid. The possibility of another entry to the substrate-binding pocket involving helices 2 and 2131,37 cannot be ruled out, however.
Structural rationalization of Substrate Specificity
As summarized in the Introduction, there are similarities as well as differences in terms of the substrate specificity for the soybean lipoxygenase isoforms. In fact, the ability of different lipoxygenase isoforms to utilize various fatty acid and lipid substrates is important for their diverse yet distinct functions. Structural similarities and differences observed in the four soybean lipoxygenases, LOX-1, LOX-3, VLXB, and VLXD may point to distinct biochemical functions for each, and highlight the importance of functional diversity between closely related lipoxygenases in plant biology. Considering the high level of sequence similarity among isoforms, subtle structural differences may account for these observed differences in their specificity of substrate and product.
The four enzymes share their ability to catalyze the hydroperoxidation of the two abundant free fatty acids in plants, linoleic and linolenic acids. Particularly in the 9/13 peroxidation of linoleic acid, LOX-1 and VLX-D produce primarily 13(S)-LOX products with a high degree of stereoselectivity, while LOX-3 and VLX-B produce a greater abundance of 9-LOX products in their racemic mixtures.10,26,32,50 In addition, VLX-D is the only isoform discussed here that utilizes triglycerides as a substrate; thus it has been hypothesized to function in the catabolism of lipid bodies in soybean leaves.26
Another distinct difference among the enzymes is their activities against esterified lipids. LOX-1 has high activity against various phospholipids including, phosphatidylcholine (PC), phosphatidylethanolamine (PE), and phosphatidylinositol (PI), which suggests a function in the degradation of biomembranes.51 Linoleate or arachidonate esterified in phosphatdylcholine are oxygenated by LOX-1 identically (at n-6) on the fatty acid chain. In contrast, the corresponding activity of LOX-3 against phospholipids has not been reported. VLX-B and VLX-D show only marginal activity against PS and PC (Sellhorn and Grimes, in preparation). Because of the extremely low activity of VLX-B and VLX-D against these two phospholipids, there is probably little physiological relevance for the observed activities.
The issues listed above, such as substrate specificity, access, orientation, oxygen pathway, and stereospecificity of the reaction have been intensely investigated by many groups, but still some controversy exists. Although solving a true lipoxygenase substrate complex would be very challenging because oxygen is the second substrate, the structures of two other complexes are available. First, the soybean LOX-3 complexed with 13(S)-hydroperoxy-9(Z),11(E)-octadecadienoic acid (13-HPOD),32 and second, the rabbit 15-LOX complexed with 3-(2-octylphenyl propanoic acid (RS75091).30 In the first of these, the carboxyl of the product hydroperoxide lies deep in the cavity and interacts with R726 (LOX-3). In the second, the carboxyl of the inhibitor is an iron ligand. Models of linoleic acid similar to each of these arrangements have been proposed.47,52 More recently, several mechanistically appealing model structures have been advanced that place the carboxyl end of the substrate near one suggested entrance to the cavity, near T259 and L541.31 These models are based on extensive mutational studies and dockings, such as coral 8R-LOX with a modeled arachidonic acid33 and soybean LOX-1 with modeled linoleic acid.37 Collectively, these have provided a structural overview of the hydrophobic substrate-binding pocket, subcavity IIa. The fact that a second oxygenation of 15-hydroperoxy arachidonic acid by LOX-1 can occur demonstrates that substrates can enter the lipoxygenase cavity in two different orientations of carboxyl and methyl ends of the chain.37 However, it is hard to reconcile either of the structures of lipoxygenase complexes with steps in the lipoxygenase mechanism. Neither structure shows how hydrogen at C-11 could be positioned for the rate limiting step in catalysis. An additional concern about the interpretation of the purple lipoxygenase structure is that the color is only seen after more than one equivalent of hydroperoxide is added to ferrous enzyme, which may mean that the purple form is a side product.53 Overall, it seems that productive and unproductive complexes of lipoxygenase with fatty acids can be formed.
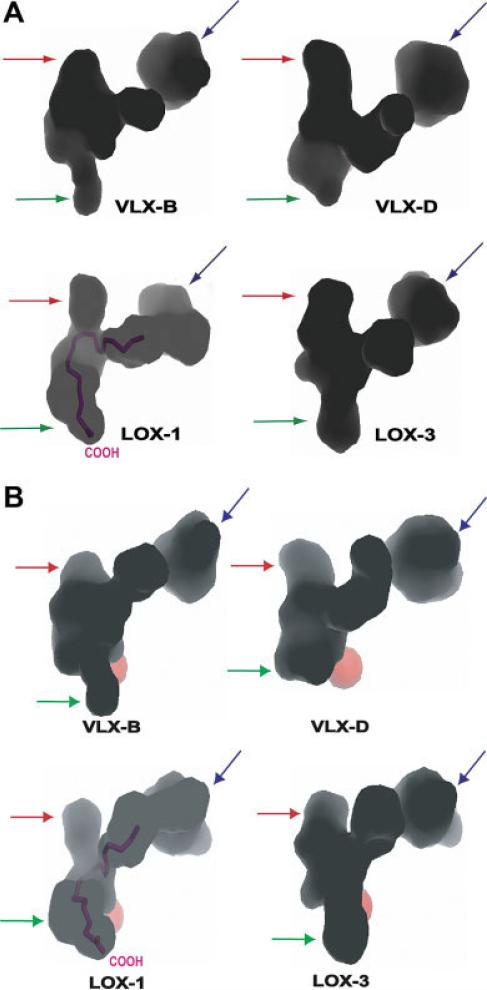
As represented in Figs. 2, 4, and 5, even though most of the residues constituting the internal cavity are highly conserved among LOX-1, LOX-3, VLX-B, and VLX-D, there are several heterogeneous residues among the sequence of these isoforms that generate slightly different shapes and volumes of each individual cavity. This subcavity IIa of both VLX-B and VLX-D is composed of roughly three branches as observed in LOX-1 and LOX-3, two of which are sharply bent near the iron and are able to accommodate the modeled linoleic acid [Figs. 4 and 5(A)]. The observed sharp bend of the cavity near the Fe2+ site is contributed by the sidechains of two residues, Q509Q521,Q495 and I552I564,I538, which are located adjacent to C-14 and C-8 of modeled linoleic acid, respectively (see Fig. 5). These two residues are conserved and superimposable among the four isoforms as shown in Figures 2 and 5.
Fig. 4.
The shape of subcavity IIa of VLX-B, VLX-D, LOX-1, and LOX-3. The O2 cavity (red arrow), extended cavity (blue arrow), and entrance site (green arrow) are indicated. The iron is depicted the by red ball (B). The view in (B) is ~45° rotated from the view in (A), along the horizontal axis. In LOX-1, the modeled linoleic acid is depicted in violet color. This figure was generated with the program PyMOL (v 0.99).
Fig. 5.
(A) Residues constituting subcavity IIa that are conserved among the four soybean lipoxygenases. The shape of subcavity IIa of VLX-B, linoleic acid (violet stick model), and iron (pink ball) are depicted. (B) The residues constituting the cavity IIa; conserved residues are represented by gray ball and sticks together with their van der Waal nets. The nonconserved residues were represented by stick model (VLX-B, orange; VLX-D, blue; LOX-1, green; LOX-3, cyan). (C) Internal cavity of VLX-B. The residues are represented in the same format as in (B).
One branch of this channel reaches the above-mentioned entry port of the molecular surface, where basic side chains may tether the fatty acid carboxylate, as suggested in other lipoxygenases.30,37 The specific activity of both VLX-B and VLX-D decreases significantly against the methyl ester of linoleic acid, which supports a role for the carboxyl group of the substrate in substrate binding (Sellhorn and Grimes, in preparation).
At the other end of this substrate-binding channel is an extended pocket (marked in blue arrow in Fig. 4) composed of A505S517,S491, T570S582,T556, F571F583,F557, G715G727,G701, I718I730,I704, R721R733,R707, T723T735,T709, D761D773,S747, L762L774,L748, I764V776,V750, I765I777,I751. This pocket has a less hydrophobic nature than the rest of the cavity in terms of the constituting residues and it also contains several water molecules observed in both VLXs. The residues in this extended cavity are less conserved compared with those constituting the central part of the cavity among soybean lipoxygenases, leading to different shapes. As shown in Figure 4, this extended cavity in LOX-1 forms a continuous channel, but the corresponding cavities observed in LOX-3, VLX-B, and VLX-D have a barrier across the channel, showing somewhat restricted access to the extended cavity compared with that of LOX-1. This feature may have some relationship to relative activities observed against linoleic acid and arachidonic acid among those soybean lipoxygenases. Only LOX-1 has a similar activity for both forms of fatty acids.54,55
The third branch of the subcavity IIa that is shorter than the other two branches intersects with the substrate-binding channel on the opposite side of the iron and has been proposed to be the O2 access channel.31 It is surrounded by residues such as Q509Q521,Q495, I510L522,L496, W514W526,W500, A556A568,A542, L560V572,L546, I561V573,I547, I567I579,I553, L578I590,V564, and L768L780,L754. In general, this channel has a hydrophobic character and some residues show heterogeneity among the four isoforms. It is apparent in Figure 4 that O2 access channel intersects the fatty acid-binding cavity with a broad area of contact in both LOX-3 and VLX-B, which is consistent with characteristic products, a mixture of 13-and 9-regio products. On the other hand, the side channels of both LOX-1 and VLX-D more specifically intersect the substrate-binding cavity near the point of the C-13 atom.
In particular, two residues L560L572,L546 and L768L780,L754 and the other residue, I567I579,I553 that intersects L560L572,L546 and Q509Q521,Q495, constitute part of this O2 channel and lie near the modeled C-11 atom and C-13, respectively (see Fig. 5). In the case of LOX-1, these residues are proposed to have a guiding role for O2 access by blocking access to the C-9 of linoleic acid.53 These residues, L560L572,L546, I567I579,I553 and L768L780,L754, are completely conserved and superimposable among the four soybean lipoxygenases.
In addition, another key residue in LOX-1, corresponding to A556A568,A542 of the VLX-B, has been noticed for its critical role in the specificity of the lipoxygenase. This residue, which resides in α11 and is positioned near the C-9 atom of the modeled linoleic acid, is completely conserved among S-lipoxygenases that includes LOX-1, LOX-3, VLX-B, and VLX-D. A556A568,A542 has been proposed to determine the stereochemistry of products by substrate orientation and flanking a pouch of O2.33,37 This alanine is replaced with glycine in other lipoxygenases such as 8R-LOX. The significance of this change may be the creation of a small pocket that can potentially accommodate O2 without creating a structural perturbation near the C-8 atom.37 Noticeably, the carbonyl oxygen of A556A568,A542 is within hydrogen bond distance to the amide nitrogens of both L560L572,L546 and I561V573,I547, which reside at the carboxy end of the same helix. Therefore, the effect of sequence heterogeneity at I561V573,I547, which is in close contact with the side chain W514W526,W500, can propagate to both L560L572,L546 and I561V573,I547. The importance of these residues to kinetic efficiency and regiospecificity has been previously noted.37,53,56,57 The Cα positions of A556A568,A542, L560L572,L546, and I561V573,I547 show maximum difference of ~0.5 Å among the four superimposed isoforms.
To model the linoleic acid into the binding pocket of VLX-B and VLX-D, the key residue in the second coordination sphere, Q509Q521,Q495, must be detached from both Q711Q723,Q697 and H513H525,H499 because of close contact with the fatty acid. A similar situation was observed in the crystal structures of the substrate-free form and the 13-HPOD complex of LOX-3.32 In addition, the side chain of the corresponding glutamine was disordered in the substrate-free form of LOX-3, which could reflect its intrinsic lack of stability or dynamic nature of the hydrogen bond. Upon ligand binding, the hydrogen bond network of the second sphere in the substrate-free form of both VLX-B and VLX-D would be partially destroyed because of dislocation of Q509Q521,Q495. Concomitantly, because of the tilted geometry of substrate-free forms, the weak N708N720,N694–Fe2+ bond would be rearranged forming a stronger bond leading to a purely 6C ligand arrangement as suggested previously.48 Therefore, this hydrogen bond network has a role of interconnecting the substrate binding and the change of iron coordination through the position of the side chain of Q509Q521,Q495. In addition, flexibility of the binding site of LOX-3, especially at this glutamine residue, has been attributed to the production of a mixture of stereoisomers (S/R: 86:14).32
CONCLUSIONS
Lipoxygenases are an important enzyme family in many biochemical and physiological processes and are found in most organisms. The multiple isoforms that are typically found in plant species, coupled with the unique biochemical properties of these isoforms, suggest distinct cellular functions. Indeed, the soybean vegetative LOXs are primarily associated with a unique cell layer, termed the paravenial mesophyll cell layer, in the leaves, and also essentially define a novel cell layer in the seed pod.25,58
One of the interesting biochemical properties found in these soybean LOXs is their substrate specificity. While all known isoforms are able to utilize the free fatty acids (18:2 and 18:3), only VLX-C and VLX-D are active against triglycerides. One rationale for the present study was to determine whether a structural basis for this disparate activity could be discerned. The crystal structures clearly show the different shapes of the substrate-binding cavity of four soybean lipoxygenases, resulting from the cumulative effect of the sequence differences near the intersection point of the three branches; especially, the noticeable shape difference of the VLX-B subcavity IIa might be connected to its apparent low activity against both linoleic acid and arachidonic acid. In addition, the unique structure of the entry port of VLX-D and extended cavity of LOX-1 might be related to their unique enzymatic capability for the triglyceride and arachidonic acid oxidation, respectively. The putative Ca2+ binding site in the N-terminal PLAT domain shows extreme heterogeneity in both sequence and conformation among four VLX-B, VLX-D, LOX-1, and LOX-3. It reaches near to the substrate entry site, and thus the membrane interaction with those flexible loops could affect the enzyme activity.
It is becoming more evident that specific LOX isoforms are often associated with specific diseases and their progression.59 Therefore, isoform specific inhibitors of LOXs can be very useful for the development of drugs to target specific LOX-related diseases while not disrupting the beneficial functions of other LOX activity.
Because of the highly conserved structure of the active site of these isoenzymes, a detailed and comparative knowledge of soybean lipoxygenase can also help in the development of LOX-based drugs that are isoform-specific inhibitors. Apparently, answering some of these questions must await a structure of true substrate-enzyme complex.
ACKNOWLEDGMENTS
This research was supported by the Murdock Charitable Trust NIH, DOE, USDA to C. Kang and H.D. Grimes and, in part, by NIH to B.J. Gaffney. We thank C. Ralston (ALS, 8.2.1).
Grant sponsor: NIH; Grant numbers: GM66173, GM65268; Grant sponsors: Murdock Charitable Trust, USDA; Grant sponsor: DOE; Grant number: DE-FG02-05ER15672.
Footnotes
B. Youn and G.E. Sellhorn have contributed equally to this work.
REFERENCES
- 1.Feussner I, Wasternack C. The lipoxygenase pathway. Annu Rev Plant Biol. 2002;53:275–297. doi: 10.1146/annurev.arplant.53.100301.135248. [DOI] [PubMed] [Google Scholar]
- 2.Kuhn H, Thiele BJ. The diversity of the lipoxygenase family. Many sequence data but little information on biological significance. FEBS Lett. 1999;449:7–11. doi: 10.1016/s0014-5793(99)00396-8. [DOI] [PubMed] [Google Scholar]
- 3.Rosahl S. Lipoxygenases in plants—their role in development and stress response. Z Naturforsch [C] 1996;51:123–138. doi: 10.1515/znc-1996-3-401. [DOI] [PubMed] [Google Scholar]
- 4.Hamberg M, Su C, Oliw E. Manganese lipoxygenase: discovery of a bis-allylic hydroperoxide as product and intermediate in a lipoyxgenase reaction. J Biol Chem. 1998;273:13080–13088. doi: 10.1074/jbc.273.21.13080. [DOI] [PubMed] [Google Scholar]
- 5.Su C, Oliw E. Manganese lipoxygenase: purification and characterization. J Biol Chem. 1998;273:13072–13079. doi: 10.1074/jbc.273.21.13072. [DOI] [PubMed] [Google Scholar]
- 6.Hornsten L, Su C, Osbourn A, Hellman U, Oliw E. Cloning of the manganese lipoxygenase gene reveals homology with the lipoxygenase gene family. Eur J Biochem. 2002;269:2609–2697. doi: 10.1046/j.1432-1033.2002.02936.x. [DOI] [PubMed] [Google Scholar]
- 7.Vance RE, Hong S, Gronert K, Serhan CN, Mekalanos JJ. The opportunistic pathogen Pseudomonas aeruginosa carries a secretable arachidonate 15-lipoxygenase. Proc Natl Acad Sci USA. 2004;101:2135–2139. doi: 10.1073/pnas.0307308101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vidal-Mas J, Busquets M, Manresa A. Cloning and expression of a lipoxygenase from Pseudomonas aeruginosa 42A2. Antonie Van Leeuwenhoek. 2005;87:245–251. doi: 10.1007/s10482-004-4021-1. [DOI] [PubMed] [Google Scholar]
- 9.Matsui K, Hijiya K, Tabuchi Y, Kajiwara T. Cucumber cotyledon lipoxygenase during postgerminative growth. Its expression and action on lipid bodies. Plant Physiol. 1999;119:1279–1287. doi: 10.1104/pp.119.4.1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feussner I, Kuhn H, Wasternack C. Lipoxygenase-dependent degradation of storage lipids. Trends Plant Sci. 2001;6:268–273. doi: 10.1016/s1360-1385(01)01950-1. [DOI] [PubMed] [Google Scholar]
- 11.Gerhardt B, Fischer K, Balkenhohl TJ, Pohnert G, Kuhn H, Wasternack C, Feussner I. Lipoxygenase-mediated metabolism of storage lipids in germinating sunflower cotyledons and β-oxidation of (9Z,11E,13S)-13-hydroxy-octadeca-9,11-dienoic acid by the cotyledonary glyoxysomes. Planta. 2005;220:919–930. doi: 10.1007/s00425-004-1408-1. [DOI] [PubMed] [Google Scholar]
- 12.Grimes HD, Tranbarger TJ, Franceschi VR. Expression and accumulation patterns of nitrogen-responsive lipoxygenase in soybeans. Plant Physiol. 1993;103:457–466. doi: 10.1104/pp.103.2.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bunker TW, Koetje DS, Stephenson LC, Creelman RA, Mullet JE, Grimes HD. Sink limitation induces the expression of multiple soybean vegetative lipoxygenase mRNAs while the endogenous jasmone acid level remains low. Plant Cell. 1995;7:1319–1331. doi: 10.1105/tpc.7.8.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stephenson LC, Bunker TW, Dubbs WE, Grimes HD. Specific soybean lipoxygenases localize to discrete subcellular compartments and their mRNAs are differentially regulated by source-sink status. Plant Physiol. 1998;116:923–933. doi: 10.1104/pp.116.3.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shah J. Lipids, lipases, and lipid-modifying enzymes in plant disease resistance. Annu Rev Phytopathol. 2005;43:229–260. doi: 10.1146/annurev.phyto.43.040204.135951. [DOI] [PubMed] [Google Scholar]
- 16.Howe GA, Lightner J, Browse J, Ryan CA. An octadecanoid pathway mutant (JL5) of tomato is compromised in signaling for defense against insect attack. Plant Cell. 1996;8:2067–2077. doi: 10.1105/tpc.8.11.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weber H, Chetelat A, Caldelari D, Farmer E. Divinyl ether fatty acid synthesis in late blight-diseased potato leaves. Plant Cell. 1999;11:485–494. doi: 10.1105/tpc.11.3.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ben-Hayyim G, Gueta-Dahan Y, Avsian-Kretchmer O, Weichert H, Feussner I. Preferential induction of a 9-lipoxygenase by salt in salt-tolerant cells of Citrus sinensis L. Osbeck. Planta. 2001;212:367–375. doi: 10.1007/s004250000397. [DOI] [PubMed] [Google Scholar]
- 19.Kolomiets MV, Hannapel DJ, Chen H, Tymeson M, Gladon RJ. Lipoxygenase is involved in the control of potato tuber development. Plant Cell. 2001;13:613–626. doi: 10.1105/tpc.13.3.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Constabel C, Bergey D, Ryan C. Systemin activates synthesis of wound-inducible tomato leaf polyphenol oxidase via the octadecanoid defense signaling pathway. Proc Natl Acad Sci USA. 1995;92:407–411. doi: 10.1073/pnas.92.2.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Porta H, Rocha-Sosa M. A Phaseolus vulgaris lipoxygenase gene expressed in nodules and in Rhizobium tropici inoculated roots. Biochim Biophys Acta. 2000;1517:139–142. doi: 10.1016/s0167-4781(00)00230-x. [DOI] [PubMed] [Google Scholar]
- 22.Hamberg M, Samuelsson B. Oxygenation of unsaturated fatty acids by the vesicular gland of sheep. J Biol Chem. 1967;242:5344–5354. [PubMed] [Google Scholar]
- 23.Egmond M, Veldink G, Vliegenthart J, Boldingh J. C-11 H-abstraction from linoleic acid, the rate-limiting step in lipoxygenase catalysis. Biochem Biophys Res Commun. 1973;54:1178–1184. doi: 10.1016/0006-291x(73)90816-4. [DOI] [PubMed] [Google Scholar]
- 24.Axelrod B, Cheesbrough TM, Laakso S. Lipoxygenase from soybeans. Methods Enzymol. 1981;71:441–451. [Google Scholar]
- 25.Fischer AM, Dubbs WE, Baker RA, Fuller MA, Stephenson LC, Grimes HD. Protein dynamics, activity and cellular localization of soybean lipoxygenases indicate distinct functional roles for individual isoforms. Plant J. 1999;19:543–554. doi: 10.1046/j.1365-313x.1999.00550.x. [DOI] [PubMed] [Google Scholar]
- 26.Fuller M, Weichert H, Fischer A, Feussner I, Grimes H. Activity of soybean lipoxygenase isoforms against esterified fatty acids indicates functional specificity. Arch Biochem Biophys. 2001;388:146–154. doi: 10.1006/abbi.2000.2269. [DOI] [PubMed] [Google Scholar]
- 27.Rickert KW, Klinman JP. Nature of hydrogen transfer in soybean lipoxygenase 1: separation of primary and secondary isotope effects. Biochemistry. 1999;38:12218–12228. doi: 10.1021/bi990834y. [DOI] [PubMed] [Google Scholar]
- 28.Pistorius E, Axelrod B, Palmer G. Evidence for participation of iron in lipoxygenase reaction from optical and electron spin resonance studies. J Biol Chem. 1976;251:7144–7148. [PubMed] [Google Scholar]
- 29.Boyington J, Gaffney B, Amzel L. The three-dimensional structure of an arachidonic acid 15-lipoxygenase. Science. 1993;260:1482–1486. doi: 10.1126/science.8502991. [DOI] [PubMed] [Google Scholar]
- 30.Gillmor SA, Villasenor A, Fletterick R, Sigal E, Browner MF. The structure of mammalian 15-lipoxygenase reveals similarity to the lipases and the determinants of substrate specificity. Nat Struct Biol. 1997;4:1003–1009. doi: 10.1038/nsb1297-1003. [DOI] [PubMed] [Google Scholar]
- 31.Minor W, Steczko J, Stec B, Otwinowski Z, Bolin JT, Walter R, Axelrod B. Crystal structure of soybean lipoxygenase L-1 at 1.4 A resolution. Biochemistry. 1996;35:10687–10701. doi: 10.1021/bi960576u. [DOI] [PubMed] [Google Scholar]
- 32.Skrzypczak-Jankun E, Bross RA, Carroll RT, Dunham WR, Funk MO., Jr. Three-dimensional structure of a purple lipoxygenase. J Am Chem Soc. 2001;123:10814–10820. doi: 10.1021/ja011759t. [DOI] [PubMed] [Google Scholar]
- 33.Oldham M, Brash A, Newcomer M. Insights from the X-ray crystal structure of coral 8R-lipoxygenase: calcium activation via A C2-like domain and a structural basis of product chirality. J Biol Chem. 2005;280:39545–39552. doi: 10.1074/jbc.M506675200. [DOI] [PubMed] [Google Scholar]
- 34.Navaza J. Implementation of molecular replacement in AMoRe. Acta Crystallogr D Biol Crystallogr. 2001;57:1367–1372. doi: 10.1107/s0907444901012422. [DOI] [PubMed] [Google Scholar]
- 35.Jones T, Zou J, Cowan S, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 36.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 37.Coffa G, Imber AN, Maguire BC, Laxmikanthan G, Schneider C, Gaffney BJ, Brash AR. On the relationships of substrate orientation, hydrogen abstraction, and product stereochemistry in single and double dioxygenations by soybean lipoxygenase-1 and its Ala542Gly mutant. J Biol Chem. 2005;280:38756–38766. doi: 10.1074/jbc.M504870200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morris GM, Goodsell DS, Halliday RS, Huey R, Hart WE, Belew RK, Olson AJ. Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J Comput Chem. 1998;19:1639–1662. [Google Scholar]
- 39.Neria E, Fischer S, Karplus S. Simulation of activation free energies in molecular systems. J Chem Phys. 1996;105:1902–1921. [Google Scholar]
- 40.Bateman A, Sandford R. The PLAT domain: a new piece in the PKD1 puzzle. Curr Biol. 1999;26:R588–R590. doi: 10.1016/s0960-9822(99)80380-7. [DOI] [PubMed] [Google Scholar]
- 41.Rizo J, Sudhof TC. C2-domains, structure and function of a universal Ca2+-binding domain. J Biol Chem. 1998;273:15879–15882. doi: 10.1074/jbc.273.26.15879. [DOI] [PubMed] [Google Scholar]
- 42.Baba A, Sakuma S, Okamoto H, Inoue T, Iwata H. Calcium induces membrane translocation of 12-lipoxygenase in rat platelets. J Biol Chem. 1989;264:15790–15795. [PubMed] [Google Scholar]
- 43.Watson A, Doherty F. Calcium promotes membrane association of reticulocyte 15-lipoxygenase. Biochem J. 1994;298:377–383. doi: 10.1042/bj2980377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Noguchi M, Miyano M, Matsumoto T, Noma M. Human 5-lipoxygenase associates with phosphatidylcholine liposomes and modulates LTA4 synthetase activity. Biochim Biophys Acta. 1994;1215:300–306. doi: 10.1016/0005-2760(94)90057-4. [DOI] [PubMed] [Google Scholar]
- 45.Tatulian S, Steczko J, Minor W. Uncovering a calcium-regulated membrane-binding mechanism for soybean lipoxygenase-1. Biochemistry. 1998;37:15481–15490. doi: 10.1021/bi981062t. [DOI] [PubMed] [Google Scholar]
- 46.Hammarberg T, Radmark O. 5-lipoxygenase binds calcium. Biochemistry. 1999;38:4441–4447. doi: 10.1021/bi9824700. [DOI] [PubMed] [Google Scholar]
- 47.Skrzypczak-Jankun E, Amzel LM, Kroa BA, Funk MO., Jr. Structure of soybean lipoxygenase L3 and a comparison with its L1 isoenzyme. Proteins: Struct Funct Genet. 1997;29:15–31. [PubMed] [Google Scholar]
- 48.Schenk G, Neidig M, Zhou J, Holman T, Solomon E. Spectroscopic characterization of soybean lipoxygenase-1 mutants: the role of second coordination sphere residues in the regulation of enzyme activity. Biochemistry. 2003;42:7294–7302. doi: 10.1021/bi027380g. [DOI] [PubMed] [Google Scholar]
- 49.Tomchick D, Phan P, Cymborowski M, Minor W, Holman T. Structural and functional characterization of second-coordination sphere mutants of soybean lipoxygenase-1. Biochemistry. 2001;40:7509–7517. doi: 10.1021/bi002893d. [DOI] [PubMed] [Google Scholar]
- 50.Fukushige H, Wang C, Simpson T, Gardner H, Hildebrand D. Purification and identification of linoleic acid hydroperoxides generated by soybean seed lipoxygenases 2 and 3. J Agric Food Chem. 2005;53:5691–5694. doi: 10.1021/jf047958o. [DOI] [PubMed] [Google Scholar]
- 51.Brash AR, Ingram CD, Harris TM. Analysis of a specific oxygenation reaction of soybean lipoxygenase-1 with fatty acids esterified in phospholipids. Biochemistry. 1987;26:5465–5471. doi: 10.1021/bi00391a038. [DOI] [PubMed] [Google Scholar]
- 52.Prigge ST, Boyington JC, Gaffney BJ, Amzel LM. Structure conservation in lipoxygenases: structural analysis of soybean lipoxygenase-1 and modeling of human lipoxygenases. Proteins: Struct Funct Genet. 1996;24:275–291. doi: 10.1002/(SICI)1097-0134(199603)24:3<275::AID-PROT1>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 53.Knapp M, Seebeck F, Klinman J. Steric control of oxygenation regiochemistry in soybean lipoxygenase-1. J Am Chem Soc. 2001;123:2931, 2932. doi: 10.1021/ja003855k. [DOI] [PubMed] [Google Scholar]
- 54.Smith WL, Lands WE. Oxygenation of unsaturated fatty acids by soybean lipoxygenase. J Biol Chem. 1972;247:1038–1047. [PubMed] [Google Scholar]
- 55.Cook HW, Lands WE. Further studies of the kinetics of oxygenation of arachidonic acid by soybean lipoxygenase. Can J Biochem. 1975;53:1220–1231. doi: 10.1139/o75-167. [DOI] [PubMed] [Google Scholar]
- 56.Gan QF, Browner M, Sloane DL, Sigal E. Defining the arachidonic acid binding site of human 15-lipoxygenase. J Biol Chem. 1996;271:25412–25418. doi: 10.1074/jbc.271.41.25412. [DOI] [PubMed] [Google Scholar]
- 57.Hughes R, Lawson D, Hornostaj A, Fairhurst S, Casey R. Muta-genesis and modelling of linoleate-binding to pea seed lipoxygenase. Eur J Biochem. 2001;268:1030–1040. doi: 10.1046/j.1432-1327.2001.01964.x. [DOI] [PubMed] [Google Scholar]
- 58.Dubbs W, Grimes H. Specific lipoxygenase isoforms accumulate in distinct regions of soybean pod walls and mark a unique cell layer. Plant Physiol. 2000;123:1269–1280. doi: 10.1104/pp.123.4.1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhao L, Funk CD. Lipoxygenase pathways in atherogenesis. Trends Cardiovasc Med. 2004;14:191–195. doi: 10.1016/j.tcm.2004.04.003. [DOI] [PubMed] [Google Scholar]