Abstract
Bacterial flagellar motors rotate, obtaining power from the membrane gradient of protons or, in some species, sodium ions. Torque generation in the flagellar motor must involve interactions between components of the rotor and components of the stator. Sites of interaction between the rotor and stator have not been identified. Mutational studies of the rotor protein FliG and the stator protein MotA showed that both proteins contain charged residues essential for motor rotation. This suggests that functionally important electrostatic interactions might occur between the rotor and stator. To test this proposal, we examined double mutants with charged-residue substitutions in both the rotor protein FliG and the stator protein MotA. Several combinations of FliG mutations with MotA mutations exhibited strong synergism, whereas others showed strong suppression, in a pattern that indicates that the functionally important charged residues of FliG interact with those of MotA. These results identify a functionally important site of interaction between the rotor and stator and suggest a hypothesis for electrostatic interactions at the rotor–stator interface.
Keywords: flagella, motility, energy transduction
Many species of bacteria are propelled by flagella, each consisting of a thin helical propeller attached, via a flexible coupling, to a rotary motor in the cell membrane (for recent reviews, see refs. 1 and 2). The energy for rotation of the flagella comes from the transmembrane gradient of protons (3–5) or, in certain marine or alkaliphilic species, sodium ions (6). The molecular mechanism of torque generation is not understood. The proteins most closely involved are MotA, MotB, and FliG. MotA and MotB are integral membrane proteins (7–11) that function in proton conduction (12–14) and that are believed to form the stator, or nonrotating part, of the motor (9, 15–17). FliG is located on the cytoplasmic side of the membrane, and is thought to be a component of the rotor (18–22).
In hypotheses for the mechanism of the flagellar motor, charged residues have often been suggested to play key roles, serving, for example, to regulate movements of the rotor or to control proton flow (5, 23–25). We recently identified charged residues essential for motor function in both the rotor protein FliG and the stator protein MotA in mutational analyses of these proteins (26, 27). In FliG, the residues Arg-281, Asp-288, and Asp-289 are of primary importance for function, whereas residues Lys-264 and Arg-297 might have secondary roles [FliG residue numbers here are higher by two than those given in the previous study, owing to recent corrections to the Escherichia coli fliG sequence (28)]. In MotA, the charged residues most important for function are Arg-90 and Glu-98, whereas Glu-150 may have a secondary role. Mutant phenotypes suggest that charge is the most important feature of these residues and that the charged residues in each protein function redundantly. Single charge-neutralizing replacements had only mild effects on function, whereas double replacements in either protein impaired function severely; mutations that reversed charge caused more severe impairments than mutations that neutralized charge; and mutations that altered side-chains but preserved charge had little effect. The precise function of these residues in torque generation is not known. Because charge appears to be their most important property, it was suggested that they might mediate essential electrostatic interactions between the rotor and stator (26, 27).
To test the hypothesis that charged residues of the rotor protein FliG interact with those of the stator protein MotA, we made and characterized double mutants with replacements of charged residues in both proteins. Many instances of strong synergism, and some instances of strong suppression, were observed in the double mutants. Cases of strong synergism or supression all involved residues shown previously to be important for torque generation. The results indicate that charged residues of the rotor and stator interact, and that these interactions are important for motor rotation. We propose a hypothesis for electrostatic interactions at the rotor–stator interface, and discuss possible roles for these interactions in torque generation by the flagellar motor.
EXPERIMENTAL PROCEDURES
Strains, Plasmids, and Materials.
A strain defective in both fliG and motA was constructed by transferring a plasmid-encoded deletion in fliG (29) into the chromosome of the motA-deficient strain MS5037, a gift from M. I. Simon (California Institute of Technology, Pasadena), by using the procedure of Hamilton et al. (30). This strain, designated DFB245, was used as host in assays of function of mutant FliG and MotA proteins.
Most of the motA and fliG mutations were described previously (26, 27). Additional site-directed mutations of fliG were made by using the Altered Sites procedure (Promega) with plasmid pSL27, a derivative of pAlter-1 (Promega) that encodes fliG. Mutations were verified by dideoxynucleotide sequencing (31) of double-stranded plasmid DNA. To allow coexpression of the mutant FliG and MotA proteins in various combinations, two compatible plasmids were used. FliG and its mutant variants were expressed from pSL27, which confers ampicillin resistance (29). MotA and its mutant variants were expressed from plasmid pJZ19, a derivative of plasmid pACYC184 (New England Biolabs) that confers chloramphenicol resistance, constructed as follows. A 2.84-kb SspI/BsmI fragment of pDFB45 (14) that encoded motA and motB, and a 2.06-kb Tth111I/HindIII fragment of pACYC184 that encoded chloramphenicol acetyltransferase, were treated with mung bean nuclease and ligated. The ligation mixture was used to transform a motA mutant strain, and a motile, chloramphenicol-resistant transformant was selected. The plasmid isolated from this strain was designated pJZ19. Its size and composition were confirmed by digestion with EcoRI and HindIII. Mutant motA genes were then transferred (from pRF4; ref. 27) into pJZ19 by using a HindIII site upstream of motA and a NsiI site in motB.
Plasmid transformations and DNA manipulations were according to the procedures of Sambrook et al. (32). Restriction enzymes were from New England Biolabs. Plasmid DNA was isolated from single colonies by using the Flexi-Prep kit from Pharmacia. Deoxyadenosine 5′-[α-[35S]thio]triphosphate was from DuPont/NEN and Sequenase was from Amersham. Deoxyoligonucleotides were synthesized at the University of Utah Protein-DNA Core Facility.
Assays of Motility and Flagellation.
Cells were cultured at 32°C with shaking in tryptone broth [1% Bacto-tryptone (Difco), 0.5% NaCl]. When appropriate, ampicillin was included in plates and liquid medium at 100 μg/ml, and chloramphenicol at 35 μg/ml.
For assays of swarming motility, 1-μl aliquots of overnight cultures were spotted onto swarm plates containing tryptone broth, 0.28% Bacto-agar (Difco), and appropriate antibiotics, and plates were incubated at 32°C. Swarm diameters were measured at regular intervals (typically once every hour) and used to compute swarming rates. Rates are reported relative to wild-type controls present on the same plates. In some cases, cell motility in liquid culture was also characterized by visual observation under a phase-contrast microscope. Assays of tethered-cell rotation were carried out as described (33). Flagella were stained and counted as described (33); values reported are averages for 50 cells.
RESULTS
A strain with chromosomal defects in both fliG and motA was constructed as described in Experimental Procedures to serve as host for the expression of mutant FliG and MotA proteins. Null mutants of fliG are nonflagellate because FliG is essential for flagellar assembly as well as for rotation. Null mutants of motA are well-flagellated but immotile (34). The fliG motA double mutant was nonflagellate, as expected. Transformation of this strain with a plasmid encoding fliG restored flagella but not motility. Cotransformation with two plasmids, one encoding fliG and another encoding motA, restored both flagella and good motility, as judged by microscopic examination of swimming cells and by swarming in soft-agar plates (Fig. 1). Strains with normal motility swarm outward rapidly from the point of inoculation on such plates, whereas those with impaired motility swarm more slowly or not at all.
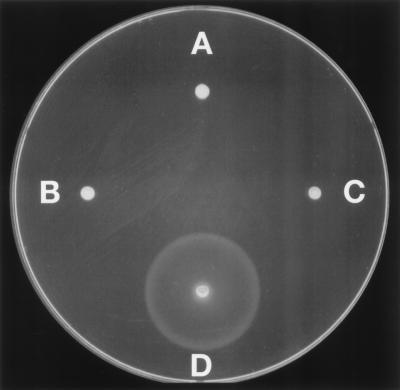
Figure 1.
Complementation of the fliG motA double-mutant strain by wild-type fliG and motA genes on plasmids. Shown are swarms in soft-agar plates of (A) strain DFB245, a fliG motA double mutant; (B) strain DFB245 transformed with plasmid pSL27, which encodes FliG; (C) strain DFB245 transformed with plasmid pJZ19, which encodes MotA; and (D) strain DFB245 transformed with both pSL27 and PJZ19. Flagellar staining showed that strains A and C were nonflagellate, whereas strains B and D had 2.4 and 4.5 flagella per cell, respectively (averages for 50 cells).
The mutant MotA and FliG proteins were coexpressed in the fliG motA double-mutant strain in various combinations by using two compatible plasmids. All of the mutations studied were in conserved charged residues, of which there are 3 in MotA (27), and 14 in the C-terminal domain of FliG that has previously been shown to function most closely in torque generation (26, 29). Motility of the double-mutant strains was assessed by measuring rates of swarming in soft-agar plates. Swarming rates of the double mutants are summarized in Table 1, and examples of swarming phenotypes are shown in Fig. 2. Because the strain is wild type for homologous recombination (rather than recA−, for example), recombination between plasmid and chromosome sequences is possible and could in principle produce wild-type copies of the gene that allow swarming. In such a case, the onset of swarming would be significantly delayed, however. This was not observed for any of the mutants, indicating that the observed swarming was caused by plasmid-encoded genes rather than by products of homologous recombination.
Table 1.
Swarming rates in soft agar of fliG motA double mutants, measured relative to wild-type controls on the same plates
| FliG mutation | MotA mutation
|
||||||
|---|---|---|---|---|---|---|---|
| WT | R90A | E98Q | E150Q | R90E | E98K | E150K | |
| WT | 1.00 | 0.71 | 0.98 | 1.09 | 0.00 | 0.00 | 0.66 |
| E224A | 1.02 | 0.69 | 0.86 | 1.07 | 0.00 | 0.00 | 0.56 |
| E237A | 1.12 | 0.73 | 0.94 | 1.14 | 0.00 | 0.00 | 0.58 |
| K264A | 0.96 | 0.09 | 0.66 | 1.03 | 0.00 | 0.00 | 0.45 |
| K264E | 0.69 | 0.00 | 0.00 | 0.66 | 0.00 | 0.00 | 0.00 |
| E272A | 1.01 | 0.71 | 0.81 | 1.04 | 0.00 | 0.00 | 0.70 |
| R281A | 0.86* | 0.00 | 0.53 | 0.91 | 0.00 | 0.00 | 0.00 |
| R281D | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| R281V | 0.04 | NM | NM | NM | NM | 0.29 | NM |
| R281W | 0.12 | NM | NM | NM | NM | 0.18 | NM |
| R287A | 1.02 | 0.59 | 0.85 | 1.06 | 0.00 | 0.00 | 0.64 |
| D288A | 0.94 | 0.10 | 0.15 | 0.97 | 0.00 | 0.00 | 0.00 |
| D289A | 0.92 | 0.72 | 0.00 | 0.91 | 0.40 | 0.00 | 0.00 |
| D289K | 0.23 | 0.58 | 0.00 | 0.15 | 0.77 | 0.00 | 0.00 |
| R297A | 1.10 | 0.11 | 0.66 | 1.03 | 0.00 | 0.00 | 0.09 |
| R297D | 0.27 | 0.00 | 0.00 | 0.34 | 0.00 | 0.00 | 0.00 |
| E302A | 0.96 | 0.34 | 0.61 | 1.00 | 0.00 | 0.00 | 0.44 |
| R313A | 1.00 | 0.54 | 0.81 | 0.84 | 0.00 | 0.00 | 0.68 |
| E320A | 1.06 | 0.72 | 0.85 | 1.14 | 0.00 | 0.00 | 0.78 |
| E327A | 0.96 | 0.40 | 0.75 | 0.95 | 0.00 | 0.00 | 0.43 |
| D328A | 1.12 | 0.41 | 0.92 | 1.07 | 0.00 | 0.00 | 0.35 |
Values are the mean of two determinations that differed by an average of 6%. The mutant FliG and MotA proteins were expressed from plasmids in the fliG motA double-mutant strain DFB245. The wild-type (WT) control was the fliG motA double-mutant strain expressing wild-type FliG and MotA from the plasmids; its swarm rate was 7.5 mm/hr. Instances of synergism are indicated by boldface type, and instances of suppression are indicated by boldface italic type. NM, not measured.
The FliG mutant R281A swarmed at only 40% of wild type in a previous mutational study of fliG (26) but faster in the present study. This difference is due to different levels of MotA in the two experiments. In the previous study, MotA was expressed from the chromosome, whereas in the present study MotA was moderately overexpressed from a plasmid. When a plasmid expressing wild-type MotA (pJZ19) was introduced into the strain used in the previous study, the swarming rate of the R281A mutant increased to about 90% of wild type.
Figure 2.
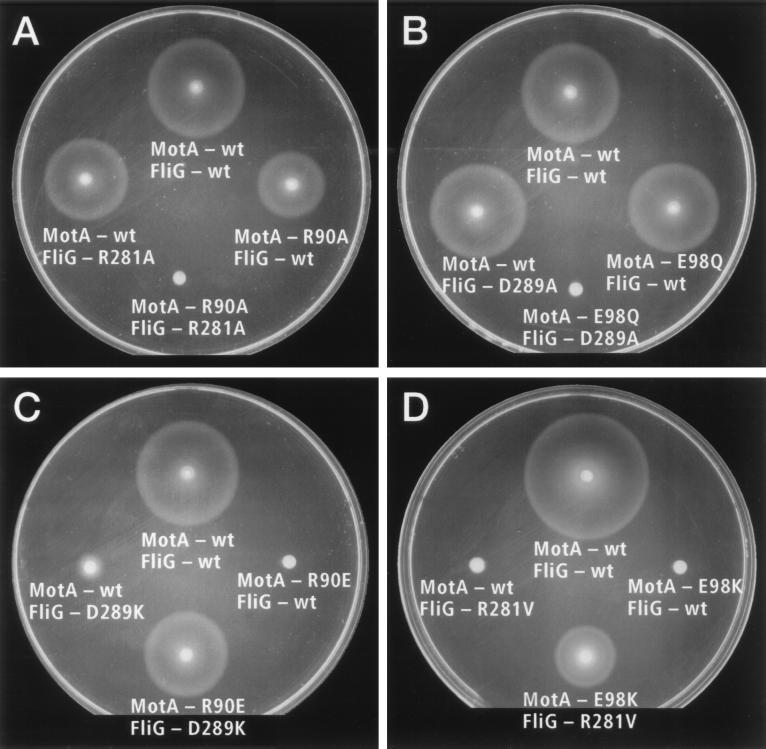
Examples of swarming phenotypes of fliG motA double mutants, illustrating cases of synergism and suppression. FliG and MotA proteins were expressed from two compatible plasmids in a fliG motA double-mutant strain. (A) Synergistic effect of combining the MotA mutation R90A with the FliG mutation R281A. (B) Synergistic effect of combining the MotA mutation E98Q with the FliG mutation D289A. (C) Mutual suppression of the mutations R90E (MotA) and D289K (FliG). (D) Mutual suppression of the mutations E98K (MotA) and R281V (FliG).
Several combinations of charge-neutralizing mutations in FliG and MotA acted synergistically, together impairing function strongly even though the individual mutations had only mild effects (Table 1). Two combinations abolished swarming completely. These were the R90A mutation of MotA together with the R281A mutation of FliG, and the E98Q mutation of MotA together with the D289A mutation of FliG. Other combinations reduced swarming to about one-tenth of normal. These were the E98Q mutation of MotA together with the D286A mutation of FliG, and the R90A mutation of MotA together with the K264A, D288A, or R297A mutations of FliG. All such cases of strong synergism thus involve residues shown previously to be functionally important, to various extents, in the mutational studies of FliG and MotA (26, 27). The residues classified previously as secondarily important (Glu-150 of MotA, and Lys-264 and Arg-297 of FliG) were also parties to strong synergistic effects. Generally, charge-reversing mutations in these residues had effects about as severe as charge-neutralizing mutations in the residues of primary importance (Table 1).
The neutral-replacement mutants that showed very strong synergism, R90A(MotA)/R281A(FliG) and E98Q(MotA)/D289A(FliG), were examined more closely to determine the nature of the impairment. When examined in the microscope, the R90A(MotA)/R281A(FliG) mutant was immotile, and the E98Q(MotA)/D289A(FliG) mutant was very weakly motile. Flagellar staining showed that both mutants were well flagellated (2.7 and 3.1 flagella per cell in the R90A/R281A and E98Q/D289A mutants, respectively, compared with 4.3 flagella per cell in a wild-type control; averages for 50 cells of each strain). When tethered to coverslips by their flagellar filaments, cells of the mutants rotated more slowly than wild-type cells. The impaired motility of these mutants is therefore caused by defects in flagellar rotation and not flagellar assembly.
Instances of strong suppression were also observed and involved the same set of residues (Table 1 and Fig. 2). The charge-reversing mutation R90E in MotA abolishes motility. This MotA defect was partially suppressed by the charge-neutralizing mutation D289A in FliG (swarming rate 40% of wild type) and was suppressed more effectively by the charge-reversing mutation D289K (swarming rate 77% of wild type). The suppression is mutual because the FliG mutant D289K swarmed slowly when MotA was wild type, faster when MotA harbored the mutation R90A, and still faster when it harbored the mutation R90E.
The charge-reversing mutation E98K in MotA also abolishes motility, but in contrast to the R90E mutation, this defect was not suppressed by any of the alanine-replacement mutations or charge-reversing mutations in FliG. As discussed below, the pattern of synergistic effects suggests that Glu-98 of MotA interacts with Arg-281 of FliG, so the failure of the FliG mutations R281A and R281D to suppress the E98K defect is most significant. This failure to suppress might reflect a requirement for residues other than alanine or aspartic acid at position 281 in FliG. To test this, residue 281 of FliG was mutagenized by using an oligonucleotide randomized at the corresponding codon, and replacements were sought that could suppress the E98K defect in MotA. Two replacements in FliG were isolated, R281V and R281W, that suppressed the E98K defect of MotA (swarming rates of 29% and 18% of wild type, respectively). The suppression is again mutual, most strongly in the case of the mutation R281V, which nearly abolished swarming when MotA was wild type (Table 1 and Fig. 2).
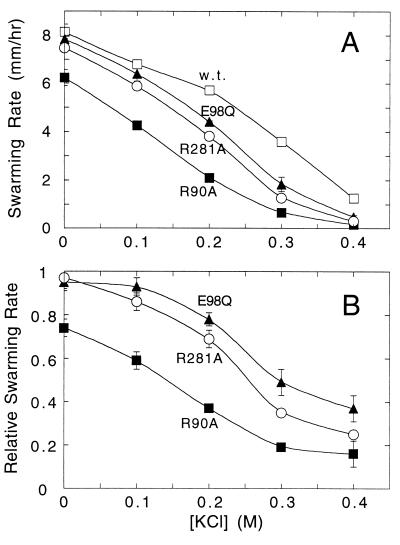
As discussed below, these results suggest that functionally important electrostatic interactions occur between rotor and stator. Increased ionic strength might therefore be expected to impair motor function, possibly more strongly in charged-residue mutants than in the wild type. To test this, we measured the effects of increased ionic strength on the motility of wild-type and mutant cells. Intracellular ionic strength can be increased to a significant extent by the addition of salts to the medium (35). Swarming rates of wild-type and selected mutant strains were measured on soft-agar plates supplemented with KCl, NaCl, choline-Cl, or sucrose (sucrose at twice the concentrations used for the salts). As shown in Fig. 3, KCl in the plates significantly slowed the swarming of all strains. More importantly, the KCl effect was stronger for charged-residue mutants more than for the wild type. Some charged-residue mutants that swarmed almost normally in standard medium (MotA E98Q; FliG R281A) swarmed at less than half the wild-type rate in the presence of 0.3–0.4 M KCl. Comparable effects were observed with NaCl, but not with the less-permeant salt choline chloride or the neutral osmolyte sucrose (data not shown).
Figure 3.
Effects of KCl on swarming motility of wild-type E. coli and strains with mutations in key charged residues. (A) Swarming rates of wild-type and mutant strains in soft-agar plates containing KCl at the concentrations indicated. Data are the average of 10 determinations for the wild type and 2 determinations for each of the mutants. (B) Swarming rates of the charged-residue mutants, measured relative to wild-type controls present on the same plates at various concentrations of KCl (mean ± SD; n = 3).
DISCUSSION
Previous mutational studies (26, 27) identified a small number of functionally important charged residues in both the rotor protein FliG and the stator protein MotA. In this study we examined the effect of combining mutations in the two proteins. Certain combinations of FliG mutations and MotA mutations acted synergistically, together abolishing swarming even though the individual mutations had only mild effects. Other combinations showed strong suppression, restoring very good motility to immotile or poorly motile mutants. All such instances of strong nonadditivity involved residues shown to be important for function in the previous mutational studies of FliG and MotA.
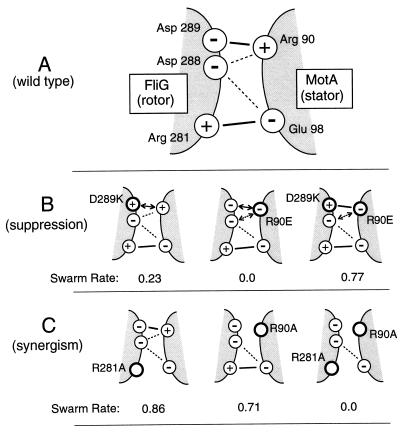
A straightforward interpretation of the synergism and suppression seen with the FliG and MotA mutations is that the charged residues of these proteins interact, and that these electrostatic interactions are important for motor function. A specific hypothesis for interactions at the rotor–stator interface is presented in Fig. 4. Assumptions of the hypothesis are that (i) the electrostatic interactions pictured function to promote an essential step(s) in torque generation; (ii) no single electrostatic interaction is, by itself, critical to motor function; and (iii) reversing the sign of an electrostatic interaction by reversing one of the charges impairs function more severely than eliminating the interaction by neutralizing a charge. As detailed below, this hypothesis can account for the pattern of synergism and suppression observed in the FliG/MotA double mutants and for the effects of single and multiple mutations in each protein as reported previously (26, 27).
Figure 4.
(A) Hypothesis for charged-residue interactions at the rotor–stator interface. The functionally important charged residues of wild-type FliG and MotA, and hypothesized electrostatic interactions between them, are shown. Solid lines indicate interactions of primary importance for motor function, and thin dashed lines indicate interactions of secondary importance. The interactions are drawn as occurring simultaneously, but might actually occur sequentially, as the rotor and stator move relative to one another or as one of the proteins undergoes a conformational change. (B) An example of strong suppression and its rationalization in the framework of the model. Mutually suppressing mutations of FliG and MotA, and their hypothesized effects on electrostatic interactions at the rotor–stator interface, are shown. Double-headed arrows indicate repulsive interactions in the mutants that take the place of attractive interactions in the wild type. (C) An example of strong synergism.
The synergistic action of certain combinations of FliG mutations with MotA mutations mirrors what is observed when multiple mutations are made within each protein. Neutralizing any one of the key charged residues in FliG or MotA has relatively little effect, but neutralizing two key residues in either protein abolishes or severely impairs function. Thus, for example, the mutation R90A in MotA allows good swarming, but swarming is abolished by additionally mutating either Glu-98 of the same MotA protein (27) or residue Arg-281 of FliG (Table 1). This suggests that Glu-98 of MotA interacts with Arg-281 of FliG. Similar reasoning suggests that Arg-90 of MotA interacts with Asp-289 of FliG: the mutation E98Q in MotA allows good swarming, but swarming is abolished by the additional mutation of either Arg-90 of MotA (27) or Asp-289 of FliG (Table 1). The instances of suppression provide additional, stronger evidence for the same interactions. The E98K defect in MotA is suppressed by mutations in Arg-281 of FliG, and the R90E defect in MotA is suppressed by mutations in Asp-289 of FliG. In both cases, suppression is mutual, and in the case of the Arg-90/Asp-289 pair, charge-reversing mutations are more effective suppressors than charge-neutralizing mutations. These results suggest a simple interpretation of the Arg-90/Asp-289 suppression: an attractive electrostatic interaction between these residues is helpful for motor function, whereas a repulsive interaction is harmful.
Other electrostatic interactions between rotor and stator probably occur in addition to the ones pictured in Fig. 4. Certain instances of synergism in Table 1 suggest that the residues previously classified as secondarily important (Lys-264 and Arg-297 of FliG, and Glu-150 of MotA) might also engage in rotor–stator interactions. Those interactions appear less important for function than the ones shown, however.
We cannot rule out the occurrence of other kinds of interactions between rotor and stator, such as hydrophobic interactions. A number of observations suggest that the electrostatic interactions hypothesized are directly important for function, however, rather than serving just to maintain the integrity of an interface for interaction by other forces. The charges of the key residues are conserved evolutionarily, which would not be expected if any kind of attractive interaction could serve. Replacements that preserve charge but otherwise alter side-chain structure (e.g., the R90K/E98D mutation in MotA) allow good motor function; this would not be expected if these residues functioned primarily to maintain complementarity of interacting surfaces. Finally, certain charge-reversing mutations in MotA and FliG disrupt function by themselves but allow good function when present together; this is expected if electrostatic interactions are of primary importance but would be unlikely if the mutations disrupted function by disrupting surface complementarity.
Electrostatic interactions are weaker in water than in a less polar milieu. They could still exert significant forces, however, particularly if the interacting groups are positioned in the motor so as to ensure that they will approach each other closely at some step(s) in motor rotation. Also, water might be partially excluded from the rotor–stator interface, which would make the interactions stronger. The effects of increased ionic strength (Fig. 3) support the proposal that electrostatic interactions, occurring between groups exposed to water, are important for motor function. We cannot rule out indirect effects of ionic strength, however. It is possible, for example, that increased intracellular salt affects the folding and/or stability of the mutant MotA and FliG proteins. The swimming speed of cells in liquid medium is also reduced by added salt, and this effect occurs quickly (within the ≈30 sec required for mixing; unpublished results). Thus, if salt-induced misfolding is the cause of reduced motor performance, it must involve the rapid inactivation of already folded, functional protein.
What might be the function of electrostatic interactions between the rotor and stator? One possibility is that electrostatic interactions between MotA and FliG trigger a needed event in the torque-generating cycle, such as a conformational change in one of the proteins. Conformational changes in the MotA/MotB complexes appear most likely because the movement of protons (or sodium ions) through these complexes must somehow be coupled to rotation. The hypothesized electrostatic interactions might ensure that a conformational change in the stator occurred only when the rotor and stator were correctly aligned, thus serving to link ion movements to rotor movements. Another possibility is that electrostatic interactions between rotor and stator act directly to control or accelerate rotor movements at some steps in rotation. A third possibility is that the charged residues at the MotA–FliG interface form a pathway for the conduction of protons. Rapid, rotation-coupled proton conduction would presumably require a fairly specific arrangement of chemical groups (more so than would conformational changes or rotor movements). This last alternative thus appears less likely given the resilience of the site toward mutation (26, 27).
The identification of a functionally important site of contact between the rotor and stator should be useful in the development of more detailed hypotheses for the mechanism of the flagellar motor. Additional physiological and structural studies will be needed to clarify the role of electrostatic interactions in torque generation and to determine the arrangement of key residues at the rotor–stator interface.
Acknowledgments
We thank Sandy Parkinson and Melvin Simon for strains, Stephanie Billings for construction of the fliG motA double-mutant strain and for flagellar staining, Carolyn Sammon and Jonathan Gully for assistance with tethering experiments, and Sandy Parkinson, David Goldenberg, and Patricia Renfranz for discussions. This work was supported by Grant 2-R01-GM46683 from the National Institutes of Health and by Grant MCB-9513486 from the National Science Foundation. S.A.L. was supported in part by Training Grant 5T32-GM08537 from the National Institutes of Health. The Protein-DNA Core Facility at the University of Utah receives support from the National Cancer Institute (5P30 CA42014).
References
- 1.Blair D F. Annu Rev Microbiol. 1995;49:489–522. doi: 10.1146/annurev.mi.49.100195.002421. [DOI] [PubMed] [Google Scholar]
- 2.Macnab R M. In: Escherichia coli and Salmonella, Cell and Molecular Biology. Neidhardt F C, editor. Washington, DC: Am. Soc. Microbiol.; 1995. pp. 123–145. [Google Scholar]
- 3.Larsen S H, Adler J, Gargus J J, Hogg R W. Proc Natl Acad Sci USA. 1974;71:1239–1243. doi: 10.1073/pnas.71.4.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Matsuura S, Shioi J-I, Imae Y. FEBS Lett. 1977;82:187–190. doi: 10.1016/0014-5793(77)80581-4. [DOI] [PubMed] [Google Scholar]
- 5.Glagolev A N, Skulachev V P. Nature (London) 1978;272:280–282. doi: 10.1038/272280a0. [DOI] [PubMed] [Google Scholar]
- 6.Hirota N, Imae Y. J Biol Chem. 1983;258:10577–10581. [PubMed] [Google Scholar]
- 7.Dean G E, Macnab R M, Stader J, Matsumura P, Burke C. J Bacteriol. 1984;159:991–999. doi: 10.1128/jb.159.3.991-999.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stader J, Matsumura P, Vacante D, Dean G E, Macnab R M. J Bacteriol. 1986;166:244–252. doi: 10.1128/jb.166.1.244-252.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chun S Y, Parkinson J S. Science. 1988;239:276–278. doi: 10.1126/science.2447650. [DOI] [PubMed] [Google Scholar]
- 10.Wilson L M, Macnab R M. J Bacteriol. 1988;170:588–597. doi: 10.1128/jb.170.2.588-597.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou J, Fazzio R T, Blair D F. J Mol Biol. 1995;251:237–242. doi: 10.1006/jmbi.1995.0431. [DOI] [PubMed] [Google Scholar]
- 12.Blair D F, Berg H C. Cell. 1990;60:439–449. doi: 10.1016/0092-8674(90)90595-6. [DOI] [PubMed] [Google Scholar]
- 13.Blair D F, Berg H C. J Mol Biol. 1991;221:1433–1442. doi: 10.1016/0022-2836(91)90943-z. [DOI] [PubMed] [Google Scholar]
- 14.Stolz B, Berg H C. J Bacteriol. 1991;173:7033–7037. doi: 10.1128/jb.173.21.7033-7037.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blair D F, Kim D-Y, Berg H C. J Bacteriol. 1991;173:4049–4055. doi: 10.1128/jb.173.13.4049-4055.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.DeMot R, Vanderleyden J. Mol Microbiol. 1994;12:333–334. doi: 10.1111/j.1365-2958.1994.tb01021.x. [DOI] [PubMed] [Google Scholar]
- 17.Garza A G, Harris-Haller L W, Stoebner R A, Manson M D. Proc Natl Acad Sci USA. 1995;92:1970–1974. doi: 10.1073/pnas.92.6.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Francis N R, Irikura V M, Yamaguchi S, DeRosier D J, Macnab R M. Proc Natl Acad Sci USA. 1992;89:6304–6308. doi: 10.1073/pnas.89.14.6304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Francis N R, Sosinsky G E, Thomas D, DeRosier D J. J Mol Biol. 1994;235:1261–1270. doi: 10.1006/jmbi.1994.1079. [DOI] [PubMed] [Google Scholar]
- 20.Zhao R, Schuster S C, Khan S. J Mol Biol. 1995;251:400–412. doi: 10.1006/jmbi.1995.0443. [DOI] [PubMed] [Google Scholar]
- 21.Garza A G, Biran R, Wohlschlegel J A, Manson M D. J Mol Biol. 1996;258:270–285. doi: 10.1006/jmbi.1996.0249. [DOI] [PubMed] [Google Scholar]
- 22.Tang H, Braun T F, Blair D F. J Mol Biol. 1996;261:209–221. doi: 10.1006/jmbi.1996.0453. [DOI] [PubMed] [Google Scholar]
- 23.Murata T, Yano M, Shimizu H. J Theor Biol. 1989;139:531–559. doi: 10.1016/s0022-5193(89)80069-4. [DOI] [PubMed] [Google Scholar]
- 24.Blair D F. Semin Cell Biol. 1990;1:75–85. [PubMed] [Google Scholar]
- 25.Berry R M. Biophys J. 1993;64:961–973. doi: 10.1016/S0006-3495(93)81462-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lloyd S A, Blair D F. J Mol Biol. 1997;266:733–744. doi: 10.1006/jmbi.1996.0836. [DOI] [PubMed] [Google Scholar]
- 27.Zhou J, Blair D F. J Mol Biol. 1997;273:428–439. doi: 10.1006/jmbi.1997.1316. [DOI] [PubMed] [Google Scholar]
- 28.Marykwas D L, Berg H C. J Bacteriol. 1996;178:1289–1294. doi: 10.1128/jb.178.5.1289-1294.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lloyd S A, Tang H, Wang X, Billings S, Blair D F. J Bacteriol. 1996;178:223–231. doi: 10.1128/jb.178.1.223-231.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hamilton C M, Aldea M, Washburn B K, Babitzke P, Kushner S R. J Bacteriol. 1989;171:4617–4622. doi: 10.1128/jb.171.9.4617-4622.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sanger F, Nicklen S, Coulson A R. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 33.Tang H, Blair D F. J Bacteriol. 1995;177:3485–3495. doi: 10.1128/jb.177.12.3485-3495.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Macnab R M. Annu Rev Genet. 1992;26:129–156. doi: 10.1146/annurev.ge.26.120192.001023. [DOI] [PubMed] [Google Scholar]
- 35.Kohno T, Roth J. Biochemistry. 1979;18:1386–1392. doi: 10.1021/bi00574a041. [DOI] [PubMed] [Google Scholar]