Abstract
Despite the prevalence of craniofacial disorders, the genetic contribution remains poorly understood. Class III malocclusion represents a specific craniofacial problem that can be handicapping, both functionally and socially. We hypothesized that the Class III phenotype is genetically linked to specific loci that regulate maxillary or mandibular growth. To determine the region linked to the Class III phenotype in four Hispanic families, we performed a genome-wide scan and linkage analysis using 500 microsatellite markers. Pedigree and linkage analyses revealed that the Class III phenotype (primarily maxillary deficiency) segregates in an autosomal-dominant manner, and that 5 loci (1p22.1, 3q26.2, 11q22, 12q13.13, and 12q23) are suggestive of linkage. Candidate genes within the 12q23 region (ZLR = 2.93) include IGF1, HOXC, and COL2A1. Chromosome 1 results (ZLR = 2.92) were similar to those reported previously in an Asian cohort with mandibular prognathism, suggesting that a common upstream genetic element may be responsible for both mandibular prognathism and maxillary deficiency.
Keywords: Class III phenotype, malocclusion, dentofacial deformity, Hispanic
Introduction
Class III malocclusion is a dentofacial phenotype characterized by negative anterior overjet (underbite), with a prevalence of 1-5% in the US (Proffit et al., 1998) and up to 23% in Asia (Susami et al., 1972; Tang, 1994). If this is the result of a significant underlying skeletal dysplasia, the individual is said to have a ‘dentofacial deformity’ (Proffit and White, 1991). Dentofacial deformities are a major problem in the US, with 1-2% of the population having a jaw deformity severe enough to be handicapping, both functionally and socially (Proffit et al., 1998). About half of these individuals are affected severely enough that orthognathic surgery is the only treatment method (Proffit and White, 1991).
In comparison with Class I and Class II (relative mandibular deficiency) anteroposterior relationships, the Class III relationship occurs least frequently in the US, but is most often cited as being an inherited trait. Despite being the least common malocclusion, the Class III phenotype can be thought of as common when compared with other craniofacial disorders (i.e., cleft lip and palate). It is defined by a disproportionately larger mandible (mandibular prognathism) [OMIM #176700], a disproportionately smaller maxilla (maxillary deficiency), or a combination of both, and can occur either as part of a syndrome or as an isolated trait. The current diagnosis and treatment of the Class III phenotype are fraught with inconsistencies in the timing, duration, and type of treatment. To date, many investigations have focused largely on treatment modalities and outcomes, with little being accomplished toward an understanding of the etiology of the Class III phenotype and the potential relationship between the genetic components or how genetic factors may influence the response to treatment.
Our rationale for investigating the genetic etiology of the Class III phenotype is logical, since considerable evidence exists to support this theory. Both human and animal studies have shown a significant genetic contribution in the development of the Class III phenotype. Human studies support an autosomal-dominant mode of inheritance in two independent studies of the Class III phenotype (El-Gheriani et al., 2003; Cruz et al., 2008). Specifically, genome-wide scan and linkage analysis of mandibular prognathism in Korean and Japanese persons revealed that there was a statistically significant, although nominal, linkage of the mandibular prognathic trait to 3 regions (Yamaguchi et al., 2005). Studies in mice also support the genetic basis of maxillary and mandibular size. Some investigators have used inbred mouse strains toconfirm the hypothesis that the D12mit7 segment on mouse chromosome12 determines maxillary growth (Oh et al., 2007), while others applied Quantitative Trait Locus (QTL) studies in inbred mice to identify specific QTLs on mouse chromosomes 10 and 11 that are correlated with the anteroposterior length of the mandible (Dohmoto et al., 2002).
We have previously shown that several subtypes exist for the broadly defined Class III phenotype (Bui et al., 2006). In this report, we focus on a specific subtype occurring in a Hispanic cohort in an effort to increase our power to detect linkage and test our hypothesis that the Class III phenotype is genetically linked to specific loci that regulate maxillary or mandibular growth.
Materials & Methods
Ascertainment of Families and Diagnosis
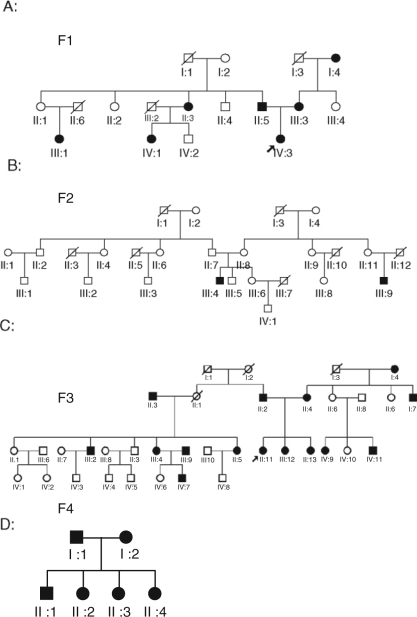
Approval for this study was granted by the Biomedical Institutional Review Board at the University of North Carolina at Chapel Hill and the Bioethics Committee of Universidad de Antioquia. Consent to participate in this study (including a release for dental records) was obtained from every adult participant or from a parental guardian in the case of minors. Through collaboration with the Universidad de Antiouqia in Medellín, Colombia, individuals from Medellín were recruited based on the presence of the Class III phenotype in at least two family members. Through subsequent interviews, the pedigrees (Fig. 1) were extended, for a total of 57 individuals from four families (28 affected and 29 unaffected). The age range was from 6 to 76 yrs, with an average age of 34.77 yrs. The family historian described the dentofacial phenotype for deceased individuals based on personal recollection. Forty-three individuals were available for pre-treatment clinical photographs, and panoramic and cephalometric radiographs following the clinical evaluation described below.
Figure 1.
Pedigrees of four families showing segregation of the Class III phenotype as indicated by darkened (affected) circles or squares. Squares indicate male; circles indicate female; diagonal line indicates deceased.
Two independent orthodontic evaluators reviewed clinical photos and/or cephalometric records to determine inclusion as affected or unaffected. A positive diagnosis of the Class III phenotype was made for all available individuals based on the evaluation above, using clinical appearance and dental relationships. Specifically, diagnostic criteria included at least 2 of the following: determination of straight or concave facial profile, overjet £ 0, Angle’s Class III molar or cuspid relationship, the Wits appraisal (useful in describing the severity of an anteroposterior jaw discrepancy without influence from the anterior cranial base angulation), or ANB angle £ 0 (Stellzig-Eisenhauer et al., 2002). The ANB angle represents the anteroposterior relationship of the maxilla to the mandible. Participants were excluded from the study if they had previous orthodontic treatment or any craniofacial anomalies (e.g., cleft lip/palate).
Cephalometric Analysis
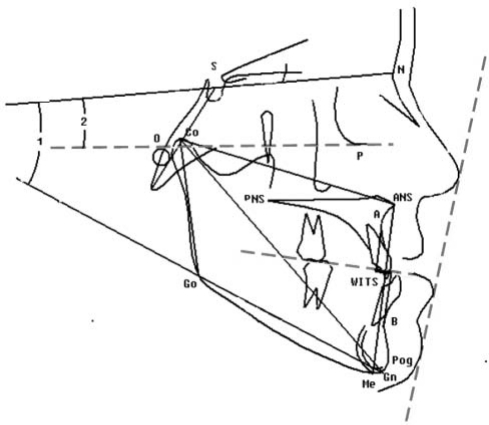
A cephalometric analysis was performed after lateral cephalometric radiographs (taken in natural head position) were scanned and digitized by one investigator using Dolphin, version 9.0 (Dolphin Imaging Systems, Chatsworth, CA, USA) to determine the extent of the skeletal and/or dental malocclusion. Sixty-seven cephalometric variables were selected and used for phenotypic characterization of the Class III phenotype. Among the measurements calculated (see Fig. 2 for selected measurements), 38 were linear, 25 angular, and 4 proportional (Riolo et al., 1975).
Figure 2.
Cephalometric tracing showing summary of selected linear and angular cephalometric measurements used in study. Co-ANS (mm), maxillary unit length; Co-Gn (mm), mandibular unit length; Co-Gn :: Co-ANS (mm), maxillary and mandibular difference; Go-Gn (mm), mandibular corpus length; S-N (mm), anteroposterior length of cranial base; ANS-Me (mm), lower facial height; Co-Go (mm), posterior facial height; Wits (mm), length of distance AO-BO (AO, intersection between perpendicular line dropped from Point A and occlusal plane; BO, intersection between perpendicular line dropped from Point B and occlusal plane); ANB, anteroposterior relation of maxilla and mandible; SNA, anteroposterior maxillary position to anterior cranial plane; SNB, anteroposterior mandibular position to anterior cranial plane; SN to GoGn (angle 1), inclination of the frontal cranial base SN to mandible plate GoGn; SN to FH (angle 2), inclination of the frontal cranial base SN to Frankfort horizontal (FH) plane.
The Method Error (ME) was calculated once all the cephalometric tracings were completed. Ten randomly selected cephalograms were traced and digitized on three occasions at two-week intervals by the same observer. The ME calculations were performed by the Intraclass Correlation (ICC) method (SPSS for Windows, version 14, Chicago, IL, USA).
Power Estimation
We calculated the power to detect linkage using the program SIMLINK in four Colombian families. Our calculation provided an expected LOD score greater than 1.0 individually (Family F3 provides an expected LOD score of 2.8), assuming an autosomal-dominant model with incomplete penetrance and a sporadic rate of 0.05. We estimate a cumulative expected LOD score of 2.9. We would have approximately 60% and 40% power to detect LODs of 2.0 and 3.0, respectively.
Genotyping and Linkage Analysis
Extraction and purification of DNA were carried out in blood (PureGene kit, Gentra Systems, Minneapolis, MN, USA) or saliva with Oragene kits (DNA Genotek, San Diego, CA, USA) for all individuals in this study. Genotyping was carried out via an automated 4 cM genome-wide microsatellite repeat scan for 57 individuals in four families (Fig. 1) of Hispanic descent, with 546 microsatellite markers (deCODE Genetics, Reykjavik, Iceland). In these four families, the Class III diagnosis and therefore affection status were made based on assignment of a dichotomous classification (i.e., Class III facial pattern or absence of Class III facial pattern). Because the definition of the Class III phenotype varies, linkage analyses were re-run with classifications based on maxillary/mandibular differences larger than one standard deviation. Map distances were taken from the deCODE map based on human genome build 36 at NCBI. Parametric and non-parametric (model-free) multipoint linkage analyses (Whittemore et al., 2005) were performed for four families with Allegro software version 2.1 (Allegro Microsystems, Inc., Worcester, MA, USA). Non-parametric linkage (NPL) analysis was run to account for the possibility of alternate modes of inheritance. NPL has been described as a powerful approach compared with the commonly used parametric methods, and has been referred to as the method of choice for pedigree studies of complex traits (Kruglyak, 1996). Since the literature strongly favors an autosomal-dominant inheritance of the Class III trait (see above), and our pedigree analyses by inspection also suggested this pattern, we assumed an autosomal-dominant inheritance with incomplete penetrance (97%) for the parametric analysis and an affected allele frequency of 0.0001. Calculations with marker allele frequencies were based on the allele frequencies generated from the Colombian families, since there were no allele frequency data for the Colombian/Hispanic population available in known databases. Linkage calculations were made at 1-cM intervals, and LOD scores were generated for both parametric and non-parametric analyses.
Results
Phenotypic Characterization (Cephalometric Analysis)
Cephalometric analysis (Fig. 2) of four families with a positive history of the Class III phenotype (where n = 41 with available cephalometric radiographs) showed that there was a tendency for one subphenotype to segregate within families. Specifically, we found that affected individuals from these families were maxillary-deficient (59.5%), with a normal or small mandible, when compared with normative cephalometric standards. This finding is an interesting contrast to reports from previous studies (El-Gheriani et al., 2003; Yamaguchi et al., 2005; Cruz et al., 2008) that focused largely on the mandibular prognathic subtype. Although mandibular unit length was normal in most affected individuals (49%), the maxillary/mandibular difference (measured in millimeters) was significantly increased in 20 individuals (48.7%) classified with maxillary/mandibular differences greater than normal, confirming the anteroposterior discrepancy. Moreover, in 76.7% individuals, the anterior cranial base (SN) was significantly smaller than normal, while the posterior cranial base was normal. Analysis of 43 cephalometric radiographs also revealed that 31 individuals (72%) were shown to have a decreased Wits number. This is particularly important in this cohort, since the anterior cranial base was greatly decreased in three out of four families (76.7%).
Genotyping and Linkage Mapping
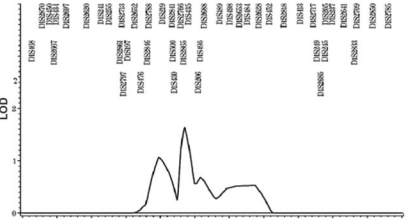
Pedigree analysis by visual inspection suggested an autosomal-dominant inheritance with incomplete penetrance. Non-parametric linkage analysis revealed that 5 loci (1p22.1, 3q26.2, 11q22, 12q13.13, and 12q23) on 4 chromosomes were suggestive of linkage (see Table), according to the criteria of an LOD threshold of 1.86 (Rao and Gu, 2001) for suggestive linkage in genome-wide linkage studies. We found that one locus identified in our report (Fig. 3), 1p22, is near previously reported loci, 1p35 and 1p36, identified by sib-pair analysis of Japanese and Korean children with mandibular prognathism (Yamaguchi et al., 2005). Although the previous study differed in ethnicity and subtype, our linkage resulted in ZLR and LOD scores that were slightly higher. We further identified candidate genes of biologic interest for 3 loci (11q22, 12q13.13, and 12q23) using bioinformatic approaches (http://www.ncbi.nlm.nih.gov/projects/omim/Homology/). This search revealed that human genes IGF1, HOXC, and COL2A1, respectively, are within these regions. HOX genes are believed to be pivotal in vertebrate craniofacial development. IGF1 is an excellent candidate gene known to influence body size in both mice and humans. COL2A1 gene (collagen, type II, alpha 1) encodes type II collagen in cartilage. Our search revealed a potential candidate on chromosome 11, matrix metallopeptidase 13 or collagenase 3, but, based on the literature, we did not consider this a high-priority candidate gene.
Table.
Multi-point Linkage Results from Parametric and Non-parametric Linkage Analysis Reveal that Regions on Chromosomes 1p, 12, and 3q are Suggestive of Linkage
| Locus* | Marker | Position from pter (cM) | LOD | ZLR | Parametric (p)/Non-parametric (npl) |
|---|---|---|---|---|---|
| 1p22.1 | D1S2865 | 114.2 | 1.8554 | 2.92 | p |
| 1p22.2 | D1S435 | 117.7 | 1.6382 | 2.74 | npl |
| 3q26.2 | D3S3041 | 178.3 | 1.9136 | 2.97 | npl |
| 11q22.2-q22.3 | D11s1886 | 105.6 | 1.9960 | 3.03 | P |
| D11s4206 | 108.7 | 1.8377 | 2.90 | P | |
| 12q23 | D12s1613 | 121.5 | 1.6971 | 2.79 | P |
| D12s1583 | 124.7 | 1.8730 | 2.93 | P | |
| D12s354 | 130.9 | 1.8412 | 2.91 | P | |
| D12s369 | 131.6 | 1.8355 | 2.91 | P | |
| 12q13.13 | D12S368 | 68.0 | 1.7820 | 2.70 | np |
Locus refers to map position as identified by Marker. Map position is indicated in centiMorgans from pter of chromosome. LOD denotes logarithm (base 10) of odds. ZLR refers to the maximum likelihood ratio z-score.
Figure 3.
Non-parametric linkage results of chromosome 1 for four families with an autosomal-dominant Class III trait. Y axis indicates LOD score; X-axis indicates map position in centi-Morgan (cM); microsatellite markers are indicated above the peak tracing.
Discussion
While previous studies have contributed to our understanding of the inheritance of the Class III phenotype, there are still significant gaps in the knowledge of the specific genetic contribution. In this report, we sought to define the Class III dentofacial phenotype in this Hispanic population in terms of its genetic and phenotypic profile. Analysis of our data supports that the Class III phenotype is an inherited trait, and that subphenotypes of the more broadly defined Class III tend to aggregate within the families studied. The craniofacial morphology leading to a Class III phenotype is clearly more complex than the relative sizes of the maxilla and mandible. Nonetheless, as a simple measure of variation, we believed that these dimensions were most useful.
We further found that 5 loci—including the intervals 1p22, 3q26.2, 11q22, 12q13.13, and 12q23—show evidence ‘suggestive’ of linkage based on previously defined criteria (Rao and Guo, 2001). Although heterogeneity is always a problem in the analysis of multiple families, the Colombian families of the Antiouquiam municipality are from a population more genetically homogenous than the US population, and have been specifically noted to show minimal genetic stratification (Bedoya et al., 2006). Our results, taken collectively with previous reports, underscore the fact that heterogeneity exists in the Class III phenotype, since different populations (Japanese/ Korean and Hispanic) reveal that differing subtypes of the Class III phenotype share linkage to loci on chromosome 1 (Yamaguchi et al., 2005). We speculate that this may point to a common upstream regulator that affects both maxillary and mandibular development.
Additional regions identified in this report include the 12q13 and 12q23 regions that correlate positional candidate genes of interest, including the Homeobox region, HOX3, IGF1, and the COL2A1 gene. The Hox families of genes are highly conserved master regulatory genes shown to play a definitive role in patterning the hindbrain and branchial regions of the developing head, up to and including structures derived from the second branchial arch. The HOX3 region contains at least 7 genes in a 160-Kb stretch of DNA, including Hoxc4, Hoxc5, Hoxc6, Hoxc8, Hoxc9, Hoxc10, Hoxc11, Hoxc12, and Hoxc13 (Trainor and Krumlauf, 2000). The COL2A1 (collagen, type II, alpha 1) gene, located between positions 12q13.11 and 12q13.2, encodes the alpha-1 chain of type II collagen found in cartilage. Previous studies in mice point to the rhizomelic effect of Col2A1 mutations in overall somatic growth, but also confirm the importance of Col2A1 in craniofacial growth (Garofalo et al., 1991).
An additional candidate gene of biologic interest, IGF1, was identified in the 12q23 region. It is an excellent candidate gene known to influence body size in both mice and humans (Baker et al., 1993; Woods et al., 1996). This is a logical candidate gene, because the GH/GHR/IGF1 system plays an important role in skeletal growth and the development (Sjogren et al., 2000) of body size in mice and humans (Sutter et al., 2007). More specifically, IGF1 receptors are found in the fibrous articular surface of the temporomandibular joint condyle (Visnapuu et al., 2001), while GHR SNPs are associated with mandibular height in Chinese and Japanese populations (Yamaguchi et al., 2001; Zhou et al., 2005).
Finally, results from mouse studies of craniofacial growth support our findings that a region on chromosome 12 is biologically relevant to craniofacial development and may be linked to the Class III phenotype. In an SMXA recombinant inbred strain of mice, the positions of the mouse chromosome 10 and chromosome 11 were determined to be responsible for mandibular length and corresponded to regions 12q21 and 2p13, respectively, in human chromosomes. These results suggest that the major gene(s) responsible for mandibular length are located in these regions (Dohmoto et al., 2002). Of particular interest is region 12q21, which is very close to 12q22 and 12q23 identified in our study.
Progress in the craniofacial genetics field toward human genetic mapping of the Class III trait is gradual but limited. The improved annotation of genetic and physical maps offers great future potential for identifying genes associated with this trait. Future studies can make use of genotyping data from human and mouse studies to understand how morphological phenotypes segregate in families, and whether these subphenotypes are linked with specific major genes such as those in the GH/GHR/IGF-1 system.
Acknowledgments
We gratefully acknowledge the support of the family and dentists who participated in this study, the assistance of Dr. James Ackerman in the preparation of this manuscript, and Debbie Price and Melody Torain in the preparation of the data.
Footnotes
This research was supported by the Southern Association of Orthodontists, the American Association of Orthodontists Foundation, University of North Carolina at Chapel Hill Faculty Development Funds to SAFB, and NIH grants 1K23RR17442 to SAFB and M01RR-00046.
References
- Baker J, Liu JP, Robertson EJ, Efstratiadis A. (1993). Role of insulin-like growth factors in embryonic and postnatal growth. Cell 75:73-82 [PubMed] [Google Scholar]
- Bedoya G, Montoya P, García J, Soto I, Bourgeois S, Carvajal L, et al. (2006). Admixture dynamics in Hispanics: a shift in the nuclear genetic ancestry of a South American population isolate. Proc Natl Acad Sci USA 103:7234-7239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bui C, King T, Proffit W, Frazier-Bowers S. (2006). Phenotypic characterization of Class III patients. Angle Orthod 76:564-569 [DOI] [PubMed] [Google Scholar]
- Cruz RM, Krieger H, Ferreira R, Mah J, Hartsfield J, Jr, Oliveira S. (2008). Major gene and multifactorial inheritance of mandibular prognathism. Am J Med Genet A 146:71-77 [DOI] [PubMed] [Google Scholar]
- Dohmoto A, Shimizu K, Asada Y, Maeda T. (2002). Quantitative trait loci on chromosomes 10 and 11 influencing mandible size of SMXA RI mouse strains. J Dent Res 81:501-504 [DOI] [PubMed] [Google Scholar]
- El-Gheriani AA, Maher BS, El-Gheriani AS, Sciote JJ, Abu-Shahba FA, Al-Azemi R, et al. (2003). Segregation analysis of mandibular prognathism in Libya. J Dent Res 82:523-527 [DOI] [PubMed] [Google Scholar]
- Garofalo S, Vuorio E, Metsaranta M, Rosati R, Toman D, Vaughan J, et al. (1991). Reduced amounts of cartilage collagen fibrils and growth plate anomalies in transgenic mice harboring a glycine-to-cysteine mutation in the mouse type II procollagen alpha 1-chain gene. Proc Natl Acad Sci USA 88:9648-9652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruglyak L. (1996). Thresholds and sample sizes. Nat Genet 14:132-133 [DOI] [PubMed] [Google Scholar]
- Oh J, Wang CJ, Poole M, Kim E, Davis RC, Nishimura I, et al. (2007). A genome segment on mouse chromosome 12 determines maxillary growth. J Dent Res 86:1203-1206 [DOI] [PubMed] [Google Scholar]
- Proffit WR, White RP. (1991). Surgical orthodontic treatment. St. Louis, MO: Mosby Year Book, p. 2 [Google Scholar]
- Proffit WR, Fields HW, Jr, Moray LJ. (1998). Prevalence of malocclusion and orthodontic treatment need in the United States: estimates from the NHANES III survey. Int J Adult Orthodont Orthognath Surg 13:97-106 [PubMed] [Google Scholar]
- Rao DC, Gu C. (2001). False positives and false negatives in genome scans. Adv Genet 42:487-498 [DOI] [PubMed] [Google Scholar]
- Riolo ML, Moyers RE, McNamara JA, Hunter WS. (1975). An atlas of craniofacial growth. Monograph #2. Craniofacial Growth Series, Ann Arbor, MI: University of Michigan, Center for Human Growth and Development [Google Scholar]
- Sjögren K, Bohlooly YM, Olsson B, Coschigano K, Törnell J, Mohan S, et al. (2000). Disproportional skeletal growth and markedly decreased bone mineral content in growth hormone receptor -/- mice. Biochem Biophys Res Commun 267:603-608 [DOI] [PubMed] [Google Scholar]
- Stellzig-Eisenhauer A, Lux CJ, Schuster G. (2002). Treatment decision in adult patients with Class III malocclusion: orthodontic therapy or orthognathic surgery? Am J Orthod Dentofacial Orthop 122:27-37 [DOI] [PubMed] [Google Scholar]
- Susami R, Asai Y, Hirose K, Hosoi T, Hayashi I. (1972). Prevalence of malocclusion in Japanese school children. 4. The frequency of mandibular overjet. J Jpn Orthod Soc 31:319-324 [PubMed] [Google Scholar]
- Sutter NB, Bustamante CD, Chase K, Gray MM, Zhao K, Zhu L, et al. (2007). A single IGF1 allele is a major determinant of small size in dogs. Science 316:112-115; erratum in Science 316:1284, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang EL. (1994). The prevalence of malocclusion amongst Hong Kong male dental students. Br J Orthod 21:57-63 [DOI] [PubMed] [Google Scholar]
- Trainor PA, Krumlauf R. (2000). Patterning the cranial neural crest: hindbrain segmentation and Hox gene plasticity. Nat Rev Neurosci 1:116-124 [DOI] [PubMed] [Google Scholar]
- Visnapuu V, Peltomaki T, Ronning O, Vahlberg T, Helenius H. (2001). Growth hormone and insulin-like growth factor I receptors in the temporomandibular joint of the rat. J Dent Res 80:1903-1907 [DOI] [PubMed] [Google Scholar]
- Whittemore AS, Halpern J, Ahsan H. (2005). Covariate adjustment in family-based association studies. Genet Epidemiol 28:244-255 [DOI] [PubMed] [Google Scholar]
- Woods KA, Camacho-Hübner C, Savage MO, Clark AJ. (1996). Intrauterine growth retardation and postnatal growth failure associated with deletion of the insulin-like growth factor I gene. N Engl J Med 335:1363-1367 [DOI] [PubMed] [Google Scholar]
- Yamaguchi T, Maki K, Shibasaki Y. (2001). Growth hormone receptor gene variant and mandibular height in the normal Japanese population. Am J Orthod Dentofacial Orthop 119:650-653 [DOI] [PubMed] [Google Scholar]
- Yamaguchi T, Park SB, Narita A, Maki K, Inoue I. (2005). Genome-wide linkage analysis of mandibular prognathism in Korean and Japanese patients. J Dent Res 84:255-259 [DOI] [PubMed] [Google Scholar]
- Zhou J, Lu Y, Gao XH, Chen YC, Lu JJ, Bai YX, et al. (2005). The growth hormone receptor gene is associated with mandibular height in a Chinese population. J Dent Res 84:1052-1056 [DOI] [PubMed] [Google Scholar]