Abstract
RNA secondary structures can be divided into helical regions composed of canonical Watson-Crick and related basepairs, as well as single-stranded regions such as hairpin loops, internal loops, and junctions. These elements function as building blocks in the design of diverse RNA molecules with various fundamental functions in the cell. To better understand the intricate architecture of three-dimensional RNAs, we analyze existing RNA 4-way junctions in terms of basepair interactions and three-dimensional configurations. Specifically, we identify nine broad junction families according to coaxial stacking patterns and helical configurations. We find that helices within junctions tend to arrange in roughly parallel and perpendicular patterns, and stabilize their conformations using common tertiary motifs like coaxial stacking, loop-helix interaction, and helix packing interaction. Our analysis also reveals a number of highly conserved basepair interaction patterns and novel tertiary motifs such as A-minor-coaxial stacking combinations and sarcin/ricin motif variants. Such analyses of RNA building blocks can ultimately help in the difficult task of RNA 3D structure prediction.
Keywords: RNA structure, 4-way junction, tertiary motifs, coaxial stacking, non-Watson-Crick-basepair
INTRODUCTION
Recent studies have demonstrated the amazing capacity of RNA to form complex tertiary structures as well as perform many surprisingly intricate cellular functions1; 2; 3. As new roles for RNAs are being discovered, the functionality of many non-coding RNAs remains unknown4.
RNA crystallography has offered unprecedented opportunities to analyze RNA tertiary (3D) structure5; 6; 7; 8; 9 and relate structure to function. RNA molecules have also been studied extensively at the secondary-structure level, where building blocks include helical stems and single-stranded regions such as hairpins, bulges, internal loops, and junctions. In particular a junction – defined as the point of connection between different helical segments10– is a common structural element found in a wide range of contexts from within small RNA structures11; 12 to the large ribosomal subunits9; 13; 14. These structural elements have well defined 3D configurations that are important in the organization of the global structure of RNA molecules. While more is known about hairpins and internal loops15, our current understanding of the more complex junction elements is limited. An advance in our knowledge of junctions is important because junctions define main architectural building blocks of RNA tertiary arrangements. In particular, to better understand how RNAs function, a quantitative analysis of these important structural elements is needed.
Experimental techniques such as NMR and crystallography have produced a number of high resolution RNA 3D structures16, allowing researchers to observe and study some structural properties of junctions such as coaxial stacking of helices and long-range tertiary interactions12; 17; 18; 19. For instance, Lilley et al.20; 21; 22 analyzed the conformations of specific examples of 3-way and 4-way junctions (junctions composed of three and four helical arms, respectively) in nucleic acids using FRET techniques, and observed transitional changes in their helical configuration under Mg2+ and Na+ concentration variations. Lescoute and Westhof23 compiled and analyzed the topology of three-way junctions in folded RNAs, specifying rules to predict coaxial stacking, which occurs when two separate helical regions stack to form coaxial helices as a pseudo-continuous helix (see Fig. 1b). Tyagi and Mathews24 also predicted coaxial stacking based on free energy minimization and concluded that non-canonical basepairs make coaxial stacking more difficult to predict. RNAJunction, a database developed by Bindewald et al.25, contains information on RNA structural elements including junctions.
Figure 1.
(a) 2D diagram of a 4-way junction element composed of four helices labeled and color-coded by H1 (red), H2 (blue), H3 (green) and H4 (magenta), and the corresponding single stranded loop regions labeled J1/2 to J4/1 with nucleotides color-coded in yellow. Helices and loop regions are labeled in a unique way according to the 5′ to 3′ orientation of the entire RNA structure, by labeling H1 as the first helix encountered, while entering the junction region, as one moves along the nucleotide chain in the 5′ to 3′ direction and so forth. Lines inside the helices represent the canonical WC basepairs G-C, A-U, and the wobble basepair G-U. (b) 3D diagrams containing two pairs of helices: H1 with H4, and H2 with H3, which are coaxial stacked. The 4-way junction illustrated corresponds to the 23S rRNA 1S72_2678 from Table 1.
Our previous work on annotation and analysis of RNA tertiary motifs19, based on a representative set of high-resolution RNA structures, showed that coaxial helices are abundant tertiary motifs that often cooperate with other long-range interactions such as A-minor to stabilize RNA’s structure. Motivated by these results, we investigate here the structure of 4-way junctions in more detail, using the currently available solved crystal structures of folded RNAs. Our long-term goal is to find sequence “signatures” and other properties that will ultimately aid the prediction of coaxial stacking patterns and helical configurations of a given RNA based solely on sequence or computationally-predicted secondary structure. Our classification of nine families of 4-way junctions here shows that helices within junctions arrange in roughly parallel or perpendicular patterns, and stabilize their conformations using common tertiary motifs. Within junctions we also encounter novel tertiary motifs such as A-minor-coaxial stacking combinations and sarcin/ricin motif variants.
RESULTS
We begin with a classification of 4-way junctions based on their coaxial stacking, parallel and perpendicular helix arrangement patterns, and configuration of their flexible helical arms. By using the Leontis and Westhof notation26; 27, we study the associated basepair interactions and describe common motifs. A helix here is required to contain at least two consecutive Watson-Crick (WC) basepairs (G-C, A-U and G-U). For convenience, we label and color code helices sequentially according to the 5′ to 3′ orientation of the entire RNA as shown in Fig. 1. The single stranded region between each pair of consecutive helices Hi and Hi+1 is labeled by Ji/i+1. The point where strands exchange is called the point of strand exchange or simply crossover. A relative rotation of one helical pair could be right handed (clockwise) or left handed (counterclockwise)22.
Our list of 62 4-way junctions (Table 1) was assembled by taking all high-resolution RNA structures from the Protein Data Bank16 as of April 2009. RNA 4-way junctions are the second most abundant junction type after 3-way junctions. Previously Lescoute and Westhof23 analyzed and divided RNA three-way junctions into three families according to their topology. As the degree of helix branching increases, the number of possible junction conformers grows rapidly, and the junctions become highly diverse in terms of possible interactions and motifs. This diversity complicates classification of RNA junctions. However, a natural way to group them is according to their coaxial stacking patterns and helical organization. Our list of 62 four-way junctions (Table 1) is divided into nine families as shown on Fig. 2 (one diagram per RNA type). Families H, cH and cL contain junctions with two coaxial stacking; families cK and π are formed by junctions with one coaxial stacking; and junctions in families cW, ψ, X, and cX contain no coaxial stacking (see name selections below). Our classification differs from that of Lilley on DNA four-way junctions conformers22 in the sense that we group related conformers into one family; however, we also distinguish between parallel and antiparallel conformers. See also comment in Discussion on the flexibility and dynamic nature of RNA junctions. We now describe each family in turn. The Leontis-Westhof notation is used in our annotation – see also inset tables at the end of Fig. 2.
Table 1.
List of RNA 3D structures containing 62 four-way junctions. The name describes the PDB code and the number of the first nucleotide of helix H1 in the junction. The nomenclature is based on 10 and the helix number correspond to the labels given as published.
| Name | RNA Type | Coaxial stacks |
Helical alignments |
Family | Nomenclature | Domain | Helix Numbers |
|---|---|---|---|---|---|---|---|
| 1U9S _78 | Ribonuclease P_A | H1H4 , H2H3 | H | 2HS1HS2H | P7-8-9-10 | ||
| 2A2E _70 | Ribonuclease P_A | H1H4 , H2H3 | H | HS13HS1 | P7-8-9-10 | ||
| 1NBS _89 | Ribonuclease P_B | H1H4 , H2H3 | H | 2HS1HS2H | P7-8-9-10 | ||
| 2A64 _90 | Ribonuclease P_B | H1H4 , H2H3 | H | 2HS1HS2H | P7-8-9-10 | ||
| 1M50 _13 | Hairpin Ribozyme | H1H4 , H2H3 | H | 4H | A-B-C-D | ||
| 1S72 _1827 | 23S rRNA | H1H4 , H2H3 | H | HS3HS3HS3HS4 | IV | H64-65-66-67 | |
| 2AW4 _1771 | 23S rRNA | H1H4 , H2H3 | H | HS3HS3HS2HS3 | IV | H64-65-66-67 | |
| 2J01 _1771 | 23S rRNA | H1H4 , H2H3 | H | HS3HS3HS2HS3 | IV | H64-65-66-67 | |
| 1KH6 _4 | HCV IRES | H1H4 , H2H3 | cH | HS22HS1H | III-IIIa-IIIb-IIIc | ||
| 2AVY _141 | 16S rRNA | H1H4 , H2H3 | cH | HS3HS7HS4HS1 | 5' | H7-8-9-10 | |
| 2J00 _141 | 16S rRNA | H1H4 , H2H3 | cH | HS1HS4HS3HS1 | 5' | H7-8-9-10 | |
| 1NKW _2621 | 23S rRNA | H1H4 , H2H3 | cH | HS2HS1HS4HS2 | VI | H94-95-96-97 | |
| 1S72 _2678 | 23S rRNA | H1H4 , H2H3 | cH | HS22HS2HS1 | VI | H94-95-96-97 | |
| 2AW4 _2642 | 23S rRNA | H1H4 , H2H3 | cH | HS2HS1HS3HS1 | VI | H94-95-96-97 | |
| 2J01 _2642 | 23S rRNA | H1H4 , H2H3 | cH | HS2HS1HS3HS1 | VI | H94-95-96-97 | |
| 3F2Q_7 | Riboswitch (FMN) | H1H4 , H2H3 | cH | HS6HS3HS1HS7 | P1-P2-X-P6 | ||
| 3F2Q_31 | Riboswitch (FMN) | H1H4 , H2H3 | cH | 2HS2HS3HS2 | X-P3-P4-P5 | ||
| 1NKW _1457 | 23S rRNA | H1H4 , H2H3 | cH | HS1HS3HS6HS4 | III | H56-57-58-59 | |
| 2AW4 _1443 | 23S rRNA | H1H4 , H2H3 | cH | 3HS3HS4 | III | H56-57-58-59 | |
| 2AVY _568 | 16S rRNA | H1H4 , H2H3 | cL | HS7HS4HS10HS1 | C | H19-20-24-25 | |
| 2J00 _568 | 16S rRNA | H1H4 , H2H3 | cL | HS7HS4HS10HS1 | C | H19-20-24-25 | |
| 1NKW _1282 | 23S rRNA | H1H4 , H2H3 | cL | HS52HS2H | III | H47A-47-48-61 | |
| 1S72 _1373 | 23S rRNA | H1H4 , H2H3 | cL | HS72HS3H | III | H47A-47-48-61 | |
| 2AW4 _1269 | 23S rRNA | H1H4 , H2H3 | cL | HS52HS2H | III | H47A-47-48-61 | |
| 2J01 _1269 | 23S rRNA | H1H4 , H2H3 | cL | HS52HS2H | III | H49A-49-50-51 | |
| 1EFW _6 | transfer RNA | H1H4 , H2H3 | cL | HS2HS1HS5H | H1-2-3-4 | ||
| 1EHZ _6 | transfer RNA | H1H4 , H2H3 | cL | HS2HS1HS5H | H1-2-3-4 | ||
| 1N78 _506 | transfer RNA | H1H4 , H2H3 | cL | HS2HS1HS4H | H1-2-3-4 | ||
| 1QRS _6 | transfer RNA | H1H4 , H2H3 | cL | HS2HS1HS5H | H1-2-3-4 | ||
| 1U08 _6 | transfer RNA | H1H4 , H2H3 | cL | HS2HS1HS4H | H1-2-3-4 | ||
| 2GIS _7 | Riboswitch (SAM I) | H1H4 , H2H3 | cL | HS6HS1HS8HS3 | P1-2A-3-4 | ||
| 2AVY _114 | 16S rRNA | H1H4 | cK | HS62HS2 | 5' | H7-11-12-13A | |
| 2J00 _114 | 16S rRNA | H1H4 | cK | HS62HS2 | 5' | H7-11-12-13A | |
| 1NKW _2263 | 23S rRNA | H3H4 | cK | HS4HS4HS1HS1 | V | H82-83-86-87 | |
| 1S72 _2318 | 23S rRNA | H3H4 | cK | HS3HS3HS1HS1 | V | H82-83-86-87 | |
| 2AW4 _2284 | 23S rRNA | H3H4 | cK | HS3HS3HS1HS1 | V | H82-83-86-87 | |
| 2J01 _2284 | 23S rRNA | H3H4 | cK | HS3HS3HS1HS1 | V | H83A-83-86-87 | |
| 1NKW _1360 | 23S rRNA | H3H4 | cK | HS1HS3HS4HS2 | III | H51-52-53-54 | |
| 1S72 _1452 | 23S rRNA | H3H4 | cK | HS1HS3HS2HS1 | III | H51-52-53-54 | |
| 2AW4 _1346 | 23S rRNA | H3H4 | cK | HS2HS3HS2HS1 | III | H49A-49-50-51 | |
| 2J01 _1347 | 23S rRNA | H3H4 | cK | HS1HS3HS2H | III | H51-52-53-54 | |
| 2AVY _18 | 16S rRNA | H1H2 | cK | HS7HS10HS1HS3 | C | H2-3-19-27 | |
| 2J00 _18 | 16S rRNA | H1H2 | cK | HS7HS10HS1HS3 | C | H2-3-19-28 | |
| 1U9S _118 | Ribonuclease P_A | H3H4 | H1H2 | π | HS12HS7HS1HS4 | P11-12-13-14 | |
| 2A2E _110 | Ribonuclease P_A | H3H4 | H1H2 | π | HS10HS7HS2HS4 | P11-12-13-14 | |
| 1NKW _1682 | 23S rRNA | H1H4 | cW | HS152HS12HS5 | IV | H61-62-63-64 | |
| 1S72 _1743 | 23S rRNA | H1H4 | cW | HS152HS12HS5 | IV | H61-62-63-64 | |
| 2AW4 _1665 | 23S rRNA | H1H4 | cW | HS152HS12HS5 | IV | H61-62-63-64 | |
| 2J01 _1665 | 23S rRNA | H1H4 | cW | HS152HS12HS5 | IV | H61-62-63-64 | |
| 1S72 _42 | 23S rRNA | H2H4 | ψ | HS6HS4HS5HS1 | I | H4-5-8-10 | |
| 1NKW _1824 | 23S rRNA | H2H4 | ψ | HS1HS2HS22HS10 | IV | H64-65-66-67 | |
| 1S72 _1888 | 23S rRNA | H2H4 | ψ | HS12HS20HS10 | IV | H67-68-69-71 | |
| 2AW4 _1832 | 23S rRNA | H2H4 | ψ | HS12HS20HS10 | IV | H64-65-66-67 | |
| 2J01 _1832 | 23S rRNA | H2H4 | ψ | HS12HS20HS10 | IV | H64-65-66-67 | |
| 1NKW _244 | 23S rRNA | H2H4 | ψ | HS5HS6HS6HS2 | I | H14-16-21-22 | |
| 2AW4 _267 | 23S rRNA | H2H4 | ψ | HS2HS8HS6HS2 | I | H14-16-21-22 | |
| 1NKW_608 | 23S rRNA | X | HS10HS9HS3HS11 | I | H27-28-29-31 | ||
| 2AW4 _600 | 23S rRNA | X | HS2HS3HS2HS5 | I | H27-28-29-31 | ||
| 2J01 _600 | 23S rRNA | X | HS2HS3HS2HS2 | I | H27-28-29-31 | ||
| 2IHX _166 | Sarcoma Virus | cX | HS3HS42HS3 | A-B-C-O3 | |||
| 2AVY _942 | 16S rRNA | cX | HS2HS5HS11HS10 | 3'M | H29-30-41-42 | ||
| 2J00 _940 | 16S rRNA | cX | HS4HS5HS11HS12 | 3'M | H29-30-41-42 | ||
Figure 2.
Network interaction diagrams for the nine families of 4-way junctions. Family H, cH and cL contains two coaxial helices; family cK and π contains one coaxial stacking; while families cW, ψ, X and cX contains no coaxial stacking. The “c” before the capital letter in the family name denotes the crossing observed at the point of strand exchange. The network symbology follows the Leontis-Westhof notation27 (see inset boxes).
Four-way junction families
4-way junctions with two coaxial stacking
Family H is characterized by two coaxial stacking roughly aligned, resembling the letter H (see Fig. 2a). The continuous strands in each coaxial helix are antiparallel to each other, resembling the DNA Holliday junction4. The coaxial helices are stabilized by their long-range interactions and, in some instances, these interactions contribute to small (left or right-handed) rotations (e.g. hairpin ribozyme and ribonuclease P A-type in Fig. 2a) similar to the X-stacked conformer in DNA 4-way junctions28.
Family cH also consists of two coaxial helices roughly aligned, but now the continuous strands at each coaxial helix runs in the same direction (Fig. 2b). When viewed from a direction perpendicular to the coaxial helix axis, the exchanging strands appear to cross at the center. A-minor interactions29 (denoted in Fig. 2 by empty and solid triangles known as Sugar-Sugar interactions) are the most conserved interactions responsible of such crossings at the point of strand exchange, as we discuss below in more detail. Note that two types of pairwise coaxial stacking patterns are observed: H1H4 with H2H3, and H1H2 with H3H4.
In family cL, the pair of coaxial stacks H1H4 and H2H3 aligns in a perpendicular fashion, making an “L” shape. The most well known structure in this family is the transfer RNA12. The “L” shape can be stabilized by a diversity of long-range interactions such as loop-loop, loop-helix, or helix packing interactions such as P-interactions30; 31 (Fig. 2c), but other factors such as ion concentrations also play a role. As in family H, A-minor interactions within the junction domain anchor single stranded regions to the end of its helices to produce crossing at the point of strand exchange. Note that the riboswitch (2GIS_7) represents a different conformer from the three examples in Fig. 2c, because the coaxial helix H2H3 is rotated relative to H1H4 so that helices H1 and H3 are sufficiently close to interact.
4-way junctions with one coaxial stack
Family cK consists of two helical arms stacked, while the third helix becomes perpendicular to the coaxial helix, and the fourth subtends an angle that depends on the number of unpaired bases and tertiary interactions (Fig. 2d). Long-range interactions help stabilize the perpendicular helical arrangement. Family cK also contains a crossing at the point of strand exchange, usually formed by adenine bases that make up A-minor interactions at the locus of the strand exchange. In addition, helix packing interactions, pseudoknots, and other types of non-canonical basepair interactions can help rotate the helical arm and produce the same perpendicular arrangement. Three types of junction conformers can be noted, each with one coaxial stacking (H1H2, H3H4, and H1H4), and one helix perpendicular to them (H4, H2 and H3 respectively).
Note that in the 16S rRNA 2AVY_114 (Fig. 2d), both H2 and H3 are perpendicular to each other and to the coaxial helix, forming a perpendicular frame in three-dimensional space.
Family π resembles family H but instead of the two coaxial stacking interactions of family H, family π has only one, with the second pair of helices aligned rather than stacked. The ribonuclease P structure (1U9S_118) uses non-canonical basepair interactions32 to reduce the instability caused by the long strands J1/2 and J2/3. Helix H2 is anchored to H3 through A-minor interactions (Fig. 2e).
4-way junctions with no stacking
Families cW, ψ, cX and X are less common and so far only observed within the large ribosomal structures 16S and 23S rRNA. They are characterized by longer single-strand elements and no coaxial stacking, but they contain at least one helical alignment or perpendicular helix interaction. Like the other families, they also contain a high degree of junction symmetry. The specific conformations depend on the tertiary interactions that form, as well as the binding of proteins. Family cW has a helical alignment between consecutive helical arms H1 and H4 (Fig. 2f). Family ψ has also a helical alignment, but is defined by the two non-consecutive helical arms H2 and H4 (Fig. 2g). Families cX and X contain 4-way junctions with helical arms in perpendicular arrangements (Fig. 2h–i). The junction in family X has a non-planar triad of helices roughly perpendicular to each other, while family cX has two pairs of helical arms arranged perpendicular to each other by helix packing interactions.
Tertiary motifs in four-way junctions
Our analysis underscores the diversity of RNA 4-way junction in structure. Still, common features such as sequence and stacking preferences, loop sizes, basepair interactions, and tertiary motifs are often preserved within and across families, as we describe next.
Coaxial stacking
Coaxial stacking is a common tertiary motif present in many junctions, as well as internal loops, and even pseudoknots and kissing hairpins18; 19. From our list of 62 4-way junctions (Table 1), which contains 75 cases of coaxial stacking, about 33 (53%) of the junctions contain two coaxial stacking interactions (Fig. 2a–c), 14 (22%) contain one coaxial stacking (Fig. 2d–e), and the remaining 17 (27%) of the junctions contain no coaxial stacking (Fig. 2f–i).
Table 2 describes the frequency of these 75 coaxial stacking cases in our dataset of 62 4-way junctions (Table 1), ordered by size of loop Ji/i+1 between the helices Hi and Hi+1 forming the stacking. A strong preference for stacking between helices with small loop size Ji/i+1 (between 0 and 1) can be observed. Similar patterns have been reported for 3-way junctions23. As the size of Ji/i+1 increases, coaxial stacking between helices becomes less likely and no coaxial stacking with Ji/i+1>7 was observed. Note that a small loop size does not guarantee coaxial stacking (see for instance the lengths of J1/2 and J3/4 for junctions on family H in Fig. 2a).
Table 2.
Frequency of coaxial stacking between helices Hi and Hi+1 joined by the size of the single stranded loop in between Ji/(i+1) ranging from 0 to 7 as performed for the 75 coaxial stacking cases within our 62 4-way junction list (Table 1).
| Size of Ji/(i+1) | 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 |
|---|---|---|---|---|---|---|---|---|
| Frequency | 25 | 26 | 7 | 8 | 5 | 0 | 2 | 2 |
Interestingly, from the list of observed coaxial helices in our dataset of junctions (Table 1), we note a strong preference for stacking between H1H4 and H2H3. A total of 30 (40%) and 28 (38%) out of 74 coaxial helices are formed between H1H4 and H2H3, respectively (the hairpin ribozyme 1M5O_13 was excluded here since this junction is formed by two strands, making it difficult to label the first helix). Furthermore, 28 (93%) out of the 30 four-way junctions with two coaxial stacking form both H1H4 and H2H3 patterns. Although the reason for these strong coaxial stacking preferences is unclear, we speculate that this is related to the right-handedness of RNA molecules.
Coaxial stacking interactions also occur in helical stems that form pseudoknots33. In fact, pseudoknots involving single stranded loops regions Ji/i+1 in junctions will facilitate coaxial stacking between helices Hi and Hi+1 as observed in 16S rRNA 2AVY_18 and 23S rRNA 1S72_1452 in Fig. 2d.
Non-canonical basepairs are frequently formed between loops Ji−1/i and Ji/i+1 next to their common helix Hi. These non-canonical basepairs stack to Hi to reduce the number unpaired nucleotides between Ji−1/i or Ji/i+1 and help promote coaxial stacking between Hi and a neighboring helix. It has been previously reported that sheared GA basepairs (trans Hoogsteen/Sugar) of cis WC GA occur often at the end of helices34; 35. Other basepairs such as the AU trans Hoogsteen/Watson are also frequent like observed in Fig. 2. In agreement with previous studies on three-way junctions36, the stability of junctions depends on the amount of unpaired nucleotides at the Ji/i+1 regions. Thus, not only is the length of Ji/i+1 important in coaxial stacking, but the non-canonical basepair formation plays also an important role as well.
Parallel and perpendicular helical configurations
A small number of helical arms align their axis without stacking forces, or arrange in roughly perpendicular configurations (Fig. 2f–i). This is not exclusive of junctions18. Parallel conformations between helices are stabilized using long-range interactions, preferably A-minor interactions as in the case of 23S rRNA 2AW4_1443 in Fig. 2b, but other basepairs such as WC GC basepairs and even base-backbone interactions are frequent. The dotted-line interactions in Fig. 2 denote one hydrogen bond or base-backbone interactions that do not fit into the base-base classification of Leontis and Westhof. Helices that arrange in perpendicular configurations are often stabilized by helix packing interactions such as the P-interaction30; 31 between WC GU wobble basepairs on a first helix and a WC basepair in a second helix (see for instance 23S rRNA 2AW4_600 in Fig. 2i). This P-interaction functions by anchoring the former helix into the minor groove of the latter. Loop-helix and loop-loop interactions that stabilize perpendicular helix configurations also occur, as in the case of the 23S rRNA 2J01_1269 in Fig. 2c and the tRNA D-loop/T-loop interaction37 (see tRNA 1EHZ_6 in Fig. 2c). Besides P-interactions, other forms of interactions are of course possible, requiring a larger dataset of junctions.
A-minor and other sugar-edge interactions
A-minor motifs are among the most abundant tertiary interactions found in RNA. In our recent annotation of a representative high-resolution set of solved RNA, A-minor interactions were observed in 37% of the tertiary motifs. A-minor motifs involve sugar-edge interactions which can be recognized in the diagrams by the small connector triangles between adenines located in single stranded regions, and the helical receptor, usually a WC (GC) basepair. We previously reported that the helical receptor of A-minor has a strong preference to lie at the end of helices rather the inside helices19. Our data here indicate that A-minor interactions within junctions form two main types of motifs.
The first and most common interaction often involves two adenines (but it could also be one or three adenines) in the loop region Ji/i+1 forming sugar-edge interactions, often A-minor (type I and II), but also cis Sugar-Hoogsteen and cis Watson-Sugar (e.g. HCV IRES domain 1KH6_4 in Fig. 2a). These adenines interact with helical elements of the junction near the end of the helix (see Fig. 3a), forming a crossing at the point of strand exchange. Several examples are found in junction families cH, cL and cK.
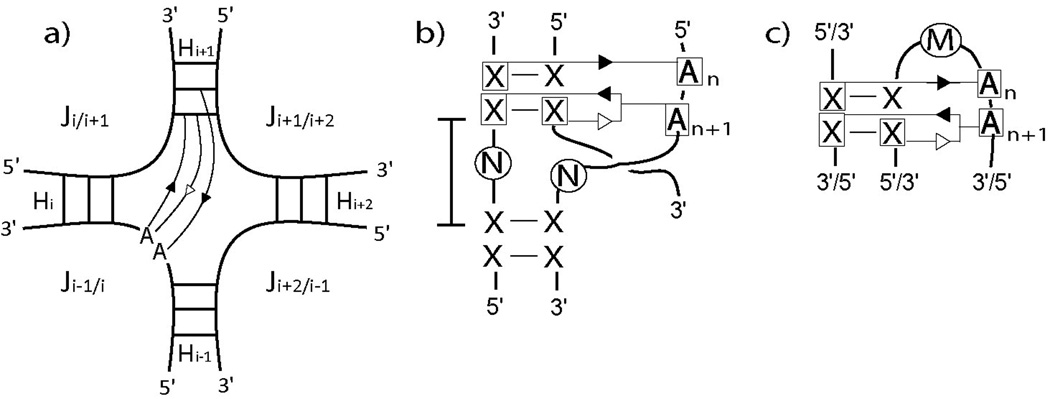
Figure 3.
A-minor interactions within junction domains. a) Secondary structure diagram for the most common interaction. b) Motif consensus for the most common interaction. c) Consensus motif for the less common interaction. N and M represent 0–3 and 2–3 nucleotides respectively. Often, a WC (GC) interaction appears next to the consecutive adenines at the n+2 position.
As was previously observed in 3-way junctions23, the right handedness of RNA implies that when a coaxial stacking between helices say Hi and Hi+1 is formed, the 5′-end strand entering Hi faces the shallow/minor groove of Hi+1, thus allowing nucleotides in Ji−1/i to interact with Hi+1 as sugar-edge interactions (see Fig. 3a). This property reflects the occurrence of the A-minor (and other sugar-edge) interactions described above. By analyzing cases of A-minor/coaxial stacking interactions across several families, we constructed a consensus diagram in Fig. 3b. Here N denotes a small number of nucleotides (0 to 3); the same number is required on both loop strands. X-X denotes standard WC basepairs (GC, AU) and the GU wobble basepair. If a pseudoknot forms between helices which appear stacked, the adenines can also interact with the helix produced by this pseudoknot (see for instance 23S rRNA 1S72_1452 in Fig. 2d). Because this pattern occurs very often, we consider it an important functional arrangement of helices. Similar interactions between pseudoknots and A-minor motif has been previously observed19.
A second and less common interaction involving A-minor occurs when either the 5′-end or the 3′-end strand leaving the helix makes a u-turn and interacts again with its starting helix (Fig. 3c). A number of nucleotides M are needed (2 to 3) to allow the u-turn. A case is observed on 16S rRNA 2J00_568 in Fig. 2c when two nucleotides in M form a pseudoknot with another RNA strand, thus reorienting the 5′-end strand back to its starting helix. A second example is found on and 23S rRNA 2J01_1832 in Fig. 2g where adenines in J4/1 interacts with helix H1.
One interesting example exists in the 23S rRNA (see Fig. 2g, 2J01_1832 in family ψ) where the direction of the A-minor interaction pattern is reversed. A pair of adenines in J3/4 interacts with helix H2 rather than H1. This interaction can be explained by the fact that RNA is for the most part a right handed molecule, but in this junction, due to the sarcin/ricin like motif inside, a portion of the loop strand J3/4 folds in a left-handed orientation, thus reversing the direction of the pattern shown in Fig. 3a. Sarcin/ricin like motifs are described in more detail next.
Sarcin/ricin like motifs
A different type of tertiary interaction resembling the sarcin/ricin motif32 occurs within the single-stranded regions of junctions, particularly for members of families π and cX. Sarcin/ricin like interactions appear on junctions where helical alignment rather than coaxial stacking is present. These interactions show a surprising similarity to the sarcin/ricin motif. However, they lack the AG (shown in Fig. 4 in green) trans Hoogsteen-Sugar or the AA trans Hoogsteen-Hoogsteen (orange in Fig. 4), as well as all UC trans Sugar-Hoogsteen basepair interactions (cyan in Fig. 4). As in sarcin/ricin motifs38, these interactions stabilize RNA-RNA conformations as shown in Fig. 4 (magenta), as well as RNA-protein interactions (red color in Fig. 4).
Figure 4.
Interactions similar to sarcin/ricin motif (left box). The interactions are part of the junctions 23S rRNA 2AW4_267 and 23S rRNA 2J01_1832 of family ψ, and 16S rRNA 2AVY_942 of family cX. The interaction in 2AW4_267 was previously observed by Leontis et al38. RNA-Protein interactions (red font) are denoted by protein name followed by amino acid type and residue number.
DISCUSSION
Annotating and analyzing is a major task in structural biology. For RNA, classification and other aspects of RNA structure and function have provided much work for many researchers under the RNA Ontology Consortium (ROC)39 (http://roc.bgsu.edu/). The notion of classes as discussed here for 4-way junctions is important for understanding common properties that members of a family share. Ultimately, such classification can help interpret RNA function.
The classification of 4-way junctions considered here is a complementary and compatible approach to the classification of RNA 3-way junctions given by Lescoute and Westhof23, which groups elements according to their topology. RNA junctions listed in the RNAJunction25 database have been classified according to standard nomenclature10 based on the size of each loop region. However, similar junctions from homologous RNAs can differ by single insertions of deletions in the loop regions, leading to different classifications under the standard nomenclature. Similarly, the SCOR40 database lists examples of coaxial helices as elements of tertiary motifs. Our work extends these definitions/classifications to all known coaxial helices encountered in four-way junctions as of October 2008. The previous classification of DNA 4–way junctions22 is only based on forms containing two coaxial helices, whereas our framework additionally includes junctions that contain one or no-coaxial stacking.
The classification presented here identifies nine major families of 4-way junctions; other conformations and families are of course theoretically possible. For each example in Fig. 2a, we observed a stacking of helices H1H4, and H2H3, but the conformer H1H2 and H3H4 might exist in nature. Although not yet observed, one can also imagine the existence of family L where pairs of coaxial stacking align in a perpendicular fashion but without the crossing of the single strands at the point of strand exchange. Similarly, one can predict the existence of a family K where the crossings at the point of strand exchange is not present. Conformations also include members in family π yet to be discovered with a high degree of rotation between the inter-helical angles of H1H2 with H3H4, instead of the almost parallel conformer of ribonuclease P 1U9S_118 as we observed in Fig. 2e.
In general, due to the conformational flexibility and dynamic character of 4-way junctions, a continuum of junction conformations might be possible. Still, current structural information suggests a preference for conformations consisting of parallel and perpendicular helical arrangements. Thus, new conformations will likely oscillate around these observed families and possibly new ones such as the families L and K that we define. We are currently extending this work to all higher order junctions available (Laing et al., in preparation41).
The data from Table 2 reveal a high frequency of coaxial stacking of helices when the size of their common single stranded loop is small; we also note certain sequence preferences and that the presence of pseudoknots can strongly induce coaxial stacking. Our analysis reveals a strong tendency for coaxial stacking between helices H1 with H4 and H2 with H3. Although the reason for this is unclear, we speculate that the right handedness of RNA has a role. Additionally, such topologies could be favored during RNA transcription because helices that form first could have a greater opportunity to stack first. Furthermore, in the large ribosomal RNA, proteins that bind to sites in the junction near the 5′-end of the starting helix may assemble earlier than those located near the 3′-end; thus, those proteins buried in the interior of junctions influence the coaxial stacking formation by enhancing or restricting conformational flexibility of the helical arms.
One advantage of grouping junctions is that it allows recognizing important repeating motifs such as the sugar-edge interactions (mostly A-minor interactions) and the sarcin/ricin like motifs. These sets of non-canonical basepairs play important roles in RNA’s structure and therefore function. For instance, it has been reported42 that mutations on the adenines in the loop regions of the 4-way junction (HCV IRES domain 1KH6_4 in Fig. 2b) in the HCV IRES RNA are lethal to the virus; thus, the sugar-edge interactions are critical elements for the correct structure of the junction. Another example showing the importance of these long-range interactions is found in the hairpin ribozyme (1M5O_13 in Fig. 2a). While this ribozyme can be active in the absence of the junction, under physiological ionic conditions the junction’s presence accelerates the ion-induced folding of the ribozyme by 500-fold43. Sarcin/ricin like motifs are important structural elements that stabilize the junctions when no coaxial stacking is present, but also serve as sites for specific RNA-RNA and RNA-protein recognition. The existence of such variants of the original sarcin/ricin motifs agrees with the idea of RNA modularity44 and the principle of structural scaffolding45, where RNA motifs are stable interactions formed by submotifs. While these submotifs are more versatile, they retain key structural tertiary interactions.
The junctions we encountered containing two coaxially-stacked elements belonging to families H, cL and cH differ in the angle between the axes of the coaxial-stacks, roughly 0°, 90° or 180° respectively. While the degree of rotation depends on the environment (e.g., ion concentration, proteins), the length of the loops forming the exchanging strands for each family also determines its final conformation. For instance, the lengths of the loops in family H are small compared to those found in the other families. In family cL, the lengths of the loops at the exchanging strands are often larger than those in family H to allow the perpendicular rotation, while avoiding steric clashes. In family cH, the lengths of the loops are slightly larger than in family H but smaller than in cL; however, as previously mentioned, the presence of sugar-edge interactions help stabilize the conformation (see Table S1).
Furthermore, A-minor or other sugar-edge interactions within junction domains are important structural elements for excluding interconversion between families such as cH and H to one another20. Correctly predicting A-minor interactions can help predict coaxial stacking patterns since loops that contain adenines involved in A-minor interactions will not form coaxial stacking with their neighboring helices. However, it is not clear whether these interactions will occur even in the presence of consecutive adenines in loop regions. Such adenines could form stacking interactions or long-range A-minor interactions with other RNA elements, or could interact with proteins.
Indeed, experiments for the hammerhead ribozyme46 and hairpin ribozyme47 have shown that loop-loop interactions act as important elements in the function of these ribozymes, by stabilizing the correct conformation of these junctions. While more data will strengthen these assertions, it clear that long-range interactions are important complementary elements in the junction domains.
Our compilation of RNA junction domains illustrates nature’s strong preferences for the arrangement of RNA helical elements in parallel and perpendicular patterns. The conformations of some 4-way junction elements also greatly resemble helical configurations of three-way junctions. For instance, in the classification of Lescoute and Westhof23, the conformation given in family C is a subset of our family cH, where in both cases a coaxial stack aligns in parallel to a third helical arm which is stabilized by A-minor interactions. Similarly, 3-way junction elements belonging to the Family A resemble the conformation observed for 4-way junctions in our family cK.
The junction 2J01_1832 in family ψ shown in Fig. 2g is also of interest. Here the loop region J3/4 interacts with H2 using A-minor interactions, while near H3, it is structured like a hairpin using the standard U-turn motif, and closed by a trans WC GC basepair. This U-turn behaves like a small extra helix or a like a cap. The resulting motifs align H3 parallel to both H2 and H4. This pattern is the characteristic signature of the 3-way junction elements of family C. Understanding such preferences for RNA’s helical conformations can greatly improve RNA 3D structure prediction. However, more work on understanding such topologies is required. Ongoing efforts will continue to analyze higher order junctions.
Our analysis underscores the notion20 that RNA junctions are composed of both rigid and flexible elements. Tertiary motifs such as coaxial stacking, pseudoknots and RNA-RNA long-range interactions are interactions responsible for maintaining the rigid parts of the junction, while flexible elements appear on helical arms with longer loop regions and are more sensitive to external forces such as proteins and ion concentration. This is consistent with the fact that loop regions involved in RNA-protein interactions are consistently longer in size48; 49 and appear on the large ribosomal subunits. FRET experiments also show changes on inter-helical angles at high or low magnesium concentrations, with coaxial stacking interactions unchanged50. Interestingly, the crystal structure of the 4-way junction HCV IRES domain solved by Kieft et al.42 (1KH6_4 in Fig. 2b) describes one conformation containing a pair of coaxial stacks parallel to each other. While only one conformer can be incorporated in the crystal lattice, studies using comparative gel electrophoresis and FRET analysis have shown that this junction exists in a dynamic equilibrium between parallel and antiparallel structural conformations51. In contrast, the junction 2AW4_1443 (Fig. 2b) contains A-minor interactions outside the junction domain which helps stabilize the parallel junction configuration; however, no long-range interactions are observed in the HCV IRES crystal structure. Similar studies on the junction obtained by removing the neighboring internal loops20 of the hairpin ribozyme (1M5O_13 from Fig. 2a) in the presence of Mg2+ have shown a continuous interconversion between parallel and antiparallel forms. These findings underscore the polymorphic and dynamic character of junctions as needed for biological function, including interactions with other molecules.
Finally, we propose in Fig. 5 what could be described as the anatomy of a 4-way junction. The idea is to build upon secondary structure features that can help predict three-dimensional shape of junctions. Coaxial stacking occurs between helical arms with a small number of intervening single stranded nucleotides. Non-canonical basepairs, preferably GA (sheared) trans Sugar-Hoogsteen, or a AU trans Watson-Hoogsteen (or GC WC basepairs) can help to reduce the number of nucleotides between helices by base stacking interactions. Also, internal basepair interactions between non-consecutive loop elements of the junctions help reduce the spatial distance between helical arms, with the most common interaction involving AU trans Watson-Hoogsteen or WC GC basepairs. Helix packing interactions such as P-interactions involving GU cis WC near the end of the helix help promote perpendicular arrangements between helices. Long-range interactions, preferably A-minor motifs, stabilize helical elements and align them in parallel; for these interactions to form, hairpin loops or internal loops must exist near the junction domain. Other types of RNA-RNA or RNA-protein interactions can occur at the single stranded regions, but this requires longer loop chains. Analysis of higher-order junctions and other RNA tertiary motifs will further help put these ideas into a growing framework of RNA architecture and ultimately function.
Figure 5.
Anatomy of a 4-way junction.
MATERIALS AND METHODS
Data of our 3D RNA junctions were collected from the RCSB Protein Data Bank16. Based on available structures as of April 2009, 554 high-resolution structures were selected with repetitions omitted by choosing the more recent structures. Junction elements were searched within these and analyzed for basepair interactions (see below).
Dataset of RNA junctions
To perform our comprehensive search of 4-way-junctions in the set of RNA structures above, we first considered the secondary structure associated with every 3D structure defined in terms of its WC basepairs (G-C, A-U and G-U) and the single stranded regions. The search for canonical WC and wobble basepairs was performed using the program FR3D52. Next we searched for sets of four distinct strands connecting in a cyclical way by at least two consecutive canonical WC basepairs (Fig. 1). For simplicity, pseudoknots were automatically removed during the search, but later re-inserted for statistical analysis. Visual inspection was also used to verify the correctness of our procedure. In addition, we compared our search outcome to data available from the RNAJunction database25, to ensure the verity of all junctions.
Our search of 20 crystal structures contained at least one 4-way junction each. The structures include the two high resolution crystal structures of the 16S (PDB 2AVY, 2J00) and four 23S rRNA (PDB 1NKW, 1S72, 2AW4, 2J01). Although the 3D shape of homologous rRNA molecules is highly conserved among species, differences are informative because they help to understand evolutionary changes that Nature allows while keeping their molecular function intact. In total, our dataset thus contains 62 four-way junctions as listed in Table 1. Additional detailed junction information such as PDB source, sequence, and residue numbers are available in Table S1 from the Supplementary Material.
Basepair Interactions and Coaxial Stacking
Non-canonical basepairing with alternate hydrogen bonding patterns occur often in RNA. A consensus between FR3D and RNAVIEW53 was considered to classify basepairs. Where discrepancies occur, we employed visual programs such as Pymol (DeLano Scientific LLC) and Swiss PDB viewer54 to clear the analysis. Additionally, the junction data were analyzed from different perspectives: sequence signatures, length of loop regions, 3D motifs, and the 3D organization of their helices. Orientation aspects such as in coaxial stacking, helices that form perpendicular inter-helical angles, and helices aligning their axis in parallel without the use of stacking forces were analyzed on the basis of inspection.
Network Interaction Diagrams
Network interaction diagrams describing basepair interactions are represented symbolically according to the Leontis and Westhof basepairing classification26; 27. The diagrams were created using S2S55, a visual aid program based on RNAVIEW. We also used the 3D visual program Pymol to classify 4-way junctions into families.
Supplementary Material
Acknowledgments
FUNDING
The work was supported by the Human Frontier Science Program (HFSP), by a joint NSF/NIGMS initiative in Mathematical Biology (DMS-0201160), by NSF EMT award # CF-0727001. Partial support by NIH (grant # R01-GM055164), NIH (grant # 1 R01 ES 012692), and NSF (grant # CCF-0727001) is also gratefully acknowledged. The authors thank Abdul Iqbal and Segun Jung for their help in figure preparation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Hannon GJ. RNA interference. Nature. 2002;418:244–251. doi: 10.1038/418244a. [DOI] [PubMed] [Google Scholar]
- 2.Moore MJ. From birth to death: the complex lives of eukaryotic mRNAs. Science. 2005;309:1514–1518. doi: 10.1126/science.1111443. [DOI] [PubMed] [Google Scholar]
- 3.Schroeder R, Barta A, Semrad K. Strategies for RNA folding and assembly. Nat Rev Mol Cell Biol. 2004;5:908–919. doi: 10.1038/nrm1497. [DOI] [PubMed] [Google Scholar]
- 4.Birney E, Stamatoyannopoulos JA, Dutta A, Guigo R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis ET, Thurman RE, Kuehn MS, Taylor CM, Neph S, Koch CM, Asthana S, Malhotra A, Adzhubei I, Greenbaum JA, Andrews RM, Flicek P, Boyle PJ, Cao H, Carter NP, Clelland GK, Davis S, Day N, Dhami P, Dillon SC, Dorschner MO, Fiegler H, Giresi PG, Goldy J, Hawrylycz M, Haydock A, Humbert R, James KD, Johnson BE, Johnson EM, Frum TT, Rosenzweig ER, Karnani N, Lee K, Lefebvre GC, Navas PA, Neri F, Parker SC, Sabo PJ, Sandstrom R, Shafer A, Vetrie D, Weaver M, Wilcox S, Yu M, Collins FS, Dekker J, Lieb JD, Tullius TD, Crawford GE, Sunyaev S, Noble WS, Dunham I, Denoeud F, Reymond A, Kapranov P, Rozowsky J, Zheng D, Castelo R, Frankish A, Harrow J, Ghosh S, Sandelin A, Hofacker IL, Baertsch R, Keefe D, Dike S, Cheng J, Hirsch HA, Sekinger EA, Lagarde J, Abril JF, Shahab A, Flamm C, Fried C, Hackermuller J, Hertel J, Lindemeyer M, Missal K, Tanzer A, Washietl S, Korbel J, Emanuelsson O, Pedersen JS, Holroyd N, Taylor R, Swarbreck D, Matthews N, Dickson MC, Thomas DJ, Weirauch MT, Gilbert J, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ban N, Nissen P, Hansen J, Moore PB, Steitz TA. The complete atomic structure of the large ribosomal subunit at 2.4 A resolution. Science. 2000;289:905–920. doi: 10.1126/science.289.5481.905. [DOI] [PubMed] [Google Scholar]
- 6.Cate JH, Gooding AR, Podell E, Zhou K, Golden BL, Kundrot CE, Cech TR, Doudna JA. Crystal structure of a group I ribozyme domain: principles of RNA packing. Science. 1996;273:1678–1685. doi: 10.1126/science.273.5282.1678. [DOI] [PubMed] [Google Scholar]
- 7.Toor N, Keating KS, Taylor SD, Pyle AM. Crystal structure of a self-spliced group II intron. Science. 2008;320:77–82. doi: 10.1126/science.1153803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wimberly BT, Brodersen DE, Clemons WM, Jr, Morgan-Warren RJ, Carter AP, Vonrhein C, Hartsch T, Ramakrishnan V. Structure of the 30S ribosomal subunit. Nature. 2000;407:327–339. doi: 10.1038/35030006. [DOI] [PubMed] [Google Scholar]
- 9.Yusupov MM, Yusupova GZ, Baucom A, Lieberman K, Earnest TN, Cate JH, Noller HF. Crystal structure of the ribosome at 5.5 A resolution. Science. 2001;292:883–896. doi: 10.1126/science.1060089. [DOI] [PubMed] [Google Scholar]
- 10.Lilley DM, Clegg RM, Diekmann S, Seeman NC, Von Kitzing E, Hagerman PJ. A nomenclature of junctions and branchpoints in nucleic acids. Nucleic Acids Res. 1995;23:3363–3364. doi: 10.1093/nar/23.17.3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Batey RT, Gilbert SD, Montange RK. Structure of a natural guanine-responsive riboswitch complexed with the metabolite hypoxanthine. Nature. 2004;432:411–415. doi: 10.1038/nature03037. [DOI] [PubMed] [Google Scholar]
- 12.Kim SH, Sussman JL, Suddath FL, Quigley GJ, McPherson A, Wang AH, Seeman NC, Rich A. The general structure of transfer RNA molecules. Proc Natl Acad Sci U S A. 1974;71:4970–4974. doi: 10.1073/pnas.71.12.4970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cate JH, Yusupov MM, Yusupova GZ, Earnest TN, Noller HF. X-ray crystal structures of 70S ribosome functional complexes. Science. 1999;285:2095–2104. doi: 10.1126/science.285.5436.2095. [DOI] [PubMed] [Google Scholar]
- 14.Noller HF. RNA structure: reading the ribosome. Science. 2005;309:1508–1514. doi: 10.1126/science.1111771. [DOI] [PubMed] [Google Scholar]
- 15.Hendrix DK, Brenner SE, Holbrook SR. RNA structural motifs: building blocks of a modular biomolecule. Q Rev Biophys. 2005;38:221–243. doi: 10.1017/S0033583506004215. [DOI] [PubMed] [Google Scholar]
- 16.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Holbrook SR. RNA structure: the long and the short of it. Curr Opin Struct Biol. 2005;15:302–308. doi: 10.1016/j.sbi.2005.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holbrook SR. Structural principles from large RNAs. Annu Rev Biophys. 2008;37:445–464. doi: 10.1146/annurev.biophys.36.040306.132755. [DOI] [PubMed] [Google Scholar]
- 19.Xin Y, Laing C, Leontis NB, Schlick T. Annotation of tertiary interactions in RNA structures reveals variations and correlations. RNA. 2008;14:2465–2477. doi: 10.1261/rna.1249208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hohng S, Wilson TJ, Tan E, Clegg RM, Lilley DM, Ha T. Conformational flexibility of four-way junctions in RNA. J Mol Biol. 2004;336:69–79. doi: 10.1016/j.jmb.2003.12.014. [DOI] [PubMed] [Google Scholar]
- 21.Lilley DM. Folding of branched RNA species. Biopolymers. 1998;48:101–112. doi: 10.1002/(SICI)1097-0282(1998)48:2<101::AID-BIP2>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 22.Lilley DM. Structures of helical junctions in nucleic acids. Q Rev Biophys. 2000;33:109–159. doi: 10.1017/s0033583500003590. [DOI] [PubMed] [Google Scholar]
- 23.Lescoute A, Westhof E. Topology of three-way junctions in folded RNAs. RNA. 2006;12:83–93. doi: 10.1261/rna.2208106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tyagi R, Mathews DH. Predicting helical coaxial stacking in RNA multibranch loops. RNA. 2007;13:939–951. doi: 10.1261/rna.305307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bindewald E, Hayes R, Yingling YG, Kasprzak W, Shapiro BA. RNAJunction: a database of RNA junctions and kissing loops for three-dimensional structural analysis and nanodesign. Nucleic Acids Res. 2008;36:D392–D397. doi: 10.1093/nar/gkm842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leontis NB, Stombaugh J, Westhof E. The non-Watson-Crick base pairs and their associated isostericity matrices. Nucleic Acids Res. 2002;30:3497–3531. doi: 10.1093/nar/gkf481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leontis NB, Westhof E. Geometric nomenclature and classification of RNA base pairs. RNA. 2001;7:499–512. doi: 10.1017/s1355838201002515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Duckett DR, Murchie AI, Diekmann S, von Kitzing E, Kemper B, Lilley DM. The structure of the Holliday junction, and its resolution. Cell. 1988;55:79–89. doi: 10.1016/0092-8674(88)90011-6. [DOI] [PubMed] [Google Scholar]
- 29.Nissen P, Ippolito JA, Ban N, Moore PB, Steitz TA. RNA tertiary interactions in the large ribosomal subunit: the A-minor motif. Proc Natl Acad Sci U S A. 2001;98:4899–4903. doi: 10.1073/pnas.081082398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gagnon MG, Steinberg SV. GU receptors of double helices mediate tRNA movement in the ribosome. RNA. 2002;8:873–877. doi: 10.1017/s135583820202602x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mokdad A, Krasovska MV, Sponer J, Leontis NB. Structural and evolutionary classification of G/U wobble basepairs in the ribosome. Nucleic Acids Res. 2006;34:1326–1341. doi: 10.1093/nar/gkl025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leontis NB, Westhof E. A common motif organizes the structure of multi-helix loops in 16 S and 23 S ribosomal RNAs. J Mol Biol. 1998;283:571–583. doi: 10.1006/jmbi.1998.2106. [DOI] [PubMed] [Google Scholar]
- 33.Aalberts DP, Hodas NO. Asymmetry in RNA pseudoknots: observation and theory. Nucleic Acids Res. 2005;33:2210–2214. doi: 10.1093/nar/gki508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Elgavish T, Cannone JJ, Lee JC, Harvey SC, Gutell RR. AA.AG@helix.ends: A:A and A:G base-pairs at the ends of 16 S and 23 S rRNA helices. J Mol Biol. 2001;310:735–753. doi: 10.1006/jmbi.2001.4807. [DOI] [PubMed] [Google Scholar]
- 35.Moazed D, Stern S, Noller HF. Rapid chemical probing of conformation in 16 S ribosomal RNA and 30 S ribosomal subunits using primer extension. J Mol Biol. 1986;187:399–416. doi: 10.1016/0022-2836(86)90441-9. [DOI] [PubMed] [Google Scholar]
- 36.Leontis NB, Kwok W, Newman JS. Stability and structure of three-way DNA junctions containing unpaired nucleotides. Nucleic Acids Res. 1991;19:759–766. doi: 10.1093/nar/19.4.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Holbrook SR, Sussman JL, Warrant RW, Kim SH. Crystal structure of yeast phenylalanine transfer RNA. II. Structural features and functional implications. J Mol Biol. 1978;123:631–660. doi: 10.1016/0022-2836(78)90210-3. [DOI] [PubMed] [Google Scholar]
- 38.Leontis NB, Stombaugh J, Westhof E. Motif prediction in ribosomal RNAs Lessons and prospects for automated motif prediction in homologous RNA molecules. Biochimie. 2002;84:961–973. doi: 10.1016/s0300-9084(02)01463-3. [DOI] [PubMed] [Google Scholar]
- 39.Leontis NB, Altman RB, Berman HM, Brenner SE, Brown JW, Engelke DR, Harvey SC, Holbrook SR, Jossinet F, Lewis SE, Major F, Mathews DH, Richardson JS, Williamson JR, Westhof E. The RNA Ontology Consortium: an open invitation to the RNA community. RNA. 2006;12:533–541. doi: 10.1261/rna.2343206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Klosterman PS, Hendrix DK, Tamura M, Holbrook SR, Brenner SE. Three-dimensional motifs from the SCOR, structural classification of RNA database: extruded strands, base triples, tetraloops and U-turns. Nucleic Acids Res. 2004;32:2342–2352. doi: 10.1093/nar/gkh537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Laing C, Jung S, Iqbal A, Schwede T. Recurrent interaction motifs direct and stabilize the 3D structure of RNA junctions. In preparation. [Google Scholar]
- 42.Kieft JS, Zhou K, Grech A, Jubin R, Doudna JA. Crystal structure of an RNA tertiary domain essential to HCV IRES-mediated translation initiation. Nat Struct Biol. 2002;9:370–374. doi: 10.1038/nsb781. [DOI] [PubMed] [Google Scholar]
- 43.Tan E, Wilson TJ, Nahas MK, Clegg RM, Lilley DM, Ha T. A four-way junction accelerates hairpin ribozyme folding via a discrete intermediate. Proc Natl Acad Sci U S A. 2003;100:9308–9313. doi: 10.1073/pnas.1233536100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lescoute A, Westhof E. The interaction networks of structured RNAs. Nucleic Acids Res. 2006;34:6587–6604. doi: 10.1093/nar/gkl963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jaeger L, Verzemnieks EJ, Geary C. The UA_handle: a versatile submotif in stable RNA architectures. Nucleic Acids Res. 2009;37:215–230. doi: 10.1093/nar/gkn911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Penedo JC, Wilson TJ, Jayasena SD, Khvorova A, Lilley DM. Folding of the natural hammerhead ribozyme is enhanced by interaction of auxiliary elements. Rna. 2004;10:880–888. doi: 10.1261/rna.5268404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Walter F, Murchie AI, Thomson JB, Lilley DM. Structure and activity of the hairpin ribozyme in its natural junction conformation: effect of metal ions. Biochemistry. 1998;37:14195–14203. doi: 10.1021/bi981513+. [DOI] [PubMed] [Google Scholar]
- 48.Klein DJ, Moore PB, Steitz TA. The roles of ribosomal proteins in the structure assembly, and evolution of the large ribosomal subunit. J Mol Biol. 2004;340:141–177. doi: 10.1016/j.jmb.2004.03.076. [DOI] [PubMed] [Google Scholar]
- 49.Zhou J, Bean RL, Vogt VM, Summers M. Solution structure of the Rous sarcoma virus nucleocapsid protein: muPsi RNA packaging signal complex. J Mol Biol. 2007;365:453–467. doi: 10.1016/j.jmb.2006.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Walter F, Murchie AI, Duckett DR, Lilley DM. Global structure of four-way RNA junctions studied using fluorescence resonance energy transfer. RNA. 1998;4:719–728. doi: 10.1017/s135583829898030x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Melcher SE, Wilson TJ, Lilley DM. The dynamic nature of the four-way junction of the hepatitis C virus IRES. Rna. 2003;9:809–820. doi: 10.1261/rna.5130703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sarver M, Zirbel CL, Stombaugh J, Mokdad A, Leontis NB. FR3D: finding local and composite recurrent structural motifs in RNA 3D structures. J Math Biol. 2008;56:215–252. doi: 10.1007/s00285-007-0110-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang H, Jossinet F, Leontis N, Chen L, Westbrook J, Berman H, Westhof E. Tools for the automatic identification and classification of RNA base pairs. Nucleic Acids Res. 2003;31:3450–3460. doi: 10.1093/nar/gkg529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Guex NaP, M C. SWISS-MODEL and the Swiss-PdbViewer: An environment for comparative protein modelling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 55.Jossinet F, Westhof E. Sequence to Structure (S2S): display, manipulate and interconnect RNA data from sequence to structure. Bioinformatics. 2005;21:3320–3321. doi: 10.1093/bioinformatics/bti504. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.