Abstract
Staphylococcus epidermidis and S. aureus are the most frequent causes of nosocomial infections and infections on indwelling medical devices, which characteristically involve biofilms. Recent advances in staphylococcal molecular biology have provided more detailed insight into the basis of biofilm formation in these opportunistic pathogens. A series of surface proteins mediate initial attachment to host matrix proteins, which is followed by the expression of a cationic glucosamine-based exopolysaccharide that aggregates the bacterial cells. In some cases, proteins may function as alternative aggregating substances. Furthermore, surfactant peptides have now been recognized as key factors involved in generating the 3-dimensional structure of a staphylococcal biofilm by cell-cell disruptive forces, which eventually may lead to the detachment of entire cell clusters. Transcriptional profiling experiments have defined the specific physiology of staphylococcal biofilms and demonstrated that biofilm resistance to antimicrobials is due to gene-regulated processes. Finally, novel animal models of staphylococcal biofilm-associated infection have given us important information on which factors define biofilm formation in vivo. These recent advances constitute an important basis for the development of anti-staphylococcal drugs and vaccines.
1. Introduction
Staphylococci are recognized as the most frequent causes of biofilm-associated infections. This exceptional status among biofilm-associated pathogens is due to the fact that staphylococci are frequent commensal bacteria on the human skin and mucous surfaces (and those of many other mammals). Thus, staphylococci are among the most likely germs to infect any medical device that penetrates those surfaces, such as when being inserted during surgery (Vuong and Otto 2002).
For a long time, research on the molecular basis of biofilm formation was focused on gram-negative pathogens, predominantly Pseudomonas aeruginosa, which is more easily accessible to molecular genetic investigation. More recently, advances in staphylococcal molecular biology have allowed researchers to determine the molecular basis of biofilm formation in staphylococci. In addition, animal models of staphylococcal biofilm-associated infection have been established. Therefore, we now find staphylococci, and particularly S. epidermidis, among the best studied clinically relevant biofilm-forming organisms.
This review will give an overview of the role of staphylococci in biofilm-associated human diseases and focus on the mechanism of biofilm development and the molecular basis of virulence in biofilm-forming S. epidermidis and S. aureus.
2. Biofilms and staphylococcal infections
The Nosocomial Infections Surveillance System (http://www.cdc.gov/ncidod/hip/NNIS/2004NNISreport.pdf) recognizes S. aureus and CoNS (coagulase-negative staphylococci, i.e. S. epidermidis and most other staphylococci other than S. aureus) as the most frequently isolated nosocomial pathogens from intensive care unit patients. An extremely high percentage of these isolates are resistant to methicillin (89% CoNS compared to 59.5% for S. epidermidis). In addition to specific antibiotic resistance, which is based on the acquisition of genetic resistance factors and may be chromosomally, or more often plasmid-encoded, staphylococci have non-specific mechanisms of resistance, of which biofilm formation is undoubtedly the most important.
2.1. S. epidermidis infections on indwelling medical devices
S. epidermidis is known as an opportunistic pathogen because it predominantly causes infection in immuno-compromised individuals such as intravenous drug abusers, AIDS patients, patients receiving immuno-suppressive therapy and premature newborns (Vadyvaloo and Otto 2005). In otherwise healthy patients, S. epidermidis causes infection only after penetration of the skin or mucous membranes, which can occur by trauma, inoculation, or implantation of medical devices. These patients may develop septicemia or endocarditis (Arber et al. 1994). As S. epidermidis makes up a significant part of the normal bacterial flora of the human skin and mucous membranes, it is probably easily introduced as a contaminant during the surgical implantation of the polymeric device. Notably, a device-related infection of S. epidermidis characteristically involves biofilm formation, which generally is considered the most important factor involved in the pathogenesis of S. epidermidis.
2.2. S. aureus biofilm-associated infection
S. aureus only colonizes a certain percentage of healthy adults permanently or transiently (van Belkum 2006). The reasons for these differences are not understood, but may involve yet undiscovered host factors that predispose for S. aureus colonization. Thus, whether indwelling medical devices are contaminated with S. aureus during insertion depends significantly on the carrier, be it the patient or health care personnel. To some extent, biofilm-associated infections with S. aureus are similar to those with S. epidermidis. However, the involvement of S. aureus usually requires more intensive care. Often, S. aureus biofilm-associated infections are difficult to treat with antibiotics and devices need to be replaced more frequently than those infected with S. epidermidis (Jones et al. 2001). In addition, they represent a reservoir of dissemination of S. aureus infection to other sites in the human body. In this regard, it is critical from a perspective of molecular pathogenesis, whether biofilm-forming S. aureus strains are genetically different from those involved in more serious infections, or – alternatively - whether they are in a different physiological status and might thus develop a more aggressive behavior when spreading within the body.
2.3. Other staphylococci
Similar to S. epidermidis, most other staphylococci have a benign relationship with their host and develop from commensals to pathogens only after damage of a natural barrier such as the skin. In comparison with S. epidermidis and S. aureus, biofilm-associated infections with other staphylococci are far less frequent. It is not known if this is due to a difference in virulence or abundance on the human skin, or – which appears most likely – a combination of both factors. CoNS found in humans colonize different parts of the human skin and mucous membranes, with each species having a certain predominance on specific parts of the body (Kloos and Schleifer 1986). Notably, every species of CoNS that has been characterized as a resident of the human body (S. epidermidis, S. capitis, S. hominis, S. haemolyticus, S. saccharolyticus, S. warneri, S. lugdunensis, S. saprophyticus, S. cohnii) has also at least once been connected to an infection. The specific sites and frequency of infection seem to be related to those of normal colonization. S. saprophyticus for example is often found in the inguinal and perineal areas and is a common cause of urinary tract infections (Kloos and Schleifer 1986). In these infections, biofilm formation is probably a crucial determinant of disease, although this remains to be investigated. In general, the specific molecular determinants of biofilm formation in CoNS may be different from S. epidermidis and S. aureus, but appear to use the same basic mechanisms.
2.4. Interaction of staphylococci with other pathogens in mixed-species medical biofilms
In contrast to many other medical biofilms, such as multi-species dental plaque formation, biofilm-associated infections with staphylococci are usually not mixed with other species (Arciola et al. 2005). In addition, it is rare to find more than one strain in an infection. A possible explanation for this phenomenon is interspecies communication by quorum-sensing signals, which in staphylococci leads to interspecies inhibition of virulence factor expression (Ji et al. 1997). Similarly, bacterial interference by quorum-sensing signals may explain why P. aeruginosa outgrows S. aureus and other bacterial pathogens in progressed lung infections (Renders et al. 2001; Qazi et al. 2006). However, these phenomena are poorly understood and there may be a simpler explanation based on the evolutionary adaptation of the bacteria to a specific environment, such as of S. epidermidis on the skin.
3. The molecular basis of biofilm formation in staphylococci
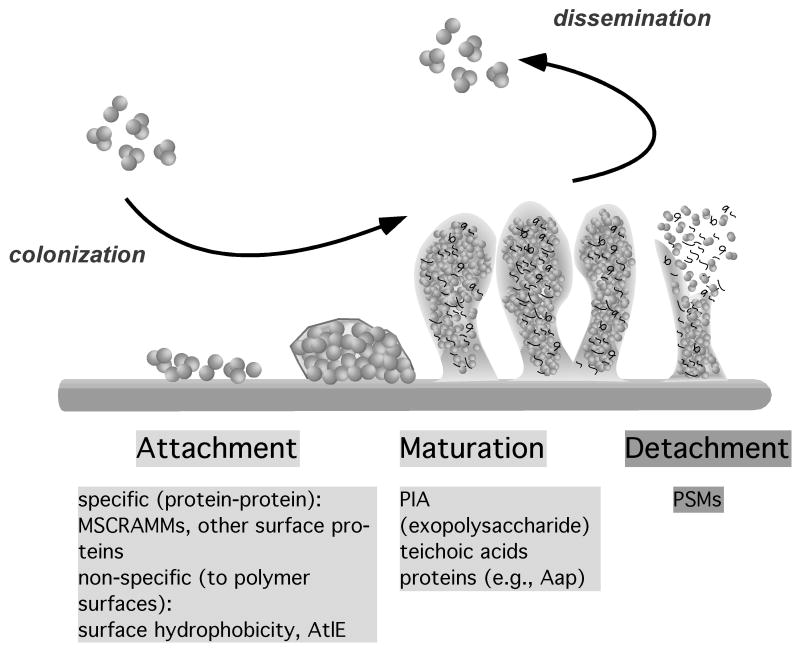
Research performed in many biofilm-forming organisms has revealed that the development of a biofilm is a 2-step process involving an initial attachment and a subsequent maturation phase, which are physiologically different from each other and require phase-specific factors. A final detachment (or dispersal) phase involves the detachment of single cells or cell clusters by various mechanisms and is believed to be crucial for the dissemination of the bacteria, in the case of pathogens to new infection sites in the human body (Fig. 1).
Fig. 1.
Phases of biofilm development in staphylococci. Biofilms form by initial attachment to a surface, which can occur on tissues or after covering of an abiotic surface by host matrix proteins in the human body (specific, protein-protein interaction), or directly to an abiotic surface (non-specific). Subsequently, biofilms grow and mature. The molecules that connect the cells in a staphylococcal biofilm are predominantly the exopolysaccharide PIA, teichoic acids, and some proteins such as the accumulation-associated protein Aap. Finally, cell clusters detach. Detachment is facilitated by expression of the surfactant-like PSM peptides, which are also important in producing the 3-dimensional structure of the biofilm. During infection, attachment is a crucial part of the colonization on host tissues or on indwelling medical devices, whereas detachment is a prerequisite for the dissemination of an infection.
3.1. Attachment
In the human body, the attachment to human matrix proteins represents the first step of biofilm formation. S. epidermidis and S. aureus express dozens of so-called MSCRAMMs (microbial surface components recognizing adhesive matrix molecules) that have the capacity to bind to human matrix proteins such as fibrinogen or fibronectin, and often combine binding capacity for several different matrix proteins (Patti et al. 1994). MSCRAMMs have a common structure that includes an exposed binding domain, a cell-wall spanning domain, which often has a repeat structure, and a domain that is responsible for the covalent or non-covalent attachment to the bacterial surface. Covalent attachment is catalyzed by a family of enzymes called sortases that link a conserved motif of the MSCRAMMS to peptidoglycan (Marraffini et al. 2006). The most important one is sortase A, which recognizes an LPXTG motif at the C-terminus of the surface protein sequence (Mazmanian et al. 1999). S. aureus strains have a wider variety of LPXTG-type MSCRAMMs (~20), compared to ~ 12 in S. epidermidis (Gill et al. 2005). The only functional equivalents between the two species appear to be several members of the serine-aspartate-repeat family (Sdr proteins). This family comprises several surface proteins that have a characteristic serine-aspartate repeat cell-wall spanning domain (McCrea et al. 2000). In addition, both species have the accumulation-associated protein Aap and several non-covalently bound surface proteins, such as the autolysin Atl, in common.
The forces that govern the attachment of non-covalently bound MSCRAMMs to the surface of staphylococci are not well understood (Navarre and Schneewind 1999). The most important examples are autolysins, which often represent some of the most abundant proteins on the staphylococcal cell surface. There is some evidence to suggest that autolysins are non-covalently attached to teichoic acids (Peschel et al. 2000). These enzymes, in addition to their primary role in cell wall turnover, also facilitate attachment to plastic surfaces and harbor binding sites for human matrix proteins (Heilmann et al. 1997; Heilmann et al. 2003). Thus, they have a crucial bi-functional importance for bacterial attachment. Similar to the autolysins, the lipase GehD has a primary catalytic role, but there is evidence to suggest that it has an additional adhesive function (Bowden et al. 2002).
Staphylococci are known for their extraordinary ability to stick to plastic surfaces. While this ability has been the basis for most of the in vitro biofilm research performed in staphylococci (and in other biofilm-forming pathogens), it is not clear if direct attachment to plastic plays a significant role in the pathogenesis of medical device-associated infection. Host matrix proteins cover the devices soon after insertion and thus, the specific interaction between these proteins and MSCRAMMs most likely is of much greater importance for colonization. The classic microtiter plate assay for biofilm formation on abiotic surfaces has been a valuable tool especially in large screens for biofilm-related factors. However, it is far from representing the detailed characteristics of biofilm-associated infection in vivo and might have led to an over-estimation of the importance of some molecules in biofilm formation. It should thus optimally be accompanied by more elaborate in vitro methods, such as flow cells and confocal laser scanning microscopy, and animal models of biofilm-associated infection. For example, subcutaneous infection models with catheter tubing (Rupp et al. 1999a) or tissue-cage models (Zimmerli et al. 1982) have been used successfully to monitor staphylococcal biofilm-associated infection.
3.2. Maturation
The maturation phase of biofilm formation is characterized by 1) intercellular aggregation that can be accomplished by a variety of molecules such as adhesive proteins or – usually polysaccharide-based - exopolymers, and 2) biofilm structuring forces that lead to the typical 3-dimensional appearance of mature biofilms with its mushroom-like cell towers surrounding fluid-filled channels.
3.2.1. Adhesive forces: aggregation
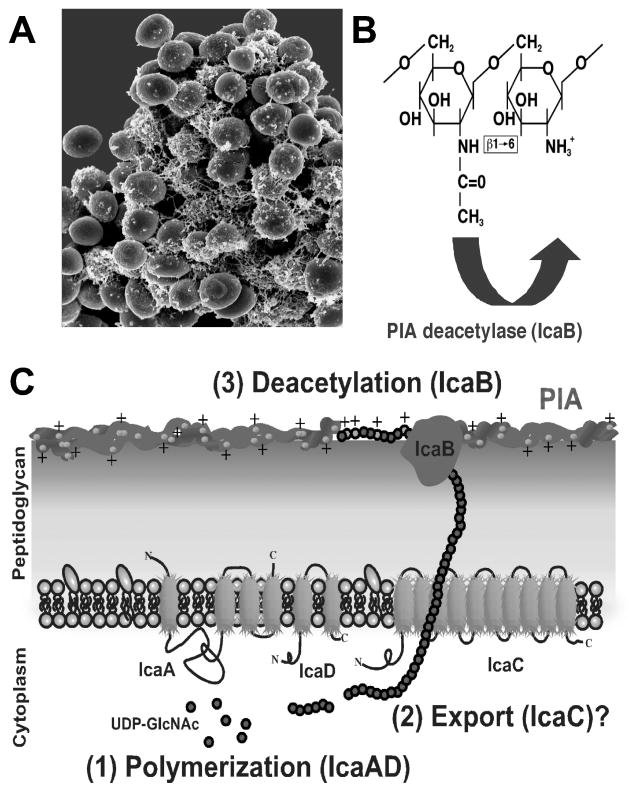
In staphylococci, the main molecule responsible for intercellular adhesion is the polysaccharide intercellular adhesin (PIA), which is also called poly-N-acetylglucosamine (PNAG) according to its chemical composition (Mack et al. 1996). It is a partially de-acetylated polymer of beta-1-6-linked N-acetylglucosamine, which together with other polymers such as teichoic acids and proteins forms the major part of what has often been called “slime”, the extracellular matrix of biofilm-forming staphylococci (Fig. 2). More recently, PIA homologs have been detected in a variety of biofilm-forming pathogens, suggesting that this polymer has a widespread function in biofilms and biofilm-associated infections (Darby et al. 2002; Kaplan et al. 2004b; Wang et al. 2004).
Fig. 2.
The biofilm exopolysaccharide PIA. A, PIA covers staphylococcal cells and sticks them together as the major component of the extracellular matrix (backscatter scanning electron microscopic picture of S. epidermidis). B, PIA is a homopolymer of beta 1-6–linked N-acetylglucosamine, of which about 10–20% of residues are de-acetylated. C, The biosynthesis of PIA in S. epidermidis occurs in 3 steps. (1) IcaA adds GlcNAc moieties from UDP-GlcNAc to the growing PIA chain. The IcaA transferase needs the presence of IcaD for full activity. (2) Presumably, the nascent PIA chain is then exported by IcaC. (3) After export, PIA is de-acetylated by the surface-attached IcaB to introduce positive charges, which are crucial for its surface location and biological function.
The de-acetylation of N-acetylglucosamine residues in PIA is of major biological importance. It introduces a positively charged character in the otherwise neutral molecule by liberating free amino groups that become charged at neutral or acid pH, such as found in the natural habitat of staphylococci, the human skin (Vuong et al. 2004a). As the bacterial cell surface is negatively charged, PIA supposedly works like glue that sticks the cells together by electrostatic interaction. Teichoic acids may represent the negatively charged molecules that interact with PIA on the cell surface. Interestingly, the relative amounts of teichoic acids and PIA are subject to environmental control – the biological role of which is not yet understood (Sadovskaya et al. 2005).
PIA biosynthesis is accomplished by the products of the ica gene locus, which comprises an N-acetylglucosamine transferase (icaA and icaD), a PIA deacetylase (icaB), a putative PIA exporter (icaC), and a regulatory gene (icaR) (Heilmann et al. 1996; Gerke et al. 1998; Vuong et al. 2004a). Expression of the ica gene locus is regulated by a variety of environmental factors and regulatory proteins (see 4.3). The production of PIA and its deacetylation have been recognized as key virulence factors in S. epidermidis (Rupp et al. 1999a; Rupp et al. 1999b; Rupp et al. 2001; Vuong et al. 2004a; Fluckiger et al. 2005). Several animal models have confirmed this key role, although some conflicting results exist (Kristian et al. 2004). However, PIA production does not seem to be of universal importance for biofilm formation and biofilm-associated infection, as PIA-independent biofilm formation has been demonstrated (Rohde et al. 2005). Furthermore, some strains isolated from biofilm-associated infection do not have the ica genes (Arciola et al. 2006). Interestingly, invasiveness of non-invasive S. epidermidis that lack the ica operon can be restored by introduction of the ica genes (Li et al. 2005).
In cases of PIA-independent biofilm formation, adhesive proteins most likely substitute for PIA. The most important protein involved in PIA-independent biofilm formation appears to be Aap (Hussain et al. 1997). In a recent study, 27% of biofilm-forming strains isolated from the infection of prosthetic joint infections formed PIA-independent biofilms, in most of which biofilm formation appeared to be mediated by Aap (Rohde et al. 2007). In this study, S. aureus biofilms, in contrast, always seemed to be dependent on PIA. Furthermore, biofilm formation was less pronounced when exclusively dependent on proteins. Thus, although PIA does not have an absolutely universal importance for staphylococcal biofilms, this study confirms its key role in staphylococcal biofilm formation.
Aap is a 220 kD protein that needs to be proteolytically cleaved to a smaller 140 kD form to induce biofilm formation (Rohde et al. 2005) and has been suggested to interact with PIA (Hussain et al. 1997). Aap may be identical to the SSP-1 and SSP-2 proteins, which have been implicated in biofilm formation but whose identity was not investigated further (Veenstra et al. 1996). Interestingly, it was shown that SSP forms protein strands on the S. epidermidis surface, thus possibly contributing to cell-cell adhesion over greater distances. This capacity could explain how proteins contribute to the aggregation step of biofilm development. A very recent publication has in fact demonstrated that the formation of fibril-like structures on the S. epidermidis surface is dependent on Aap (Banner et al. 2007).
In S. aureus isolates from animals suffering from mastitis, a cell wall bound surface protein named biofilm associated protein, Bap, is involved in adherence to a polystyrene surface, intercellular adhesion and biofilm formation (Cucarella et al. 2001). There is evidence for the significance of Bap during infection of bovine mammary glands (Cucarella et al. 2004). A homolog of bap named bhp occurs in human strains of S. epidermidis (Zhang et al. 2003; Gill et al. 2005). Bap homologs are also found in other bacteria, suggesting that the Bap family of surface proteins may have widespread importance in biofilm formation (Latasa et al. 2005; Lasa and Penades 2006).
S. aureus and S. epidermidis contain teichoic acids (TA), which are commonly found in many Gram-positive bacteria (Hussain et al. 1992; Hussain et al. 1993). TA can be linked to the cell wall in which case they are referred to as cell wall TA (WTA), or they can be linked to the cell membrane via a lipid anchor, known as lipoteichoic acid (LTA). TA consist of (1,3)-linked poly (glycerol/ribitol phosphate), substituted with glucose, N-acetylglucosamine, D-alanine, or 6-alanyl glucose at the position 2 of the glycerol residue. S. epidermidis TA significantly increase adhesion to fibronectin-coated surfaces, suggesting a probable role for TA in S. epidermidis virulence (Hussain et al. 2001). Furthermore, the importance of the D-alanylation of S. aureus TA in biofilm formation has been demonstrated (Gross et al. 2001).
3.2.2. Disruptive forces: biofilm structuring
A mature biofilm has a specific 3-dimensional structure, which has been described to consist of “towers” or “mushrooms” (Costerton et al. 1995). In between those towers, there are fluid-filled channels that are believed to have a vital function in delivering nutrients to cells in deeper biofilm layers. The mechanisms that lead to channel formation and biofilm structuring are far less well understood than those governing intercellular adhesion. Findings primarily achieved in P. aeruginosa indicate the involvement of cell-to-cell signaling, e.g. by quorum-sensing systems (Davies et al. 1998). The quorum-sensing controlled expression of the surfactant rhamnolipid appears to be the major mechanism for biofilm structuring in P. aeruginosa (Davey et al. 2003; Boles et al. 2005). In staphylococci, differential expression of the biofilm exopolysacharide PIA might to some degree contribute to biofilm structuring. In contrast, enzymatic degradation of PIA, which appears to occur in other bacteria that express PIA homologs (Kaplan et al. 2003), is very likely not present in staphylococci.
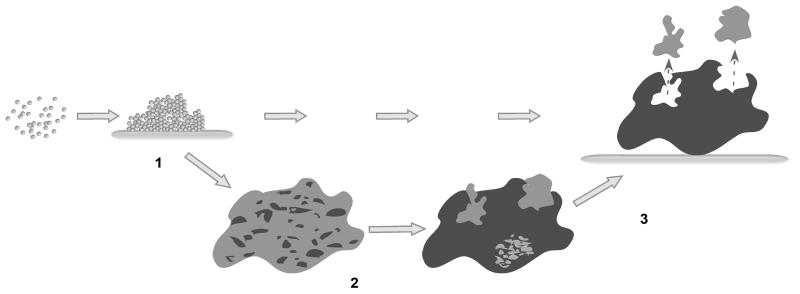
Recent findings in my laboratory suggest that staphylococci use quorum-sensing controlled surfactant peptides to structure biofilms in a mechanism similar to P. aeruginosa, but based on chemically different effector molecules. Phenol-soluble modulins (PSMs) are a class of peptides that have first been described as pro-inflammatory agents in S. epidermidis (Mehlin et al. 1999). All PSMs have a pronounced amphipathic alpha-helical character and thus, strong surfactant-like properties. They can be subdivided in two classes: those with a length of ~ 20 amino acids (alpha type) and those with a length of ~ 40–45 amino acids (beta type). Notably, under biofilm conditions, PSM expression is shifted to the beta type of PSM peptides, which are encoded in an operon (Yao et al. 2005). Recently, we found that expression of PSM beta peptides has a key role in biofilm development in S. epidermidis. During dynamic S. epidermidis biofilm formation in flow cells, expression of the PSM beta peptides leads to the detachment of cell clusters (unpublished results). This likely leads to the formation of “holes” in early biofilms and thereby to biofilm structuring (Fig. 3). Consistently, a PSM beta operon deletion strain forms a more compact biofilm than the isogenic wild-type strain. PSM homologs also occur in S. aureus and other staphylococci (unpublished results). Whether they have the same role in biofilm development needs to be determined.
Fig. 3.
Model of PSM function in biofilm structuring. (1) Cells actively expressing PSM beta peptides attach to a surface. (2) Later on, some cell clusters discontinue PSM beta expression for yet unknown reasons, possibly due to limited oxygen concentration. (3) Cell clusters with active PSM beta expression detach, leaving gaps in the biofilm, which ultimately leads to the typical structure of a biofilm with cell towers and fluid-filled channels.
3.3. Detachment
Biofilm detachment is crucial for the dissemination of bacteria to other colonization sites. It may occur by the detachment of single cells or larger cell clusters. Several factors may contribute to detachment: 1) mechanical forces, such as flow in a blood vessel, 2) cessation of the production of biofilm building material, such as exopolysaccharide, and 3) detachment factors sensu strictu, such as enzymes that destroy the matrix, or surfactants. For all that we know, the latter factors are not different from those discussed as biofilm structuring agents. When produced at a high rate, these factors will cause detachment, especially at the biofilm surface area. In fact, controlled detachment maintains a certain biofilm thickness and governs a specific rate of biofilm dissemination. In staphylococci, this mechanism is controlled by the quorum-sensing system agr (see below).
3.4. Cell death and extracellular DNA
Some more recent publications claim that controlled cell death in staphylococci contributes to biofilm development. While the phenomenon of controlled cell death in bacteria is still a controversial issue (Rice and Bayles 2003), an increased degree of cell lysis clearly appears to influence biofilm formation. Several regulators that control autolysis have been shown to affect biofilms, such as CidR (Yang et al. 2006). In the case of the CidA murein hydrolase regulator, the release of DNA, a process naturally involved in cell lysis, contributes to biofilm development (Rice et al. 2007). In fact, DNA has recently been frequently implicated in biofilm formation. As a polyanionic molecule, DNA has the capacity to link other molecules together in the biofilm matrix in a way similar to teichoic acids, notably including cationic polymers such as the genuine biofilm polymer PIA discussed above. Due to the conserved nature of the DNA molecule, it is to be expected that autolysis in general will have a similar impact on biofilm formation by that mechanism, which may also in part be responsible for observations made with the Atl type of autolysins (Heilmann et al. 1997; Heilmann and Gotz 1998).
4. Regulation of biofilm formation in staphylococci
Biofilms are the common way of growth for a multitude of microorganisms. Thus, it is to be expected that biofilm growth is under the influence of a vast variety of regulatory mechanisms, just as planktonic growth. However, we lack knowledge on the specific metabolism of biofilms. Regulatory influences on biofilm factors sensu strictu will be discussed here, whereas our current knowledge on the physiology of staphylococcal biofilms as determined by transcriptional profiling will be discussed in 5. We will focus on regulators, for which a mechanism for the influence on biofilm formation has been described in more detail. There are several regulatory systems described in the literature, such as the rbf (regulator of biofilm formation) (Lim et al. 2004), for which this is still elusive. In addition, very recent reports suggest that the effect that has been described for the Trap regulator, allegedly affecting biofilm formation in response to a peptide called RIP (Balaban et al. 2003; Balaban et al. 2007), is not genuine but due to a second site mutation, most likely in the agr system (Shaw et al. 2007; Tsang et al. 2007).
4.1. Environmental influences
In the earlier literature, when the composition of the staphylococcal “slime” matrix was not yet known, one can find many reports on the influence of environmental changes on slime formation and biofilm formation as a whole. From a biological point of view, the biofilm-increasing influence of oxygen and iron limitation, and high osmolarity, appear to be the most crucial. More recently, knowledge of slime composition and the availability of reporter gene constructs have given a clearer picture of what controls specific biofilm factors.
4.2. Regulation of attachment factors
The classical notion of quorum-sensing regulation in S. aureus comprises the up-regulation of adhesion factors such as MSCRAMMs when the cell density as low, a situation encountered during the beginning of a staphylococcal infection. After colonization has been accomplished, increasing activity of the agr quorum-sensing system is believed to abolish the expression of the no longer needed colonization factors (Novick 2003). Consistently, many MSCRAMMs are under negative regulation by agr in S. aureus (Patti et al. 1994). Real-time monitoring of agr activity during S. aureus infection using bioluminescence has provided a better understanding of quorum-sensing regulation in vivo (Wright et al. 2005), but results from biofilm-associated infection monitored in real-time are not yet available. In S. epidermidis, our knowledge of colonization factors and their regulation is more limited. Results obtained by transcriptional profiling (Yao et al. 2006) and measurement of MSCRAMM expression (Bowden et al. 2005) suggest that several MSCRAMMs do not follow the classical notion of agr down-regulation.
Other attachment factors may be controlled by completely different regulation. For example, the autolysins are in general mainly expressed during times of high cell wall turnover, as this is their primary function (Giesbrecht et al. 1976). It is not known how this influences biofilm development.
4.3. Regulation of exopolysaccharide (PIA) synthesis
The regulation of PIA expression is probably the best studied among the regulatory influences on staphylococcal biofilm formation. Many previously found regulatory influences on biofilm formation as a whole could be attributed to a change of PIA expression, after tools to pinpoint the regulated targets had become available, such as PIA-specific antisera or ica-reporter gene fusion constructs. However, somewhat rashly, many researchers have equated the staphylococcal biofilm matrix with PIA, which as we now know is not completely valid. Thus, there is some confusion in the literature as to which factors have clearly been shown to influence specifically the production of PIA.
A clearly demonstrated influence on PIA expression has been shown for N-acetylglucosamine and glucosamine, the building blocks of PIA, probably as these molecules are readily available substrates for the biosynthesis of PIA (Gerke et al. 1998). Furthermore, anaerobiosis significantly increases PIA expression (Cramton et al. 2001). This represents a very important finding for biofilm physiology, as oxygen concentration thus would limit biofilm formation in the oxygen-loaded arterial bloodstream. Also, it would lead to increased expression of PIA in an established biofilm, in which oxygen concentration decreases significantly with increasing depth. Finally, subinhibitory concentrations of specific antibiotics increase ica transcription in S. epidermidis (Rachid et al. 2000), a factor to be taken into account during therapy of staphylococcal biofilm-associated infection.
In S. aureus or in S. epidermidis, several global regulators have been shown to regulate ica transcription or PIA expression: the DNA-binding protein SarA and the alternative sigma factor SigB up-regulate whereas the quorum-sensing system luxS down-regulates (Knobloch et al. 2001; Tormo et al. 2005; Xu et al. 2006). In contrast, agr does not regulate PIA expression (Vuong et al. 2000; Vuong et al. 2003). The exact mechanism of sarA and sigB influence on ica transcription is complicated. Briefly, whereas the influence of sigB appears to occur via repression of icaR transcription (Knobloch et al. 2004), which in turn represses transcription of icaADBC (Conlon et al. 2002), sarA regulates the icaA promoter independently of icaR (Tormo et al. 2005).
The widespread insertion element IS256 can integrate in the ica genes, thus abolishing PIA production (Ziebuhr et al. 1999; Conlon et al. 2004). It has been speculated that IS256 thereby contributes to virulence by a mechanism of adaptation to changing environments during infection. The integration of IS256 in the agr operon might have a very similar function of environmental adaptation (Vuong et al. 2004b). Strains with IS256 integrated into ica and agr genes have been isolated from infection (Kozitskaya et al. 2004; Vuong et al. 2004b). In addition, the presence of IS256 appears to be correlated with the invasiveness of S. epidermidis strains (Gu et al. 2005; Kozitskaya et al. 2005). However, although IS256 might have a genuine role in the adaptation of the bacterial population to a different ecological niche, and thus to bacterial versatility, one can probably not call it a true regulator. The action of IS256 appears to be final – it has not been demonstrated to excise from a gene thus re-establishing its function in vivo.
4.4. Regulation of phenol-soluble modulin expression: agr
We have discussed how the quorum-sensing system agr represses surface protein expression after the initial attachment phase. The major agr-dependent control of biofilm development is however likely accomplished by the strict regulation of PSM expression. Expression of agr in a biofilm is limited to surface-exposed areas, where it is probably the key regulator that controls biofilm detachment by up-regulating the expression of the PSM effector molecules (Vuong et al. 2004b). Yarwood et al. have used gfp expression to measure agr activity in S. aureus biofilms over time and have proposed a model for agr-dependent biofilm maintenance (Yarwood et al. 2004). We have speculated earlier that staphylococcal delta-toxin, one of the PSMs, is a major effector of agr-controlled biofilm detachment in S. aureus (Vuong et al. 2000). However, recent research on S. epidermidis in our laboratory suggests that the PSMs of the beta type are more important in that regard (see above), at least in this species.
As a consequence of the influence of agr on PSM expression, agr mutants from a thicker and more compact biofilm in vitro compared to isogenic wild-type strains (Vuong et al. 2000; Vuong et al. 2003). Furthermore, agr mutants occur naturally and can be isolated from biofilm-associated infections at a rate of about 25% (Vuong et al. 2004b). Most likely, the permanent disabling of agr regulation and the consequent excessive biofilm formation are of advantage to bacterial survival in specific stages or types of infection. Notably, mutations that produce agr-negative phenotypes are common and can also be seen in vitro where they occur at a high rate (Somerville et al. 2002).
5. Physiology of staphylococcal biofilms: lessons from transcriptional profiling
After complete genome sequences of S. aureus, S. epidermidis, and other staphylococci had become available, transcriptional profiling of biofilm gene expression was soon initiated. Three transcriptional profiling-based manuscripts have been published, two on S. aureus (Beenken et al. 2004; Resch et al. 2005) and one on S. epidermidis (Yao et al. 2005), and in addition, proteomics were used to confirm results obtained by the microarray experiments (Resch et al. 2006). The general lessons learned from these studies are comparable although differences exist that originate most likely from different experimental setups. In addition, it has to be taken into consideration that two studies (Resch et al. 2005; Resch et al. 2006) were performed in the SA113 strain of S. aureus, which is a natural agr mutant.
First and foremost, staphylococcal biofilms have a physiological status that is characterized by a general down-regulation of active cell processes, such as protein, DNA, and cell wall biosynthesis, which is typical of slow growing cells. Other metabolic changes can be interpreted as a switch to fermentative processes such as acetoin metabolism, resulting from the low oxygen concentration in biofilms. Finally, the up-regulation of urease and the arginine deiminase pathway, which ultimately produce ammonia compounds, has been explained as a switch to limit the deleterious effects of the reduced pH associated with anaerobic growth conditions (Beenken et al. 2004). In general, although similarities exist, a crucial finding of these experiments was that biofilms are physiologically different from planktonic cells in stationary growth phase. In addition, specific resistance mechanisms were found to be up-regulated in staphylococcal biofilms (Yao et al. 2005). Thus, gene-regulatory effects add to the intrinsic structure-based resistance that biofilms have to antibiotics and other antibacterial agents (see 6).
6. The molecular basis of biofilm resistance to host defenses and antibiotics
It has long been recognized that biofilms have dramatically increased resistance to antibiotics, and to key mechanisms of innate host defense, such as antimicrobial peptides (AMPs) and neutrophil phagocytosis (Costerton et al. 1999). However, the molecular basis of this phenomenon has only recently been further investigated. Two main mechanisms contribute to biofilm resistance: 1) prevention of the antibacterial substance from reaching its target, e.g. by limited diffusion or repulsion, and 2) the specific physiology of a biofilm, which limits the efficacy of antibiotics, mainly of those that target active cell processes and which may also include specific subpopulations of resistant cells (“persisters’) (Keren et al. 2004).
Limited diffusion of antibiotics through the extracellular biofilm matrix may be the mechanism of resistance to some antibiotics, such as ciprofloxacin in P. aeruginosa (Walters et al. 2003), whereas several others (e.g. rifampicin and vancomycin) have been shown to break through the exopolysaccharide layer of S. epidermidis (Dunne et al. 1993). A major role in preventing an antibacterial agent from reaching its target (often, the cytoplasm, the cytoplasmic membrane, or the peptidoglycan layer) is electrostatic repulsion or sequestration by surface polymers. Interestingly, PIA protects from cationic and anionic AMPs, and thus may use either mechanism for differently charged molecules (Vuong et al. 2004c). Similarly, the exopolymer poly-gamma-glutamic acid, which is present in S. epidermidis and a variety of other CoNS and is up-regulated during biofilm formation, contributes to resistance to AMPs of either charge (Kocianova et al. 2005) and unpublished results; for a review on biofilm resistance to AMPs see (Otto 2006).
Phagocytosis, mainly by neutrophils, is a major mechanism by which the innate immune system eliminates invading microorganisms. It has been known for a long time that neutrophil phagocytosis is impaired against staphylococci in a biofilm. More recently, we could show that the exopolysaccharide PIA, and the exopolymer PGA, are specific molecules that shield cells from neutrophil phagocytosis, thus significantly contributing to biofilm resistance from elimination by innate host defense (Vuong et al. 2004c; Kocianova et al. 2005).
7. Possible anti-biofilm therapeutics
Biofilms are involved in a multitude of different infections and often contribute significantly to the difficulties encountered in treatment. Developing anti-biofilm drugs aims to combine these drugs with conventional antibiotics, thus restoring the efficacy that the latter show to bacteria in a non-biofilm status.
7.1. Interfering with essential staphylococcal biofilm factors
An ideal anti-biofilm drug in staphylococci would inactivate a factor that is indispensable for every case of staphylococcal biofilm-associated infection. However, such a factor very likely does not exist, because staphylococcal biofilm formation, as we now know, is multi-factorial. Still, targeting the biosynthesis of a factor that appears to be involved in at least the majority of staphylococcal biofilm-associated infection, such as PIA, seems worth considering. Interestingly, some bacteria produce a PIA-degrading enzyme, which – although not present in staphylococci – can degrade staphylococcal PIA and destroy staphylococcal biofilms (Kaplan et al. 2004a). This PIAse, named dispersin B, has first been found in Actinobacillus actinomycetemcomitans, and appears to have potential as an anti-biofilm drug (Kaplan et al. 2003). Similarly, although not biofilm-specific, the peptidoglycan-degrading enzyme lysostaphin is being evaluated for therapeutic use against biofilm-forming staphylococci (Wu et al. 2003).
7.2. Altering adhesive features of indwelling medical devices
The surface of indwelling medical devices can be altered in attempts to decrease bacterial adhesion. However, staphylococci show great versatility and can still adhere to the modern polymers that are in use now. As an additional or complimentary approach, it has been proposed to coat indwelling medical devices with antibiotics or other antibacterial substances. These approaches had limited success, with one problem being plasmid-encoded resistance that is widespread in staphylococci. It is evident that due to the difficulties that staphylococci present to antibacterial therapy, considerable efforts need to be made in both the alteration of device surfaces and the molecular approaches to control staphylococcal biofilm formation.
7.3. Vaccination
Whether vaccination against staphylococcal infection is a promising means to control staphylococcal diseases is controversial. However, many antisera that have been raised for example to PIA (Kelly-Quintos et al. 2006) and several surface proteins, such as fibronectin-binding protein (Rennermalm et al. 2001), have proven effective in animal infection models. Nevertheless, many of these vaccines still need to be tested for their usefulness against biofilm-associated infections.
8. Conclusions and outlook
Recent advances in our understanding of staphylococcal biofilm development have demonstrated that there are some key structural and regulatory factors that determine the form and physiology of Staphylococcus biofilms. Although not all staphylococcal biofilms depend on the expression of the exopolysaccharide PIA, it is by far the most crucial determinant that we know for biofilm-associated infection in staphylococci, and possibly a variety of other pathogens. We are only beginning to comprehend the physiological role of the surfactant PSM peptides in biofilm structuring and it is to be expected that we will soon know better how these peptides contribute to the formation of biofilm structure. Additionally, the more recent development of real time-monitoring of biofilm-associated infection using animal models with bioluminescent bacteria will yield a better understanding of the detailed roles of biofilm factors during biofilm-associated infection in vivo. Finally, an even more intensive use of genome-wide approaches to understand biofilm physiology in greater detail will be a key step in our efforts to establish the molecular basis for the development of anti-staphylococal drugs and vaccines.
References
- Arber N, et al. Pacemaker endocarditis. Report of 44 cases and review of the literature. Medicine (Baltimore) 1994;73:299–305. doi: 10.1097/00005792-199411000-00003. [DOI] [PubMed] [Google Scholar]
- Arciola CR, An YH, Campoccia D, Donati ME, Montanaro L. Etiology of implant orthopedic infections: a survey on 1027 clinical isolates. Int J Artif Organs. 2005;28:1091–1100. doi: 10.1177/039139880502801106. [DOI] [PubMed] [Google Scholar]
- Arciola CR, et al. Detection of biofilm formation in Staphylococcus epidermidis from implant infections. Comparison of a PCR-method that recognizes the presence of ica genes with two classic phenotypic methods. J Biomed Mater Res A. 2006;76:425–430. doi: 10.1002/jbm.a.30552. [DOI] [PubMed] [Google Scholar]
- Balaban N, et al. Treatment of Staphylococcus aureus Biofilm Infection by the Quorum-Sensing Inhibitor RIP. Antimicrob Agents Chemother. 2007;51:2226–2229. doi: 10.1128/AAC.01097-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balaban N, et al. Use of the quorum-sensing inhibitor RNAIII-inhibiting peptide to prevent biofilm formation in vivo by drug-resistant Staphylococcus epidermidis. J Infect Dis. 2003;187:625–630. doi: 10.1086/345879. [DOI] [PubMed] [Google Scholar]
- Banner MA, et al. Localized tufts of fibrils on Staphylococcus epidermidis NCTC 11047 are comprised of the accumulation-associated protein. J Bacteriol. 2007;189:2793–2804. doi: 10.1128/JB.00952-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beenken KE, et al. Global gene expression in Staphylococcus aureus biofilms. J Bacteriol. 2004;186:4665–4684. doi: 10.1128/JB.186.14.4665-4684.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boles BR, Thoendel M, Singh PK. Rhamnolipids mediate detachment of Pseudomonas aeruginosa from biofilms. Mol Microbiol. 2005;57:1210–1223. doi: 10.1111/j.1365-2958.2005.04743.x. [DOI] [PubMed] [Google Scholar]
- Bowden MG, et al. Identification and preliminary characterization of cell-wall-anchored proteins of Staphylococcus epidermidis. Microbiology. 2005;151:1453–1464. doi: 10.1099/mic.0.27534-0. [DOI] [PubMed] [Google Scholar]
- Bowden MG, Visai L, Longshaw CM, Holland KT, Speziale P, Hook M. Is the GehD lipase from Staphylococcus epidermidis a collagen binding adhesin? J Biol Chem. 2002;277:43017–43023. doi: 10.1074/jbc.M207921200. [DOI] [PubMed] [Google Scholar]
- Conlon KM, Humphreys H, O’Gara JP. icaR encodes a transcriptional repressor involved in environmental regulation of ica operon expression and biofilm formation in Staphylococcus epidermidis. J Bacteriol. 2002;184:4400–4408. doi: 10.1128/JB.184.16.4400-4408.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conlon KM, Humphreys H, O’Gara JP. Inactivations of rsbU and sarA by IS256 represent novel mechanisms of biofilm phenotypic variation in Staphylococcus epidermidis. J Bacteriol. 2004;186:6208–6219. doi: 10.1128/JB.186.18.6208-6219.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costerton JW, Lewandowski Z, Caldwell DE, Korber DR, Lappin-Scott HM. Microbial biofilms. Annu Rev Microbiol. 1995;49:711–745. doi: 10.1146/annurev.mi.49.100195.003431. [DOI] [PubMed] [Google Scholar]
- Costerton JW, Stewart PS, Greenberg EP. Bacterial biofilms: a common cause of persistent infections. Science. 1999;284:1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- Cramton SE, Ulrich M, Gotz F, Doring G. Anaerobic conditions induce expression of polysaccharide intercellular adhesin in Staphylococcus aureus and Staphylococcus epidermidis. Infect Immun. 2001;69:4079–4085. doi: 10.1128/IAI.69.6.4079-4085.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cucarella C, Solano C, Valle J, Amorena B, Lasa I, Penades JR. Bap, a Staphylococcus aureus surface protein involved in biofilm formation. J Bacteriol. 2001;183:2888–2896. doi: 10.1128/JB.183.9.2888-2896.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cucarella C, et al. Role of biofilm-associated protein bap in the pathogenesis of bovine Staphylococcus aureus. Infect Immun. 2004;72:2177–2185. doi: 10.1128/IAI.72.4.2177-2185.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darby C, Hsu JW, Ghori N, Falkow S. Caenorhabditis elegans: plague bacteria biofilm blocks food intake. Nature. 2002;417:243–244. doi: 10.1038/417243a. [DOI] [PubMed] [Google Scholar]
- Davey ME, Caiazza NC, O’Toole GA. Rhamnolipid surfactant production affects biofilm architecture in Pseudomonas aeruginosa PAO1. J Bacteriol. 2003;185:1027–1036. doi: 10.1128/JB.185.3.1027-1036.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies DG, Parsek MR, Pearson JP, Iglewski BH, Costerton JW, Greenberg EP. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science. 1998;280:295–298. doi: 10.1126/science.280.5361.295. [DOI] [PubMed] [Google Scholar]
- Dunne WM, Jr, Mason EO, Jr, Kaplan SL. Diffusion of rifampin and vancomycin through a Staphylococcus epidermidis biofilm. Antimicrob Agents Chemother. 1993;37:2522–2526. doi: 10.1128/aac.37.12.2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fluckiger U, et al. Biofilm formation, icaADBC transcription, and polysaccharide intercellular adhesin synthesis by staphylococci in a device-related infection model. Infect Immun. 2005;73:1811–1819. doi: 10.1128/IAI.73.3.1811-1819.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerke C, Kraft A, Sussmuth R, Schweitzer O, Gotz F. Characterization of the N-acetylglucosaminyltransferase activity involved in the biosynthesis of the Staphylococcus epidermidis polysaccharide intercellular adhesin. J Biol Chem. 1998;273:18586–18593. doi: 10.1074/jbc.273.29.18586. [DOI] [PubMed] [Google Scholar]
- Giesbrecht P, Wecke J, Reinicke B. On the morphogenesis of the cell wall of staphylococci. Int Rev Cytol. 1976;44:225–318. doi: 10.1016/s0074-7696(08)61651-4. [DOI] [PubMed] [Google Scholar]
- Gill SR, et al. Insights on evolution of virulence and resistance from the complete genome analysis of an early methicillin-resistant Staphylococcus aureus strain and a biofilm-producing methicillin-resistant Staphylococcus epidermidis strain. J Bacteriol. 2005;187:2426–2438. doi: 10.1128/JB.187.7.2426-2438.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross M, Cramton SE, Gotz F, Peschel A. Key role of teichoic acid net charge in Staphylococcus aureus colonization of artificial surfaces. Infect Immun. 2001;69:3423–3426. doi: 10.1128/IAI.69.5.3423-3426.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu J, et al. Bacterial insertion sequence IS256 as a potential molecular marker to discriminate invasive strains from commensal strains of Staphylococcus epidermidis. J Hosp Infect. 2005;61:342–348. doi: 10.1016/j.jhin.2005.04.017. [DOI] [PubMed] [Google Scholar]
- Heilmann C, Gotz F. Further characterization of Staphylococcus epidermidis transposon mutants deficient in primary attachment or intercellular adhesion. Zentralbl Bakteriol. 1998;287:69–83. doi: 10.1016/s0934-8840(98)80149-7. [DOI] [PubMed] [Google Scholar]
- Heilmann C, Hussain M, Peters G, Gotz F. Evidence for autolysin-mediated primary attachment of Staphylococcus epidermidis to a polystyrene surface. Mol Microbiol. 1997;24:1013–1024. doi: 10.1046/j.1365-2958.1997.4101774.x. [DOI] [PubMed] [Google Scholar]
- Heilmann C, Schweitzer O, Gerke C, Vanittanakom N, Mack D, Gotz F. Molecular basis of intercellular adhesion in the biofilm-forming Staphylococcus epidermidis. Mol Microbiol. 1996;20:1083–1091. doi: 10.1111/j.1365-2958.1996.tb02548.x. [DOI] [PubMed] [Google Scholar]
- Heilmann C, Thumm G, Chhatwal GS, Hartleib J, Uekotter A, Peters G. Identification and characterization of a novel autolysin (Aae) with adhesive properties from Staphylococcus epidermidis. Microbiology. 2003;149:2769–2778. doi: 10.1099/mic.0.26527-0. [DOI] [PubMed] [Google Scholar]
- Hussain M, Hastings JG, White PJ. Comparison of cell-wall teichoic acid with high-molecular-weight extracellular slime material from Staphylococcus epidermidis. J Med Microbiol. 1992;37:368–375. doi: 10.1099/00222615-37-6-368. [DOI] [PubMed] [Google Scholar]
- Hussain M, Heilmann C, Peters G, Herrmann M. Teichoic acid enhances adhesion of Staphylococcus epidermidis to immobilized fibronectin. Microb Pathog. 2001;31:261–270. doi: 10.1006/mpat.2001.0469. [DOI] [PubMed] [Google Scholar]
- Hussain M, Herrmann M, von Eiff C, Perdreau-Remington F, Peters G. A 140-kilodalton extracellular protein is essential for the accumulation of Staphylococcus epidermidis strains on surfaces. Infect Immun. 1997;65:519–524. doi: 10.1128/iai.65.2.519-524.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain M, Wilcox MH, White PJ. The slime of coagulase-negative staphylococci: biochemistry and relation to adherence. FEMS Microbiol Rev. 1993;10:191–207. doi: 10.1111/j.1574-6968.1993.tb05867.x. [DOI] [PubMed] [Google Scholar]
- Ji G, Beavis R, Novick RP. Bacterial interference caused by autoinducing peptide variants. Science. 1997;276:2027–2030. doi: 10.1126/science.276.5321.2027. [DOI] [PubMed] [Google Scholar]
- Jones SM, Morgan M, Humphrey TJ, Lappin-Scott H. Effect of vancomycin and rifampicin on meticillin-resistant Staphylococcus aureus biofilms. Lancet. 2001;357:40–41. doi: 10.1016/S0140-6736(00)03572-8. [DOI] [PubMed] [Google Scholar]
- Kaplan JB, Ragunath C, Ramasubbu N, Fine DH. Detachment of Actinobacillus actinomycetemcomitans biofilm cells by an endogenous beta-hexosaminidase activity. J Bacteriol. 2003;185:4693–4698. doi: 10.1128/JB.185.16.4693-4698.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan JB, Ragunath C, Velliyagounder K, Fine DH, Ramasubbu N. Enzymatic detachment of Staphylococcus epidermidis biofilms. Antimicrob Agents Chemother. 2004a;48:2633–2636. doi: 10.1128/AAC.48.7.2633-2636.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan JB, et al. Genes involved in the synthesis and degradation of matrix polysaccharide in Actinobacillus actinomycetemcomitans and Actinobacillus pleuropneumoniae biofilms. J Bacteriol. 2004b;186:8213–8220. doi: 10.1128/JB.186.24.8213-8220.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly-Quintos C, Cavacini LA, Posner MR, Goldmann D, Pier GB. Characterization of the opsonic and protective activity against Staphylococcus aureus of fully human monoclonal antibodies specific for the bacterial surface polysaccharide poly-N-acetylglucosamine. Infect Immun. 2006;74:2742–2750. doi: 10.1128/IAI.74.5.2742-2750.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keren I, Kaldalu N, Spoering A, Wang Y, Lewis K. Persister cells and tolerance to antimicrobials. FEMS Microbiol Lett. 2004;230:13–18. doi: 10.1016/S0378-1097(03)00856-5. [DOI] [PubMed] [Google Scholar]
- Kloos W, Schleifer KH. Staphylococcus. In: PHAS, SM, MES, JGH, editors. Bergey’s Manual of Systematic Bacteriology. Williams & Wilkins; Baltimore: 1986. [Google Scholar]
- Knobloch JK, Bartscht K, Sabottke A, Rohde H, Feucht HH, Mack D. Biofilm formation by Staphylococcus epidermidis depends on functional RsbU, an activator of the sigB operon: differential activation mechanisms due to ethanol and salt stress. J Bacteriol. 2001;183:2624–2633. doi: 10.1128/JB.183.8.2624-2633.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knobloch JK, Jager S, Horstkotte MA, Rohde H, Mack D. RsbU-dependent regulation of Staphylococcus epidermidis biofilm formation is mediated via the alternative sigma factor sigmaB by repression of the negative regulator gene icaR. Infect Immun. 2004;72:3838–3848. doi: 10.1128/IAI.72.7.3838-3848.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocianova S, et al. Key role of poly-gamma-DL-glutamic acid in immune evasion and virulence of Staphylococcus epidermidis. J Clin Invest. 2005;115:688–694. doi: 10.1172/JCI23523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozitskaya S, Cho SH, Dietrich K, Marre R, Naber K, Ziebuhr W. The bacterial insertion sequence element IS256 occurs preferentially in nosocomial Staphylococcus epidermidis isolates: association with biofilm formation and resistance to aminoglycosides. Infect Immun. 2004;72:1210–1215. doi: 10.1128/IAI.72.2.1210-1215.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozitskaya S, Olson ME, Fey PD, Witte W, Ohlsen K, Ziebuhr W. Clonal analysis of Staphylococcus epidermidis isolates carrying or lacking biofilm-mediating genes by multilocus sequence typing. J Clin Microbiol. 2005;43:4751–4757. doi: 10.1128/JCM.43.9.4751-4757.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristian SA, et al. The ability of biofilm formation does not influence virulence of Staphylococcus aureus and host response in a mouse tissue cage infection model. Microb Pathog. 2004;36:237–245. doi: 10.1016/j.micpath.2003.12.004. [DOI] [PubMed] [Google Scholar]
- Lasa I, Penades JR. Bap: a family of surface proteins involved in biofilm formation. Res Microbiol. 2006;157:99–107. doi: 10.1016/j.resmic.2005.11.003. [DOI] [PubMed] [Google Scholar]
- Latasa C, et al. BapA, a large secreted protein required for biofilm formation and host colonization of Salmonella enterica serovar Enteritidis. Mol Microbiol. 2005;58:1322–1339. doi: 10.1111/j.1365-2958.2005.04907.x. [DOI] [PubMed] [Google Scholar]
- Li H, et al. Conversion of Staphylococcus epidermidis strains from commensal to invasive by expression of the ica locus encoding production of biofilm exopolysaccharide. Infect Immun. 2005;73:3188–3191. doi: 10.1128/IAI.73.5.3188-3191.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim Y, Jana M, Luong TT, Lee CY. Control of glucose- and NaCl-induced biofilm formation by rbf in Staphylococcus aureus. J Bacteriol. 2004;186:722–729. doi: 10.1128/JB.186.3.722-729.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mack D, et al. The intercellular adhesin involved in biofilm accumulation of Staphylococcus epidermidis is a linear beta-1,6-linked glucosaminoglycan: purification and structural analysis. J Bacteriol. 1996;178:175–183. doi: 10.1128/jb.178.1.175-183.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marraffini LA, Dedent AC, Schneewind O. Sortases and the art of anchoring proteins to the envelopes of gram-positive bacteria. Microbiol Mol Biol Rev. 2006;70:192–221. doi: 10.1128/MMBR.70.1.192-221.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazmanian SK, Liu G, Ton-That H, Schneewind O. Staphylococcus aureus sortase, an enzyme that anchors surface proteins to the cell wall. Science. 1999;285:760–763. doi: 10.1126/science.285.5428.760. [DOI] [PubMed] [Google Scholar]
- McCrea KW, et al. The serine-aspartate repeat (Sdr) protein family in Staphylococcus epidermidis. Microbiology. 2000;146 (Pt 7):1535–1546. doi: 10.1099/00221287-146-7-1535. [DOI] [PubMed] [Google Scholar]
- Mehlin C, Headley CM, Klebanoff SJ. An inflammatory polypeptide complex from Staphylococcus epidermidis: isolation and characterization. J Exp Med. 1999;189:907–918. doi: 10.1084/jem.189.6.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarre WW, Schneewind O. Surface proteins of gram-positive bacteria and mechanisms of their targeting to the cell wall envelope. Microbiol Mol Biol Rev. 1999;63:174–229. doi: 10.1128/mmbr.63.1.174-229.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick RP. Autoinduction and signal transduction in the regulation of staphylococcal virulence. Mol Microbiol. 2003;48:1429–1449. doi: 10.1046/j.1365-2958.2003.03526.x. [DOI] [PubMed] [Google Scholar]
- Otto M. Bacterial evasion of antimicrobial peptides by biofilm formation. Curr Top Microbiol Immunol. 2006;306:251–258. doi: 10.1007/3-540-29916-5_10. [DOI] [PubMed] [Google Scholar]
- Patti JM, Allen BL, McGavin MJ, Hook M. MSCRAMM-mediated adherence of microorganisms to host tissues. Annu Rev Microbiol. 1994;48:585–617. doi: 10.1146/annurev.mi.48.100194.003101. [DOI] [PubMed] [Google Scholar]
- Peschel A, Vuong C, Otto M, Gotz F. The D-alanine residues of Staphylococcus aureus teichoic acids alter the susceptibility to vancomycin and the activity of autolytic enzymes. Antimicrob Agents Chemother. 2000;44:2845–2847. doi: 10.1128/aac.44.10.2845-2847.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qazi S, et al. N-acylhomoserine lactones antagonize virulence gene expression and quorum sensing in Staphylococcus aureus. Infect Immun. 2006;74:910–919. doi: 10.1128/IAI.74.2.910-919.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rachid S, Ohlsen K, Witte W, Hacker J, Ziebuhr W. Effect of subinhibitory antibiotic concentrations on polysaccharide intercellular adhesin expression in biofilm-forming Staphylococcus epidermidis. Antimicrob Agents Chemother. 2000;44:3357–3363. doi: 10.1128/aac.44.12.3357-3363.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renders N, Verbrugh H, Van Belkum A. Dynamics of bacterial colonisation in the respiratory tract of patients with cystic fibrosis. Infect Genet Evol. 2001;1:29–39. doi: 10.1016/s1567-1348(01)00004-1. [DOI] [PubMed] [Google Scholar]
- Rennermalm A, et al. Antibodies against a truncated Staphylococcus aureus fibronectin-binding protein protect against dissemination of infection in the rat. Vaccine. 2001;19:3376–3383. doi: 10.1016/s0264-410x(01)00080-9. [DOI] [PubMed] [Google Scholar]
- Resch A, et al. Comparative proteome analysis of Staphylococcus aureus biofilm and planktonic cells and correlation with transcriptome profiling. Proteomics. 2006;6:1867–1877. doi: 10.1002/pmic.200500531. [DOI] [PubMed] [Google Scholar]
- Resch A, Rosenstein R, Nerz C, Gotz F. Differential gene expression profiling of Staphylococcus aureus cultivated under biofilm and planktonic conditions. Appl Environ Microbiol. 2005;71:2663–2676. doi: 10.1128/AEM.71.5.2663-2676.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice KC, Bayles KW. Death’s toolbox: examining the molecular components of bacterial programmed cell death. Mol Microbiol. 2003;50:729–738. doi: 10.1046/j.1365-2958.2003.t01-1-03720.x. [DOI] [PubMed] [Google Scholar]
- Rice KC, et al. The cidA murein hydrolase regulator contributes to DNA release and biofilm development in Staphylococcus aureus. Proc Natl Acad Sci U S A. 2007;104:8113–8118. doi: 10.1073/pnas.0610226104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohde H, et al. Polysaccharide intercellular adhesin or protein factors in biofilm accumulation of Staphylococcus epidermidis and Staphylococcus aureus isolated from prosthetic hip and knee joint infections. Biomaterials. 2007;28:1711–1720. doi: 10.1016/j.biomaterials.2006.11.046. [DOI] [PubMed] [Google Scholar]
- Rohde H, et al. Induction of Staphylococcus epidermidis biofilm formation via proteolytic processing of the accumulation-associated protein by staphylococcal and host proteases. Mol Microbiol. 2005;55:1883–1895. doi: 10.1111/j.1365-2958.2005.04515.x. [DOI] [PubMed] [Google Scholar]
- Rupp ME, Fey PD, Heilmann C, Gotz F. Characterization of the importance of Staphylococcus epidermidis autolysin and polysaccharide intercellular adhesin in the pathogenesis of intravascular catheter-associated infection in a rat model. J Infect Dis. 2001;183:1038–1042. doi: 10.1086/319279. [DOI] [PubMed] [Google Scholar]
- Rupp ME, Ulphani JS, Fey PD, Bartscht K, Mack D. Characterization of the importance of polysaccharide intercellular adhesin/hemagglutinin of Staphylococcus epidermidis in the pathogenesis of biomaterial-based infection in a mouse foreign body infection model. Infect Immun. 1999a;67:2627–2632. doi: 10.1128/iai.67.5.2627-2632.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupp ME, Ulphani JS, Fey PD, Mack D. Characterization of Staphylococcus epidermidis polysaccharide intercellular adhesin/hemagglutinin in the pathogenesis of intravascular catheter-associated infection in a rat model. Infect Immun. 1999b;67:2656–2659. doi: 10.1128/iai.67.5.2656-2659.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadovskaya I, Vinogradov E, Flahaut S, Kogan G, Jabbouri S. Extracellular carbohydrate-containing polymers of a model biofilm-producing strain, Staphylococcus epidermidis RP62A. Infect Immun. 2005;73:3007–3017. doi: 10.1128/IAI.73.5.3007-3017.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw LN, Jonnson I-M, Singh VK, Tarkowski A, Stewart GC. Inactivation of traP has no effect on the Agr quorum sensing system or virulence of Staphylococcus aureus. Infect Immun. 2007 doi: 10.1128/IAI.00491-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somerville GA, et al. In vitro serial passage of Staphylococcus aureus: changes in physiology, virulence factor production, and agr nucleotide sequence. J Bacteriol. 2002;184:1430–1437. doi: 10.1128/JB.184.5.1430-1437.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tormo MA, et al. SarA Is an Essential Positive Regulator of Staphylococcus epidermidis Biofilm Development. J Bacteriol. 2005;187:2348–2356. doi: 10.1128/JB.187.7.2348-2356.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang LH, Daily ST, Weiss EC, Smeltzer MS. Mutation of traP in Staphylococcus aureus has no impact on expression of agr or biofilm formation. Infect Immun. 2007 doi: 10.1128/IAI.00603-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vadyvaloo V, Otto M. Molecular genetics of Staphylococcus epidermidis biofilms on indwelling medical devices. Int J Artif Organs. 2005;28:1069–1078. doi: 10.1177/039139880502801104. [DOI] [PubMed] [Google Scholar]
- van Belkum A. Staphylococcal colonization and infection: homeostasis versus disbalance of human (innate) immunity and bacterial virulence. Curr Opin Infect Dis. 2006;19:339–344. doi: 10.1097/01.qco.0000235159.40184.61. [DOI] [PubMed] [Google Scholar]
- Veenstra GJ, Cremers FF, van Dijk H, Fleer A. Ultrastructural organization and regulation of a biomaterial adhesin of Staphylococcus epidermidis. J Bacteriol. 1996;178:537–541. doi: 10.1128/jb.178.2.537-541.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuong C, Gerke C, Somerville GA, Fischer ER, Otto M. Quorum-sensing control of biofilm factors in Staphylococcus epidermidis. J Infect Dis. 2003;188:706–718. doi: 10.1086/377239. [DOI] [PubMed] [Google Scholar]
- Vuong C, et al. A crucial role for exopolysaccharide modification in bacterial biofilm formation, immune evasion, and virulence. J Biol Chem. 2004a;279:54881–54886. doi: 10.1074/jbc.M411374200. [DOI] [PubMed] [Google Scholar]
- Vuong C, Kocianova S, Yao Y, Carmody AB, Otto M. Increased colonization of indwelling medical devices by quorum-sensing mutants of Staphylococcus epidermidis in vivo. J Infect Dis. 2004b;190:1498–1505. doi: 10.1086/424487. [DOI] [PubMed] [Google Scholar]
- Vuong C, Otto M. Staphylococcus epidermidis infections. Microbes Infect. 2002;4:481–489. doi: 10.1016/s1286-4579(02)01563-0. [DOI] [PubMed] [Google Scholar]
- Vuong C, Saenz HL, Gotz F, Otto M. Impact of the agr quorum-sensing system on adherence to polystyrene in Staphylococcus aureus. J Infect Dis. 2000;182:1688–1693. doi: 10.1086/317606. [DOI] [PubMed] [Google Scholar]
- Vuong C, et al. Polysaccharide intercellular adhesin (PIA) protects Staphylococcus epidermidis against major components of the human innate immune system. Cell Microbiol. 2004c;6:269–275. doi: 10.1046/j.1462-5822.2004.00367.x. [DOI] [PubMed] [Google Scholar]
- Walters MC, 3rd, Roe F, Bugnicourt A, Franklin MJ, Stewart PS. Contributions of antibiotic penetration, oxygen limitation, and low metabolic activity to tolerance of Pseudomonas aeruginosa biofilms to ciprofloxacin and tobramycin. Antimicrob Agents Chemother. 2003;47:317–323. doi: 10.1128/AAC.47.1.317-323.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Preston JFI, Romeo T. The pgaABCD locus of Escherichia coli promotes the synthesis of a polysaccharide adhesin required for biofilm formation. J Bacteriol. 2004;186:2724–2734. doi: 10.1128/JB.186.9.2724-2734.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright JS, 3rd, Jin R, Novick RP. Transient interference with staphylococcal quorum sensing blocks abscess formation. Proc Natl Acad Sci U S A. 2005;102:1691–1696. doi: 10.1073/pnas.0407661102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu JA, Kusuma C, Mond JJ, Kokai-Kun JF. Lysostaphin disrupts Staphylococcus aureus and Staphylococcus epidermidis biofilms on artificial surfaces. Antimicrob Agents Chemother. 2003;47:3407–3414. doi: 10.1128/AAC.47.11.3407-3414.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, et al. Role of the luxS quorum-sensing system in biofilm formation and virulence of Staphylococcus epidermidis. Infect Immun. 2006;74:488–496. doi: 10.1128/IAI.74.1.488-496.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang SJ, Dunman PM, Projan SJ, Bayles KW. Characterization of the Staphylococcus aureus CidR regulon: elucidation of a novel role for acetoin metabolism in cell death and lysis. Mol Microbiol. 2006;60:458–468. doi: 10.1111/j.1365-2958.2006.05105.x. [DOI] [PubMed] [Google Scholar]
- Yao Y, Sturdevant DE, Otto M. Genomewide analysis of gene expression in Staphylococcus epidermidis biofilms: insights into the pathophysiology of S. epidermidis biofilms and the role of phenol-soluble modulins in formation of biofilms. J Infect Dis. 2005;191:289–298. doi: 10.1086/426945. [DOI] [PubMed] [Google Scholar]
- Yao Y, et al. Characterization of the Staphylococcus epidermidis Accessory-Gene Regulator Response: Quorum-Sensing Regulation of Resistance to Human Innate Host Defense. J Infect Dis. 2006;193:841–848. doi: 10.1086/500246. [DOI] [PubMed] [Google Scholar]
- Yarwood JM, Bartels DJ, Volper EM, Greenberg EP. Quorum sensing in Staphylococcus aureus biofilms. J Bacteriol. 2004;186:1838–1850. doi: 10.1128/JB.186.6.1838-1850.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YQ, et al. Genome-based analysis of virulence genes in a non-biofilm-forming Staphylococcus epidermidis strain (ATCC 12228) Mol Microbiol. 2003;49:1577–1593. doi: 10.1046/j.1365-2958.2003.03671.x. [DOI] [PubMed] [Google Scholar]
- Ziebuhr W, Krimmer V, Rachid S, Lossner I, Gotz F, Hacker J. A novel mechanism of phase variation of virulence in Staphylococcus epidermidis: evidence for control of the polysaccharide intercellular adhesin synthesis by alternating insertion and excision of the insertion sequence element IS256. Mol Microbiol. 1999;32:345–356. doi: 10.1046/j.1365-2958.1999.01353.x. [DOI] [PubMed] [Google Scholar]
- Zimmerli W, Waldvogel FA, Vaudaux P, Nydegger UE. Pathogenesis of foreign body infection: description and characteristics of an animal model. J Infect Dis. 1982;146:487–497. doi: 10.1093/infdis/146.4.487. [DOI] [PubMed] [Google Scholar]