Abstract
In an effort to discover genes important for human development, we have ascertained patients with congenital anomalies and cytogenetically balanced chromosomal rearrangements. Herein, we report a four year-old girl with profound deafness, a history of feeding difficulties, dysmorphism, strabismus, developmental delay, and an apparently balanced de novo paracentric chromosome 5 inversion, inv(5)(q15q33.2). Molecular cytogenetic analysis of the inversion revealed the presence of microdeletions of approximately 400-500 kb at or near both breakpoints. The 5q15 microdeletion completely removes the nuclear receptor NR2F1 (COUP-TFI) from the inverted chromosome 5. We propose haploinsufficiency of NR2F1 to be the cause of the patient's deafness and many of the other associated anomalies based on striking similarity with the Nr2f1 null mouse. Additionally, this study further highlights the need for high resolution analysis of clinical samples with chromosomal rearrangements as associated deletions may be primarily responsible for the clinical features of these patients.
Keywords: NR2F1, deafness, chromosomal inversion, microdeletion, FISH, 5q
INTRODUCTION
Approximately 1 in 2000 newborns has a de novo balanced chromosomal rearrangement [Warburton, 1991]. The risk for a congenital anomaly in this population is estimated at 6.1% for translocations and 9.4% for inversions [Warburton, 1991]. Overall, the risk of a congenital anomaly in individuals with a de novo balanced rearrangement is two to three times higher than that observed in an unselected population of newborns, suggesting that the phenotype may be due to deletion, disruption, or dysregulation of gene(s) in the breakpoint regions. Through the Developmental Genome Anatomy Project (DGAP, http://dgap.harvard.edu) we have been studying patients with de novo balanced chromosomal rearrangements and congenital anomalies. The goal of DGAP is to identify genes at the chromosomal breakpoints that are critical for human development. Recent studies of patients with phenotypic abnormalities and chromosomal rearrangements using high resolution molecular cytogenetic techniques, such as array comparative genomic hybridization (aCGH), have revealed that many presumably balanced rearrangements include cryptic microdeletions at or near the breakpoints [Astbury et al., 2004; Gribble et al., 2005; De Gregori et al., 2007; Higgins et al., 2008; Fantes et al., 2008; Baptista et al., 2008]. While the majority of patients we have studied have balanced rearrangements, detailed molecular mapping has also revealed some rearrangements with cryptic microdeletions [Higgins et al., 2008]. High resolution characterization of these microdeletions is essential for identifying candidate genes responsible for the patient's phenotype and for distinguishing haploinsufficiencies with clinical consequences from nonpathogenic copy number polymorphisms.
Herein, we describe a 4-year-old girl (designated DGAP169) with profound sensorineural deafness, feeding difficulties, dysmorphism, strabismus, developmental delay, and an apparently balanced de novo paracentric inversion of chromosome 5 identified by GTG-banded analysis of metaphase chromosomes. Detailed FISH mapping of the inversion breakpoints revealed two microdeletions of ~400-500 kb, one at the 5q33 breakpoint and the other a short distance from the 5q15 breakpoint. While several genes are completely or partially deleted due to the rearrangement, NR2F1 is a compelling positional candidate gene for the patient's phenotype.
MATERIALS AND METHODS
Clinical details
DGAP169 was ascertained as part of the Developmental Genome Anatomy Project. The human study protocol for this project has been reviewed and approved by the Partners HealthCare System Human Research Committee.
DGAP169 was born at term to healthy, unrelated parents following an uneventful pregnancy via planned cesarean due to prior cesarean delivery. Her G3P2SAB1 mother was 26 years old at the time of delivery and has one healthy son. At birth, DGAP169 was immediately noticed to have malformed ears. Major sucking and swallowing difficulties further complicated her neonatal course. GTG-banded chromosomal analysis of peripheral blood lymphocytes revealed the presence of an apparently balanced chromosome 5 inversion. Her karyotype was reported as 46,XX,inv(5)(q14q34). Both parents were subsequently determined to have normal karyotypes.
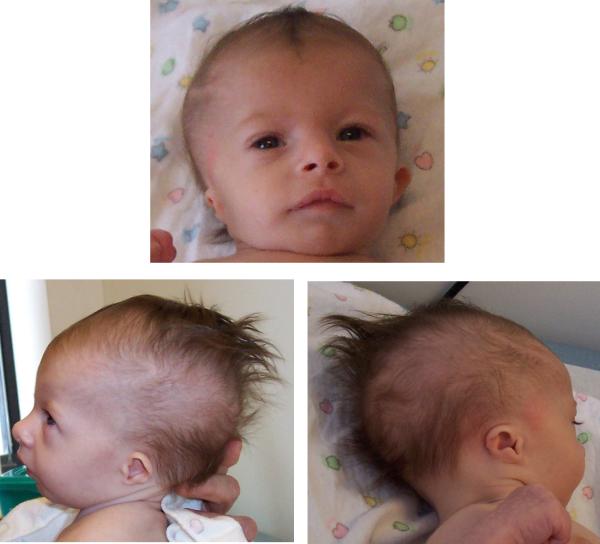
DGAP169 was evaluated at Children's Hospital Boston at 11 weeks of age. Her weight of 3.22 kg had barely increased over her birth weight because of poor sucking and suspected uncoordinated sucking and swallowing. Swallow study confirmed the uncoordinated sucking and swallowing. Her scalp hair pattern was unusual in that the direction of hair growth was toward the midline resembling a “comb” that extended backward along the line of the sagittal suture, with very sparse temporal hair (Fig 1). Her face was somewhat asymmetric with her eyes positioned at slightly different levels. She had upslanted palpebral fissures and anteverted nares with a broad tip of the nose. Remarkable micrognathia was noted with no associated cleft palate. Her ear pinnae were low-set, small, and significantly malformed (Fig 1) but her external auditory canals were patent. Her neurological examination was notable for significant axial hypotonia with decreased deep tendon reflexes. Auditory brainstem response (ABR) performed to test her hearing showed no air conduction response for 1-4 kHz even at 100 dB and no bone conduction response for 0.5-1.5 kHz at 65 dB indicating bilateral profound sensorineural deafness.
Figure 1.
Front and side views of DGAP169 at 11 weeks of age. Note the low-set, small, malformed ears, unusual pattern of hair growth, upslanting palpebral fissures, broad nasal tip, anteverted nares, and micrognathia. The face is also asymmetric with the eyes positioned at slightly different levels. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
Temporal bone CT revealed malformations of the middle and inner ear, and suggested an abnormality of the cochlear division of the vestibulocochlear nerve. The cochlea was malformed bilaterally with only one to one and a quarter turns. The vestibular aqueduct was enlarged and the position of the lateral semicircular canal was abnormal. All three ossicles, particularly the stapes, were malformed and the incus and malleus appeared to be fused (Figs 2a and 2b). Additionally, the oval window was stenotic. The canal for the cochlear nerve was small suggesting that the cochlear nerve is either small or absent and the course of the facial nerve was bilaterally anomalous in the tympanic and mastoid segments (Figs 2c and 2d). A follow-up MRI to visualize the facial and vestibulocochlear nerve was performed. The facial nerve was visible but very small; however, the cochlear division of the vestibulocochlear nerve could not be seen due to suboptimal quality of the MRI. DGAP169 received a cochlear implant at 28 months with the hope that she would gain some functional hearing ability. Two years after implantation she still has no significant hearing. Although she shows some response to sound during conditioned play audiometry testing, it appears to be a vibrotactile response rather than a true auditory response. Continued implant use and continued adjustment of the programming of the implant will reveal if it is providing any meaningful auditory input.
Figure 2.

CT images of DGAP169 temporal bone. a) The malformed stapes is indicated by the arrow. b) The malformed malleus is indicated by the arrow. c) The asterisk marks the underdeveloped middle and apical turns of the cochlea, while the arrow marks the abnormal path of the facial nerve through the mastoid bone. d) The abnormal course of the facial nerve through the middle ear is indicated by the arrow.
Recent follow-up of DGAP169 at four years of age has uncovered several additional abnormal features. Her feeding has improved significantly with age and her caloric intake has been consistently above her estimated needs for growth catch-up; however, she still shows poor growth, weighing only 28 lbs (3rd-5th centile). As an infant, she stopped breathing whenever her arms were elevated. The cause of this condition is unknown but seems to persist as she is still unwilling to lift her arms upward. Otherwise, she moves and uses her arms normally. Strabismus was noted at 18 months and persists at four years. She continues to have sparse temporal hair at four years of age, although this feature is less obvious since the rest of her hair has been grown long. Evaluation of developmental milestones at 28 months showed significant delay in gross motor skills (18-21 month level), cognitive skills (15.5-18 month level), and communication (5.5-7 month level), although she had recently started walking. By 35 months, she engaged in limited signing, was able to follow simple commands, could stack blocks, and enjoyed toys. At four years, she climbs stairs with alternating feet and has started toilet training.
Fluorescence in situ hybridization (FISH) analysis
After informed consent was obtained, a peripheral blood sample was collected from the child. A lymphoblastoid cell line was generated from the blood sample at the Massachusetts General Hospital Cell Transformation Core using standard protocols. Based on the reported karyotype, BAC clones spanning the breakpoint regions were selected for FISH mapping using the University of California Santa Cruz Genome Browser (http://genome.ucsc.edu). BACs from the RP11 library were acquired from Children's Hospital Oakland Research Institute (Oakland, CA) and BACs from the CTD library were acquired from Invitrogen (Carlsbad, CA). Metaphase chromosome spreads were prepared from the lymphoblastoid cells using standard cytogenetic protocols and FISH performed as previously described [Ney et al., 1993]. BACs were labeled directly with either Spectrum Orange or Spectrum Green conjugated dUTP using a nick translation kit (Vysis, Downers Grove, IL) and differentially labeled pairs were hybridized overnight to metaphase chromosome preparations. After washing, chromosomes were counterstained with DAPI and analyzed with a Zeiss Axioskop microscope (Thornwood, NY) and Applied Imaging CytoVision software (Santa Clara, CA). At least 10 metaphases were scored per BAC probe.
To interpret accurately the results of the child's FISH experiments, peripheral blood samples were obtained from the biological parents. The blood samples were cultured, lymphocytes harvested, and metaphase spreads prepared according to standard cytogenetic protocols. FISH was then performed as described above.
Sequencing of NR2F1
DNA was isolated from the DGAP169 lymphoblastoid cell line using the PureGene system (Gentra, Minneapolis, MN). M13-tagged primer pairs were used to amplify the three exons of NR2F1. PCR reactions contained 50 ng DNA, 0.4 uM each primer, 2 mM MgSO4, 0.4 mM dNTP, 1X High Fidelity PCR Buffer, 2.5% DMSO, and 2.5 units Platinum High Fidelity Taq DNA polymerase (Invitrogen). PCR was performed with an initial denaturation of 97°C for 5 minutes, 35 cycles of 97°C for 30 seconds, 55°C for 30 seconds, 72°C for 45 seconds, and a final extension of 72°C for 7 minutes. PCR products were purified using the QIAquick PCR Purification kit (Qiagen, Valencia, CA) and sequenced at the Dana-Farber/Harvard Cancer Center DNA Resource Core.
Reverse transcriptase-polymerase chain reaction (RT-PCR) of NR2F1
RNA was isolated from the DGAP169 lymphoblastoid cell line using Trizol (Invitrogen). NR2F1 was reverse transcribed and amplified from 1 ug of RNA using SuperScript One-Step RT-PCR with Platinum Taq (Invitrogen).
RESULTS
To map the inversion breakpoints, we performed FISH on metaphase chromosomes from patient lymphoblastoid cells. Based on the reported karyotype, inv(5)(q14q34), BACs localized to 5q13-q15 and 5q33-q35 on the UCSC website were selected for FISH. Sequential FISH experiments were performed to narrow the breakpoint regions. Results for a subset of BACs used for each breakpoint are listed in Table I. BAC clones RP11-108e6 and RP11-176p18 from 5q15 hybridized to 5q15 on the normal chromosome 5 and to both 5q15 and 5q33 on the inverted chromosome 5, narrowing the breakpoint region to about 135 kb (Fig 3a). The ends of a BAC centromeric to the breakpoint, RP11-213a8, and a BAC telomeric to the breakpoint, RP11-297g19, slightly overlap but neither shows a split hybridization signal, indicating that the breakpoint likely occurs within the 20 kb of overlapping sequence. Based on this localization, the 5q15 breakpoint disrupts a large gene, C5ORF21, which spans about 500 kb of genomic sequence (Fig 4a).
Table I.
FISH results for BACs mapping to the 5q15 and 5q33 breakpoint regions
| 5q15 BAC | BAC position on chromosome 5* | FISH result for inv(5) |
|---|---|---|
| RP11-60g8 | 90,227,590-90,372,004 | 5q15 |
| RP11-92c19 | 92,593,206-92,749,220 | 5q15 |
| CTD-3236h3 | 92,612,533-92,691,223 | Deleted |
| RP11-829m2 | 92,620,518-92,801,910 | Deleted |
| RP11-608g16 | 92,784,673-92,972,632 | Deleted |
| RP11-65f13 | 92,894,088-93,074,143 | 5q15 |
| RP11-521h6 | 93,023,218-93,207,639 | 5q15 |
| RP11-213a8 | 93,168,124-93,308,711 | 5q15 |
| RP11-108e6 | 93,196,412-93,376,816 | 5q15 and 5q33 |
| RP11-176p18 | 93,241,892-93,406,803 | 5q15 and 5q33 |
| RP11-297g19 | 93,288,262-93,463,665 | 5q33 |
| RP11-33a7 | 93,746,809-93,916,228 | 5q33 |
| RP11-86c20 | 152,618,661-152,788,737 | 5q15 |
| RP11-343m7 | 153,138,586-153,319,561 | 5q15 |
| CTD-2270b22 | 153,311,373-153,523,768 | 5q15 |
| CTD-2338o23 | 153,390,771-153,495,446 | Deleted |
| CTD-2336d9 | 153,409,619-153,523,782 | Deleted |
| RP11-635k22 | 153,436,850-153,605,891 | Deleted |
| CTD-2314a16 | 153,557,982-153,723,774 | Deleted |
| RP11-314p8 | 153,647,251-153,820,842 | Deleted |
| RP11-108c12 | 153,647,252-153,844,258 | Deleted |
| RP11-790o1 | 153,693,987-153,872,038 | 5q33 |
| RP11-960p1 | 153,880,910-154,059,745 | 5q33 |
| RP11-1149b7 | 154,371,494-154,551,424 | 5q33 |
BAC position based on March 2006 genome assembly
Figure 3.
FISH results on DGAP169 metaphase chromosomes using chromosome 5 BAC probes. a) BAC RP11-108e6 (red) maps to 5q15 and spans the 5q15 inversion breakpoint. b) BAC CTD-3236h3 (red) maps to 5q15 and is deleted from the inv(5). BAC RP11-790o1 (green) at 5q33.2 marks chromosome 5. C) BAC RP11-635k22 (green) maps to 5q33.2 and is deleted from the inv(5). BAC RP11-521h6 (red) at 5q15 marks chromosome 5. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
Figure 4.
Maps of the inv(5)(q15q33.2) breakpoint regions from the UCSC genome browser. a) Map of 5q15 showing the breakpoint region and the deleted region centromeric to the breakpoint. b) Map of 5q33.2 showing the region deleted by the breakpoint. The red portion of the line at the bottom of each section shows the minimal deleted regions. The grey portions of the line show the maximal deleted regions. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
While mapping the 5q15 breakpoint, a cryptic deletion of 360-480 kb was discovered approximately 220-330 kb centromeric to the breakpoint region. Three BAC clones, CTD-3236h3, RP11-829m2, and RP11-608g16, produced hybridization signals only on the normal chromosome 5, while the flanking BAC clones, RP11-92c19 and RP11-65f13, produced hybridization signals on both the normal 5 and inv(5) (Fig 3b). As a result, NR2F1 and AK124699 are completely deleted and C5ORF21 may be partially deleted from the inv(5) (Fig 4a).
A cryptic deletion of 450-560 kb was also detected at the 5q33.2 breakpoint. All BACs centromeric to the 5q33.2 deletion showed a hybridization signal at 5q33.2 on the normal chromosome 5 and at 5q15 on the inv(5). All BACs telomeric to the 5q33.2 deletion showed a hybridization signal at 5q33.2 on both the normal 5 and the inv(5). These results indicate that the deletion marks the inversion breakpoint. Six BAC clones, CTD-2338o23, CTD-2336d9, RP11-635k22, CTD-2314a16, RP11-314p8, and RP11-108c12, produced hybridization signals only on the normal chromosome 5, while the flanking BAC clones, CTD-2270b22 and RP11-790o1, produced hybridization signals on the normal 5 and the inv(5) (Figure 3c). As a result of this deletion, five additional genes, C5ORF3, MFAP3, GALNT10, SAP30L, and HAND1, are completely or partially deleted from the inv(5) (Fig 4b).
To determine if these deletions occurred de novo in the child, FISH was performed on chromosomes from both parents using two BAC probes within the 5q15 deleted region and two BAC probes within the 5q33.2 deleted region. Two chromosome 5 signals were detected for both parents with all BACs tested, indicating that the microdeletions are only present in the child.
To determine if any of the deleted genes could be candidate deafness genes, we queried our human fetal cochlear cDNA library [Skvorak et al., 1999] for transcripts from all deleted genes. Four of the eight possibly deleted genes, NR2F1, C5ORF21, AK124699, and MFAP3, are expressed in fetal cochlea with NR2F1 among the top 50 most highly expressed cochlear transcripts. Because NR2F1 is highly expressed in the cochlea and Nr2f1 knockout mice have profound sensorineural deafness by three weeks of age [Wenzel et al., 2004], we investigated this gene further as a possible candidate for deafness in DGAP169. We sequenced the remaining NR2F1 allele in DGAP169 and found no additional mutations suggesting that any phenotype attributed to the gene would likely be due to haploinsufficiency. We also attempted to assess expression levels of NR2F1 in the DGAP169 lymphoblastoid cell line to determine whether the deletion leads to reduced NR2F1 expression. Using reverse transcriptase-polymerase chain reaction (RT-PCR) we were unable to detect NR2F1 expression in either control or patient lymphoblastoid cell lines indicating that the gene is not expressed in this cell type. The RT-PCR results are consistent with the NR2F1 profile in the UniGene database (http://www.ncbi.nlm.nih.gov) in which no expression was detected in blood.
DISCUSSION
Molecular cytogenetic analysis of an inv(5)(q15q33.2)dn in a female with syndromic deafness revealed microdeletions of about 400-500 kb in the vicinity of both inversion breakpoints. The two microdeletions result in partial or complete deletion of approximately eight genes, and haploinsufficiency for one or more of these genes is the likely etiology of the child's phenotype. Copy number gains within the centromeric portion of the 5q15 deletion overlapping AK124699 have been reported in normal individuals, but no additional copy number polymorphisms overlapping the other deleted genes have been reported (Database of Genomic Variants, http://projects.tcag.ca/variation/; USCS Genome Browser, http://genome.ucsc.edu; Ensembl Genome Browser, http://www.ensembl.org). Pathogenic interstitial deletions including bands 5q15 and 5q33.2 have been reported in the literature; however, all reported patients have much larger deletions than detected in DGAP169 and only a few of these deletions have been mapped molecularly making it difficult to compare this patient to those reports. Hearing defects have been noted in two patients with interstitial deletions involving 5q15 [Harprecht-Beato et al., 1983; Harreus and Issing, 2002], but it is unclear whether these deletions overlap with that of DGAP169. Only conductive hearing loss has been noted in patients with deletions involving 5q33.2 [Giltay et al., 1997].
We propose haploinsufficiency for NR2F1 as the primary cause of the patient's syndrome. NR2F1 is an orphan receptor of the steroid/thyroid hormone receptor superfamily, which consists of ligand inducible transcription factors. It plays an important role in neurogenesis and neural crest cell differentiation [Qiu et al., 1997]. Studies of mouse embryos show that Nr2f1 is expressed in the otic vesicle, forebrain, midbrain, hindbrain, branchial arches, and neural tube at E9.0 [Qiu et al., 1997; Jonk et al., 1994]. At later time points, Nr2f1 is expressed in the cochlea, middle ear, restricted brain regions, spinal cord, tongue, and other tissues [Jonk et al., 1994; Pereira et al., 1995]. Most Nr2f1 knockout mice die between 8 and 36 hours of birth due to inability to feed [Qiu et al., 1997]. Similar to DGAP169, the mice have difficulty swallowing. Nr2f1 null mice that survive to P20 are profoundly deaf displaying no response when tested for auditory brainstem response (ABR) or distortion product otoacoustic emissions (DPOAE) [Wenzel et al., 2004]. Null mice have a shortened cochlear duct and malformed vestibular chambers [Tang et al., 2006]. The null mice also have axonal guidance and projection defects. Specifically, the paths of some of the cranial nerves are abnormal, both the saccule and organ of Corti of the inner ear are poorly innervated, and misguided axonal projections are present in the cochlear duct [Qiu et al., 1997; Pereira et al., 2001]. DGAP169 seems to have similar axonal guidance defects. Imaging revealed an abnormal course of the facial nerve bilaterally, and the limited response to cochlear implantation may result from poor innervation of the cochlea or other defects in the auditory nerve. Additionally, the patient's strabismus could be due to defects in cranial nerves III, IV, or VI. Nr2f1 null mice that survived to three weeks are described as ataxic, suggesting there could also be problems with connections of other motor neurons [Qiu et al., 1997]. Likewise, the hypotonia and delay in motor skills seen in DGAP169 suggest deficits in motor neurons. While no abnormalities in hair patterning were reported for Nr2f1 null mice, Nr2f1 can regulate Notch signaling [Tang et al., 2006], which is important for proper hair patterning [Uyttendaele et al., 2004; Vauclair et al., 2005]. Because of the strikingly similar phenotypic features observed in DGAP169 and Nr2f1 null mice, we propose that haploinsufficiency for NR2F1 is the primary cause of the DGAP169 phenotype.
While mice haploinsufficient for Nr2f1 appear to be normal [Qiu et al., 1997], this does not exclude our hypothesis that NR2F1 is primarily responsible for the DGAP169 phenotype, as humans seem to be more sensitive than mice to dosage differences in certain genes. For example, holoprosencephaly type 3 is caused by haploinsufficiency for Sonic Hedgehog (SHH). Shh null mice generally display an embryonic lethal phenotype with CNS abnormalities and midline defects, while Shh haploinsufficient mice appear completely normal [Chiang et al., 1996]. Additionally, mice haploinsufficient for Tbx1 show only cardiac abnormalities, while Tbx1 null mice exhibit most of the common features seen in haploinsufficient DiGeorge/Velocardiofacial syndrome patients, including cardiac abnormalities, hypoplasia of the thymus, abnormal facial structures, abnormal vertebrae, and cleft palate [Jerome and Papaioannou, 2001]. Consistent with our hypothesis, the complete absence of Nr2f1 in the null mouse causes a more severe phenotype than seen in DGAP169 who presumably has some residual NR2F1 function. Furthermore, while some differences in phenotypic manifestations between species are expected, the tissues and systems affected in the Nr2f1 null mice and in DGAP169 are quite similar and, perhaps, a mouse model homozygous for an Nr2f1 hypomorphic mutation would mimic even more closely the human phenotype.
While NR2F1 is the most attractive positional candidate gene, we cannot conclusively exclude the other deleted genes from playing a role in the patient's phenotype. Most of the deleted genes are widely expressed; however, HAND1 has a more restricted expression pattern. HAND1 is the only deleted gene other than NR2F1 that has been well studied, and it is a transcription factor essential for proper development of the placenta and the heart [Firulli et al., 1998; Riley et al., 1998]. MFAP3 localizes to microfibrils and could play a role in microfibril diseases [Abrams et al., 1995]. Genes encoding other microfibril components cause Marfan syndrome, congenital contractural arachnodactyly, familial mitral valve prolapse syndrome, annuloaortic ectasia, and familial ectopia lentis. SAP30L interacts with several components of the Sin3A corepressor complex and can induce transcriptional repression probably through recruitment of histone deacetylases [Viiri et al., 2006]. GALNT10 is an enzyme that catalyzes the first step of O-linked protein glycosylation [Nelson et al., 2002]. Very little is known about C5ORF3, C5ORF21, and AK124699, only that C5ORF3 contains an ATP/GTP binding site [Boultwood et al., 2000] and C5ORF21 has an endoplasmic reticulum targeting sequence [Simpson et al., 2000]. MFAP3, C5ORF21, and AK124699 appear to be weakly expressed in fetal cochlea, although NR2F1 is the more compelling candidate human deafness gene.
Several recent reports have noted that individuals with congenital anomalies due to apparently balanced chromosomal rearrangements commonly have deletions near the rearrangement breakpoints [Astbury et al., 2004; Gribble et al., 2005; De Gregori et al, 2007; Higgins et al., 2008; Baptista et al., 2008; Fantes et al., 2008]. These additional deletions will presumably affect more genes than the rearrangement alone increasing the chances that the individual will have an abnormal phenotype. As of yet, none of these studies have assessed if there are differences in the rate of cryptic deletions associated with inversions versus translocations. Three of these reports [Astbury et al., 2004; Higgins et al., 2008; Fantes et al., 2008] contain data on patients whose congenital anomalies are associated with inversions. Combining the data on patients with congenital anomalies studied in these reports reveals that 5 of 11 (45%) inversions and 18 of 78 (23%) simple reciprocal translocations have associated deletions. These data suggest that patients with inversions may be more likely to have cryptic deletions. So far, the number of reported inversion patients whose rearrangements have been studied at high resolution is very small and, therefore, the difference between the frequency of deletions associated with inversions versus translocations does not reach statistical significance. Consequently, additional studies with larger numbers of patients may be warranted to investigate further this preliminary observation. Interestingly, the increased number of deletions seen in these inversion patients correlates with Warburton's study [Warburton, 1991] which estimates the risk of congenital anomaly in individuals with apparently balanced inversions to be 1.5 times higher than in individuals with apparently balanced translocations. Perhaps, a higher incidence of associated deletions contributes to the increased risk seen for inversion patients.
While we cannot definitively determine the causal gene from a single patient, we suggest that haploinsufficiency for NR2F1 could be etiologic for all or at least most of the anomalies observed in DGAP169, including deafness, feeding difficulties, abnormal course of the facial nerve, strabismus, hypotonia, and developmental delay. Genetic analysis of other individuals with a similar phenotype will be necessary to confirm this prediction. Additionally, we recommend high resolution screening for imbalances in patients with congenital anomalies and chromosomal rearrangements as associated deletions seem to be a common occurrence.
ACKNOWLEDGMENTS
We would like to express our sincere gratitude to the patient and her family for making this study possible. We would also like to thank Heather Ferguson for assistance with patient enrollment, the Massachusetts General Hospital Cell Transformation Core for assistance with EBV transformation, the Brigham and Women's Hospital Clinical Cytogenetics Laboratory for assistance with processing the parental blood samples, and the Dana-Farber/Harvard Cancer Center (DF/HCC) DNA resource core (P30CA06516) for assistance with DNA sequencing. This work was supported by grants from the National Institutes of Health, P01 GM061354 (to C.C.M.) and F31 DC007540 (to K.K.B).
REFERENCES
- Abrams WR, Ma RI, Kucich U, Bashir MM, Decker S, Tsipouras P, McPherson JD, Wasmuth JJ, Rosenbloom J. Molecular cloning of the microfibrillar protein MFAP3 and assignment of the gene to human chromosome 5q32-q33.2. Genomics. 1995;26:47–54. doi: 10.1016/0888-7543(95)80081-v. [DOI] [PubMed] [Google Scholar]
- Astbury C, Christ LA, Aughton DJ, Cassidy SB, Kumar A, Eichler EE, Schwartz S. Detection of deletions in de novo “balanced” chromosome rearrangements: further evidence for their role in phenotypic abnormalities. Genet Med. 2004;6:81–89. doi: 10.1097/01.gim.0000117850.04443.c9. [DOI] [PubMed] [Google Scholar]
- Baptista J, Mercer C, Prigmore E, Gribble SM, Carter NP, Maloney V, Thomas NS, Jacobs PA, Crolla JA. Breakpoint mapping and array CGH in translocations: comparison of a phenotypically normal and an abnormal cohort. Am J Hum Genet. 2008;82:927–936. doi: 10.1016/j.ajhg.2008.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boultwood J, Fidler C, Strickson AJ, Watkins F, Kostrzewa M, Jaju RJ, Muller U, Wainscoat JS. Transcription mapping of the 5q- syndrome critical region: cloning of two novel genes and sequencing, expression, and mapping of a further six novel cDNAs. Genomics. 2000;66:26–34. doi: 10.1006/geno.2000.6193. [DOI] [PubMed] [Google Scholar]
- Chiang C, Litingtung Y, Lee E, Young KE, Corden JL, Westphal H, Beachy PA. Cyclopia and defective axial patterning in mice lacking Sonic hedgehog gene function. Nature. 1996;383:407–413. doi: 10.1038/383407a0. [DOI] [PubMed] [Google Scholar]
- De Gregori M, Ciccone R, Magini P, Pramparo T, Gimelli S, Messa J, Novara F, Vetro A, Rossi E, Maraschio P, Bonaglia MC, Anichini C, Ferrero GB, Silengo M, Fazzi E, Zatterale A, Fischetto R, Previdere C, Belli S, Turci A, Calabrese G, Bernardi F, Meneghelli E, Riegel M, Rocchi M, Guerneri S, Lalatta F, Zelante L, Romano C, Fichera M, Mattina T, Arrigo G, Zollino M, Giglio S, Lonardo F, Bonfante A, Ferlini A, Cifuentes F, Van Esch H, Backx L, Schinzel A, Vermeesch JR, Zuffardi O. Cryptic deletions are a common finding in “balanced” reciprocal and complex chromosome rearrangements: a study of 59 patients. J Med Genet. 2007;44:750–762. doi: 10.1136/jmg.2007.052787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fantes JA, Boland E, Ramsay J, Donnai D, Splitt M, Goodship JA, Stewart H, Whiteford M, Gautier P, Harewood L, Holloway S, Sharkey F, Maher E, van Heyningen V, Clayton-Smith J, Fitzpatrick DR, Black GC. FISH mapping of de novo apparently balanced chromosome rearrangements identifies characteristics associated with phenotypic abnormality. Am J Hum Genet. 2008;82:916–926. doi: 10.1016/j.ajhg.2008.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firulli AB, McFadden DG, Lin Q, Srivastava D, Olson EN. Heart and extra-embryonic mesodermal defects in mouse embryos lacking the bHLH transcription factor Hand1. Nat Genet. 1998;18:266–270. doi: 10.1038/ng0398-266. [DOI] [PubMed] [Google Scholar]
- Giltay JC, Gerssen-Schoorl KB, Luitse GH, Dauwerse HG. A case of de novo interstitial deletion of chromosome 5(q33q34) Clin Genet. 1997;52:173–176. doi: 10.1111/j.1399-0004.1997.tb02539.x. [DOI] [PubMed] [Google Scholar]
- Gribble SM, Prigmore E, Burford DC, Porter KM, Ng BL, Douglas EJ, Fiegler H, Carr P, Kalaitzopoulos D, Clegg S, Sandstrom R, Temple IK, Youings SA, Thomas NS, Dennis NR, Jacobs PA, Crolla JA, Carter NP. The complex nature of constitutional de novo apparently balanced translocations in patients presenting with abnormal phenotypes. J Med Genet. 2005;42:8–16. doi: 10.1136/jmg.2004.024141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harprecht-Beato W, Kaiser P, Steuber E, Reinhard W. Interstitial deletion in the long arm of chromosome No. 5. Clin Genet. 1983;23:167–171. doi: 10.1111/j.1399-0004.1983.tb01867.x. [DOI] [PubMed] [Google Scholar]
- Harreus UA, Issing WJ. [Chromosome 5q-syndrome-ENT pathologies] Laryngorhinootologie. 2002;81:565–567. doi: 10.1055/s-2002-33361. [DOI] [PubMed] [Google Scholar]
- Higgins AW, Alkuraya FS, Bosco AF, Brown KK, Bruns GA, Donovan DJ, Eisenman R, Fan Y, Farra CG, Ferguson HL, Gusella JF, Harris DJ, Herrick SR, Kelly C, Kim H-G, Kishikawa S, Korf BR, Kulkarni S, Lally E, Leach NT, Lemyre E, Lewis J, Ligon AH, Lu W, Maas RL, MacDonald ME, Moore SD, Peters RE, Quade BJ, Quintero-Rivera F, Saadi I, Shen Y, Shendure J, Williamson RE, Morton CC. Characterization of apparently balanced chromosomal rearrangements from the developmental genome anatomy project. Am J Hum Genet. 2008;82:712–722. doi: 10.1016/j.ajhg.2008.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jerome LA, Papaioannou VE. DiGeorge syndrome phenotype in mice mutant for the T-box gene, Tbx1. Nat Genet. 2001;27:286–291. doi: 10.1038/85845. [DOI] [PubMed] [Google Scholar]
- Jonk LJ, de Jonge ME, Pals CE, Wissink S, Vervaart JM, Schoorlemmer J, Kruijer W. Cloning and expression during development of three murine members of the COUP family of nuclear orphan receptors. Mech Dev. 1994;47:81–97. doi: 10.1016/0925-4773(94)90098-1. [DOI] [PubMed] [Google Scholar]
- Nelson PA, Sutcliffe JG, Thomas EA. A new UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase mRNA exhibits predominant expression in the hypothalamus, thalamus and amygdala of mouse forebrain. Brain Res Gene Expr Patterns. 2002;1:95–99. doi: 10.1016/s1567-133x(01)00019-9. [DOI] [PubMed] [Google Scholar]
- Ney PA, Andrews NC, Jane SM, Safer B, Purucker ME, Weremowicz S, Morton CC, Goff SC, Orkin SH, Nienhuis AW. Purification of the human NF-E2 complex: cDNA cloning of the hematopoietic cell-specific subunit and evidence for an associated partner. Mol Cell Biol. 1993;13:5604–5612. doi: 10.1128/mcb.13.9.5604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira FA, Qiu Y, Tsai MJ, Tsai SY. Chicken ovalbumin upstream promoter transcription factor (COUP-TF): expression during mouse embryogenesis. J Steroid Biochem Mol Biol. 1995;53:503–508. doi: 10.1016/0960-0760(95)00097-j. [DOI] [PubMed] [Google Scholar]
- Pereira FA, Xu J, Price SD, Shope CD, Brownell WE, Lysakowski A, Tsai M-J. Vestibular and auditory system defects in COUP-TFI mutants. Paper presented at the 24th Annual Mid-Winter Research Meeting of the Association for Research in Otolaryngology; St. Petersburg, FL. 2001. [Google Scholar]
- Qiu Y, Pereira FA, DeMayo FJ, Lydon JP, Tsai SY, Tsai MJ. Null mutation of mCOUPTFI results in defects in morphogenesis of the glossopharyngeal ganglion, axonal projection, and arborization. Genes Dev. 1997;11:1925–1937. doi: 10.1101/gad.11.15.1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley P, Anson-Cartwright L, Cross JC. The Hand1 bHLH transcription factor is essential for placentation and cardiac morphogenesis. Nat Genet. 1998;18:271–275. doi: 10.1038/ng0398-271. [DOI] [PubMed] [Google Scholar]
- Simpson JC, Wellenreuther R, Poustka A, Pepperkok R, Wiemann S. Systematic subcellular localization of novel proteins identified by large-scale cDNA sequencing. EMBO Rep. 2000;1:287–292. doi: 10.1093/embo-reports/kvd058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skvorak AB, Weng Z, Yee AJ, Robertson NG, Morton CC. Human cochlear expressed sequence tags provide insight into cochlear gene expression and identify candidate genes for deafness. Hum Mol Genet. 1999;8:439–452. doi: 10.1093/hmg/8.3.439. [DOI] [PubMed] [Google Scholar]
- Tang LS, Alger HM, Pereira FA. COUP-TFI controls Notch regulation of hair cell and support cell differentiation. Development. 2006;133:3683–3693. doi: 10.1242/dev.02536. [DOI] [PubMed] [Google Scholar]
- Uyttendaele H, Panteleyev AA, de Berker D, Tobin DT, Christiano AM. Activation of Notch1 in the hair follicle leads to cell-fate switch and Mohawk alopecia. Differentiation. 2004;72:396–409. doi: 10.1111/j.1432-0436.2004.07208006.x. [DOI] [PubMed] [Google Scholar]
- Vauclair S, Nicolas M, Barrandon Y, Radtke F. Notch1 is essential for postnatal hair follicle development and homeostasis. Dev Biol. 2005;284:184–193. doi: 10.1016/j.ydbio.2005.05.018. [DOI] [PubMed] [Google Scholar]
- Viiri KM, Korkeamaki H, Kukkonen MK, Nieminen LK, Lindfors K, Peterson P, Maki M, Kainulainen H, Lohi O. SAP30L interacts with members of the Sin3A corepressor complex and targets Sin3A to the nucleolus. Nucleic Acids Res. 2006;34:3288–3298. doi: 10.1093/nar/gkl401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburton D. De novo balanced chromosome rearrangements and extra marker chromosomes identified at prenatal diagnosis: clinical significance and distribution of breakpoints. Am J Hum Genet. 1991;49:995–1013. [PMC free article] [PubMed] [Google Scholar]
- Wenzel GI, Pereira FA, Oghalai JS. Cochlear Dysfunction in COUP-TFI Mutant Mice. Paper presented at The 27th Annual Mid-Winter Research Meeting of the Association for Research in Otolaryngology; Daytona Beach, FL. 2004. [Google Scholar]