Abstract
Antimicrobial peptides (AMPs) form a crucial part of human innate host defense, especially in neutrophil phagosomes and on epithelial surfaces. Bacteria have a variety of efficient resistance mechanisms to human AMPs, such as efflux pumps, secreted proteases, and alterations of the bacterial cell surface that are aimed to minimize attraction of the typically cationic AMPs. In addition, bacteria have specific sensors that activate AMP resistance mechanisms when AMPs are present. AMP resistance mechanisms and AMP sensors may differ significantly between Gram-positive and Gram-negative bacteria. The prototypical Gram-negative PhoP/PhoQ and the Gram-positive Aps AMP sensing systems have been first described and investigated in Salmonella typhimurium and Staphylococcus epidermidis, respectively. Both include a classical bacterial two-component sensor/regulator system, but show many significant structural, mechanistic, and functional differences. At least the staphylococcal Aps prototype contains a third essential component of unknown function termed ApsX. Furthermore, the extracellular loop that interacts with AMPs is extremely short (9 amino acids) in the ApsS sensor in Staphylococcus, while it contains 145 amino acids in PhoQ of Salmonella. Moreover, while the PhoP/PhoQ regulon controls a variety of genes not necessarily limited to AMP resistance mechanisms, but apparently aimed to combat innate host defense on a broad scale, the staphylococcal Aps system predominantly up-regulates AMP resistance mechanisms, namely the D-alanylation of teichoic acids, inclusion of lysyl-phosphatidylglycerol in the cytoplasmic membrane, and expression of the putative VraFG AMP efflux pump. Notably, both systems are crucial for virulence and represent possible targets for antimicrobial therapy.
Introduction: Role of antimicrobial peptides in innate host defense
The first line of defense against invading microorganisms is formed by the human innate immune system. This system reacts swiftly long before the acquired host defense, which is dependent on the production of antibodies, becomes effective. It comprises phagocytes such as neutrophils that recognize invariant structures on the microorganism’s surface (pathogen-associated molecular patterns, PAMPs) and kill microorganisms by the combined activities of reactive oxygen species and antimicrobial proteins and peptides (12, 25). Additionally, antimicrobial peptides (AMPs) are secreted by epithelial cells and other cell types (22). They form an especially important part of host defense on epithelial and mucosal surfaces such as in the gut and on the skin, and contribute significantly to balancing the composition of the colonizing microflora. While AMPs are evolutionarily ancient and in lower organisms often represent the only available mechanism of immune defense, their important role in higher organisms, including humans, has been recognized more recently. In addition to their microbicidal activities, they may also function as signaling molecules, connecting the innate and acquired immune systems by the activation of immune cell types such as T cells (8).
There are three major AMP classes in humans: the defensins, cathelicidins, and thrombocidins. Defensins have a β-sheet structure and 3 disulfide bridges. They are further classified by their disulfide bridging pattern, distinguishing α and β defensins. The only cathelicidin produced in humans is the α-helical LL-37. Finally, while defensins and cathelicidins are produced by a variety of cell types that include neutrophils, the thrombocidins are released from platelets. All these AMPs are cationic (CAMPs), which is a typical feature of most AMPs and believed to have evolved to interact with the negatively charged bacterial surface (48). However, there are also anionic AMPs. For example, the anionic peptide DCD-1L, a proteolytic product of dermcidin, is found in human sweat (52).
Mechanisms of bacterial resistance to cationic antimicrobial peptides
As a consequence of the long interplay between bacteria and other microorganisms with host AMPs during evolution, bacteria have invented a series of mechanisms to combat AMPs (48). These mechanisms can be classified in mechanisms aimed at (1) destruction of AMPs, (2) change of the AMP target to make it less susceptible to AMPs, and (3) removal of AMPs from their site of action.
Many AMPs can be efficiently inactivated via proteolytic digestion by secreted bacterial proteases. However, during evolution hosts have learned to produce AMPs that are less susceptible to this relatively non-specific bacterial mechanism of evading AMP activity. The frequent presence of multiple disulfide bridges in AMPs is believed to be due to providing resistance to proteolytic digestion by bacterial proteases (48).
Most AMPs function by forming pores in the cytoplasmic membrane of bacteria. As membrane integration of the AMP is dependent on the physico-chemical properties of the membrane, resistance can be achieved by altering those properties, for example by altering fluidity of the phospholipid bilayer via introduction of fatty acids with different chain lengths (30).
Mechanisms of AMP removal from the site of action are the most frequently found mechanisms of AMP resistance. They comprise 2 major mechanisms, efflux pumps and alteration of the bacterial cell surface composition. Further, the streptococcal SIC proteins (streptococcal inhibitors of complement) and the Staphylococcus aureus staphylokinase interact specifically with AMPs to prevent them from reaching the cytoplasmic membrane (2, 29).
Efflux pumps such as the MtrCE system of Neisseria gonorrhoeae and QacA of S. aureus work by energy-dependent constant export of toxic substances out of the cell and away from the membrane (21, 51). Although they primarily export hydrophobic drugs, they have relatively low specificity and may also accept a subset of AMPs as substrates (34, 55). However, in the case of S. aureus QacA, it has been shown that resistance to the platelet AMP, tPMP-1 is not mediated by efflux pump activity, exemplifying that real transport phenomena should always be shown by biochemical studies to rule out that low-level resistance is mediated by mere binding effects (6).
Mechanisms based on alteration of the bacterial surface are often differ between Gram-positive and Gram-negative bacteria owing to the different composition of the Gram-positive and Gram-negative cell surface. In the Gram-negative Salmonella typhimurium, AMP resistance is achieved by modification of lipid A either by acylation (PagP) (20) or addition of an aminoarabinose moiety (Pmr system) (18). In the Gram-positive S. aureus, substitution of teichoic acids with D-alanine confers AMP resistance by decreasing the negative net charge of teichoic acids, which leads to diminished attraction of CAMPs (47). This mechanism is catalyzed by enzymes encoded in the dlt locus and contributes to CAMP resistance in many Gram-positive bacteria, for example Streptococcus pneumonia and Listeria monocytogenes (1, 32). Further, formation and integration into the membrane of lysyl-phosphatidylglycerol (L-PG), a phospholipid that is distinguished by a positive net charge not found in other common bacterial membrane phospholipids, equally leads to reduced attraction of CAMPs to the cytoplasmic membrane (46) and inhibits killing by neutrophils (33). The transmembrane enzyme that catalyzes formation and likely integration of L-PG into the membrane, MprF, is present in many bacteria and thus represents a widespread mechanism of AMP resistance. It is important to stress that these latter mechanisms are specific for CAMPs. The presence of anionic AMPs in human sweat likely represents a response of the human innate immune system to circumvent those mechanisms.
Resistance mechanisms against anionic AMPs have only been investigated more recently. Interestingly, in Staphylococcus epidermidis the cationic exopolymer polysaccharide intercellular adhesin (PIA) and the anionic poly-γ-glutamic acid (PGA) have been shown to protect from cationic and anionic AMPs (31, 57, 59). These results indicate that charged bacterial exoploymers do not work exclusively by electrostatic repulsion such as in the cases of teichoic acid D-alanylation. Alternative mechanisms that may explain the findings with PIA and PGA include charge-independent mechanical exclusion and charge-dependent sequestration of AMPs, a mechanism also proposed for example for the cationic antibiotic tobramycin that is sequestered by the anionic Pseudomonas aeruginosa exopolymer alginate (40). Alginate and PIA are typical components of the extracellular matrix in bacterial biofilms, and possibly main contributors to the observed high resistance of biofilms to AMPs.
Finally, some yet poorly understood mechanisms of AMP resistance may be unique to some bacteria. For example, Shigella species appear to inhibit production of LL-37 and human β-defensin 1 in human rectal epithelial cells (28).
The Gram-negative PhoP/PhoQ antimicrobial peptide sensor
The PhoP/PhoQ system is a virulence regulator originally described in S. typhimurium (16, 42). It regulates a series of genes and cellular activities (15), such as Mg2+ transport (mgtA and mgtCB genes) (54), survival in macrophages (mgtCB genes) (53), modification of lipopolysaccharide (LPS, pagP gene)(7, 20), and also activates a further two-component regulatory system, pmrA/pmrB, by activation of pmrD transcription (19). The pmrA/pmrB system also up-regulates genes involved in LPS modification and resistance to polymyxin B (pbgP, pbgE, ugd) (19).
PhoP/PhoQ is a classical and one of the most intensively studied bacterial two-component signal transduction systems (16, 42). PhoQ is the histidine kinase sensor part that is present as a dimer in the bacterial cytoplasmic membrane. It has two transmembrane helices, one extracellular (periplasmic) loop of 145 amino acid length, and a cytoplasmic domain that contains a conserved autophosphorylation and an ATP binding site. Upon stimulation, PhoQ trans-autophosphorylates within the dimer, and a phosphate is transferred to a conserved aspartate residue on PhoP. PhoP is the response regulator DNA-binding protein of the system that interacts with target gene promoters (Fig. 1).
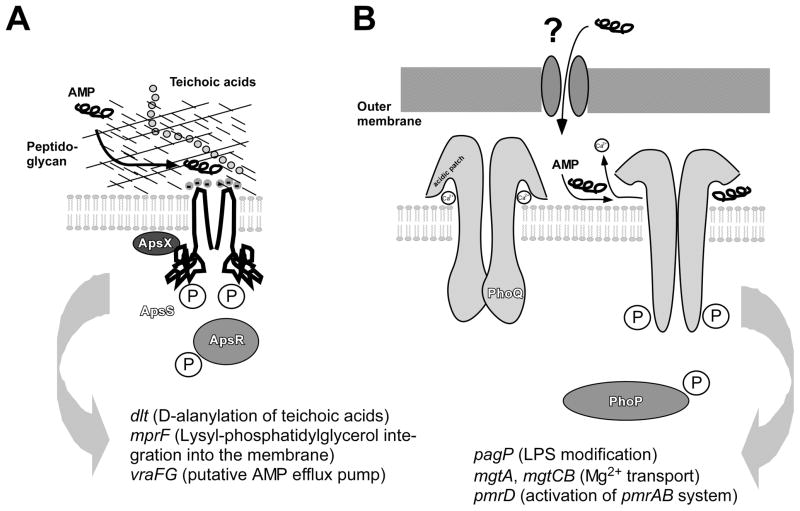
Fig. 1. The Gram-negative and Gram-positive AMP sensors.
A, The Staphylococcus aps system. The Gram-positive cell surface, containing a thick layer of peptidoglycan and anionic teichoic acids, is shown schematically. Binding of a cationic AMP leads to activation of the system in a mechanistically yet poorly understood fashion. The extracellular loop of the ApsS sensor is extremely short, carrying three negative charges that presumably are crucial for the interaction with the cationic AMP. A phosphorylation cascade as typical for two-component systems is believed to activate target gene transcription, with the ApsX protein participating in signal transduction in an unknown way. It is not known whether the ApsS sensor forms a dimer, but dimer formation of sensor proteins is common. B, The Salmonella phoP/phoQ system. Activation by CAMPs, which penetrate the Gram-negative outer membrane in a yet undefined manner, occurs by deplacement of divalent cations in an acidic patch of the PhoQ sensor. The PhoQ acidic region is much larger than the acidic extracellular loop of ApsS. This leads to structural rearrangements including notably two α-helices that move away from the membrane, and ultimately auto-phosphorylation of ApsS and subsequent phosphorylation of PhoP. Main regulatory targets of the systems are shown at the bottom. While both systems control many targets, those involved in CAMP resistance are the dlt, mprF, and vraFG loci in Staphylococcus, and the pagP locus in Salmonella. Further, the pbgPE operon, which is also involved in CAMP resistance by lipid A modification, is regulated by pmrAB, which is under control of phoPQ.
Three stimulants of the PhoQ sensor have been described: low concentration of divalent cations (13), low pH (3), and CAMPs (5). Recent studies have shown that PhoQ is repressed in the presence of divalent cations such as Mg2+ or Ca2+ (5). By solving the crystal structure of PhoQ in the presence of Ca2+, it was found that these divalent cations bind to a set of acidic residues termed the acidic patch that is adjacent to two α-helices (10). Binding of Ca2+ ions alleviates some of the charge repulsion between the acidic patch and the membrane that can be predicted from the crystal structure and which is believed to be important during the activation process. NMR studies have shown that apparently, there is no difference in the structure of PhoQ with Mg2+ versus Ca2+, indicating that the type of divalent ion is not critical for keeping the PhoQ dimer in a repressed state (10).
Removing repression of PhoQ by lowering the concentration of divalent cations in principle would lead to activation of the system. However, this is unlikely to represent a signal that occurs in vivo, because metal ions are abundant in host tissues and fluids. Rather, activation of the PhoP/PhoQ system is believed to be triggered in vivo by low pH and CAMPs. Bader et al. reconstituted PhoQ in membrane vesicles and showed that CAMPs lead to increased levels of phosphorylated PhoP and target gene activation in that system (5). Activation by CAMPs can be competed out by increasing concentrations of Mg2+, indicating that CAMPs and divalent cations bind to the same site in PhoQ. Based on these studies, a model was developed in which CAMPs substitute divalent cations at the acidic patch and owing to their larger size, thereby force two α-helices of the PhoQ protein away from the membrane, ultimately leading to PhoQ activation (5, 49).
Similarly to CAMPs, PhoQ can be activated by low pH. This was also shown using reconstituted PhoQ in membrane vesicles (5). It is important to stress that the effects of pH and CAMPs on PhoQ phosphorylation are additive, suggesting a different mechanism for the interaction of these two signals with the sensor protein. Interestingly, activation by low pH does not involve loss of capacity to bind divalent cations, which would represent a hypothetical mechanism for pH-dependent activation of PhoQ, but leads to increased flexibility of the PhoQ protein. This increased flexibility would allow the two α-helices to move in a model similar to the one proposed for PhoQ activation by CAMPs.
Activation by CAMPs and pH occurs in the presence of ~ 1 mM Mg2+, which is about the divalent cation concentration in a neutrophil phagosome (5). Thus, these two signals are good candidates for stimuli that activate PhoQ in vivo. Further, the additive effects of pH and CAMPs on PhoQ indicate that for maximal activation, both signals need to be present, limiting the expression of PhoQ-regulated CAMP resistance mechanisms to situations that are relevant for pathogen survival in vivo, notably in the phagosome and the acidic intestinal microenvironment around Paneth cells, which secrete the CAMP HD-5 (human α-defensin 5).
While only investigated in detail in Salmonella, PhoQ homologues have been found in a variety of Gram-negative pathogens. In some of those such as Shigella flexneri, Yersinia pestis, the insect pathogen Photorhabdus luminescens, and the plant pathogen Erwinia chrysanthemi, there are results that suggest that PhoQ is also involved in sensing CAMPs and low pH and affects virulence (11, 39, 43, 45, 50). In contrast to these eukaryote-associated bacteria, the acidic patch crucial for CAMP recognition is missing in Pseudomonas aeruginosa, which may cause infection, but is primarily a bacterium found in soil and water. Sensing CAMPs with strongly varying structure is much more essential for eukaryote-associated pathogens, indicating that either the acidic patch in PhoQ has evolved during the interaction with the host, or has been lost in organisms in which CAMP sensing is not as crucial for survival (49).
Despite the evidence outlined so far, the role of the PhoP/PhoQ system in CAMP and pH sensing is debatable. It has been stressed that the PhoP regulon comprises many more genes than those activated by the CAMP polymyxin B (17). Furthermore, PmrA/PmrB appears to interact with PhoP/PhoQ post-translationally via PmrD and is primarily involved in low pH sensing. Finally, it has been argued that most CAMPs are larger than porins in the outer membrane of Gram-negative bacteria and thus, would cause perturbation that may be sensed by other systems before PhoQ can be reached. For example, polymyxin B promoted the expression of genes regulated by RcsB and the alternative sigma factor RpoS. Taken together, it appears that while the interaction of CAMPs with PhoQ has been studied on a very detailed molecular level, there may be more systems involved in CAMP sensing in a complicated regulatory network.
The Gram-positive Aps antimicrobial peptide sensor
While the Gram-negative PhoP/PhoQ system as a regulator of virulence and CAMP resistance has been known for a while and its role in CAMP sensing has been described about 2 years ago (5), a Gram-positive sensor with an equivalent function in CAMP sensing and regulation of Gram-positive CAMP resistance mechanisms has remained elusive. (The PhoR/PhoP system described in some Gram-positive bacteria to respond to inorganic phosphate and control virulence (27) must not be confused with the Gram-negative PhoP/PhoQ sensor.) Recently, during studies aimed to determine genome-wide responses to the human AMP human β-defensin 3 (hBD3) in the nosocomial pathogen and skin colonizer S. epidermidis, using transcriptional profiling with microarrays, my group identified a Gram-positive CAMP sensor/gene regulator (38). The AMP human β-defensin 3 (hBD3), the only β-defensin that maintains anti-staphylococcal activity at physiological salt concentration as found in the natural habitat of skin-colonizing staphylococci (24), induced expression of the major staphylococcal mechanisms involved in CAMP resistance: the dlt operon for D-alanylation of teichoic acids and the mprF gene for formation of L-PG. In addition, several transporters were up-regulated that may function to remove CAMP from the cytoplasmic membrane. Exceptionally strong up-regulation was detected for the VraFG transporter, previously described to confer resistance to the glycopeptide antibiotic vancomycin (35). The vraFG genes are located in the S. epidermidis and S. aureus genome adjacent to an apparent operon consisting of genes coding for a two-component system and a third gene without striking similarity to genes with known function. Based on the fact that in bacteria, regulatory systems are often involved in the regulation of adjacent genes, we constructed deletion mutants in each of the three genes and characterized genome-wide changes in gene expression compared to the wild-type strain. The results showed that this system, which we termed aps for antimicrobial peptide sensor, regulates the major CAMP resistance mechanisms, dlt and mprF, in addition to vraFG. Notably, all three aps components were essential for gene regulation, indicating that this system represents an unusual three-component regulatory system consisting of a classical two-component system with a sensor histidine kinase and a DNA-binding response regulator, whereas the third component has a yet undefined role in signal transduction. The apsS and apsR genes have also been called graS and graR due to their involvement in resistance to glycopeptide antibiotics (41, 44). We suggest using the aps name, because it is based on the presumably natural role of this system and publications on graRS have failed to recognize the three-component nature of the regulatory system.
Similar to results achieved in Salmonella with the PhoP/PhoQ system (15), analysis of the aps regulon using deletion mutants revealed genes that were not found to be under regulation of CAMPs (38). In addition, differential expression of autolysins was observed in all three aps gene deletion mutants. However, it needs to be stressed that the deletion of a gene represents an unnatural situation that is not likely to be achieved in this extreme fashion by differential phosphorylation of the ApsS/ApsR/(ApsX), or the PhoP/PhoQ regulatory cascades. Thus, these results do not necessarily contradict the notion that the primary tasks of these systems are the sensing of CAMPs and regulation of CAMP resistance mechanisms.
The ApsS sensor is a transmembrane protein with two predicted transmembrane helices and an extracellular acidic loop, features that are reminiscent of the PhoQ architecture. However, there is no significant sequence similarity and most strikingly, the ApsS extracellular loop is only 9 amino acids long and thus much smaller than the acidic patch of PhoQ (Fig. 1). Interaction of this 9 amino acid loop with CAMPs has been shown using specific antibodies developed against the loop epitope that blocked up-regulation of aps target genes (38).
In contrast to the S. epidermidis aps system, which was activated by any CAMP tested, we found activation of the S. aureus aps system only with specific CAMPs, including indolicidin and LL-37, for example, but not using hBD3 and several other peptides (37). Using complementation vectors that expressed heterologous S. epidermidis or hybrid proteins consisting of the S. aureus apsS gene with an S. epidermidis loop inserted in an apsS deletion mutant of S. aureus, we could demonstrate that substrate specificity of the system is due to amino acid sequence differences in the loop between S. aureus and S. epidermidis. The biological significance of the fact that S. aureus is more discriminatory in accepting only some CAMPs as aps stimuli is not known. Further mechanistic details on AMP sensing by ApsS are not known yet. However, divalent cations do not appear to have a significant AMP-dependent impact on aps-dependent gene regulation (Li and Otto, unpublished data), suggesting that the regulators involved in divalent cation-dependent gene expression are different from aps and the mechanism of ApsS activation differs from that used by PhoQ.
In addition to regulating the dlt, mprF, and vraFG loci (with a somewhat differing relative emphasis compared to S. epidermidis), the aps regulon in S. aureus comprises genes involved in the biosynthesis of lysine (37). This is in accordance with the increased need for lysine when L-PG production via MprF is stimulated. Thus, basic metabolic adaptation supports the specific up-regulation of CAMP resistance mechanisms during stimulation of the aps regulon by CAMPs. It needs to be stressed that these results achieved in the hyper-virulent community-associated MRSA strain strain MW2 (4, 56) are contradictory to those achieved by other authors, who did not find the mprF and lysine biosynthesis genes in the graRS regulon of S. aureus strain SA113 (26), which is a mutant in the global regulatory system agr (58). Thus, there might be strain-specific differences in the aps regulon in S. aureus.
Sensing CAMPs and responding with dedicated resistance mechanisms is supposed to be important for staphylococcal virulence. In a peritoneal mouse infection model, increased cfu counts of the wild-type strain in comparison to the apsS deletion mutant strain in the kidneys indicated a role for the aps system in S. aureus infection (37).
Homologues of the aps system are found in a multitude of Gram-positive pathogens, including Clostridium difficile, Listeria monocytogenes, Bacillus anthracis, S. haemolyticus, and Streptococcus pneumonia (38). Interestingly, homologues of the ApsX third component are only found in staphylococcal species. Thus, determining the role of ApsX in the staphylococcal Aps system and those of the ApsS homologues in other bacteria will be an important task for future research.
Anionic antimicrobial peptides and non-specific sensing
The recently discovered human anionic AMP dermcidin does not activate the aps system due to its negative net charge and thus represents a valuable means to investigate gene regulatory responses to an AMP not mediated via that sensor (38). In S. epidermidis, dermcidin (in its proteolytically processed, active form called DCD-1) led to an up-regulation of the gene coding for the secreted protease SepA, and increased extracellular proteolytic activity (36). SepA had a major role in degrading DCD-1, while it only had marginal effects on hBD3 and LL-37, which may be explained by the secondary and tertiary structures of these AMPs. Global regulatory systems, namely the agr, saeRS, and sarA regulators were changed in a fashion to promote increased production of secreted proteases including SepA. Notably, these regulatory changes were also achieved by incubation with hBD3, indicating that they are independent of AMP charge. Interestingly, it has been found recently that the saeRS system responds to a series of phagocyte-derived signals (14) in addition to AMPs, suggesting a key role in the regulation of immune evasion mechanisms in S. aureus. These results indicate that while anionic AMPs may be interpreted as an adaptation of human innate host defense to bacterial AMP resistance mechanisms specific for CAMPs, bacteria still have less specific ways to combat those peptides, because the respective sensing mechanisms do not discriminate between positively and negatively charged AMPs. While the detailed mechanism of the staphylococcal adaptive response to dermcidin and other AMPs in an aps-independent manner needs to be further analyzed, the involvement of global regulatory systems is reminiscent of the Gram-negative rpoS-dependent response to polymyxin B and indicates a broad gene-regulatory response most likely caused by membrane perturbations.
A potential target for antimicrobial therapeutics?
In a time of desperate search for new antimicrobial agents and targets, it has been frequently proposed to develop antimicrobial therapeutics based on AMPs (23). While the commonly bactericidal mode of action of AMPs is a clear advantage, pre-existing bacterial resistance mechanisms and AMP sensors such as those discussed herein, in addition to the sensitivity of AMPs to proteolytic digestion, make this approach appear problematic. Nevertheless, due to the empty pipeline in antibiotic development and strongly increasing antibiotic resistance in premier pathogens such as S. aureus, a closer look at AMPs as potential therapeutics is certainly warranted. In that context, targeting AMP sensor mechanisms involved in regulating bacterial resistance to AMPs may form a valuable part (9). While the therapeutic benefit from targeting a regulatory system is debatable for many reasons, the fact that AMP sensors are fairly conserved among Gram-negative or Gram-positive bacteria is encouraging in that regard.
Acknowledgments
This work was supported by the Intramural Program of the National Institute of Allergy and Infectious Diseases, NIH.
References
- 1.Abachin E, Poyart C, Pellegrini E, Milohanic E, Fiedler F, Berche P, Trieu-Cuot P. Formation of D-alanyl-lipoteichoic acid is required for adhesion and virulence of Listeria monocytogenes. Mol Microbiol. 2002;43:1–14. doi: 10.1046/j.1365-2958.2002.02723.x. [DOI] [PubMed] [Google Scholar]
- 2.Akesson P, Sjoholm AG, Bjorck L. Protein SIC, a novel extracellular protein of Streptococcus pyogenes interfering with complement function. J Biol Chem. 1996;271:1081–8. doi: 10.1074/jbc.271.2.1081. [DOI] [PubMed] [Google Scholar]
- 3.Alpuche Aranda CM, Swanson JA, Loomis WP, Miller SI. Salmonella typhimurium activates virulence gene transcription within acidified macrophage phagosomes. Proc Natl Acad Sci U S A. 1992;89:10079–83. doi: 10.1073/pnas.89.21.10079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baba T, Takeuchi F, Kuroda M, Yuzawa H, Aoki K, Oguchi A, Nagai Y, Iwama N, Asano K, Naimi T, Kuroda H, Cui L, Yamamoto K, Hiramatsu K. Genome and virulence determinants of high virulence community-acquired MRSA. Lancet. 2002;359:1819–27. doi: 10.1016/s0140-6736(02)08713-5. [DOI] [PubMed] [Google Scholar]
- 5.Bader MW, Sanowar S, Daley ME, Schneider AR, Cho U, Xu W, Klevit RE, Le Moual H, Miller SI. Recognition of antimicrobial peptides by a bacterial sensor kinase. Cell. 2005;122:461–72. doi: 10.1016/j.cell.2005.05.030. [DOI] [PubMed] [Google Scholar]
- 6.Bayer AS, Kupferwasser LI, Brown MH, Skurray RA, Grkovic S, Jones T, Mukhopadhay K, Yeaman MR. Low-level resistance of Staphylococcus aureus to thrombin-induced platelet microbicidal protein 1 in vitro associated with qacA gene carriage is independent of multidrug efflux pump activity. Antimicrob Agents Chemother. 2006;50:2448–54. doi: 10.1128/AAC.00028-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Belden WJ, Miller SI. Further characterization of the PhoP regulon: identification of new PhoP-activated virulence loci. Infect Immun. 1994;62:5095–101. doi: 10.1128/iai.62.11.5095-5101.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bowdish DM, Davidson DJ, Scott MG, Hancock RE. Immunomodulatory activities of small host defense peptides. Antimicrob Agents Chemother. 2005;49:1727–32. doi: 10.1128/AAC.49.5.1727-1732.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brodsky IE, Gunn JS. A bacterial sensory system that activates resistance to innate immune defenses: potential targets for antimicrobial therapeutics. Mol Interv. 2005;5:335–7. doi: 10.1124/mi.5.6.4. [DOI] [PubMed] [Google Scholar]
- 10.Cho US, Bader MW, Amaya MF, Daley ME, Klevit RE, Miller SI, Xu W. Metal bridges between the PhoQ sensor domain and the membrane regulate transmembrane signaling. J Mol Biol. 2006;356:1193–206. doi: 10.1016/j.jmb.2005.12.032. [DOI] [PubMed] [Google Scholar]
- 11.Derzelle S, Turlin E, Duchaud E, Pages S, Kunst F, Givaudan A, Danchin A. The PhoP-PhoQ two-component regulatory system of Photorhabdus luminescens is essential for virulence in insects. J Bacteriol. 2004;186:1270–9. doi: 10.1128/JB.186.5.1270-1279.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Faurschou M, Borregaard N. Neutrophil granules and secretory vesicles in inflammation. Microbes Infect. 2003;5:1317–27. doi: 10.1016/j.micinf.2003.09.008. [DOI] [PubMed] [Google Scholar]
- 13.Garcia Vescovi E, Soncini FC, Groisman EA. Mg2+ as an extracellular signal: environmental regulation of Salmonella virulence. Cell. 1996;84:165–74. doi: 10.1016/s0092-8674(00)81003-x. [DOI] [PubMed] [Google Scholar]
- 14.Geiger T, Goerke C, Mainiero M, Kraus D, Wolz C. The Virulence Regulator Sae of Staphylococcus aureus: Promoter Activities and Response to Phagocytosis-Related Signals. J Bacteriol. 2008 doi: 10.1128/JB.01927-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Groisman EA. The pleiotropic two-component regulatory system PhoP-PhoQ. J Bacteriol. 2001;183:1835–42. doi: 10.1128/JB.183.6.1835-1842.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Groisman EA, Chiao E, Lipps CJ, Heffron F. Salmonella typhimurium phoP virulence gene is a transcriptional regulator. Proc Natl Acad Sci U S A. 1989;86:7077–81. doi: 10.1073/pnas.86.18.7077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Groisman EA, Mouslim C. Sensing by bacterial regulatory systems in host and non-host environments. Nat Rev Microbiol. 2006;4:705–9. doi: 10.1038/nrmicro1478. [DOI] [PubMed] [Google Scholar]
- 18.Gunn JS, Lim KB, Krueger J, Kim K, Guo L, Hackett M, Miller SI. PmrA-PmrB-regulated genes necessary for 4-aminoarabinose lipid A modification and polymyxin resistance. Mol Microbiol. 1998;27:1171–82. doi: 10.1046/j.1365-2958.1998.00757.x. [DOI] [PubMed] [Google Scholar]
- 19.Gunn JS, Miller SI. PhoP-PhoQ activates transcription of pmrAB, encoding a two-component regulatory system involved in Salmonella typhimurium antimicrobial peptide resistance. J Bacteriol. 1996;178:6857–64. doi: 10.1128/jb.178.23.6857-6864.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guo L, Lim KB, Poduje CM, Daniel M, Gunn JS, Hackett M, Miller SI. Lipid A acylation and bacterial resistance against vertebrate antimicrobial peptides. Cell. 1998;95:189–98. doi: 10.1016/s0092-8674(00)81750-x. [DOI] [PubMed] [Google Scholar]
- 21.Hagman KE, Pan W, Spratt BG, Balthazar JT, Judd RC, Shafer WM. Resistance of Neisseria gonorrhoeae to antimicrobial hydrophobic agents is modulated by the mtrRCDE efflux system. Microbiology. 1995;141(Pt 3):611–22. doi: 10.1099/13500872-141-3-611. [DOI] [PubMed] [Google Scholar]
- 22.Hancock RE, Diamond G. The role of cationic antimicrobial peptides in innate host defences. Trends Microbiol. 2000;8:402–10. doi: 10.1016/s0966-842x(00)01823-0. [DOI] [PubMed] [Google Scholar]
- 23.Hancock RE, Sahl HG. Antimicrobial and host-defense peptides as new anti-infective therapeutic strategies. Nat Biotechnol. 2006;24:1551–7. doi: 10.1038/nbt1267. [DOI] [PubMed] [Google Scholar]
- 24.Harder J, Bartels J, Christophers E, Schroder JM. Isolation and characterization of human beta -defensin-3, a novel human inducible peptide antibiotic. J Biol Chem. 2001;276:5707–13. doi: 10.1074/jbc.M008557200. [DOI] [PubMed] [Google Scholar]
- 25.Heine H, Ulmer AJ. Recognition of bacterial products by toll-like receptors. Chem Immunol Allergy. 2005;86:99–119. doi: 10.1159/000086654. [DOI] [PubMed] [Google Scholar]
- 26.Herbert S, Bera A, Nerz C, Kraus D, Peschel A, Goerke C, Meehl M, Cheung A, Gotz F. Molecular basis of resistance to muramidase and cationic antimicrobial peptide activity of lysozyme in staphylococci. PLoS Pathog. 2007;3:e102. doi: 10.1371/journal.ppat.0030102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hulett FM. The signal-transduction network for Pho regulation in Bacillus subtilis. Mol Microbiol. 1996;19:933–9. doi: 10.1046/j.1365-2958.1996.421953.x. [DOI] [PubMed] [Google Scholar]
- 28.Islam D, Bandholtz L, Nilsson J, Wigzell H, Christensson B, Agerberth B, Gudmundsson G. Downregulation of bactericidal peptides in enteric infections: a novel immune escape mechanism with bacterial DNA as a potential regulator. Nat Med. 2001;7:180–5. doi: 10.1038/84627. [DOI] [PubMed] [Google Scholar]
- 29.Jin T, Bokarewa M, Foster T, Mitchell J, Higgins J, Tarkowski A. Staphylococcus aureus resists human defensins by production of staphylokinase, a novel bacterial evasion mechanism. J Immunol. 2004;172:1169–76. doi: 10.4049/jimmunol.172.2.1169. [DOI] [PubMed] [Google Scholar]
- 30.Jing W, Prenner EJ, Vogel HJ, Waring AJ, Lehrer RI, Lohner K. Headgroup structure and fatty acid chain length of the acidic phospholipids modulate the interaction of membrane mimetic vesicles with the antimicrobial peptide protegrin-1. J Pept Sci. 2005;11:735–43. doi: 10.1002/psc.702. [DOI] [PubMed] [Google Scholar]
- 31.Kocianova S, Vuong C, Yao Y, Voyich JM, Fischer ER, DeLeo FR, Otto M. Key role of poly-gamma-DL-glutamic acid in immune evasion and virulence of Staphylococcus epidermidis. J Clin Invest. 2005;115:688–94. doi: 10.1172/JCI23523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kovacs M, Halfmann A, Fedtke I, Heintz M, Peschel A, Vollmer W, Hakenbeck R, Bruckner R. A functional dlt operon, encoding proteins required for incorporation of d-alanine in teichoic acids in gram-positive bacteria, confers resistance to cationic antimicrobial peptides in Streptococcus pneumoniae. J Bacteriol. 2006;188:5797–805. doi: 10.1128/JB.00336-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kristian SA, Durr M, Van Strijp JA, Neumeister B, Peschel A. MprF-mediated lysinylation of phospholipids in Staphylococcus aureus leads to protection against oxygen-independent neutrophil killing. Infect Immun. 2003;71:546–9. doi: 10.1128/IAI.71.1.546-549.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kupferwasser LI, Skurray RA, Brown MH, Firth N, Yeaman MR, Bayer AS. Plasmid-mediated resistance to thrombin-induced platelet microbicidal protein in staphylococci: role of the qacA locus. Antimicrob Agents Chemother. 1999;43:2395–9. doi: 10.1128/aac.43.10.2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kuroda M, Kuwahara-Arai K, Hiramatsu K. Identification of the up- and down-regulated genes in vancomycin-resistant Staphylococcus aureus strains Mu3 and Mu50 by cDNA differential hybridization method. Biochem Biophys Res Commun. 2000;269:485–90. doi: 10.1006/bbrc.2000.2277. [DOI] [PubMed] [Google Scholar]
- 36.Lai Y, Villaruz AE, Li M, Cha DJ, Sturdevant DE, Otto M. The human anionic antimicrobial peptide dermcidin induces proteolytic defence mechanisms in staphylococci. Mol Microbiol. 2007;63:497–506. doi: 10.1111/j.1365-2958.2006.05540.x. [DOI] [PubMed] [Google Scholar]
- 37.Li M, Cha DJ, Lai Y, Villaruz AE, Sturdevant DE, Otto M. The antimicrobial peptide-sensing system aps of Staphylococcus aureus. Mol Microbiol. 2007;66:1136–1147. doi: 10.1111/j.1365-2958.2007.05986.x. [DOI] [PubMed] [Google Scholar]
- 38.Li M, Lai Y, Villaruz AE, Cha DJ, Sturdevant DE, Otto M. Gram-positive three-component antimicrobial peptide-sensing system. Proc Natl Acad Sci U S A. 2007;104:9469–9474. doi: 10.1073/pnas.0702159104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Llama-Palacios A, Lopez-Solanilla E, Rodriguez-Palenzuela P. Role of the PhoP-PhoQ system in the virulence of Erwinia chrysanthemi strain 3937: involvement in sensitivity to plant antimicrobial peptides, survival at acid Hh, and regulation of pectolytic enzymes. J Bacteriol. 2005;187:2157–62. doi: 10.1128/JB.187.6.2157-2162.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mah TF, Pitts B, Pellock B, Walker GC, Stewart PS, O’Toole GA. A genetic basis for Pseudomonas aeruginosa biofilm antibiotic resistance. Nature. 2003;426:306–10. doi: 10.1038/nature02122. [DOI] [PubMed] [Google Scholar]
- 41.Meehl M, Herbert S, Gotz F, Cheung A. Interaction of the GraRS two-component system with the VraFG ABC transporter to support vancomycin-intermediate resistance in Staphylococcus aureus. Antimicrob Agents Chemother. 2007;51:2679–89. doi: 10.1128/AAC.00209-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miller SI, Kukral AM, Mekalanos JJ. A two-component regulatory system (phoP phoQ) controls Salmonella typhimurium virulence. Proc Natl Acad Sci U S A. 1989;86:5054–8. doi: 10.1073/pnas.86.13.5054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moss JE, Fisher PE, Vick B, Groisman EA, Zychlinsky A. The regulatory protein PhoP controls susceptibility to the host inflammatory response in Shigella flexneri. Cell Microbiol. 2000;2:443–52. doi: 10.1046/j.1462-5822.2000.00065.x. [DOI] [PubMed] [Google Scholar]
- 44.Neoh HM, Cui L, Yuzawa H, Takeuchi F, Matsuo M, Hiramatsu K. Mutated Response Regulator graR Is Responsible for Phenotypic Conversion of Staphylococcus aureus from Heterogeneous Vancomycin-Intermediate Resistance to Vancomycin-Intermediate Resistance. Antimicrob Agents Chemother. 2008;52:45–53. doi: 10.1128/AAC.00534-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Oyston PC, Dorrell N, Williams K, Li SR, Green M, Titball RW, Wren BW. The response regulator PhoP is important for survival under conditions of macrophage-induced stress and virulence in Yersinia pestis. Infect Immun. 2000;68:3419–25. doi: 10.1128/iai.68.6.3419-3425.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Peschel A, Jack RW, Otto M, Collins LV, Staubitz P, Nicholson G, Kalbacher H, Nieuwenhuizen WF, Jung G, Tarkowski A, van Kessel KP, van Strijp JA. Staphylococcus aureus resistance to human defensins and evasion of neutrophil killing via the novel virulence factor MprF is based on modification of membrane lipids with l-lysine. J Exp Med. 2001;193:1067–76. doi: 10.1084/jem.193.9.1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Peschel A, Otto M, Jack RW, Kalbacher H, Jung G, Gotz F. Inactivation of the dlt operon in Staphylococcus aureus confers sensitivity to defensins, protegrins, and other antimicrobial peptides. J Biol Chem. 1999;274:8405–10. doi: 10.1074/jbc.274.13.8405. [DOI] [PubMed] [Google Scholar]
- 48.Peschel A, Sahl HG. The co-evolution of host cationic antimicrobial peptides and microbial resistance. Nat Rev Microbiol. 2006;4:529–36. doi: 10.1038/nrmicro1441. [DOI] [PubMed] [Google Scholar]
- 49.Prost LR, Miller SI. The Salmonellae PhoQ sensor: mechanisms of detection of phagosome signals. Cell Microbiol. 2008;10:576–82. doi: 10.1111/j.1462-5822.2007.01111.x. [DOI] [PubMed] [Google Scholar]
- 50.Rebeil R, Ernst RK, Gowen BB, Miller SI, Hinnebusch BJ. Variation in lipid A structure in the pathogenic yersiniae. Mol Microbiol. 2004;52:1363–73. doi: 10.1111/j.1365-2958.2004.04059.x. [DOI] [PubMed] [Google Scholar]
- 51.Rouch DA, Cram DS, DiBerardino D, Littlejohn TG, Skurray RA. Efflux-mediated antiseptic resistance gene qacA from Staphylococcus aureus: common ancestry with tetracycline- and sugar-transport proteins. Mol Microbiol. 1990;4:2051–62. doi: 10.1111/j.1365-2958.1990.tb00565.x. [DOI] [PubMed] [Google Scholar]
- 52.Schittek B, Hipfel R, Sauer B, Bauer J, Kalbacher H, Stevanovic S, Schirle M, Schroeder K, Blin N, Meier F, Rassner G, Garbe C. Dermcidin: a novel human antibiotic peptide secreted by sweat glands. Nat Immunol. 2001;2:1133–7. doi: 10.1038/ni732. [DOI] [PubMed] [Google Scholar]
- 53.Smith RL, Kaczmarek MT, Kucharski LM, Maguire ME. Magnesium transport in Salmonella typhimurium: regulation of mgtA and mgtCB during invasion of epithelial and macrophage cells. Microbiology. 1998;144(Pt 7):1835–43. doi: 10.1099/00221287-144-7-1835. [DOI] [PubMed] [Google Scholar]
- 54.Soncini FC, Garcia Vescovi E, Solomon F, Groisman EA. Molecular basis of the magnesium deprivation response in Salmonella typhimurium: identification of PhoP-regulated genes. J Bacteriol. 1996;178:5092–9. doi: 10.1128/jb.178.17.5092-5099.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tzeng YL, Ambrose KD, Zughaier S, Zhou X, Miller YK, Shafer WM, Stephens DS. Cationic antimicrobial peptide resistance in Neisseria meningitidis. J Bacteriol. 2005;187:5387–96. doi: 10.1128/JB.187.15.5387-5396.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Voyich JM, Braughton KR, Sturdevant DE, Whitney AR, Said-Salim B, Porcella SF, Long RD, Dorward DW, Gardner DJ, Kreiswirth BN, Musser JM, DeLeo FR. Insights into mechanisms used by Staphylococcus aureus to avoid destruction by human neutrophils. J Immunol. 2005;175:3907–19. doi: 10.4049/jimmunol.175.6.3907. [DOI] [PubMed] [Google Scholar]
- 57.Vuong C, Kocianova S, Voyich JM, Yao Y, Fischer ER, DeLeo FR, Otto M. A crucial role for exopolysaccharide modification in bacterial biofilm formation, immune evasion, and virulence. J Biol Chem. 2004;279:54881–6. doi: 10.1074/jbc.M411374200. [DOI] [PubMed] [Google Scholar]
- 58.Vuong C, Saenz HL, Gotz F, Otto M. Impact of the agr quorum-sensing system on adherence to polystyrene in Staphylococcus aureus. J Infect Dis. 2000;182:1688–93. doi: 10.1086/317606. [DOI] [PubMed] [Google Scholar]
- 59.Vuong C, Voyich JM, Fischer ER, Braughton KR, Whitney AR, DeLeo FR, Otto M. Polysaccharide intercellular adhesin (PIA) protects Staphylococcus epidermidis against major components of the human innate immune system. Cell Microbiol. 2004;6:269–75. doi: 10.1046/j.1462-5822.2004.00367.x. [DOI] [PubMed] [Google Scholar]