Abstract
Monoparental parthenotes represent a potential source of histocompatible stem cells that should be isogenic with the oocyte donor and therefore suitable for use in cell or tissue replacement therapy. We generated five rhesus monkey parthenogenetic embryonic stem cell (PESC) lines with stable, diploid female karyotypes that were morphologically indistinguishable from biparental controls, expressed key pluripotent markers, and generated cell derivatives representative of all three germ layers following in vivo and in vitro differentiation. Interestingly, high levels of heterozygosity were observed at the majority of loci that were polymorphic in the oocyte donors. Some PESC lines were also heterozygous in the major histocompatibility complex region, carrying haplotypes identical to those of the egg donor females. Expression analysis revealed transcripts from some imprinted genes that are normally expressed from only the paternal allele. These results indicate that limitations accompanying the potential use of PESC-derived phenotypes in regenerative medicine, including aberrant genomic imprinting and high levels of homozygosity, are cell line-dependent and not always present. PESC lines were derived in high enough yields to be practicable, and their derivatives are suitable for autologous transplantation into oocyte donors or could be used to establish a bank of histocompatible cell lines for a broad spectrum of patients.
Keywords: Embryonic stem cells, Parthenogenetic, Meiotic recombination, Imprinting, Histocompatible
INTRODUCTION
Embryonic stem cells (ESCs) are pluripotent cells derived from preimplantation-stage embryos that carry the potential to differentiate into lineages representing the three major germ layers. Furthermore, their differentiated derivatives have the potential to replace defective cells by cell or tissue replacement therapy. However, there remain concerns about ESC-derived tissue transplantation in humans because of safety issues and the potential for spontaneous or uncontrolled cellular proliferation. Therefore, therapeutic studies of ESCs in clinically relevant animal models such as nonhuman primates are warranted. Old World macaques provide one such model wherein ESCs are available [1] and have recently been used to generate dopaminergic neurons that function in animals with induced Parkinsonian symptoms [2].
Human ESCs derived from spare in vitro fertilized embryos will inevitably be genetically divergent from any patient requiring tissue transplantation and, as such, will likely incite an immune response resulting in rejection unless an immunosuppression regimen is used. Nuclear transfer, intended to produce ESCs genetically identical to the patient by somatic cell nuclear transfer, may avoid immune rejection and has recently been achieved in primates [3]. However, the anticipated high cost and low efficiency of nuclear transfer provide compelling reasons to explore alternative sources of histocompatible, pluripotent cells. We have shown that diploid monkey parthenogenetic embryos can be generated by artificial activation of metaphase II (MII)-arrested oocytes followed by retention of genetic material in the second polar body [4]. Such embryos represent a potential source of pluripotent cells that would be isogenic with the oocyte donor [5, 6]. Diploid parthenotes are capable of completing preimplantation development and implanting but are unable to form viable fetuses [7, 8]. This limited developmental potential bypasses most ethical concerns regarding the derivation of human ESCs because it obviates the question of destruction of potential human life.
On the other hand, several safety issues accompany the potential use of parthenogenetic embryonic stem cells (PESCs) in regenerative medicine, including aberrant genomic imprinting and high levels of homozygosity. Disruption or inappropriate expression of imprinted genes is associated with severe clinical syndromes and carcinogenesis in humans [9]. Here, we demonstrate that rhesus monkey PESCs are genetically and epigenetically variable and that some lines carry attributes that attenuate many of these safety concerns.
MATERIALS AND METHODS
Ovarian Stimulation, Recovery of Rhesus Monkey Oocytes, Parthenogenetic Activation, and Embryo Culture
Controlled ovarian stimulation and oocyte recovery was conducted as described previously [1, 10]. Cumulus-oocyte complexes were collected from anesthetized animals by laparoscopic follicular aspiration (28–29 hours post-hCG) and placed in Hepes-buffered modified Tyrode solution with albumin, lactate and pyruvate containing 0.3% bovine serum albumin (TH3) at 37°C. Unless indicated otherwise, all reagents were from Sigma-Aldrich (St. Louis, http://www.sigmaaldrich.com). Oocytes, stripped of cumulus cells by mechanical pipetting after brief exposure (<1 minute) to hyaluronidase (0.5 mg/ml), were placed in chemically defined, protein-free hamster embryo culture medium (HECM)-9 medium [11] at 37°C in 5% CO2, 5%O2, and 90% N2 until further use. Mature MII oocytes were activated by exposure to 5 µM ionomycin (Calbiochem, San Diego, http://www.emdbiosciences.com) for 5 minutes followed by a 5-hour incubation in 2 mM 6-dimethylaminopurine. After activation, oocytes were placed in four-well dishes (Nalge Nunc International, Naperville, IL, http://www.nalgenunc.com) containing HECM-9 medium and cultured at 37°C in 5% CO2, 5% O2, and 90% N2. Embryos at the 8–16-cell stage were transferred to fresh plates of HECM-9 medium supplemented with 5% fetal bovine serum (FBS; HyClone, Logan, UT, http://www.hyclone.com) and cultured for a maximum of 9 days, with medium change every other day.
PESC Isolation, Propagation, and Differentiation
Zonae pellucidae of expanded blastocysts were removed with brief protease (0.5%) treatment, and inner cell masses (ICMs) were isolated using immunosurgery [1]. ICMs were plated onto Nunc four-well dishes containing mitotically inactivated mouse embryonic fibroblasts and ESC culture medium consisting of Dulbecco’s modified Eagle’s medium/Ham’s F-12 medium supplemented with 1% nonessential amino acids, 2 mM l-glutamine, 0.1 mM β-mercaptoethanol (all from Invitrogen, Carlsbad, CA, http://www.invitrogen.com), and 15% FBS. ICMs that attached to the feeder layer and initiated outgrowth were manually dissociated into small-cell clumps with a microscalpel and replated onto new mEFs. After the first passage, colonies with ESC-like morphology were selected for further propagation, characterization, and low-temperature storage. Medium was changed daily, and ESC colonies were split every 5–7 days by manual disaggregation and replating collected cells onto dishes with fresh feeder layers. Cultures were maintained at 37°C in 3% CO2, 5% O2, and 92% N2. The cardiomyocyte differentiation protocol involved transfer of embryoid bodies (EBs) onto collagen-coated culture dishes. Following attachment, EBs spontaneously developed into clusters of contracting cardiomyocytes. For studies on teratoma formation, 3–5 million undifferentiated ESCs from each cell line were harvested and injected into the hind leg muscle or subcutaneously into 4-week-old SCID, beige, male mice using an 18-gauge needle. Six to 7 weeks after injection, mice were sacrificed, and teratomas were dissected, sectioned, and histologically characterized for the presence of representative tissues of all three germ layers.
Immunocytochemical Procedures and Reverse Transcription-Polymerase Chain Reaction
Immunocytochemistry and reverse transcription (RT)-polymerase chain reaction (PCR) were performed as described previously [1]. Transcripts of all imprinted genes were confirmed by direct sequencing of PCR products eluted from gel bands. These sequences were used to design primers and probes for quantitative RT-PCR. Information regarding sequences and annealing temperatures for each primer can be found in supplemental online Table 1. Quantitative real-time polymerase chain reaction (qPCR) was performed on total RNA isolated from each PESC line and biparental Oregon Rhesus Monkey Embryonic Stem (ORMES)-22 [1]. Total RNA was treated with RNase-free DNase (Invitrogen) to eliminate any traces of genomic DNA. Absence of DNA was further confirmed by standard −RT control reaction. The RNA concentrations were determined by Nanodrop ND1000 spectrophotometer, and quality was analyzed using an Agilent Labchip Bioanalyzer (Agilent Technologies, Palo Alto, CA, http://www.agilent.com). The cDNAs were synthesized from 800 ng of total RNA sample with Super-Script III reverse transcriptase (200 U/µl) (Invitrogen) using oligo(dT) primers. TaqMan probes and primers were designed on cDNA sequences using ABI Primer Express, and probes were synthesized by Applied Biosystems (Foster City, CA, http://www.appliedbiosystems.com). The real-time PCR primers were synthesized by Integrated DNA Technologies (Coralville, IA, http://www.idtdna.com/Home/Home.aspx) (supplemental online Table 1). qPCR was performed on an ABI 7500 Fast Real-time PCR System with the SDS 1.4.0 program and using the ABI TaqMan Fast Universal PCR master mix (Applied Biosystems). To eliminate the possibility of contamination with genomic DNA, all qPCRs included a pilot −RT control with GAPDH probes and primer set. The final concentration of the real-time primers was 300 nM, and the final concentration of the real-time probes was 250 nM. Initially, we examined the OCT4 qPCR results obtained for a fivefold dilution series for ORMES-22 cDNA to determine the optimum primer dilution for future reactions. The control sample consisted of equal amounts of ORMES-22 cell line. All reactions were carried out in duplicate and repeated three times. For each reaction, we included duplicate fivefold dilutions of GAPDH and each gene as standard curves. The cycling profile for each run was 95°C for 20 seconds and 40 cycles of 95°C for 3 seconds followed by 60°C for 30 seconds, using the default ramp rate. During the amplification cycles, changes in fluorescence emissions were monitored by the ABI Prism sequence. The number of amplification cycles required for the fluorescence signal to reach a determined threshold level (CT) was recorded for every sample and an internal standard curve. The internal standard curve, used for relative mRNA quantifications, was generated from five fivefold dilutions of control biparental ORMES-22. CT values for unknown samples were used to extrapolate the amount of RNA equivalents from the internal standard curve. The RNA equivalent values for SNRPN, NDN, PLAGL1, PEG10, SGCE, IGF2, DIRAS3, UBE3A, and H19 were calculated using standard curve method followed by normalization with endogenous housekeeping GAPDH equivalent values derived from the same internal standard curve [12].
Methylation Analysis by Southern Blot
For Southern blot analysis, 4 µg of genomic DNA (gDNA) from each sample was digested with EcoNI and the CpG methylation-blocked enzymes BsaHI (H19/IGF2) and AgeI (SNRPN) (New England Biolabs, Ipswich, MA, http://www.neb.com). The samples were electrophoresed through a 1% agarose gel and transferred to a nylon membrane. The blots were then hybridized with a probe whose template was generated by PCR of gDNA from the muscle control tissue using the following primers: H19 forward, 5′–3′, AGTGCAGGCTCACACATCATAGTC; H19 reverse, 5′–3′, TAGTCTCTGAGCAAGTAGCGCATC (629 base pairs [bp]); SNRPN forward, 5′–3′, TCTGTAAAGCCCAGGACACCCAT; and SNRPN reverse, 5′–3′, CATGCCTTTGGACTCAGGCTCTTT (707 bp).
The probes were produced by random priming (Megaprime DNA Labeling Systems; Amersham Biosciences, Piscataway, NJ, http://www.amersham.com) and labeled with 32P-dCTP (PerkinElmer Life and Analytical Sciences, Boston, http://www.perkinelmer.com). The hybridization was carried out at 65°C overnight in Rapid-hyb buffer (Amersham Biosciences). The blots were washed for 30 minutes in 2× SSC/0.1% SDS and then for 20 minutes in 0.1× SSC/0.1% SDS at 65°C followed by exposure to the autoradiography film for 3 days.
Methylation Analysis by Bisulfite Sequencing
gDNA was extracted using Puregene DNA Purification system (Gentra Systems, Minneapolis, www.gentra.com) according to the manufacturer’s instructions. Two micrograms of gDNA was modified by bisulfite treatment using a CpG Genome Modification Kit (Chemicon, Temecula, CA, http://www.chemicon.com) according to the manufacturer’s protocol. Primers for bisulfite sequencing were designed using online software (http://www.urogene.org/methprimer) as described elsewhere [13]. The sequence, annealing temperature, and cycle number for H19 primer pair are available in supplemental online Table 1, and SNRPN primers were previously reported [14]. PCRs were carried out in a 50-µl volume containing 1.5 mM MgCl2, 0.2 mM dNTP mixture, 0.2 µM each primer, 400 ng of template DNA, and 5 U of Platinum Taq DNA polymerase (Invitrogen). Amplicons were electrophoresed through 1.6% Tris-Borate-EDTA agarose gels stained with ethidium bromide and visualized on a UV transilluminator. PCR products were recovered from stained gels (QIAquick Gel Extraction Kit; Qiagen, Hilden, Germany, http://www.qiagen.com), ligated with plasmid vector (TOPO TA Cloning Kit pCR4.0 TOPO Vector for Sequencing; Invitrogen), and cloned according to the manufacturer’s protocol. Individual bacterial colonies were transferred to LB/Amp medium and cultured overnight with shaking. Cultures were then processed with Qiaprep Spin Mini-prep Kit (Qiagen) according to the manufacturer’s protocol, resulting in a single cloned PCR species per plasmid. Restriction digestion with EcoRI was carried out for all cloned plasmids to confirm the correct insert. Individual clones were then sequenced using T7 primer with an ABI 3100 capillary genetic analyzer (Applied Biosystems) using BigDye terminator sequencing chemistry [15]. Sequencing results were analyzed using Sequencher software (Gene Codes Corp., Ann Arbor, MI, http://www.genecodes.com). Unmodified genomic DNA from each sample was also amplified and sequenced to identify the presence of previously described single-nucleotide polymorphisms (SNPs) [14].
Cytogenetic Analysis
Mitotically active PESCs in log phase were incubated with 120 ng/ml ethidium bromide for 40 minutes at 37°C in 5% CO2, followed by 120 ng/ml colcemid treatment for 20–40 minutes. Cells were dislodged with 0.25% trypsin and centrifuged at 200g for 8 minutes. The cell pellet was gently resuspended in 0.075 M KCl solution and incubated for 20 minutes at 37°C followed by fixation with methanol/glacial acetic acid (3:1) solution. Fixed cells were dropped on wet slides, air dried, and baked at 90°C for 1 hour. G-banding was performed using trypsin-EDTA and Leishman stain (GTL) by immersing slides in 1 × trypsin-EDTA with 2 drops of 0.4 M Na2HPO4 for 20–30 seconds. Slides were rinsed in distilled water, stained with Leishman stain for 1.5 minutes, rinsed in distilled water again, and air dried. For GTL-banding analysis, 20 metaphases were fully karyotyped under an Olympus BX40 microscope equipped with ×10 and ×100 plan-Apo objectives (Olympus, Tokyo, http://www.olympus-global.com). Images were then captured and cells karyotyped using a CytoVision digital imaging system (Applied Imaging Corporation, San Jose, CA, http://www.aicorp.com).
Microsatellite Analysis
For microsatellite or short tandem repeat (STR) genotyping, DNA was extracted from blood or cultured cells using commercial kits (Gentra Systems). Six multiplexed PCRs were set up for the amplification of 41 markers representing 25 autosomal loci, 1 X-linked marker (DXS22685), and 15 autosomal, major histocompatibility complex (MHC)-linked loci. Based on the published rhesus monkey linkage map [16], these markers are distributed in approximately 19 chromosomes. Two of the markers included in the panel, MFGT21 and MFGT22 [17], were developed from Macaca fuscata and do not have a chromosome assignment. PCRs were set up in 25-µl reactions containing 30 – 60 ng of DNA, 2.5 mM MgCl2, 200 µM dNTPs, 1 × PCR buffer II, 0.5 U of Amplitaq (Applied Biosystems) and fluorescence-labeled primers in concentrations ranging from 0.06 to 0.9 µM, as required for each multiplex PCR. Cycling conditions consisted of four cycles of 1 minute at 94°C, 30 seconds at 58°C, and 30 seconds at 72°C; followed by 25 cycles of 45 seconds at 94°C, 30 seconds at 58°C, and 30 seconds at 72°C; and a final extension at 72°C for 30 minutes. PCR products were separated by capillary electrophoresis on ABI 3730 DNA Analyzer (Applied Biosystems) according to the manufacturer’s instructions. Fragment size analysis and genotyping was done with the computer software STRand (available at http://www.vgl.ucdavis.edu/informatics/strand.php). Primer sequences for MHC-linked STRs 9P06, 246K06, 162B17 (A and B), 151L13, 268P23, and 222I18 were designed from the corresponding rhesus monkey BAC clone sequences deposited in GenBank (accession numbers AC148662, AC148696, AC148683, AC148682, AC148698, and AC148689, respectively). Loci identified by the letter D prefix were amplified using heterologous human primers.
Single-Nucleotide Polymorphism Analysis
SNP genotyping was performed as reported previously [18] using iPLEX reagents and protocols for multiplex PCR, single-base primer extension (SBE), and generation of mass spectra, as per the manufacturer’s instructions (complete details given in iPLEX Application Note; Sequenom, San Diego, http://www.sequenom.com). Four multiplexed assays containing 28, 17, 8, and 7 SNPs were included. Initial multiplexed PCRs were performed in 5-µl reactions on 384-well plates containing 5 ng of genomic DNA. Reactions contained 0.5 U of HotStar Taq polymerase (Qiagen), 100 nM primers, 1.25 × HotStar Taq buffer, 1.625 mM MgCl2, and 500 µM dNTPs. Following enzyme activation at 94°C for 15 minutes, DNA was amplified with 45 cycles of 94°C for 20 seconds, 56°C for 30 seconds, and 72°C for 1 minute, followed by a 3-minute extension at 72°C. Unincorporated dNTPs were removed using shrimp alkaline phosphatase (0.3 U; Sequenom). Single-base extension was carried out by addition of SBE primers at concentrations from 0.625 to 1.25 µM using iPLEX enzyme and buffers (Sequenom). SBE products were measured using the MassARRAY Compact system, and mass spectra were analyzed using TYPER software (Sequenom).
RESULTS
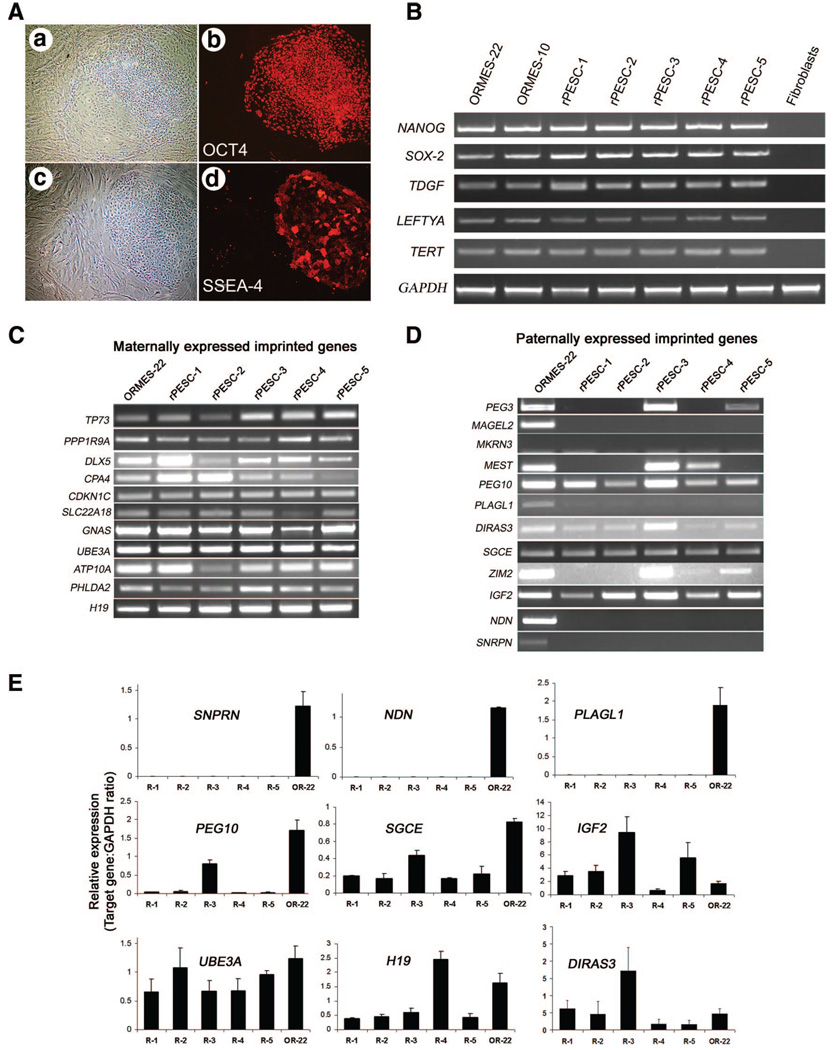
Following artificial activation of mature, MII-arrested oocytes, 38% of cleaved parthenotes developed to the expanded blastocyst-like stage with efficiency statistically similar to that of sperm-fertilized embryos (Table 1). Oocyte activation with ionomycin and DMAP first initiates a calcium flux and then inhibits protein phosphorylation, thus inducing pronuclear formation without completion of meiosis [4, 19]. The parthenotes generated carry a diploid set of chromosomes (42XX) since second polar body extrusion is inhibited. Five karyotypically normal female PESC lines (supplemental online Fig. 1) were derived from 17 blastocyst-like parthenotes for an efficiency of 30%, comparable to our yield with sperm-fertilized, biparental ESCs [1]. Interestingly, three of the five PESC lines derived originated from oocytes harvested from a single stimulation cycle in one female, suggesting that derivation of patient-specific PESCs could be a cost-effective medical option for autologous transplantation based on oocyte requirements, rates of successful development, and overall PESC isolation efficiency. Rhesus parthenogenetic embryonic stem cell (rPESC) lines were morphologically indistinguishable from biparental ESC controls (ORMES lines [1]), expressed key pluripotent markers (including OCT4, SSEA-3 and -4, and TRA-1–60 and -1– 81, as detected immunohistochemically), and produced transcripts of several genes characteristic of pluripotent cells: NANOG, SOX-2, TDGF, LEFTYA, and TERT (Fig. 1A, 1B).
Table 1.
In vitro development of monkey sperm-fertilized embryos and parthenotes and ESC isolation efficiency
| Embryo origin | n | Cleaved (%) |
Eight-cell (%) | Morula (%) |
Blastocyst (%) | Blastocysts used for ESC isolation |
PESC lines isolated (%) |
|---|---|---|---|---|---|---|---|
| Conventional IVF | 71 | 50 (70) | 47 (94) | 38 (76) | 27 (54)a | 0 | NA |
| Intracytoplasmic sperm injection | 40 | 38 (95) | 32 (84) | 29 (76) | 19 (50)a | 0 | NA |
| Parthenotes | 104 | 100 (96) | 96 (96) | 93 (93) | 38 (38)a | 17 | 5 (29) |
Percentages of eight-cell, morulae, compact morulae, and blastocysts are calculated based on the number of cleaved embryos.
Treatments with similar superscripts within a column are not significantly different (p > .05).
Abbreviations: IVF, in vitro fertilization; NA, not applicable; PESC, parthenogenetic embryonic stem cell.
Figure 1.
Pluripotency determination and imprinted gene expression in rPESCs. (A): Expression of pluripotent markers detected by immunocytochemistry. (Aa, Ac): Phase-contrast micrographs of rPESC colonies growing on feeder layers. (Ab, Ad): The same colonies as in (Aa) and (Ac), labeled with antibodies against OCT4 and SSEA-4 (in red). (B): Reverse transcription-polymerase chain reaction (PCR) detection of stemness markers. (C, D): Expression panel of known maternally and paternally expressed imprinted genes. (E): Quantitative PCR analyses of selected paternally and maternally expressed genes in parthenogenetic and biparental ESCs. The x-axis represents the cell lines analyzed (R1 and OR-22), and the y-axis shows the relative expression levels of each imprinted gene as determined by comparison with the expression level of housekeeping control GAPDH (imprinted gene/GAPDH ratio). The mean expression level was calculated using a standard curve method followed by normalization with housekeeping GAPDH. Data represent the mean ± SEM (n = 6). Abbreviations: OR, Oregon Rhesus Monkey Embryonic Stem; ORMES, Oregon Rhesus Monkey Embryonic Stem; R, rhesus parthenogenetic embryonic stem cell; rPESC, rhesus parthenogenetic embryonic stem cell.
Differentiation Potential
The absence of expression of paternal alleles of imprinted genes in parthenotes could result in limited differentiation potential, based on evidence from the mouse where PESCs did not contribute efficiently to mesodermal lineages, particularly to muscle tissues, in chimeric animals and teratomas [20]. However, our rPESC lines gave rise to cell lineages representative of all three embryonic germ layers upon injection into SCID mice, similar to results in the cynomolgus macaque [5, 6]. All rPESC lines efficiently generated teratomas whose histological composition was indistinguishable from that of the biparental controls (supplemental online Fig. 2), containing both mixed cystic and solid regions. The cysts were lined by a simple low cuboidal epithelium and columnar-type epithelium. The solid areas contained an admixture of mostly mature tissues derived from all three germ layers scattered haphazardly throughout the tumors. Ectoderm was represented by stratified squamous epithelium, neural tissue, and ganglion cells; mesodermal derivatives included cartilage, bone, muscle, and fibrous connective tissue; and endodermal tissue was represented mostly by enteric-type columnar epithelium. Immature neuroepithelium arranged in tubules and rosettes with few mitoses and blastema-like tissue was also found in teratomas derived from both ORMES and rPESC lines. We also induced mesodermal differentiation of rPESC lines in vitro by embryoid body production in suspension culture for 5–7 days, followed by plating onto collagen-coated dishes for adherent culture. Approximately 7–14 days after plating, attachment, and further differentiation, spontaneously contracting cell aggregates were observed in all rPESC lines. Analysis of these aggregates by RT-PCR revealed expression of markers specific for cardiomyocytes and muscle tissue (supplemental online Table 2). Our results, along with those of others [5, 6], indicate that diploid primate PESC lines can efficiently give rise to a wide variety of cell types and tissues representing all three germ layers.
Heterozygosity of rPESCs
Since the diploid genome of rPESC lines is achieved by retention of the second polar body following artificial activation, each homologous chromosome pair is a couplet of identical sister chromatids and, as such, might be expected to display high levels of homozygosity in daughter cells. However, we hypothesized that due to homologous crossing-over and recombination during meiosis I, heterozygosity in primate PESCs could occur, a phenomenon recently observed in mouse and human PESCs [21–23]. We conducted microsatellite (STR) analysis and compared results to those of gDNA isolated from the peripheral blood of egg donors. The analysis of 29 STR loci confirmed that rPESC-1, -2, and -3 were derived from the same egg donor and that rPESC-4 and -5 were from two other unrelated females (Table 2). The egg donor for rPESC-1, -2, and -3 (female 22184) exhibited 21 heterozygous loci out of the 29 analyzed. rPESC-1, -2, and -3 showed the presence of both alleles at a majority of these loci: 16, 17, and 13 heterozygous loci out of 21, respectively (Table 2). Similarly, rPESC-4 inherited 9 heterozygous loci out of 18 present in the egg donor, and rPESC-5 exhibited 16 out of 20 heterozygous STRs. To eliminate the possibility that rPESCs were unique, we examined polymorphic loci in the cynomolgus monkey PESC line, Cyno-1, and the egg donor female [5]. This line displayed 13 heterozygous loci out of 24 informative STRs present in the egg donor female. For corroboration, a panel of 60 SNPs localized to the 3′ ends of specific rhesus monkey genes were monitored [18]. Of the 13 informative SNPs detected in egg donor 22184, her rPESC lines-1, -2, and -3 inherited both alleles at 8, 8, and 7 loci, respectively (Table 3). rPESC-4 displayed 10 heterozygous loci out of 18 present in the egg donor (female 16715) and rPESC-5 exhibited 10 SNPs out of 16 existing in the donor (female 18178). Based on an analysis of 28 heterozygous microsatellite loci and 32 informative SNPs present in the egg donors, diploid rPESCs restored heterozygosity at 64% of the examined loci on average.
Table 2.
Short tandem repeat genotypes in monkey oocyte donors and rPESC lines
| Locus | Egg donor 22184 |
rPESC-1 | rPESC-2 | rPESC-3 | Egg donor 16715 |
rPESC-4 | Egg donor 18178 |
rPESC-5 |
|---|---|---|---|---|---|---|---|---|
| D1S548 | 190/206a | 206/206 | 190/206a | 190/190 | 190/206a | 190/206a | 199/202a | 199/202a |
| D2S1333 | 277/297a | 277/297a | 277/297a | 277/297a | 289/293a | 289/289 | 277/277 | 277/277 |
| D3S1768 | 205/221a | 205/221a | 205/221a | 205/221a | 201/217a | 201/217a | 215/273a | 215/273a |
| D4S2365 | 283/287a | 283/287a | 283/287a | 287/287 | 279/283a | 283/283 | 273/281a | 273/281a |
| D4S413 | 131/131 | 131/131 | 131/131 | 131/131 | 129/131a | 129/129 | 129/141a | 129/141a |
| D5S1457 | 132/132 | 132/132 | 132/132 | 132/132 | 132/136a | 136/136 | 128/136a | 128/128 |
| D6S276 | 215/225a | 215/225a | 225/225 | 215/225a | 211/225a | 211/225a | 239/241a | 239/241a |
| D6S291 | 206/208a | 206/208a | 206/206 | 208/208 | 208/208 | 208/208 | 205/217a | 205/217a |
| D6S501 | 180/184a | 180/180 | 180/184a | 180/184a | 180/180 | 180/180 | 176/180 | 176/180 |
| D6S1691 | 197/197 | 197/197 | 197/197 | 197/197 | 203/203 | 203/203 | 175/195a | 175/195a |
| D7S513 | 209/239a | 209/239a | 209/239a | 209/239a | 189/199a | 199/199 | 190/203a | 190/203a |
| D7S794 | 124/132a | 124/132a | 124/132a | 124/132a | 128/128 | 128/128 | 127/127 | 127/127 |
| D8S1106 | 148/160a | 148/160a | 148/160a | 148/160a | 144/156a | 144/156a | 140/152a | 140/152a |
| D9S921 | 187/195a | 187/195a | 187/195a | 187/195a | 179/199a | 179/199a | 187/195a | 187/195a |
| D10S1412 | 157/157 | 157/157 | 157/157 | 157/157 | 157/157 | 157/157 | 155/155 | 155/155 |
| D11S2002 | 252/252 | 252/252 | 252/252 | 252/252 | 252/260a | 260/260 | 260/260 | 260/260 |
| D11S925 | 308/330a | 308/330a | 308/330a | 308/308 | 308/312a | 308/312a | 305/327a | 305/327a |
| D12S364 | 290/290 | 290/290 | 290/290 | 290/290 | 280/290a | 280/290a | 286/288a | 286/288a |
| D12S67 | 117/212a | 117/212a | 117/212a | 117/212a | 113/121a | 113/113 | 125/188a | 188/188 |
| D13S765 | 224/232a | 224/232a | 232/232 | 224/232a | 224/228a | 224/228a | 222/222 | 222/222 |
| D15S823 | 333/357a | 357/357 | 333/333 | 333/333 | 353/353 | 353/353 | 369/369 | 369/369 |
| D16S403 | 162/164a | 162/164a | 162/164a | 162/164a | 152/156a | 156/156 | 163/165a | 163/163 |
| D17S1300 | 248/260a | 248/260a | 248/260a | 248/260a | 232/248a | 232/232 | 255/255 | 255/255 |
| D18S537 | 162/174a | 162/174a | 162/174a | 162/162 | 174/174 | 174/174 | 166/174a | 166/174a |
| D18S72 | 308/322a | 308/308 | 308/322a | 322/322 | 308/308 | 308/308 | 312/312 | 312/312 |
| D22S685 | 319/319 | 319/319 | 319/319 | 319/319 | 315/323a | 315/323a | 315/319a | 315/315 |
| DXS2506 | 262/262 | 262/262 | 262/262 | 262/262 | 262/262 | 262/262 | 262/274a | 262/274a |
| MFGT21 | 111/115a | 111/115a | 111/115a | 111/115a | 119/119 | 119/119 | 108/118a | 108/118a |
| MFGT22 | 110/120a | 110/110 | 110/120a | 110/110 | 104/104 | 104/104 | 92/100a | 92/100a |
Heterozygous loci.
Abbreviation: rPESC, rhesus parthenogenetic embryonic stem cell.
Table 3.
SNP analysis of monkey oocyte donors and rPESC lines
| SNP | Egg donor 22184 |
rPESC-1 | rPESC-2 | rPESC-3 | Egg donor 16715 |
rPESC-4 | Egg donor 18178 |
rPESC-5 |
|---|---|---|---|---|---|---|---|---|
| BCHE:76 | C | C | C | C | GCa | G | C | C |
| BCHE:447 | AGa | A | G | G | AGa | A | AGa | A |
| CCL8:516 | G | G | G | G | AGa | A | AGa | G |
| CCR1:641 | G | G | G | G | G | G | G | G |
| CCR9:315 | C | C | C | C | C | C | CTa | CTa |
| CD40LG:738 | G | G | G | G | G | G | AGa | AGa |
| CD44:471 | T | T | T | T | T | T | T | T |
| CD69:294 | CTa | CTa | CTa | CTa | CTa | CTa | C | C |
| CD74:213 | C | C | C | C | C | C | CTa | CTa |
| CD74:344 | CTa | CTa | CTa | CTa | CTa | CTa | C | C |
| CFTR:796 | AGa | AGa | G | A | G | G | AGa | AGa |
| CX3CR1:593 | AGa | AGa | AGa | AGa | AGa | AGa | G | G |
| CXCL12:173 | C | C | C | C | T | T | CTa | C |
| CYP11A1:150 | G | G | G | G | AGa | G | AGa | G |
| FAS:135 | A | A | A | A | AGa | A | A | A |
| FSHR:784 | CGa | CGa | CGa | C | CGa | CGa | C | C |
| HTATSF1:636 | C | C | C | C | C | C | CTa | CTa |
| IL1:755 | ATa | ATa | ATa | ATa | ATa | ATa | ATa | ATa |
| IL2RA:124 | C | C | C | C | CTa | CTa | C | C |
| INHBB:131 | C | C | C | C | TCa | TCa | C | C |
| ITGA:321 | A | A | A | A | A | A | AGa | AGa |
| LRP8:647 | T | T | T | T | C | C | CTa | CTa |
| NDN:135 | G | G | G | G | AGa | AGa | G | G |
| NOS1:216 | AGa | ND | AGa | AGa | AGa | AGa | ND | ND |
| NR3C1:458 | T | T | T | T | T | T | ATa | ATa |
| PYY:151 | T | T | T | T | TCa | TCa | T | T |
| SIRT1:277 | GTa | GTa | GTa | GTa | G | G | G | G |
| SLC5A7:79 | G | G | G | G | G | G | G | G |
| SLC6A4:132 | GCa | G | C | C | C | C | GCa | G |
| SNCA:394 | TCa | ND | TCa | C | C | C | ND | ND |
| STAR:522 | GTa | G | G | GTa | GTa | G | GTa | T |
| TLR4:735 | T | T | T | T | TCa | C | T | T |
| TLR5:389 | TCa | T | C | C | TCa | T | C | C |
| TNF:82 | T | T | T | T | T | T | CTa | CTa |
| XCL1:320 | C | C | C | C | T | T | T | T |
Heterozygous loci.
Abbreviations: ND, not determined; rPESC, rhesus parthenogenetic embryonic stem cell; SNP, single-nucleotide polymorphism.
Imprinting in rPESCs
Expression of imprinted genes normally silenced by passage through the female germline would not be expected in parthenotes, since both alleles are of maternal origin. We tested this expectation in undifferentiated rPESC lines by conducting expression analysis of several known imprinted genes. Transcripts of 11 imprinted genes normally expressed predominantly from the maternal allele were detected in both rPESCs and biparental ESCs (Fig. 1C). Control reactions (−RT) with DNased RNA samples in all qualitative and quantitative RT-PCRs were negative. Expression levels of maternally expressed UBE3A in rPESC lines were similar to those of biparental ORMES-22, whereas H19 gene activity varied between rPESC lines (Fig. 1E). Unexpectedly, amplified RT-PCR products of several imprinted genes normally expressed from the paternal allele, including PEG10, DIRAS3, SGCE, and IGF2, were also detected in rPESCs (Fig. 1D). Real-time PCR results confirmed significant levels of DIRAS3, SGCE, and IGF2 transcripts (Fig. 1E), whereas mRNA levels of PEG10 were significantly downregulated. In addition, PEG3, MEST, and ZIM2 transcripts were detected in some rPESC lines but not in others, whereas transcripts from PLAGL1, MAGEL2, MKRN3, SNRPN, and NDN were absent in all rPESCs but expressed in biparental ESCs (Fig. 1D, 1E). Thus, rPESCs expressed all maternally and at least some paternally imprinted genes.
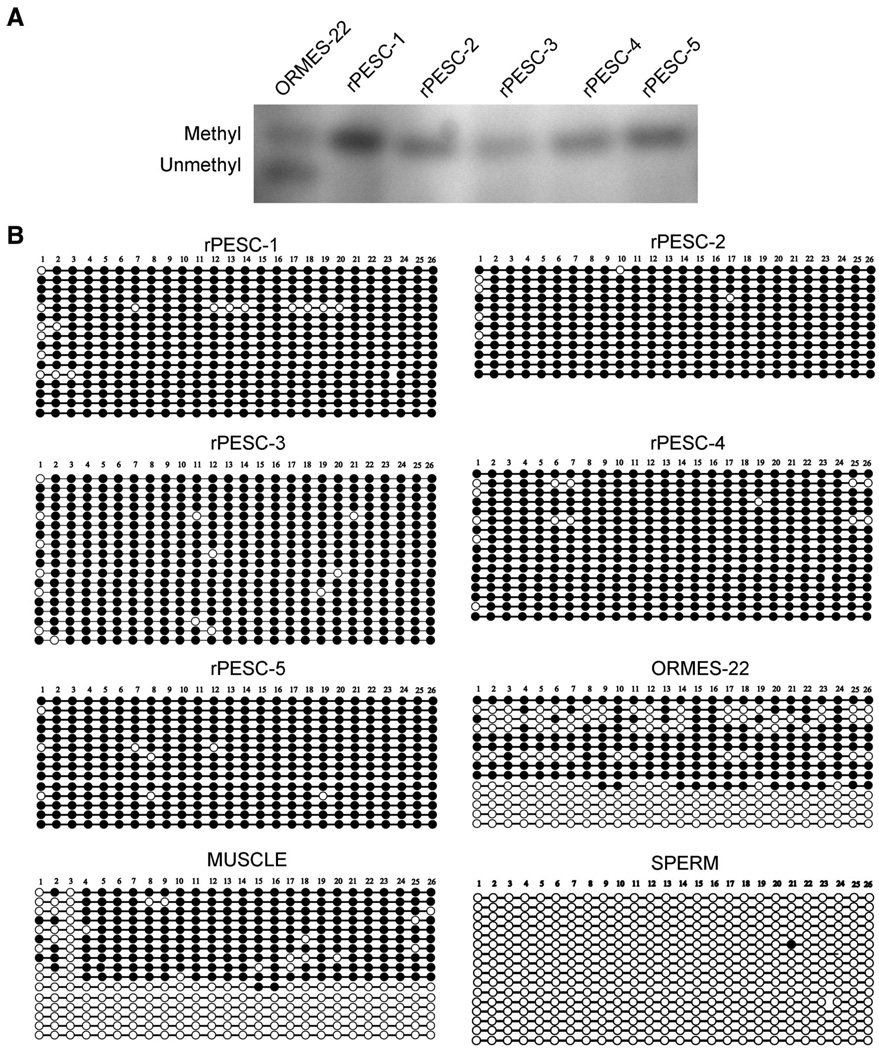
DNA methylation of CpG dinucleotides is generally associated with the regulation of allele-specific expression of imprinted genes. Parent-of-origin-dependent methylation imposed during gametogenesis within so-called imprinting centers (ICs) facilitates discrimination between maternal and paternal alleles in cells of offspring, resulting in monoallelic expression at these loci. The imprinted expression of the adjacent IGF2 and H19 genes is reciprocally controlled by a common IC located upstream of H19, harboring a CpG island that is methylated on the paternal allele and unmethylated on the maternal allele [14]. In primate MII oocytes, methylation marks are absent from both alleles within the IGF2/H19 IC, whereas both alleles in mature monkey sperm are heavily methylated [14]. Upon fertilization, biparental, differentially methylated patterns are normally restored in the zygote and retained throughout development. Based on their oocyte-specific heritage, PESCs should possess only unmethylated alleles at this IC. To test this assumption, we first analyzed the methylation status of this region by methylation-sensitive Southern blot. Digestion of skeletal muscle and biparental ORMES-22 with the methylation-sensitive BsaHI restriction enzyme produced similar results, reflecting the presence of both methylated and unmethylated alleles (Fig. 2A). Unexpectedly, rPESC lines also showed both uncut (methylated) and cut (unmethylated) fragments, although the level of methylated fragments appeared reduced compared with that observed in biparental controls. For validation, we performed independent methylation analysis of the IGF2/H19 IC by bisulfite sequencing and confirmed sporadic hypermethylation in rPESC lines (Fig. 2B). In contrast, the SNURF/SNRPN IC, which is normally methylated on the maternal alleles, was completely methylated in rPESC lines (Fig. 3). These results are consistent with the notion that primate PESCs, similar to their sperm-fertilized counterparts, are epigenetically unstable, particularly within the IGF2/H19 IC, perhaps accounting for the relaxed imprinting observed at the IGF2 locus in rPESCs [14]. Although the underlying molecular mechanisms remain unclear, culture conditions used to isolate and propagate rPESCs may alter the normal maintenance of methylation imprints.
Figure 2.
Methylation analysis of the IGF2/H19 IC in rPESCs. (A): Southern blot analysis. The larger (upper) band corresponds to a 1.3-kilobase (kb) uncut Methyl fragment, and the smaller (lower) band corresponds to the 1.07-kb digested Unmethyl fragment. Occurrence of both bands represents the presence of both Methyl and Unmethyl alleles in the same sample. (B): Methylation analysis by genomic bisulfite sequencing. Each circle represents an individual CpG site; Methyl CpG dinucleotides are depicted by black circles and the Unmethyl sites by open circles. The boxed area corresponds to the CTCF-6 core binding site. As expected, sperm DNA was completely Methyl, and control muscle maintained Unmethyl Mat and Methyl Pat alleles (separated by the presence of single-nucleotide polymorphism). In contrast, biparental ORMES-22 and rPESC lines were hypermethylated as a result of sporadic methylation of Mat alleles. Abbreviations: Mat, maternal; Methyl, methylated; ORMES, Oregon Rhesus Monkey Embryonic Stem; Pat, paternal; rPESC, rhesus parthenogenetic embryonic stem cell; Unmethyl, unmethylated.
Figure 3.
Methylation status of the SNURF/SNURPN IC involved in the regulation of paternal imprints in the Prader-Willi syndrome region in rPESC lines. (A): Southern blot analysis. The larger (upper) band corresponds to a 5.7-kilobase (kb) uncut Methyl fragment, and the smaller (lower) band corresponds to the 4.66-kb digested Unmethyl fragment. (B): Bisulfite sequencing demonstrating that oocyte-derived rPESC lines were heavily Methyl, as expected, whereas all sequenced clones of mature sperm were completely Unmethyl. In biparental ORMES-22 and muscle DNA, both Unmethyl and Methyl clones were observed. Abbreviations: Methyl, methylated; ORMES, Oregon Rhesus Monkey Embryonic Stem; rPESC, rhesus parthenogenetic embryonic stem cell; Unmethyl, unmethylated.
Histocompatibility of PESCs
To investigate whether rPESCs re-establish heterozygosity within the MHC region, as observed in the mouse [21], we genotyped rPESC lines and oocyte donors using 15 polymorphic microsatellite markers mapped within or near the MHC locus [24]. Microsatellite genotyping provides an alternative method for the rapid and accurate prediction of immunocompatibility and is widely used for MHC analysis in humans for tissue matching and donor screening [25, 26]. STR sequencing and comparisons with the complete rhesus monkey MHC genomic map indicated exact positioning of these markers within the 5.3-megabase MHC region on chromosome 6, with D6S291 located at the centromeric end and D6S276/D6S1691 at the telomeric end [24]. Comparisons of genotypes in egg donors and their corresponding rPESC lines revealed that in four cell lines, STR alleles were organized together into tightly linked haplotype blocks during meiotic recombination, resulting in either complete homozygosity (rPESC-2; Table 4) or heterozygosity (rPESC-1, -4, and -5). rPESC-1, -4, and -5 showed recombination upstream of the MHC region that fully restored heterozygosity and resulted in genotypes across the MHC region that were identical to those of the egg donors (Table 4). Therefore, these cell lines can be considered isogenic. The tight genetic linkage between MHC haplotypes appears to preclude meiotic recombination in rPESC-1, -2, -4, and -5, but rPESC-3 displayed the presence of a single meiotic crossover between major histocompatibility complex class I chain-related A and 246K06 loci (Table 4). The Cyno-1 cell line was homozygous within all 15 MHC-linked STRs analyzed.
Table 4.
Histocompatibility analysis of monkey egg donors and their rPESC lines based on MHC-linked short tandem repeat analysis
| MHC locus |
Egg donor 22184 |
rPESC-1 | rPESC-2 | rPESC-3 | Egg donor 16715 |
rPESC-4 | Egg donor 18178 |
rPESC-5 |
|---|---|---|---|---|---|---|---|---|
| D6S291 | 206/208a | 206/208a | 206/206 | 208/208 | 208/208 | 208/208 | 205/217a | 205/217a |
| D6S2741 | 269/273a | 269/273a | 273/273 | 269/269 | 277/281a | 277/281a | 259/269a | 259/269a |
| D6S2876 | 209/219a | 209/219a | 209/209 | 219/219 | 209/209 | 209/209 | 195/210a | 195/210a |
| 9P06 | 185/191a | 185/191a | 185/185 | 191/191 | 165/175a | 165/175a | 177/189a | 177/189a |
| DRA | 112/136a | 112/136a | 136/136 | 112/112 | 136/136 | 136/136 | 188/126a | 188/126a |
| MICA | 194/200a | 194/200a | 194/194 | 200/200 | 194/200a | 194/200a | 200/200 | 200/200 |
| 246K06 | 275/279a | 275/279a | 275/275 | 275/279a | 275/277a | 275/277a | 276/290a | 276/290a |
| 162B17A | 238/242a | 238/242a | 238/238 | 238/242a | 238/238 | 238/238 | 238/242a | 238/242a |
| 162B17B | 309/309 | 309/309 | 309/309 | 309/309 | 307/311a | 307/311a | 281/231a | 281/231a |
| 151L13 | 303/309a | 303/309a | 303/303 | 303/309a | 303/309a | 303/309a | 300/304a | 300/304a |
| MOGCA | 121/123a | 121/123a | 121/121 | 121/123a | 123/123 | 123/123 | 188/188 | 188/188 |
| 268P23 | 148/150a | 148/150a | 148/148 | 148/150a | 150/150 | 150/150 | 147/147 | 147/147 |
| 222I18 | 167/167 | 167/167 | 167/167 | 167/167 | 161/173a | 161/173a | 167/173a | 167/173a |
| D6S276 | 215/225a | 215/225a | 225/225 | 215/225a | 211/225a | 211/225a | 239/241a | 239/241a |
| D6S1691 | 197/197 | 197/197 | 197/197 | 197/197 | 203/203 | 203/203 | 175/195a | 175/195a |
Heterozygous loci.
Abbreviations: MHC, major histocompatibility complex; rPESC, rhesus parthenogenetic embryonic stem cell.
DISCUSSION
We have demonstrated the ability to isolate, in high yield, pluripotent cell lines from monkey parthenotes. These new PESC lines appear similar to existing ESC lines derived from biparental sperm-fertilized embryos with respect to expression of common pluripotency markers and possession of other attributes of primate ESCs, including self-renewal and the capacity to generate cell derivatives representative of all three germ layers in vivo and in vitro [1]. We found that monkey parthenotes can be produced and cultured to the blastocyst-like stage and used as a source of pluripotent cell lines as efficiently as embryos produced by natural conception or in vitro fertilization. Moreover, the establishment of three separate PESC lines from a single ovarian stimulation cycle in a single animal attests to the feasibility of deriving PESCs as a source of pluripotent cells for the treatment of degenerative diseases in a large population of adult females. Indeed, a recent study demonstrated that human parthenogenetic ESCs can also be generated with high efficiency, where six PESC lines were derived from 44 oocytes donated by four patients [23].
Since disruption or inappropriate expression of imprinted genes is associated with severe clinical syndromes and carcinogenesis typically manifested during gestation or early childhood [9, 27], the potential utility of PESCs will be dependent, in part, on the degree to which epigenetic abnormalities influence differentiation or cellular function in vivo following engraftment. However, although aberrant growth phenotypes have been seen during embryonic and fetal development in chimeras generated with mouse PESCs [20], this observation may be irrelevant to the normal functional requirements of imprinted genes in differentiated derivatives of ESCs or PESCs in the context of therapeutic engraftment, where these genes may not be expressed or required. Thus, a case can be made that relaxed imprinting need not be detrimental. Indeed, monoparental, parthenogenetic, and androgenetic ESCs in the mouse have been differentiated into transplantable hematopoietic progenitors that were capable of long-term multilineage reconstitution when engrafted into lethally irradiated adult mice [28]. Moreover, in some cases, postnatal monoparental chimeras were able to alleviate imprinting-related defects [29].
Epigenetic instability is seen in the mouse [30, 31], although human ESCs appear relatively stable, at least at low passage numbers [32]. In monkey ESCs, we reported normal imprinting in NDN and SNRPN but relaxed imprinting for IGF2 and H19 [14]. Here, we found that rPESC lines are variable, with the unexpected expression of some imprinted genes that are normally silenced by passage through the maternal germ line. The degree to which abnormal imprinting affects the therapeutic use of ESCs/PESCs must await the outcome of transplantation studies.
The high levels of genomic homozygosity anticipated in diploid, monoparental PESCs represents another safety issue in the context of therapeutic engraftment, since widespread loss of heterozygosity is the most common genetic alteration in human cancers [33]. However, based on our results here in the rhesus monkey and recent reports in mouse and human PESCs [21–23], the majority of loci in PESCs are heterozygous, having undergone meiotic recombination prior to derivation.
Immunological responses to tissue and organ transplants will vary depending on the extent of MHC antigen matching. Our analysis in the monkey provided clear evidence indicating that tight linkage of maternal MHC haplotypes typically leads to either complete homozygosity or heterozygosity of this region in PESCs, depending on whether or not this region was subject to meiotic recombination in the particular oocyte that gave rise to each PESC line. Heterozygous lines that carry haplotypes identical to the egg donors should support autologous transplantation of PESC derivatives with no or limited rejection. Conversely, homozygous PESC lines could also be used for autologous transplantation; however, the presence of only one of the two egg donor MHC haplotypes could incite rejection by natural killer cells [21]. Therefore, the creation of PESC lines heterozygous at MHC loci is a more attractive outcome for generating fully histocompatible cells. It is also important to note that minor histocompatibility antigens that scattered across the genome could also provoke an immune reaction. Hence, the latest evidence demonstrating that creation of fully matched, genetically identical primate ESCs can be achieved using somatic cell nuclear transfer or direct reprogramming represent major advances in efforts to generate patient-specific cells and tissues for therapeutic purposes [3, 34, 35].
Meiotic recombination is an important evolutionary force that plays a vital role in shaping genomes and creating diversity. Recent studies suggest that meiotic recombination events tend to happen in certain regions of the genome, leading to the concept of recombination hot spots [36]. Direct analysis of fine-scale recombination events is not feasible by pedigree analysis. The only information on recombination in primates comes from single-molecule PCR analysis of sperm. However, recombination frequencies are also sex-dependent. Crossing over is estimated to occur approximately 55 times during meiosis in males and approximately 75 times in females [36]. We present here a novel system that allows detailed analysis of recombination in the primate female. Each PESC line represents the recombination events that occurred within a single oocyte. Thus, primate PESCs, in combination with powerful genetic and cytological assays, could be valuable in vitro models with which to study increasingly complex aspects of meiosis, chromosome behavior, and recombination. It is generally accepted that exchange during meiotic recombination occurs more frequently at distal telomeric regions of chromosomes, with the frequency of recombination decreasing near the centromeric region. In murine PESC lines, evidence has been presented that the recombination rate is directly proportional to the distance from the centromere [21]. However, the location of the chromosomal centromeres is not currently known in the rhesus monkey, precluding analysis of recombination frequencies in this study.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge the Division of Animal Resources and the Endocrine Services Cores at the Oregon National Primate Research Center for assistance and technical services. We are grateful to Cathy Ramsey, Michelle Sparman, Joy Woodward, and Thea Ward for technical assistance and to Dr. John Fanton and Darla Jacobs for laparoscopic oocyte retrieval. We are indebted to Dr. Warren Sanger and Marilu Nelson for karyotyping services and to Drs. John Hennebold and Xuemei Wu for help with real-time PCR and Southern analysis. We thank Drs. Richard Stouffer, Markus Grompe, and John McCarrey for helpful discussions and critical reading of the manuscript. This study was supported by funds from the Oregon National Primate Research Center, the Stem Cell Research Foundation and NIH Grants NS044330 (to S.M.), HD18185 (to R. Stouffer), and RR00163 (to D. Dorsa). J.B. is currently affiliated with the Stanford Institute for Stem Cell Biology and Regenerative Medicine, Stanford University, Palo Alto, CA. S.G. is currently affiliated with the Department of Pathology, Roger Williams Medical Center, Providence, RI.
Footnotes
DISCLOSURE OF POTENTIAL CONFLICTS OF INTEREST
The authors indicate no potential conflicts of interest.
See www.StemCells.com for supplemental material available online.
REFERENCES
- 1.Mitalipov S, Kuo HC, Byrne J, et al. Isolation and characterization of novel rhesus monkey embryonic stem cell lines. Stem Cells. 2006;24:2177–2186. doi: 10.1634/stemcells.2006-0125. [DOI] [PubMed] [Google Scholar]
- 2.Takagi Y, Takahashi J, Saiki H, et al. Dopaminergic neurons generated from monkey embryonic stem cells function in a Parkinson primate model. J Clin Invest. 2005;115:102–109. doi: 10.1172/JCI21137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Byrne JA, Pedersen DA, Clepper LL, et al. Producing primate embryonic stem cells by somatic cell nuclear transfer. Nature. 2007;450:497–502. doi: 10.1038/nature06357. [DOI] [PubMed] [Google Scholar]
- 4.Mitalipov SM, Nusser KD, Wolf DP. Parthenogenetic activation of rhesus monkey oocytes and reconstructed embryos. Biol Reprod. 2001;65:253–259. doi: 10.1095/biolreprod65.1.253. [DOI] [PubMed] [Google Scholar]
- 5.Cibelli JB, Grant KA, Chapman KB, et al. Parthenogenetic stem cells in nonhuman primates. Science. 2002;295:819. doi: 10.1126/science.1065637. [DOI] [PubMed] [Google Scholar]
- 6.Vrana KE, Hipp JD, Goss AM, et al. Nonhuman primate parthenogenetic stem cells. Proc Natl Acad Sci U S A. 2003;100 suppl 1:11911–11916. doi: 10.1073/pnas.2034195100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barton SC, Surani MA, Norris ML. Role of paternal and maternal genomes in mouse development. Nature. 1984;311:374–376. doi: 10.1038/311374a0. [DOI] [PubMed] [Google Scholar]
- 8.Surani MA, Barton SC, Norris ML. Development of reconstituted mouse eggs suggests imprinting of the genome during gametogenesis. Nature. 1984;308:548–550. doi: 10.1038/308548a0. [DOI] [PubMed] [Google Scholar]
- 9.Nakagawa H, Chadwick RB, Peltomaki P, et al. Loss of imprinting of the insulin-like growth factor II gene occurs by biallelic methylation in a core region of H19-associated CTCF-binding sites in colorectal cancer. Proc Natl Acad Sci U S A. 2001;98:591–596. doi: 10.1073/pnas.011528698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zelinski-Wooten MB, Hutchison JS, Hess DL, et al. Follicle stimulating hormone alone supports follicle growth and oocyte development in gonadotrophin-releasing hormone antagonist-treated monkeys. Hum Reprod. 1995;10:1658–1666. doi: 10.1093/oxfordjournals.humrep.a136151. [DOI] [PubMed] [Google Scholar]
- 11.McKiernan SH, Bavister BD. Culture of one-cell hamster embryos with water soluble vitamins: Pantothenate stimulates blastocyst production. Hum Reprod. 2000;15:157–164. doi: 10.1093/humrep/15.1.157. [DOI] [PubMed] [Google Scholar]
- 12.Byrne JA, Mitalipov SM, Clepper L, et al. Transcriptional profiling of rhesus monkey embryonic stem cells. Biol Reprod. 2006;75:908–915. doi: 10.1095/biolreprod.106.053868. [DOI] [PubMed] [Google Scholar]
- 13.Li LC, Dahiya R. MethPrimer: Designing primers for methylation PCRs. Bioinformatics. 2002;18:1427–1431. doi: 10.1093/bioinformatics/18.11.1427. [DOI] [PubMed] [Google Scholar]
- 14.Mitalipov S, Clepper L, Sritanaudomchai H, et al. Methylation status of imprinting centers for H19/IGF2 and SNURF/SNRPN in primate embryonic stem cells. Stem Cells. 2007;25:581–588. doi: 10.1634/stemcells.2006-0120. [DOI] [PubMed] [Google Scholar]
- 15.Wen L. Two-step cycle sequencing improves base ambiguities and signal dropouts in DNA sequencing reactions using energy-transfer-based fluorescent dye terminators. Mol Biotechnol. 2001;17:135–142. doi: 10.1385/MB:17:2:135. [DOI] [PubMed] [Google Scholar]
- 16.Rogers J, Garcia R, Shelledy W, et al. An initial genetic linkage map of the rhesus macaque (Macaca mulatta) genome using human microsatellite loci. Genomics. 2006;87:30–38. doi: 10.1016/j.ygeno.2005.10.004. [DOI] [PubMed] [Google Scholar]
- 17.Domingo-Roura X, Lopez-Giraldez T, Shinohara M, et al. Hypervariable microsatellite loci in the Japanese macaque (Macaca fuscata) conserved in related species. Am J Primatol. 1997;43:357–360. doi: 10.1002/(SICI)1098-2345(1997)43:4<357::AID-AJP7>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 18.Ferguson B, Street SL, Wright H, et al. Single nucleotide polymorphisms (SNPs) distinguish Indian-origin and Chinese-origin rhesus macaques (Macaca mulatta) BMC Genomics. 2007;8:43. doi: 10.1186/1471-2164-8-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Susko-Parrish JL, Leibfried-Rutledge ML, Northey DL, et al. Inhibition of protein kinases after an induced calcium transient causes transition of bovine oocytes to embryonic cycles without meiotic completion. Dev Biol. 1994;166:729–739. doi: 10.1006/dbio.1994.1351. [DOI] [PubMed] [Google Scholar]
- 20.Allen ND, Barton SC, Hilton K, et al. A functional analysis of imprinting in parthenogenetic embryonic stem cells. Development. 1994;120:1473–1482. doi: 10.1242/dev.120.6.1473. [DOI] [PubMed] [Google Scholar]
- 21.Kim K, Lerou P, Yabuuchi A, et al. Histocompatible embryonic stem cells by parthenogenesis. Science. 2007;315:482–486. doi: 10.1126/science.1133542. [DOI] [PubMed] [Google Scholar]
- 22.Kim k, Ng k, Rugg-Gunn P, et al. Recombination signatures distinguish embryonic stem cells derived by parthenogenesis and somatic cell nuclear transfer. Cell Stem Cell. 2007;1:346–352. doi: 10.1016/j.stem.2007.07.001. [DOI] [PubMed] [Google Scholar]
- 23.Revazova ES, Turovets NA, Kochetkova OD, et al. Patient-specific stem cell lines derived from human parthenogenetic blastocysts. Cloning Stem Cells. 2007;9:432–449. doi: 10.1089/clo.2007.0033. [DOI] [PubMed] [Google Scholar]
- 24.Penedo MC, Bontrop RE, Heijmans CM, et al. Microsatellite typing of the rhesus macaque MHC region. Immunogenetics. 2005;57:198–209. doi: 10.1007/s00251-005-0787-1. [DOI] [PubMed] [Google Scholar]
- 25.Carrington M, Wade J. Selection of transplant donors based on MHC microsatellite data. Hum Immunol. 1996;51:106–109. doi: 10.1016/s0198-8859(96)00268-6. [DOI] [PubMed] [Google Scholar]
- 26.Foissac A, Fort M, Clayton J, et al. Microsatellites in the HLA region: HLA prediction and strategies for bone marrow donor registries. Transplant Proc. 2001;33:491–492. doi: 10.1016/s0041-1345(00)02107-2. [DOI] [PubMed] [Google Scholar]
- 27.Cui H, Onyango P, Brandenburg S, et al. Loss of imprinting in colorectal cancer linked to hypomethylation of H19 and IGF2. Cancer Res. 2002;62:6442–6446. [PubMed] [Google Scholar]
- 28.Eckardt S, Leu NA, Bradley HL, et al. Hematopoietic reconstitution with androgenetic and gynogenetic stem cells. Genes Dev. 2007;21:409–419. doi: 10.1101/gad.1524207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mann JR, Gadi I, Harbison ML, et al. Androgenetic mouse embryonic stem cells are pluripotent and cause skeletal defects in chimeras: Implications for genetic imprinting. Cell. 1990;62:251–260. doi: 10.1016/0092-8674(90)90363-j. [DOI] [PubMed] [Google Scholar]
- 30.Dean W, Bowden L, Aitchison A, et al. Altered imprinted gene methylation and expression in completely ES cell-derived mouse fetuses: Association with aberrant phenotypes. Development. 1998;125:2273–2282. doi: 10.1242/dev.125.12.2273. [DOI] [PubMed] [Google Scholar]
- 31.Humpherys D, Eggan K, Akutsu H, et al. Epigenetic instability in ES cells and cloned mice. Science. 2001;293:95–97. doi: 10.1126/science.1061402. [DOI] [PubMed] [Google Scholar]
- 32.Rugg-Gunn PJ, Ferguson-Smith AC, Pedersen RA. Epigenetic status of human embryonic stem cells. Nat Genet. 2005;37:585–587. doi: 10.1038/ng1556. [DOI] [PubMed] [Google Scholar]
- 33.Cavenee WK. Loss of heterozygosity in stages of malignancy. Clin Chem. 1989;35:B48–B52. [PubMed] [Google Scholar]
- 34.Yu J, Vodyanik MA, Smuga-Otto K, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 35.Takahashi K, Tanabe K, Ohnuki M, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 36.Jeffreys AJ, Neumann R. Factors influencing recombination frequency and distribution in a human meiotic crossover hotspot. Hum Mol Genet. 2005;14:2277–2287. doi: 10.1093/hmg/ddi232. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.