Abstract
The role of Panton-Valentine leukocidin (PVL) in Staphylococcus aureus pathogenesis is controversial. Here, we show that an unintended point mutation in the agr P2 promoter of S. aureus caused the phenotypes in gene regulation and murine pneumonia attributed to PVL by Labandeira-Rey et al. (Science 315:1130-3, 2007). In agreement with previous studies that failed to detect similar effects of PVL using community-associated methicillin-resistant S. aureus strains, we found no significant impact of PVL on gene expression or pathogenesis after we repaired the mutation. These findings further contribute to the idea that PVL does not have a major impact on S. aureus pathogenesis. Moreover, our results demonstrate that a single nucleotide polymorphism in an intergenic region can dramatically impact bacterial physiology and virulence. Finally, our work emphasizes the need to frequently evaluate the integrity of the S. aureus agr locus.
Keywords: Staphylococcus aureus, MRSA, Community-associated MRSA, Panton-Valentine Leukocidin, agr
Introduction
Methicillin-resistant Staphylococcus aureus (MRSA) is a major problem worldwide and kills more people annually in the United States than AIDS [1]. This is in part due to the evolving epidemic of community-associated MRSA (CA-MRSA), which are also the leading cause of skin and soft tissue infections reporting to the emergency department in the United States [2]. However, the molecular determinants responsible for the ability of CA-MRSA strains to spread and infect otherwise healthy people outside of the hospital setting have remained poorly defined [3]. Cytolytic peptides, the α-type phenol-soluble modulins (PSMs), and α-toxin have a crucial impact on the development of CA-MRSA disease in mice [4, 5]. However, these two virulence factors are not exclusively produced in CA-MRSA. Possibly, differences in expression of these genes can partially explain the enhanced virulence of CA-MRSA strains [4]. In contrast, many CA-MRSA strains have the lukSF-PV genes encoding the Panton-Valentine leukocidin (PVL), while lukSF-PV are present less frequently in other strains [3]. Therefore, PVL has been suspected to represent a major factor contributing to CA-MRSA virulence, and a correlation between presence of the lukSF-PV genes and specific types of skin and soft tissue infections has been noted previously [6].
Several recent studies have investigated the role of PVL in S. aureus pathogenesis using animal infection models. Using isogenic lukSF-PV deletion mutants in the most prominent CA-MRSA strains, Voyich et al. and Bubeck Wardenburg et al. found no difference in murine skin and soft tissue, bacteremia, and pneumonia models compared with the corresponding parental strains. Based upon these results, the authors concluded that PVL is not a major virulence factor in CA-MRSA disease [5, 7]. Diep et al. found only a transient positive effect of PVL in a CA-MRSA bacteremia model in rabbits [8]. This effect was limited to colonization of the kidneys and may be explained by PVL initially priming the host innate immune system.
In contrast, Labandeira-Rey et al. reported that PVL is a significant factor contributing to murine pneumonia [9]. Of note, by using laboratory strains of S. aureus with the lukSF-PV genes introduced on the PVL-encoding phage ϕSLT, Labandeira-Rey et al. [9] did not directly address the function of PVL in CA-MRSA, but tested its role solely as an S. aureus virulence factor in general. Subsequently, Bubeck Wardenburg et al. ruled out the possibility that the differential results with regard to the role of PVL in murine pneumonia were due to the use of different strains of mice [10].
In addition to the murine pneumonia results, Labandeira-Rey et al. reported a global gene regulatory effect of PVL reminiscent of that caused by the accessory gene regulator (agr) in S. aureus [9, 11]. In contrast, Diep et al. failed to find a gene regulatory effect of PVL using isogenic lukSF-PV deletion mutants in CA-MRSA strains [8]. Thus, key results obtained by Labandeira-Rey et al. are at variance with those achieved by other groups, which prompted us to re-evaluate the strains used by these authors as a potential source for the different experimental outcomes.
Methods
Bacterial strains and growth conditions
RN6390 is a laboratory S. aureus strain and the parental strain of all other strains used herein (table 1). Strains LUG855, LUG776, and LUG862 were kindly provided by M. Gabriela Bowden (Texas A&M University System Health Science Center) [9]. LUG855 contains the ϕSLT phage carrying among other genes the lukSF genes encoding PVL [9]; LUG776 contains ϕSLT in which the lukSF genes were deleted [9]; LUG862 is LUG776 carrying a plasmid with the lukSF genes (pLUG534) [9] and was grown with addition of 10 μg/ml chloramphenicol. All strains were kept in glycerol stocks after being received and were not passaged. Tryptic soy broth (TSB) was used for all experiments.
Table 1.
Strains used in this study1.
| Strain | Description | agr | lukSF (encoding PVL) | Reference |
|---|---|---|---|---|
| RN6390 | S. aureus laboratory strain | + | − | [16] |
| LUG855 | RN6390 ϕPVL+ | − (point mutation) | + | [9] |
| LUG855r | RN6390*ϕPVL+ | + (point mutation repaired) | + | This study |
| LUG776 | RN6390 ϕPVL− | + | − (inactivated on phage) | [9] |
| LUG862 | RN6390 ϕPVL− pPVL+ | + | + (plasmid) | [9] |
1 *, point mutation in agr; ϕPVL+, lysogenic phage SLT carrying lukSF genes; ϕPVL−, lysogenic phage SLT carrying inactivated lukSF genes; pPVL+, plasmid carrying lukSF genes.
Oligonucleotides and quantitative RT-PCR (qRT-PCR)
qRT-PCR was performed in triplicate using gyrB RNA as a control as described [4]. All oligonucleotides were synthesized by Sigma (table 2).
Table 2.
Oligonucleotides used in this study.
| Name | Sequence |
|---|---|
| For allelic replacement: | |
| GBattl | GGGGACAAGTTTGTACAAAAAAGCAGGCTGCCGCGAGCTTGGGAGGGGCTCACGACC |
| GBatt2 | GGGGACCACTTTGTACAAGAAAGCTGGGTGTTAATATGATTAAGGACGCGCTATCAAAC |
| For EMSAs: | |
| P2 | TACATTTAACAGTTAAGTATTTATTTCCTACAGTTAGGCAATATAATG |
| P2 Comp | CATTATATTGCCTAACTGTAGGAAATAAATACTTAACTGTTAAATGTA |
| P2 Mut | TACATTTAATAGTTAAGTATTTATTTCCTACAGTTAGGCAATATAATG |
| P2 MutComp | CATTATATTGCCTAACTGTAGGAAATAAATACTTAACTATTAAATGTA |
| For qRT-PCR: | |
| psmα | |
| Pl | TATCAAAAGCTTAATCGAACAATTC |
| P2 | CCCCTTCAAATAAGATGTTCATATC |
| Probe | AAAGACCTCCTTTGTTTGTTATGAAATCTTATTTACCAG |
| RNAIII | |
| Pl | GTGATGGAAAATAGTTGATGAGTTGTTT |
| P2 | GAATTTGTTCACTGTGTCGATAATCC |
| Probe | TGCACAAGATATCATTTCAACAATCAGTGACTTAGTAAAA |
| clfB | |
| Pl | TTCCAATGCGCAAGGAACTAG |
| P2 | CAGCATTTACTACAGGTTCAGCAACT |
| Probe | AGACTACGTACAGCTCTCGTTCTAACACTT |
| sdrD | |
| Pl | TCAGATGAGCAAGCTTCACCAA |
| P2 | TTGGTTGAGCATTTACCACTGATT |
| Probe | ATTCTCTTGCAAATCAGGTTGTAACGCTTCTTG |
| hla | |
| Pl | AAAAAACTGCTAGTTATTAGAACGAAAGG |
| P2 | GGCCAGGCTAAACCACTTTTG |
| Probe | CCTTCTTCGCTATAAACTCTATATTGACCAGCAAT |
| spa | |
| Pl | CAGCAAACCATGCAGATGCTA |
| P2 | GCTAATGATAATCCACCAAATACAGTTG |
| Probe | CATTACCAGAAACTGGTGAAGAAAATCCATTCATTG |
| For agr sequencing: | |
| 2803-3859F | GAATCCGCAGATATTTTGACTGTA |
| 2803-3859R | CCTCACTGTCATTATACGATTTAGT |
| 3653-4609F | CACATCTCTGTGATCTAGTTAT |
| 3653-4609R | TCCACCTACTATCACACTCT |
| 4351-5315F | AAAGAGCCATTTGCCCAATT |
| 4351-5315R | GAACTTGCGCATTTCGTTGT |
| 5108-6146F | TTCACAAATAAACTCGGATG |
| 5108-6146R | CTTACGAATTTCACTGCCTA |
| 5924-6927F | TTTCATTTGCGAAGACGATC |
| 5924-6927R | GATTCACGGAGTAGGAAATT |
| 6701-7710F | CAAATGCACTGTATAGCTGGCTT |
| 6701-7710R | GAATGAAGCAAACACTGCGT |
Immunoblotting
Immunoblots were performed using 10% Tris/glycine SDS-PAGE gels and blotting on nitrocellulose. Horse radish peroxidase conjugates were used as second antibodies and immune reactions were made visible using enhanced chemiluminescence.
DNA sequencing and manipulation
DNA encompassing the entire agr locus was sequenced in all strains at the Genomics Unit, Research Technologies Branch, NIAID. The mutation identified in LUG855 was repaired using the allelic replacement method with plasmid pKOR1 as described [12]. To that end, a ~ 1.6 kb fragment containing the site of the mutation was amplified from RN6390 genomic DNA using primers GBatt1 and GBatt2 introducing att1 and att2 sites, respectively, and cloned in pKOR1. The resulting plasmid was used for allelic replacement. The entire agr system of the resulting, repaired clone was sequenced and the sequence was found to be exactly the same as in RN6390.
Purification of AgrA
Purification of AgrA was performed as described previously [13]. Obtained AgrA was analyzed by SDS-PAGE, N-terminal sequencing, and analytical RP-HPLC/ESIMS, and found to be correct and pure.
Electrophoretic mobility shift assays (EMSAs)
EMSAs were performed as previously described [13]. Final DNA concentrations were 25 pM. Binding reactions were performed in 20 μl, incubated at room temperature for 30 min and run on an 8% non-denaturing 0.5 × TBE gel.
Measurement of δ-toxin concentration
Determination of δ-toxin production in S. aureus culture filtrates was performed in triplicate using RP-HPLC/ESI-MS as described [4].
Murine pneumonia model
For murine lung infections examining weight loss, lung CFU recovery and histopathology, S. aureus strains were prepared as described by Labandeira-Rey et al. [9]. Briefly, overnight cultures grown in TSB were refreshed 1:100 in media and grown with shaking to an OD600 of 1.0. Bacteria were sedimented by centrifugation, washed in PBS, and suspended at a concentration of 4-6×107 CFUs per 20-μl volume of PBS for intranasal inoculation. 7 week old mice (BALB/cAnNHsd) were anesthetized prior to inoculation of the S. aureus suspension into the left nare as previously described [14]. Microbiologic and pathologic correlates of disease were assessed 48 hours post-infection, also as previously described [14]. Animal experiments were reviewed, approved and supervised by the IACUC at the University of Chicago.
RNA isolation, transcriptional profiling, and quantitative RT-PCR
RNA isolation and cDNA preparation from cultures grown to early stationary growth phase (7 h) were performed as previously described [15]. Biotinylated S. aureus cDNA was hybridized to custom Affymetrix GeneChips (RMLChip 7) with 100% coverage of chromosomal genes from USA300 and scanned according to standard GeneChip protocols (Affymetrix). Each experiment was replicated 3 times. Affymetrix GeneChip Operating Software (GCOS v1.4, http://www.affymetrix.com) was used to perform the preliminary analysis of the custom GeneChips at the probe-set level. Subsequent data analysis was performed as described [15]. Briefly, each comparison gene list was filtered first using the 0.05 significance level, false discovery rate corrected p-values resulting from a two-way ANOVA. Second, a filter was placed upon fold change (2X), then call consistency and finally signal above background to produce the final gene list for each comparison. The complete set of microarray data was deposited in NCBIs Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/) and is accessible through GEO Series accession number GSE14394.
Results
The hemolysis-negative phenotype of LUG855 cannot be complemented by lukSF-PV
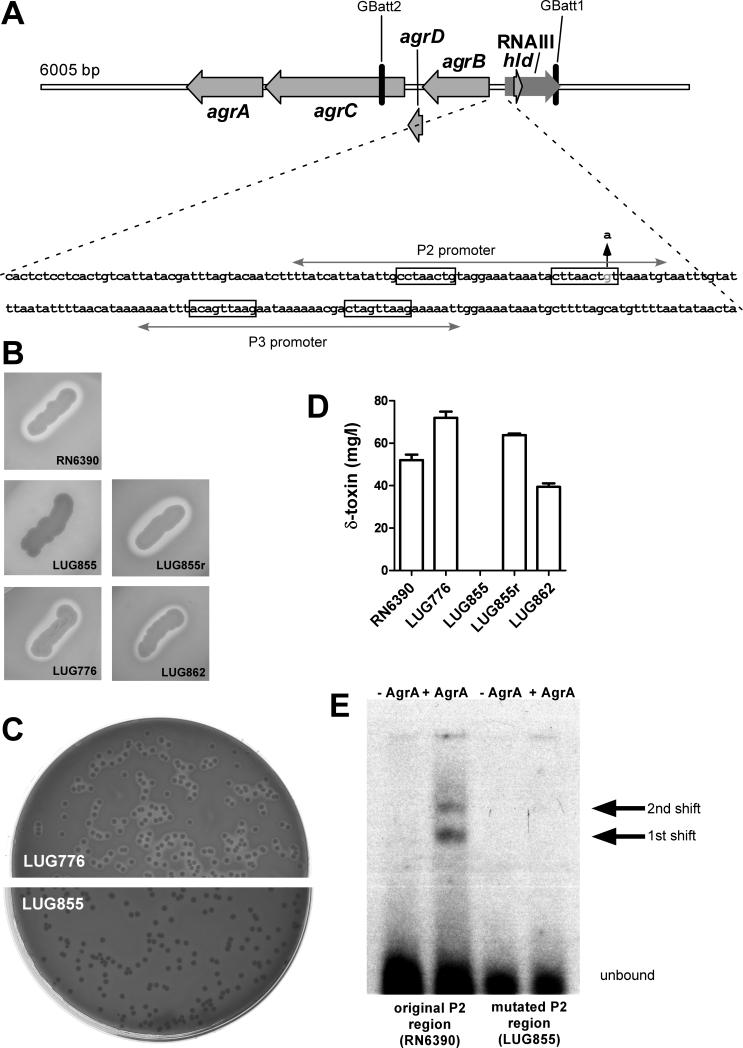
Strains LUG855 and LUG776 (table 1), represent the only isogenic strain pair compared in the Labandeira-Rey et al. study [9]. Therefore, we focused our evaluation on those strains, the respective complementation strain LUG862, and the background strain RN6390. First, we determined β-hemolysis, which is dependent on simultaneous expression of δ-toxin and other agr-regulated toxins, and thus a common and simple readout for agr functionality. While LUG855 was non-β-hemolytic, both LUG776 and LUG862 strains were β-hemolytic, even though the lukSF genes were re-introduced into LUG862 (figure 1B). Restoration of a β-hemolysis-negative phenotype in LUG862 would be expected if PVL expression were responsible for the β-hemolysis-negative phenotype of LUG855. Notably, all tested cultures showed a consistent β-hemolysis phenotype, indicating that there were no contaminations or mixed cultures (shown for LUG776 and LUG855 in figure 1C). Furthermore, we determined δ-toxin production by HPLC/MS (figure 1D). Production of this toxin is a direct readout of agr functionality, as the δ-toxin encoding gene hld is embedded within the region encoding the intracellular effector molecule of agr, RNAIII [16]. The results confirmed those achieved by measuring β-hemolysis, inasmuch as all strains were clearly δ-toxin-positive, whereas LUG855 was δ-toxin-negative. These findings indicated that the β-hemolysis- and δ-toxin-negative phenotype observed in LUG855 could not be restored by genetic complementation, lending support to the hypothesis that the phenotype of LUG855 reported by Labandeira-Rey et al. [9] was not due to PVL.
Figure 1.
Mutation in the accessory gene regulator (agr) system of S. aureus in strain LUG855. (A), The agr locus of LUG855. Locations of primers used for allelic replacement with pKOR1 (GBatt1, GBatt2) are shown. The DNA sequence of the intergenic region between the agrBDCA operon and RNAIII is at the bottom, with locations of the P2 and P3 promoters, the corresponding AgrA consensus binding sequences (in boxes), and the site and nature of the mutation observed in LUG855 (up arrow). (B), hemolysis on sheep blood agar plates of streaked cultures. (C), hemolysis of LUG776 and LUG855 in stock cultures showing a homogenous phenotype. (D), δ-toxin expression by RP-HPLC/ESI-MS. Samples were taken from cultures inoculated from pre-cultures and grown for 7 h. (E) EMSA analysis showing dramatic decrease of AgrA binding to the mutated P2 promoter present in LUG855 compared to the original promoter of RN6390 (same sequence as in LUG776 and the other tested strains except LUG855). AgrA concentration was 10 nMol/L. First and second shifts, as previously observed for AgrA binding by Koenig et al. [17], are marked.
LUG855 contains a point mutation in the agr P2 promoter that strongly impairs binding of AgrA
To test whether the observed phenotypes were rather caused by a spontaneous mutation in the agr locus, we sequenced the entire agr locus in all strains (from position 2092803 to position 2097710 in the genome sequence of S. aureus strain NCTC 8325). The agr sequences of RN6390, LUG776, and LUG862 exactly matched that published for the RN6390 progenitor strain, NCTC 8325 (NC_007795). However, strain LUG855 had a one-base pair mutation (G to A) in the intergenic region between the RNAIII and AgrB encoding regions. The mutation mapped to one of the consensus sequences in the binding site of the AgrA response regulator protein in the P2 promoter [17], which drives transcription of the agrBDCA operon [18] (figure 1A). This auto-regulatory interaction is crucial for agr function. To confirm that this one-base pair mutation affects binding of AgrA to the P2 promoter, we performed EMSAs with purified AgrA protein. We found that AgrA binding to the mutated P2 region of LUG855 was dramatically decreased compared to the corresponding region in LUG776 (figure 1E).
Gene and protein expression analysis of an agr-repaired LUG855 indicates that the LUG855 phenotype is due to the agr mutation and not PVL
To determine whether the identified mutation in the agr locus of LUG855 is responsible for the agr-negative phenotype and differences in gene expression reported by Labandeira-Rey et al. [9], we repaired the mutation in strain LUG855 using the allelic replacement strategy according to Bae and Schneewind [12]. The entire agr system of the resulting clone LUG855r was sequenced and found to be correct, exactly matching that of RN6390 and the other S. aureus strains.
We first determined global gene expression using microarray analysis. Table 3 shows the total number of differentially regulated genes and figure 2 shows selected differentially regulated virulence factors (see GEO, http://www.ncbi.nlm.nih.gov/geo/, GSE14394 for detailed results). Comparing strains LUG855 and LUG776, we confirmed the global changes in gene expression (277 genes differentially-expressed) that are characteristic of genes controlled by agr [11] and were in general described by Labandeira-Rey et al. [9] (figure 2). However, there were only 10 genes up- or down-regulated in the repaired LUG855 compared with LUG776. Several of these 10 differentially-expressed genes were either only slightly above the 2-fold cutoff or represented homologues of lukS-PV or lukF-PV, which may be explained by cross-hybridization. In addition, only expression of spa (encoding protein A) and clpB were considerably different between the strains, an effect that is not easily explained, but may be caused by differential integration of the phage. Notably, the observed change in spa expression level was opposite that described by Labandeira-Rey et al. [9]. In addition, there was very pronounced similarity in genes differentially expressed between LUG855/LUG776 and LUG855/LUG855r comparisons (figure 2), further indicating that the explanation for differential gene expression in LUG776/LUG855 is identical to that in LUG855/LUG855r – namely, the mutation in the agr locus. The results of the LUG855/LUG776 and LUG855/LUG855r comparisons are consistent with the observed decrease in AgrA–P2 promoter interaction (figure 1E); i.e., a pronounced decrease, yet not complete absence of agrA, agrB, agrC, and agrD transcripts in LUG855. Also, these results are in good agreement with the values for agr genes obtained by Labandeira-Rey et al. in their LUG855/LUG776 comparison [9]. The overall results of the gene expression analyses using the complementation strain LUG862 were consistent, as there was a much greater number of differentially expressed genes when comparing this strain to the agr-mutated LUG855 than to LUG855r or LUG776, confirming that the extensive changes in gene expression are due to a difference in agr rather than lukSF-PV (Table 3). Furthermore, this analysis revealed that the comparison of LUG862 with LUG776 or LUG855 by Labandeira-Rey et al. was not appropriate to investigate the impact of PVL on virulence, as our results demonstrate a significant influence of the plasmid background (> 100 differentially expressed genes were identified by comparing LUG862 and LUG855r) and Labandeira-Rey et al. did not compare to a strain with a control plasmid. In summary, our results demonstrate that the global gene regulatory effect described in strain LUG855 by Labandeira-Rey et al. was not caused by PVL. In contrast, our results strongly suggest that they were caused by an unintended mutation in the agr locus of LUG855.
Table 3.
Number of genes with significantly differential expression in microarray experiments.
| Comparison | number of genes passing significance tests |
|---|---|
| 8551 vs 776 | 277 |
| 855 vs 855r | 296 |
| 855r vs 776 | 10 |
| 862 vs 855 | 386 |
| 862 vs 776 | 65 |
| 862 vs 855r | 103 |
| 776 vs 6390 | 26 |
855, LUG855; 855r, LUG855r; 776, LUG776; 6390, RN6390
Figure 2.
Microarray analysis of differential gene expression in LUG776, LUG855, and the agr-repaired LUG855r. Samples for microarray analysis were harvested from cultures grown to early stationary phase (7 h) and prepared as described in Methods. Results for selected genes involved in virulence are shown. The microarray used was based on the genome for USA300 to include the PVL phage in the analysis.
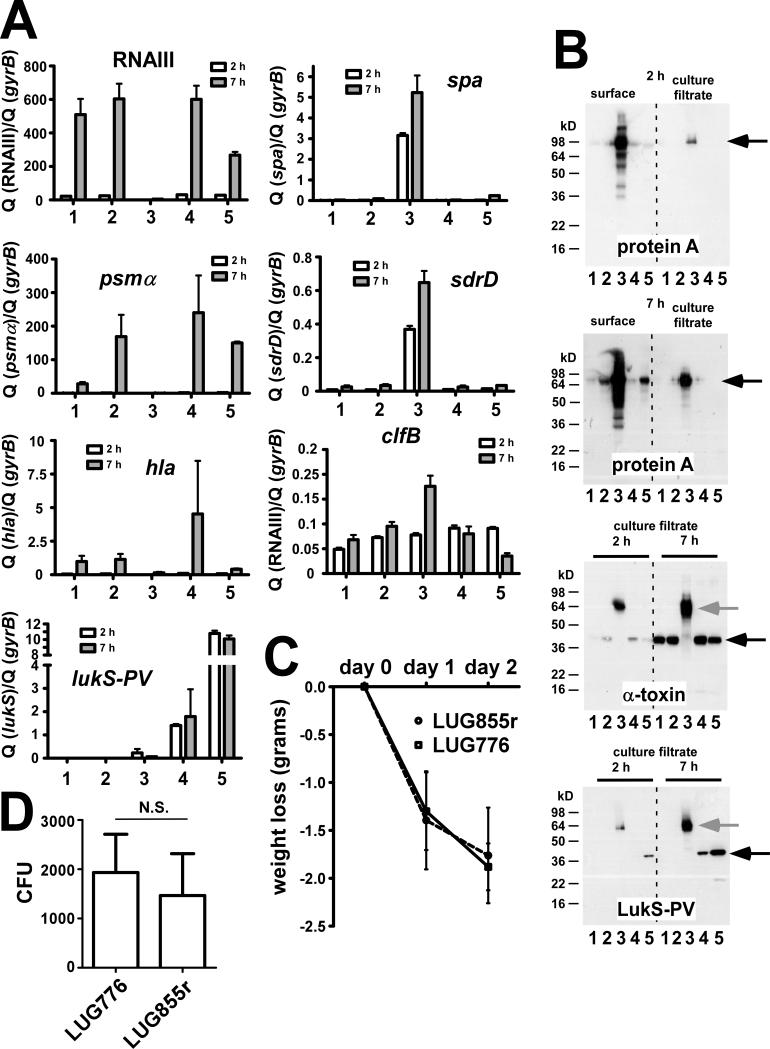
To further confirm this idea, we determined gene expression of major virulence determinants by qRT-PCR (figure 3A). Notably, we analyzed spa encoding protein A that was suggested by Labandeira-Rey et al. to underlie the ability of PVL to promote S. aureus necrotizing pneumonia [9]. Furthermore, we tested transcription of the genes encoding the surface proteins ClfB and SdrD, which were also suggested to contribute to pathogenesis via the reported gene regulatory effect of PVL [9], and α-toxin and α-type PSMs, the only factors determined so far to impact pathogenesis of CA-MRSA [4, 5]. Since the β-hemolysis data suggested that agr was defective in the original unrepaired LUG855 strain, we also tested expression of RNAIII transcripts (figure 1D) as a direct readout for agr activity. Compared to LUG776, there were no significant differences in the levels of these transcripts in the repaired LUG855r strain (figure 3A). Protein expression analyses of α-toxin and protein A using immunoblots confirmed these results on a translational level (figure 3B). These results contrast those of the original, mutated LUG855 strain, which showed a phenotype dramatically different than those of all other strains (figure 3A,B). In agreement with the microarray results, these data further indicate that the agr mutation but not PVL was the underlying cause of the differences reported between LUG855 and LUG776 [9].
Figure 3.
Gene and protein expression of various key virulence factors and mouse pneumonia model. (A), Transcription levels by qRT-PCR. lukS-PV: lukS component of PVL. (B), Immunoblots of protein A, α-toxin, and the LukS component of PVL, using specific antisera (protein A, α-toxin: commercially available from Sigma; LukS-PV: produced by GenScript). Culture filtrate samples were directly used for SDS-PAGE; surface protein samples were obtained by lysostaphin digestion. Black arrow, detected protein, grey arrow, protein A reaction with IgG Fc part. (A,B) 1, RN6390; 2, LUG776; 3, LUG855 (original); 4, LUG855 (repaired); 5, LUG862. (C,D) Animal model of murine pneumonia with strains LUG776 and the isogenic, repaired LUG855r. The model was performed as described by Labandeira-Rey et al. [9], using the same mouse strain, inocula, and experimental conditions. Fifteen mice were used for each strain. Data for weight loss (C) and CFU/g in lung tissue samples (D) are shown. Statistical analysis was by unpaired Student's t-test. Error bars depict SEM.
Additionally, we found that expression of PVL in LUG855 was very low (figure 3A,B), most likely owing to strong agr control of lukSF-PV [13], which was defective in LUG855. These observations provide a possible explanation for our inability to detect a difference in lukSF-PV expression in the microarray experiment that compared LUG776 with LUG855, whereas there was a significant difference in lukSF-PV transcript levels when we compared LUG855r with LUG776, as expected from a correct, repaired LUG855r strain expressing large amounts of PVL. Therefore, the experiments of Labandeira-Rey et al. using LUG855 were based on an S. aureus strain that produced little PVL, further supporting the idea that the phenotypic differences observed in those experiments were not due to PVL.
Use of the repaired LUG855 demonstrates that PVL does not impact murine pneumonia
Our finding demonstrating that the LUG855 strain contained an unintended mutation in agr, which dramatically changed gene expression, suggested that the virulence phenotype described by Labandeira-Rey et al. using the same strain was influenced by the agr mutation and not PVL. To evaluate this hypothesis, we repeated the murine pneumonia model of Labandeira-Rey et al. [9] using the same mouse strain, experimental conditions, and experimental readouts with the repaired LUG855r and corresponding isogenic lukSF-PV-negative LUG776. There was no statistically significant difference in weight loss (figure 3C). Furthermore, the histopathology in all lung sections of mice infected with the 2 strains was similar, inasmuch as the severity of pneumonia was similar in all tissue sections and was interpreted as moderate to severe (data not shown). Moreover, survival rates of mice infected with LUG855r and LUG776 were the same, as all mice survived the experiment. Notably, Labandeira-Rey et al. also failed to detect differences in mouse survival in this model when comparing LUG855 and LUG776 (100% survival for both strains) [9]. They only detected significant differences in survival rates when PVL was over-expressed from a plasmid and using a non-isogenic comparison. In addition, we measured the concentration of CFUs in lung tissue, which were not significantly different between the 2 groups of mice (figure 3D). Together, these findings demonstrate that PVL does not have an effect on S. aureus pathogenesis in this murine model of S. aureus pneumonia.
Introduction of the PVL phage itself impacts gene expression
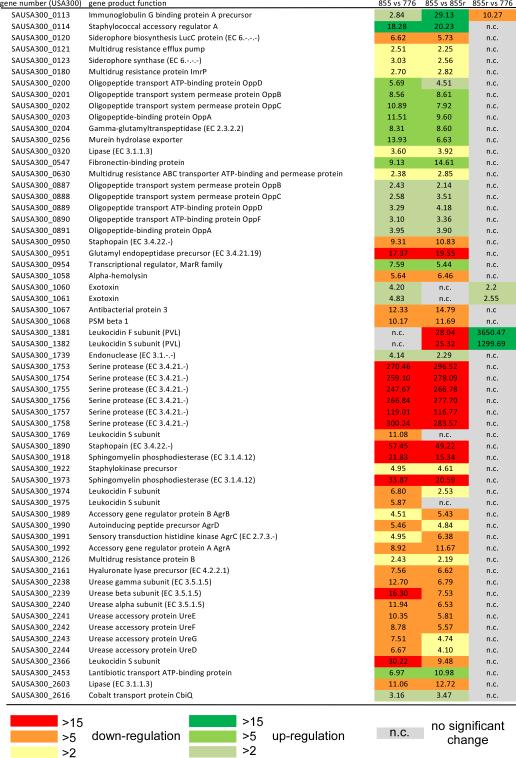
Labandeira-Rey et al. also compared PVL-positive and -negative strains produced in the S. aureus SH1000 background, which like RN6390 is a derivative of the 8325-4 laboratory strain with a repaired rsbU gene [19]. However, this strain pair was not isogenic, as the control strain did not contain the ϕSLT phage with deleted lukSF-PV genes, such as LUG855. To investigate whether the phage itself, rather than lukSF-PV, has an effect on gene expression, we compared the LUG776 (containing the ϕSLT phage without lukSF-PV) to the RN6390 background strain. We found that one group of genes with unknown function (homologues of the USA300 1377, 1378, 1379, and 1380 genes), located adjacent to the PVL phage, showed drastically increased expression in RN6390 (table 4). Furthermore, we found that clpB and spa genes were differentially regulated, lending support to our hypothesis that the differential expression of spa and clpB observed in the LUG855r versus LUG776 comparison may have been caused by differential phage integration. Notably, this analysis showed that the ϕSLT phage background without lukSF-PV has a significant impact on gene expression, emphasizing that comparisons of ϕSLT phage-containing with non-ϕSLT phage-containing strains are not valid to investigate regulatory or phenotypic effects caused by lukSF-PV.
Table 4.
Gene expression changes in RN6390 versus LUG776.
| RN6390 vs LUG776 | |||
|---|---|---|---|
| Gene number (USA300) | Gene product function | up-regulated | down-regulated |
| SAUSA300_0085 | Hypothetical protein | 2.61 | |
| SAUSA300_0113 | Immunoglobulin G binding protein A precursor | 3.66 | |
| SAUSA300_0118 | Cysteine synthase (EC 2.5.1.47) | 2.96 | |
| SAUSA300_0125 | Diaminopimelate decarboxylase (EC 4.1.1.20) | 2.88 | |
| SAUSA300_0409 | Hypothetical exported protein | 2.26 | |
| SAUSA300_0508 | ClpC ATPase | 3.24 | |
| SAUSA300_0509 | Arginine kinase (EC 2.7.3.3) | 3.16 | |
| SAUSA300_0877 | ClpB protein | 5.29 | |
| SAUSA300_1377 | Hypothetical protein | 26.38 | |
| SAUSA300_1378 | Hypothetical cytosolic protein | 22.18 | |
| SAUSA300_1379 | Hypothetical cytosolic protein | 142.65 | |
| SAUSA300_1380 | Hypothetical protein | 159.30 | |
| SAUSA300_1383 | N-acetylmuramoyl-L-alanine amidase (EC 3.5.1.28)* | 6.58 | |
| SAUSA300_1390 | Hypothetical protein* | 16.36 | |
| SAUSA300_1393 | Phage protein* | 11.25 | |
| SAUSA300_1397 | Major tail protein* | 163.08 | |
| SAUSA300_1403 | Portal protein* | 81.55 | |
| SAUSA300_1407 | Transcriptional activator RinA* | 45.76 | |
| SAUSA300_1437 | Hypothetical protein* | 742.69 | |
| SAUSA300_1438 | DNA integration/recombination/inversion protein* | 116.55 | |
| SAUSA300_2366 | Leukocidin S subunit | 3.23 | |
| SAUSA300_2367 | Hypothetical protein | 3.25 | |
| SAUSA300_2493 | Hypothetical protein | 2.45 | |
phage ϕSLT region.
Discussion
The S. aureus global gene regulator agr controls expression of multiple virulence factors and metabolic genes and is reportedly prone to spontaneous mutation [20]. As (i) the alteration in global gene expression attributed to PVL by Labandeira-Rey et al. [9] was reminiscent of that caused by mutations in agr, and (ii) control experiments in that study were not sufficient to rule out the possibility that the observed effects might have originated from a spontaneous mutation in another gene locus, we hypothesized that an unintended mutation in agr may have caused the PVL-associated phenotype reported by Labandeira-Rey et al. Failure to rule out spontaneous mutation in agr by appropriate genetic controls previously led to erroneous reports describing regulatory functions for the xprA [21], svrA [22], and traP [23-25] genes.
In the present study we demonstrated that the gene expression differences and the virulence phenotype in experimental murine pneumonia described by Labandeira-Rey et al. as attributed to PVL [9] were caused by a mutation in agr. This conclusion is based on two main observations. First, we detected an unintended agr mutation in the LUG855 strain used by Labandeira-Rey et al. that caused decreased binding of AgrA to the agr P2 promoter and dramatically decreased agr activity. Second, we could not reproduce the aforementioned results of Labandeira-Rey et al. by using a corrected LUG855 strain in which the agr mutation was repaired.
We are aware of the fact that the conclusions by Labandeira-Rey et al. were also based on other strain comparisons that revealed differences in virulence. However, these comparisons were not made between isogenic strains and may thus be influenced by factors other than PVL. In support of this, we showed that there are gene expression differences comparing LUG862 with the repaired LUG855r, and RN6390 with LUG776, demonstrating the influence of the plasmid or phage, respectively, independently of lukSF-PV.
Several reports indicate that PVL has no or only a minor role for S. aureus pathogenesis when PVL is expressed under natural conditions [5, 7, 8, 10]. Only one recent report from the same group that authored the Labandeira-Rey et al. study is at variance with these results [26]. Furthermore, there is an increasing frequency of CA-MRSA infections by strains lacking lukSF genes [3, 27, 28]. Thus, although the role of PVL for CA-MRSA disease will certainly need to be investigated further, experimental and epidemiological evidence indicates that it has been greatly overestimated.
Mutations in agr occur frequently in vitro [20] and can often be isolated from infections, particularly chronic and biofilm-associated infections [29, 30]. Most of these spontaneous mutations are found in the agrC gene. To our knowledge, this report is the first to identify a mutation in the agr promoter region causing dramatic changes in agr activity, thereby identifying a nucleotide position with extreme significance for binding of the regulatory AgrA DNA-binding protein. Our findings demonstrate that a single nucleotide change even outside an open reading frame can have a dramatic and global impact on bacterial gene expression. Furthermore, our finding underscores the importance of appropriate genetic validation and highlights the need to evaluate and re-evaluate S. aureus strains in vitro, owing to the frequent spontaneous mutations that occur in the agr locus.
Acknowledgments
The authors thank Dr. Bowden for sending strains RN6390, LUG776, LUG855, and LUG862.
This work was supported by the intramural research program of the NIAID (National Institute of Allergy and Infectious Diseases), National Institutes of Health (to F.R.D. and M.O.) and the Departments of Pediatrics and Microbiology at the University of Chicago (to J.B.W.).
Footnotes
The authors have declared that no conflict of interest exists.
References
- 1.Klevens RM, Morrison MA, Nadle J, et al. Invasive methicillin-resistant Staphylococcus aureus infections in the United States. Jama. 2007;298:1763–71. doi: 10.1001/jama.298.15.1763. [DOI] [PubMed] [Google Scholar]
- 2.Moran GJ, Krishnadasan A, Gorwitz RJ, et al. Methicillin-resistant S. aureus infections among patients in the emergency department. N Engl J Med. 2006;355:666–74. doi: 10.1056/NEJMoa055356. [DOI] [PubMed] [Google Scholar]
- 3.Diep BA, Otto M. The role of virulence determinants in community-associated MRSA pathogenesis. Trends Microbiol. 2008;16:361–9. doi: 10.1016/j.tim.2008.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang R, Braughton KR, Kretschmer D, et al. Identification of novel cytolytic peptides as key virulence determinants for community-associated MRSA. Nat Med. 2007;13:1510–4. doi: 10.1038/nm1656. [DOI] [PubMed] [Google Scholar]
- 5.Wardenburg JB, Bae T, Otto M, DeLeo FR, Schneewind O. Poring over pores: alpha-hemolysin and Panton-Valentine leukocidin in Staphylococcus aureus pneumonia. Nat Med. 2007;13:1405–6. doi: 10.1038/nm1207-1405. [DOI] [PubMed] [Google Scholar]
- 6.Lina G, Piemont Y, Godail-Gamot F, et al. Involvement of Panton-Valentine leukocidin-producing Staphylococcus aureus in primary skin infections and pneumonia. Clin Infect Dis. 1999;29:1128–32. doi: 10.1086/313461. [DOI] [PubMed] [Google Scholar]
- 7.Voyich JM, Otto M, Mathema B, et al. Is Panton-Valentine Leukocidin the Major Virulence Determinant in Community-Associated Methicillin-Resistant Staphylococcus aureus Disease? J Infect Dis. 2006;194:1761–70. doi: 10.1086/509506. [DOI] [PubMed] [Google Scholar]
- 8.Diep BA, Palazzolo-Ballance AM, Tattevin P, et al. Contribution of Panton-Valentine leukocidin in community-associated methicillin-resistant Staphylococcus aureus pathogenesis. PLoS ONE. 2008;3:e3198. doi: 10.1371/journal.pone.0003198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Labandeira-Rey M, Couzon F, Boisset S, et al. Staphylococcus aureus Panton-Valentine leukocidin causes necrotizing pneumonia. Science. 2007;315:1130–3. doi: 10.1126/science.1137165. [DOI] [PubMed] [Google Scholar]
- 10.Wardenburg JB, Palazzolo-Ballance AM, Otto M, Schneewind O, DeLeo FR. Panton-Valentine Leukocidin Is Not a Virulence Determinant in Murine Models of Community-Associated Methicillin-Resistant Staphylococcus aureus Disease. J Infect Dis. 2008;198:1166–1170. doi: 10.1086/592053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dunman PM, Murphy E, Haney S, et al. Transcription profiling-based identification of Staphylococcus aureus genes regulated by the agr and/or sarA loci. J Bacteriol. 2001;183:7341–53. doi: 10.1128/JB.183.24.7341-7353.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bae T, Schneewind O. Allelic replacement in Staphylococcus aureus with inducible counter-selection. Plasmid. 2006;55:58–63. doi: 10.1016/j.plasmid.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 13.Queck SY, Jameson-Lee M, Villaruz AE, et al. RNAIII-Independent Target Gene Control by the agr Quorum-Sensing System: Insight into the Evolution of Virulence Regulation in Staphylococcus aureus. Mol Cell. 2008;32:150–8. doi: 10.1016/j.molcel.2008.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bubeck Wardenburg J, Patel RJ, Schneewind O. Surface proteins and exotoxins are required for the pathogenesis of Staphylococcus aureus pneumonia. Infect Immun. 2007;75:1040–4. doi: 10.1128/IAI.01313-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li M, Lai Y, Villaruz AE, Cha DJ, Sturdevant DE, Otto M. Gram-positive three-component antimicrobial peptide-sensing system. Proc Natl Acad Sci U S A. 2007;104:9469–9474. doi: 10.1073/pnas.0702159104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Novick RP, Ross HF, Projan SJ, Kornblum J, Kreiswirth B, Moghazeh S. Synthesis of staphylococcal virulence factors is controlled by a regulatory RNA molecule. Embo J. 1993;12:3967–75. doi: 10.1002/j.1460-2075.1993.tb06074.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koenig RL, Ray JL, Maleki SJ, Smeltzer MS, Hurlburt BK. Staphylococcus aureus AgrA binding to the RNAIII-agr regulatory region. J Bacteriol. 2004;186:7549–55. doi: 10.1128/JB.186.22.7549-7555.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Novick RP, Projan SJ, Kornblum J, et al. The agr P2 operon: an autocatalytic sensory transduction system in Staphylococcus aureus. Mol Gen Genet. 1995;248:446–58. doi: 10.1007/BF02191645. [DOI] [PubMed] [Google Scholar]
- 19.Horsburgh MJ, Aish JL, White IJ, Shaw L, Lithgow JK, Foster SJ. sigmaB modulates virulence determinant expression and stress resistance: characterization of a functional rsbU strain derived from Staphylococcus aureus 8325-4. J Bacteriol. 2002;184:5457–67. doi: 10.1128/JB.184.19.5457-5467.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Somerville GA, Beres SB, Fitzgerald JR, et al. In vitro serial passage of Staphylococcus aureus: changes in physiology, virulence factor production, and agr nucleotide sequence. J Bacteriol. 2002;184:1430–7. doi: 10.1128/JB.184.5.1430-1437.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McNamara PJ, Iandolo JJ. Genetic instability of the global regulator agr explains the phenotype of the xpr mutation in Staphylococcus aureus KSI9051. J Bacteriol. 1998;180:2609–15. doi: 10.1128/jb.180.10.2609-2615.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen J, Novick RP. svrA, a multi-drug exporter, does not control agr. Microbiology. 2007;153:1604–8. doi: 10.1099/mic.0.2007/006247-0. [DOI] [PubMed] [Google Scholar]
- 23.Adhikari RP, Arvidson S, Novick RP. A nonsense mutation in agrA accounts for the defect in agr expression and the avirulence of Staphylococcus aureus 8325-4 traP::kan. Infect Immun. 2007;75:4534–40. doi: 10.1128/IAI.00679-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shaw LN, Jonnson I-M, Singh VK, Tarkowski A, Stewart GC. Inactivation of traP has no effect on the Agr quorum sensing system or virulence of Staphylococcus aureus. Infect Immun. 2007 doi: 10.1128/IAI.00491-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tsang LH, Daily ST, Weiss EC, Smeltzer MS. Mutation of traP in Staphylococcus aureus has no impact on expression of agr or biofilm formation. Infect Immun. 2007 doi: 10.1128/IAI.00603-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brown EL, Dumitrescu O, Thomas D, et al. The Panton-Valentine leukocidin vaccine protects mice against lung and skin infections caused by Staphylococcus aureus USA300. Clin Microbiol Infect. 2008 doi: 10.1111/j.1469-0691.2008.02648.x. DOI 10.1111/j.1469-0691.2008.02648.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Otter JA, French GL. The emergence of community-associated methicillin-resistant Staphylococcus aureus at a London teaching hospital, 2000-2006. Clin Microbiol Infect. 2008;14:670–6. doi: 10.1111/j.1469-0691.2008.02017.x. [DOI] [PubMed] [Google Scholar]
- 28.Zhang K, McClure JA, Elsayed S, Tan J, Conly JM. Coexistence of Panton-Valentine leukocidin-positive and -negative community-associated methicillin-resistant Staphylococcus aureus USA400 sibling strains in a large Canadian health-care region. J Infect Dis. 2008;197:195–204. doi: 10.1086/523763. [DOI] [PubMed] [Google Scholar]
- 29.Traber KE, Lee E, Benson S, et al. agr function in clinical Staphylococcus aureus isolates. Microbiology. 2008;154:2265–74. doi: 10.1099/mic.0.2007/011874-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vuong C, Kocianova S, Yao Y, Carmody AB, Otto M. Increased colonization of indwelling medical devices by quorum-sensing mutants of Staphylococcus epidermidis in vivo. J Infect Dis. 2004;190:1498–505. doi: 10.1086/424487. [DOI] [PubMed] [Google Scholar]