Abstract
Computational methods were used to predict the amino acid sequences and gene locations for mammalian lactate dehydrogenase (LDH) genes and proteins using genome sequence databanks. Human LDHA, LDHC and LDH6A genes were located in tandem on chromosome 11, while LDH6B and LDH6C genes were on chromosomes 15 and 12, respectively. Opossum LDHC and LDH6B genes were located in tandem with the opossum LDHA gene on chromosome 5 and contained 7 (LDHA and LDHC) or 8 (LDH6B) exons. An amino acid sequence prediction for the opossum LDH6B subunit gave an extended N-terminal sequence, similar to the human and mouse LDH6B sequences, which may support the export of this enzyme into mitochondria. The platypus genome contained at least 3 LDH genes encoding LDHA, LDHB and LDH6B subunits. Phylogenetic studies and sequence analyses indicated that LDHA, LDHB and LDH6B genes are present in all mammalian genomes examined, including a monotreme species (platypus), whereas the LDHC gene may have arisen more recently in marsupial mammals.
Keywords: Mammals, amino acid sequence, genomics, lactate dehydrogenase, opossum, platypus, data mining, sequence analyses
1. Introduction
Mammalian lactate dehydrogenase (LDH; E.C.1.1.1.27) comprises three major families of conserved enzymes that catalyse the reversible interconversion of pyruvate and lactate, a key metabolic step in glycolysis and other metabolic pathways (Everse & Kaplan, 1973) At least five LDH tetrameric isozymes are reported in somatic mammalian tissues, comprising LDHA and LDHB subunits, whereas LDHC4 is found only in mature testis and spermatozoa (Goldberg & Hawtrey, 1967; Goldberg, 1973; Li et al., 1989), where it is required for male fertility (Odet etal., 2008). The LDHA, LDHB and LDHC families of mammalian LDH genes and subunits have been extensively investigated, with human and mouse LDHA and LDHC genes located in tandem on chromosomes 11 and 7 respectively (Edwards et al., 1989), as compared with the LDHB gene, on chromosomes 12 (human) and 6 (mouse) (Takeno & Li, 1989a). Phylogenetic studies have indicated that the LDHC gene has arisen from independent gene duplication events during vertebrate evolution, including separate LDHB gene duplications in fish and birds (pigeon) (Zinkham et al., 1969; Markert et al., 1975; Hiraoka et al., 1990; Quattro et al., 1993; Mannen et al., 1997), and an LDHA gene duplication during mammalian evolution (Millan et al., 1987).
Transcription studies have reported two other human LDHA-like genes, designated as LDH6A and LDH6B, which are expressed in brain and testis respectively, and located on chromosome 11 (LDH6A in tandem with human LDHA and LDHC genes) (Ota et al., 2004) and chromosome 15 (LDH6B, an intronless gene) (Wang et al., 2005). In this study, we have identified and characterized in silico new forms of mammalian LDHs and described predicted amino acid sequences, protein secondary structures, gene locations and exonic structures for human (LDH6C), mouse (LDH6B), opossum (LDHA; LDHB; LDHC; and LDH6B) and platypus (LDHA, LDHB and LDH6B) genes and proteins, as well as the phylogenetic relationships for mammalian LDH gene families. Evidence is also presented for N-terminal extensions of LDH6B subunit sequences which may support mitochondrial export and location of human, mouse and opossum LDH6B.
2. Materials and Methods
2.1 Mammalian LDH gene and protein identification
BLAST (Basic Local Alignment Search Tool) studies were undertaken using web tools from the National Center for Biotechnology Information (NCBI) (http://blast.ncbi.nlm.nih.gov/Blast.cgi) (Altschul et al, 1997). Protein BLAST analyses used previously reported human LDHA (Tsujibo et al., 1985), LDHB (Takeno and Li, 1989a), LDHC (Millan et al., 1987) and LDH6B (Ota et al., 2004) amino acid sequences. Non-redundant protein sequence databases for several mammalian genomes were examined using the blastp algorithm, including the human (International Human Genome Sequencing Consortium, 2001); mouse (Mus musculus) (Mouse Sequencing Consortium, 2002); opossum (Mikkelsen et al., 2007); and platypus (Platypus Genome Sequencing Consortium, 2008). This procedure produced multiple BLAST ‘hits’ for each of the protein databases which were individually examined and retained in FASTA format, and a record kept of the sequences for predicted mRNAs and encoded CES-like proteins. These records were derived from annotated genomic sequences using the gene prediction method: GNOMON and predicted sequences with high similarity scores for mammalian LDH. With some exceptions, predicted LDHA, LDHB, LDHC and LDH6B protein subunit sequences were obtained in each case and subjected to computational analyses of predicted protein and gene structures. Other LDH sequences were obtained following BLAT (BLAST-Like Alignment Tool) analysis using the human LDHA, LDHB, LDHC and LDH6B sequences to interrogate human, mouse, opossum and platypus genome sequences using the UC Santa Cruz gene browser [http://genome.ucsc.edu/cgi-bin/hgBlat] (Kent et al. 2003) with the default settings to obtain Ensembl generated protein sequences by applying the method of Hubbard et al (2007) (http://www.ensembl.org/index.html).
BLAT analyses were subsequently undertaken for each of the predicted LDH amino acid sequences using the UC Santa Cruz gene browser [http://genome.ucsc.edu/cgi-bin/hgBlat] (Kent et al. 2003) with the default settings to obtain the predicted locations for each of the mammalian LDH genes, including predicted exon boundary locations and gene sizes.
2.2 Predicted Structures and Properties for Mammalian LDH Subunits
Predicted secondary structures for human and other mammalian LDH-like subunits were obtained using the PSIPRED v2.5 web site tools provided by Brunel University [http://bioinf.cs.ucl.ac.uk/psipred/psiform.html] (McGuffin et al. 2000).
Theoretical isoelectric points and molecular weights for mammalian LDH subunits were obtained using Expasy web tools (http://au.expasy.org/tools/pi_tool.html). Prediction of an LDH N-terminal protein region that may support a mitochondrial targeting sequence and the identification of a potential cleavage site was conducted using MITOPROT web based methods (Claros and Vincens, 1996) (ftp://ftp.biologie.ens.fr/pub/molbio).
2.3 Phylogenetic Studies and Sequence Divergence
Phylogenetic trees were constructed using an amino acid alignment from a ClustalW-derived alignment of CES protein sequences, obtained with default settings and corrected for multiple substitutions (Chenna et al 2003; Larkin et al. 2007) [http://www.ebi.ac.uk/clustalw/]. An alignment score was calculated for each aligned sequence by first calculating a pairwise score for every pair of sequences aligned. The alignment ambiguous amino-terminus region was excluded prior to phylogenetic analysis yielding alignments of 332 residues for comparisons of mammalian LDHA, LDHB, LDHC and LDH6B sequences with chicken LDHA and LDHB sequences, which served as ‘outgroup’ sequences (see Table 1). Sequence identities for mammalian LDH subunits were determined using the SIM-Alignment tool for Protein Sequences [http://au.expasy.org/tools/sim-prot.html] (Schwede et al. 2003).
Table 1. Mammalian and chicken lactate dehydrogenase (LDH) genes and enzymes examined.
GenBank mRNA (or cDNA) IDs identify previously reported sequences (see http://www.ncbi.nlm.nih.gov/Genbank/); 1N-scan and 2SGP IDs identify gene predictions using gene structure prediction software provided by the Computational Genomics Lab at Washington University in St. Louis, MO, USA (see http://genome.ucsc.edu); UNIPROT refers to UniprotKB/Swiss-Prot IDs for individual LDH subunits (see http://kr.expasy.org); 3Mitochondrial export probabilities and predicted signal peptides were based on MITOPROT web based tools (see Methods); Contig ID for platypus genome sequences; Prediction software based ENSOANT IDs; Sources for LDH sequences were provided by the above sources.
| Species | LDH Gene |
GenBank ID | UNIPROT ID |
NCBI RefSeq ID |
Chromosome location |
Strand | Amino Acids |
Gene size kbs |
Exons | pI | Subunit MW |
3Mitochondrial Export Probability (Residues) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Human | LDHA | BC067223 | P00338 | NP005557 | 11: 18,374,966-18,385,401 | positive | 332 | 10,436 | 7 | 8.4 | 36,689 | 0.04 (Nil) |
| LDHB | BC071860 | P07195 | NP002291 | 12: 21,679,746-21,698,872 | negative | 334 | 19,127 | 7 | 5.7 | 36,638 | 0.05 (Nil) | |
| LDHC | BC064388 | P07864 | NM002301 | 11: 18,390,841-18,429,247 | positive | 332 | 38,407 | 7 | 7.1 | 36,311 | 0.07 (Nil) | |
| LDH6A | BC014340 | NP659409 | 11: 18,434,804-18,456,990 | positive | 332 | 22,187 | 7 | 6.5 | 36,507 | 0.14 (Nil) | ||
| LDH6B | BC022034 | Q9BYZ2 | NP149972 | 15: 57,286,432-57,287,574 | positive | 381 | 1,143 | 1 | 8.9 | 41,943 | 0.92 (1–33) | |
| LDH6C | 1SGP.12.853.1 | 12: 61,683,600-61,684,723 | positive | 373 | 1,124 | 1 | 8.6 | 41,157 | 0.78 (1–33) | |||
| Mouse | LDHA | BC004639 | P06151 | NP034829 | 7: 54,102,990-54,110,508 | positive | 332 | 7,519 | 7 | 7.6 | 36,499 | 0.02 (Nil) |
| LDHB | BC046755 | P16125 | NP032518 | 6: 142,438,960-142,454,060 | negative | 334 | 15,101 | 7 | 5.7 | 36,572 | 0.09 (Nil) | |
| LDHC | BC049602 | Q548Z6 | NP038608 | 7: 54,117,140-54,133,244 | positive | 332 | 16,105 | 7 | 8.4 | 35,912 | 0.1 (Nil) | |
| LDH6B | BC019420 | NP780558 | 17: 5,417,512-5,418,657 | negative | 382 | 1,146 | 1 | 9.3 | 42,049 | 0.98 (1–37) | ||
| Opossum | LDHA | AF070996 | Q9XT87 | NP1028147 | 5: 242,665,392-242,674,081 | negative | 332 | 8,690 | 7 | 7.1 | 36,358 | 0.02 (Nil) |
| LDHB | AF070997 | Q9XT86 | 2chr8.557.a | 8: 93,264,241-93,287,176 | positive | 334 | 22,936 | 7 | 5.7 | 36,537 | 0.06 (Nil) | |
| LDHC | 2chr5.25.018 | XP1378365 | 5: 242,633,611-242,658,159 | negative | 331 | 24,549 | 7 | 6.8 | 36,303 | 0.1 (Nil) | ||
| LDH6B | 2chr5.25.016 | XP1378357 | 5: 242,565,407-242,601,987 | negative | 381 | 36,581 | 8 | 8.7 | 41,871 | 0.79 (1–50) | ||
| Platypus | LDHA | AF545182 | 411958: 1024-4697; 43118: 2244-4946 | negative | 332 | 6,377 | 7 | 8.2 | 36,451 | 0.04 (Nil) | ||
| LDHB | 5ENSOANT16632 | 459108: 2671-2799; 48353: 5168-27343 | positive | 335 | 22,305 | 7 | 7.1 | 36,525 | 0.08 (Nil) | |||
| LDH6B | 5ENSOANT13298 | 43118: 2264-26601 | positive | 385 | 17,338 | 8 | 8.8 | 41,920 | 0.27 (Nil) | |||
| Chicken | LDHA | P00340 | NP990615 | 5: 13,645,367-13,649,740 | positive | 332 | 4,373 | 7 | 7.8 | 36,514 | 0.01 (Nil) | |
| LDHB | P00337 | NP989508 | 1: 69,204,825-69,213,883 | positive | 333 | 9,059 | 7 | 7.1 | 36,318 | 0.08 (Nil) |
3. Results and Discussion
3.1 Alignments of human LDHA, LDHB, LDHC, LDH6A, LDH6B and LDH6C amino acid sequences
The amino acid sequences for human LDHA (Tsujibo et al., 1985), LDHB (Takeno and Li, 1989a), LDHC (Millan et al., 1987; Takeno and Li, 1989b) and LDH6B (Ota et al., 2004) and the computation derived LDH6A and LDH6C human subunits are aligned in Figure 1 (see Table 1). Human LDH A, B, C and 6B subunits showed 71–75% sequence identities, indicating extensive conservation in amino acid sequences for these enzymes (data not shown). Major differences were observed however at the N-termini for the human LDH6B and LDH6C subunits, which showed an extension of 49 residues. MITOPROT computer based analyses of these sequences predicted a high probability for LDH6B and LDH6C subunit export into mitochondria (0.92 and 0.78, respectively), as well as a potential cleavage site at residue 31, in each case (Table 1; see Figure 1). The predicted mitochondrial N-terminal sequences were positively charged, with excess basic amino acid residues (3 and 2 respectively for LDH6B and LDH6C), contained no acidic residues and revealed a predicted amphiphilic α-helix, which are common features for mitochondrial leader sequences (Hanmen and Weiner, 1998). Key LDH catalytic residues were present in all six human LDH subunits, including the active site proton acceptor (His193), as well as coenzyme (Arg99 and Asn138) and substrate (Arg106; Arg169; Thr248) binding residues (Figure 1) (Read et al., 2001).
Figure 1. Amino acid sequence alignments for human LDHA, LDHB, LDHC, LDH6A, LDH6B and LDH6C sequences.
See Table 1 for sources of LDH sequences; * shows identical residues; Residues identified by MITOPROT as high probability mitochondrial leader sequences; conserved active site residues Arg99 and 106; Asn138; Arg169; His193; and Thr248 Helix (Human LDHA and LDHB or predicted helix); Sheet (Human LDHA and LDHB or predicted sheet). Bold underlined font shows known or predicted exon junctions (|). A, B, C, 6A, 6B and 6C refer to the corresponding human LDH subunits.
3.2 Alignments of mammalian LDHA, LDHB, LDHC and LDH6B amino acid sequences
The amino acid sequences for predicted mouse LDH6B, opossum LDHA, LDHB, LDHC and LDH6B, and platypus LDHA, LDHB and LDH6B subunits are aligned with previously reported sequences for the corresponding human and mouse subunits (Tsujibo et al., 1985; Takeno and Li, 1989a; Millan et al., 1987; Fukasawa and Li, 1987; Sakai et al., 1987; Hiraoka et al., 1990) (Figure 2; see Table 1). The predicted opossum and platypus LDH sequences showed higher levels of identity with homologue sequences from human and mouse sources, particularly for the LDHA and LDHB sequences, which were 89–93% identical and 80–97% identical, respectively. Mammalian LDHC and LDH6B sequences, however, exhibited lower levels of identity, showing 65–74% identity for human, mouse and opossum LDHC sequences and 59–75% for human, mouse and platypus LDH6B sequences, respectively (data not shown). Mammalian LDH6B sequences showed evidence of N-terminus extensions for the predicted mouse, opossum and platypus subunits in comparison with LDHA, LDHB and LDHC sequences for all species examined (Figure 2). MITOPROT computer based analyses of these sequences predicted high probabilities for mouse and opossum LDH6B subunit export into mitochondria (0.98 and 0.79, respectively), as well as potential cleavage sites at residues 36 (mouse LDH6B) and 49 (opossum LDH6B) (Table 1; Figure 2). The platypus LDH6B sequence, however, differed significantly in this property, with the 53 residue N-terminus extension showing a lower probability as a mitochondrial signal peptide (0.27) (Table 1; Figure 2). Mitochondrial LDH (Brooks et al, 1999) has been previously proposed to play a role in the intracellular lactate shuttle and in lactate clearance by mitochondria, however the responsible LDH isozyme(s) have not been conclusively identified. The identification of a mitochondrial leader sequence for human, mouse and opossum LDH6B subunits may assist further investigations concerning a potential role for mammalian LDH in mitochondrial lactate clearance.
Figure 2. Amino acid sequence alignments for human, mouse, opossum, platypus and chicken LDH sequences.
See Table 1 for sources of LDH sequences; * shows identical residues; Residues identified by MITOPROT as high probability mitochondrial leader sequences; conserved active site residues Arg99 and 106; Asn138; Arg169; His193; and Thr248 Helix (Human LDHA and LDHB or predicted helix); Sheet (Human LDHA and LDHB or predicted sheet). Bold underlined font shows known or predicted exon junctions (|). LDHs examined included human (hu); mouse (mo); opossum (op); platypus (pl); and chicken (ch).A, B, C and 6B refer to the corresponding LDH subunits.
Each of the predicted mouse (LDH6B), opossum (LDHA; LDHB; LDHC; and LDH6B) and platypus (LDHA; LDHB; and LDH6B) sequences aligned closely with the corresponding human and mouse sequences, and all subunits (with one exception), showed sequence identity for the key active site residues previously described for human LDH subunits (see Read et al., 2001). The predicted platypus LDH6B sequence, however, contained an Arg residue in place of the key LDHA coenzyme binding residue (Asn138), which may significantly alter the kinetic properties for this enzyme.
Differences in the theoretical isoelectric points (pI) for opossum and platypus LDHA and LDHB subunits were observed, with LDHA showing higher pI values (7.1 and 8.2) than for the LDHB subunits (5.7 and 7.1), which is consistent with pI differences observed for other mammalian LDHs (Table 1). LDH6B subunits showed higher pI values than for the LDHA and LDHB subunits, which may be explained by the high basic amino acid content for the N-terminus peptide extensions, whereas theoretical pI values for mammalian LDHC subunits were intermediate between LDHB (lower pI) and LDHA/LDH6B (higher pI). Human, mouse and opossum LDHA, LDHB and LDHC subunits examined contained 331–334 amino acid sequence residues, whereas LDH6B subunits contained 381–385 amino acids due to the N-terminus extensions in each case.
3.3 Comparative Mammalian LDH Genomics
Figure 1 shows the locations of the intron-exon boundaries for the mammalian LDH gene products examined, and compares them with previously reported human and mouse LDH gene structures (Chung et al., 1985; Fusakawa and Li, 1987; Takeno and Li, 1989a,b) and their positioning within the aligned amino acid sequences. The mammalian LDHA, LDHB and LDHC genes examined, and the predicted human LDH6A gene, contained 7 exons in each case, with intron-exon boundaries in identical or comparable positions. In contrast, the human and mouse LDH6B genes were without intronic sequences, confirming a report for the human LDH6B gene (Wang et al., 2005), for which expression was observed in human testis. The predicted LDH6B genes in the opossum and platypus genomes, however, contained 8 exons, with the first exon encoding the predicted N-terminus extensions for these gene products, whereas the other 7 exons were localized in similar or identical positions to other mammalian LDH genes.
Table 1 describes the predicted locations for the mammalian LDH genes examined which showed that human, mouse and opossum LDHA and LDHC genes are located together within respective genomes on chromosomes 11, 7 and 5, respectively. The human and mouse LDHA and LDHC genes are very closely located together being separated by < 7 kilobases of DNA. The predicted human LDH6A gene is also part of this gene cluster on chromosome 11, as is the opossum LDH6B gene on chromosome 5 of the opossum genome. In addition, the platypus LDHA and LDH6B genes are apparently located on or near the same contiguous piece of DNA (Contig3116) suggesting that these genes are also closely located on the platypus genome. In contrast, the human, mouse and opossum LDHB genes are on a separate chromosome to that of the LDHA-like gene cluster (Table 1).
3.4 Predicted Secondary Structures for Mammalian (and Chicken) LDH Sequences
Figure 1 and Figure 2 show the secondary structures previously reported for human LDHA and LDHB (Read et al., 2001) and for mouse LDHC (Hogrefe et al., 1987) or predicted for mammalian LDHA, LDHB, LDHC and LDH6B subunit sequences, together with human LDH6A and LDH6C sequences. Predicted secondary structures for chicken LDHA and LDHB sequences were also examined as these were used as ‘outgroup’ LDH sequences for comparative analyses of mammalian LDH gene and protein structures. Similar α-helix β-sheet structures were observed for all mammalian and chicken LDH subunits examined, particularly near key residues or functional domains, including active site residues such as the active site proton acceptor (His193), as well as coenzyme (Arg99 and Asn138) and substrate (Arg106; Arg169; Thr248) binding residues (Read et al., 2001; Hogrefe et al., 1987). The obvious major difference in mammalian LDH secondary structure related to the N-terminus extensions for human LDH6B and LDH6C, and for mouse and opossum LDH6B, which contained an additional amphiphilic α-helix at the amino terminus, which may support being exported into mitochondria via these potential mitochondrial leader sequences (see Table 1). Although the platypus LDH6C N-terminal sequence contained a predicted α-helix, this did not extend into regions containing basic amino acid residues which may explain the lower probability for this sequence as a mitochondrial signal peptide (Table 1; Figure 2). It is apparent from these predictions that LDHA, LDHB, LDHC and LDH6B subunits are highly conserved in mammals, and it is likely that LDH subunits in the opossum will resemble the corresponding LDHs in human.
3.5 Evolution of Mammalian LDH Genes and Proteins
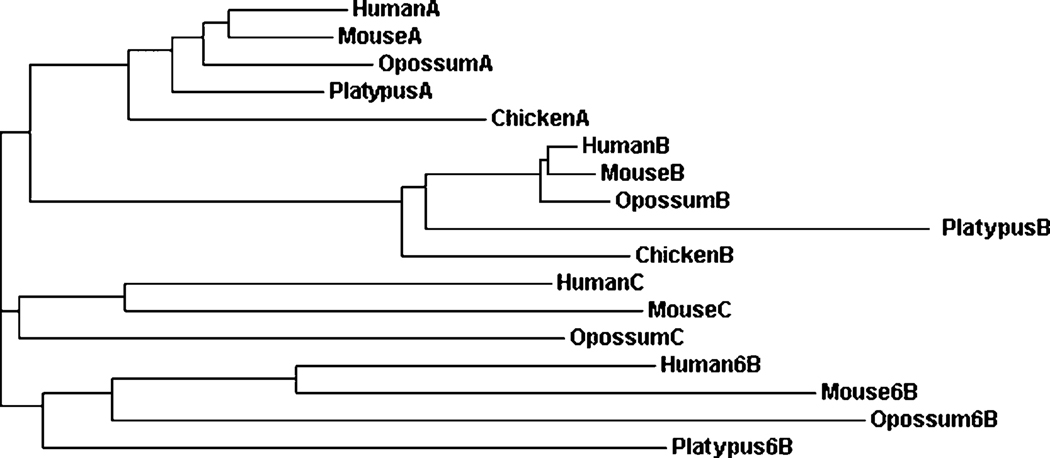
A phylogenetic tree (Figure 3) was calculated by the progressive alignment of human LDHA, LDHB, LDHC and LDH6B amino acid sequences with the corresponding LDH sequences from mouse, opossum and the platypus. Chicken LDHA and LDHB sequences were also included and served as an ‘outgroup’ for this analysis of mammalian LDHs. Four major clusters of mammalian and chicken LDHs were observed: the mammalian (and chicken) LDHA and LDHB gene clusters; the LDHC gene cluster of human, mouse and opossum; and the LDH6B cluster of human, mouse, opossum and platypus. This is consistent with the existence of four distinct mammalian LDH gene families: LDHA, encoding the major skeletal muscle isozyme; LDHB, encoding the major heart isozyme (Markert et al., 1975); LDHC, encoding the testis and sperm specific isozyme (Millan et al., 1987); and LDH6B, which awaits more detailed investigation. LDHA and LDHB have been described in all vertebrates examined and may be considered as the ‘ancestral’ genes for this enzyme (Holmes, 1972; Markert et al., 1975). In contrast, the LDHC gene has arisen independently from the LDHB gene in both teleost fish (Quattro et al., 1993) and in some birds (eg. pigeon) (Zinkham et al., 1969; Mannen et al., 1997), while in mammals, the LDHC gene has been apparently formed from an LDHA gene duplication event (Millan et al., 1987; Mannen et al., 1997). Biochemical studies have previously shown that LDHA, LDHB and LDHC isozymes are present in several Australian marsupials examined, including the pretty-faced wallaby (Macropus parryi), the koala (Phascolarctos cincereus) and the brush-tailed possum (Trichosurus vulpecula) (Holmes et al., 1973) whereas LDHC is apparently absent in monotreme mammals, the echidna (Tachyglossus aculeatus) and the platypus (Ornithorhynchus anatinus) (Baldwin and Temple-Smith, 1973). This study of LDH genes and proteins predicted from the South American gray short-tailed opossum (Monodelphis domestica) genome lends support to the distribution of LDHA, LDHB and LDHC genes and proteins among marsupials from both Australia and South America. The absence of an LDHC-like gene in the monotreme (platypus) genome, however, suggests that the proposed LDH-A gene duplication event leading to the appearance of the marsupial LDHC gene may have occurred following the separation of marsupial and monotreme common ancestors. In contrast, the mammalian LDH6B gene is apparently present throughout eutherian, marsupial and monotreme mammalian evolution but is apparently absent in the chicken genome (Table 1; Figure 3). A further LDHA gene duplication event is proposed forming the ancestral LDH6B gene at an earlier stage of mammalian evolution, prior to the separation of monotremes from the marsupial and eutherian mammalian common ancestors. This is supported by the higher levels of sequence identities observed for LDHA and LDH6B subunits (65–71%) as compared with LDHB and LDH6B subunits (57–62%), and the close locations observed for LDHA and LDH6B genes for the mammalian genomes examined.
Figure 3. Phylogenetic tree of mammalian CES6 and of human CES1, CES2, CES3 and CES5 sequences.
The tree is labeled with the LDH gene family number and the species name. Note the separation of the LDH genes into four LDH family clusters: LDHA; LDHB; LDHC; and LDH6B.
In summary, computer based predictions are presented of amino acid sequences, structures and gene locations for LDH genes and proteins of four mammalian species, the human, mouse, opossum (a South American marsupial) and platypus (an Australian monotreme). Opossum LDHC and LDH6B genes were located in tandem with the opossum LDHA gene on chromosome 5 and contained 7 (LDHA and LDHC) or 8 (LDH6B) exons. The predicted amino acid sequence for the opossum LDH6B subunit yielded an extended N-terminal sequence, similar to the human and mouse LDH6B sequences, which are proposed to support the export of these enzymes into mitochondria. The platypus genome contained at least 3 LDH genes encoding LDHA, LDHB and LDH6B subunits. Phylogenetic studies analyses indicated that LDHA, LDHB and LDH6B genes are present in all mammalian genomes examined, including a monotreme (platypus), whereas the LDHC gene may have arisen more recently in marsupial mammals prior to the appearance of eutherian mammals.
Acknowledgements
This project was supported in part by NIH HD05863 to E.G.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1997;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Baldwin J, Temple-Smith P. Distribution of LDHX in mammals: presence in marsupials and absence in the monotremes platypus and echidna. Comp. Biochem. Physiol. B. 1973;46:805–811. doi: 10.1016/0305-0491(73)90124-7. [DOI] [PubMed] [Google Scholar]
- Brooks GA, Dubouchaud H, Brown M, Sicurello JP, Butz CE. Role of mitochondrial lactate dehydrogenase and lactate oxidation in the intracellular lactate shuttle. Proc. Natl. Acad. Sci. USA. 1999;96:1129–1134. doi: 10.1073/pnas.96.3.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DJ, Thompson JD. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 2003;31:3497–3500. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung FZ, Tsujibo H, Bhattacharyya U, Sharief FS, Li SS-L. Genomic organization of the human lactate dehydrogenase-A gene. Biochem. J. 1985;231:537–541. doi: 10.1042/bj2310537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claros MG, Vincens P. Computational method to predict mitochondrially imported proteins and their targeting sequences. Eur. J. Biochem. 1996;241:779–786. doi: 10.1111/j.1432-1033.1996.00779.x. [DOI] [PubMed] [Google Scholar]
- Edwards Y, West L, Van Heyningen V, Cowell J, Goldberg E. Regional localization of the sperm specific lactate dehydrogenase, LDHC, gene on human chromosomal 11. Ann. Human Genet. 1989;53:215–219. doi: 10.1111/j.1469-1809.1989.tb01787.x. [DOI] [PubMed] [Google Scholar]
- Everse J, Kaplan NO. Lactate dehydrogenases: structure and function. Adv. Enzym. Mol. Biol. 1973;37:61–133. doi: 10.1002/9780470122822.ch2. [DOI] [PubMed] [Google Scholar]
- Fukasawa KM, Li SS-L. Complete nucleotide sequence of the mouse lactate dehydrogenase-A functional gene: comparison of the exon-intron organization of dehydrogenase genes. Genetics. 1987;116:99–105. doi: 10.1093/genetics/116.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg R, Hawtrey C. The ontogeny of sperm specific lactate dehydrogenase in mice. J. Exp. Zool. 1967;164:309–316. doi: 10.1002/jez.1401640302. [DOI] [PubMed] [Google Scholar]
- Goldberg E. Molecular basis of multiple forms of LDH-X. J. Exp. Zool. 1973;186:273–278. doi: 10.1002/jez.1401860306. [DOI] [PubMed] [Google Scholar]
- Gue N, Peitsch MC. SWISS-MODEL and the Swiss-Pdb viewer. An environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- Hanmen PK, Weiner H. Mitochondrial leader sequence: structural similarities and sequence differences. J. Exp. Zool. 1998;282:280–283. [PubMed] [Google Scholar]
- Hiraoka KA, Sharief FS, Yamg YW, Li WH, Li SS-L. The cDNA and protein sequences of mouse lactate dehydrogenase B. Molecular evolution of vertebrate lactate dehydrogenase genes A (muscle), B (heart) and C (testis) Eur. J. Biochem. 1990;189:215–220. doi: 10.1111/j.1432-1033.1990.tb15479.x. [DOI] [PubMed] [Google Scholar]
- Hogrefe HH, Griffith JP, Rossmann MG, Goldberg E. Characterization of the antigenic sites on the refined 3-A resolution structure of mouse testicular lactate dehydrogenase C4. J.Biol.Chem. 1987;262:13155–13162. [PubMed] [Google Scholar]
- Holmes RS. Evolution of lactate dehydrogenase genes. FEBS Letters. 1972;28:51–55. doi: 10.1016/0014-5793(72)80675-6. [DOI] [PubMed] [Google Scholar]
- Holmes RS, Cooper DW, VandeBerg JL. Marsupial and monotreme lactate dehydrogenase isozymes: phylogeny, ontogeny and homology with eutherian mammals. J. Exp Zool. 1973;184:127–148. doi: 10.1002/jez.1401840109. [DOI] [PubMed] [Google Scholar]
- Hubbard TJP, Aken BL, Beal1 K, Ballester B, Caccamo M, Chen Y, Clarke L, Coates G, Cunningham F, Cutts T, Down T, Dyer SC, Fitzgerald S, Fernandez-Banet J, Graf S, Haider S, Hammond M, Herrero J, Holland R, Howe K, Johnson N, Kahari A, Keefe D, Kokocinski F, Kulesha E, Lawson D, Longden I, Melsopp C, Megy K, Meidl P, Overduin B, Parker A, Prlic A, Rice S, Rios D, Schuster M, Sealy I, Severin J, Slater G, Smedley D, Spudich G, Trevanion S, Vilella A, Vogel J, White S, Wood M, Cox T, Curwen V, Durbin F, Fernandez-Suarez XP, Flicek P, Kasprzyk A, Proctor G, Searle S, Smith J, Ureta-Vidal A, Birney E. Ensembl. Nucleic Acids Res. 2007;35:D610–D617. doi: 10.1093/nar/gkl996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:994–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopp J, Schwede T. The SWISS-MODEL repository of three dimensional protein structure homology models. Nucl. Acids res. 2004;32:D230–D234. doi: 10.1093/nar/gkh008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. ClustalW2 and ClustalX version 2. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Li SS, O’Brien DA, How EW, Versola J, Rockett DL, Eddy EM. Differential activity and synthesis of lactate dehydrogenase isozymes A (muscle), B (heart) and C (testis) in mouse spermatogenic cells. Biol. Reprod. 1989;40:173–180. doi: 10.1095/biolreprod40.1.173. [DOI] [PubMed] [Google Scholar]
- McGuffin LJ, Bryson K, Jones DT. The PSIPRED protein structure prediction server. Bioinformatics. 2000;16:404–405. doi: 10.1093/bioinformatics/16.4.404. [DOI] [PubMed] [Google Scholar]
- Mannen H, Tsoi SC-M, Krushkal JS, Li W-H, Li SS-L. The cDNA cloning and molecular evolution of reptile and pigeon lactate dehydrogenase isozymes. Mol. Biol. Evol. 1997;14:1081–1087. doi: 10.1093/oxfordjournals.molbev.a025717. [DOI] [PubMed] [Google Scholar]
- Markert CL, Shaklee JB, Whitt GS. Evolution of a gene. Multiple genes for LDH isozymes provide a model of the evolution of gene structure, function and regulation. Science. 1975;189:102–114. doi: 10.1126/science.1138367. [DOI] [PubMed] [Google Scholar]
- Mikkelsen TS, Wakefield MJ, Aken B, Amemiya CT, Chang JL, Duke S, Garber M, Gentles AJ, Goodstadt L, Heger A, Jurka J, Kamal M, Mauceli E, Searle SMJ, Sharpe T, Baker ML, Batzer MA, Benos PV, Belov K, Clamp M, Cook A, Cuff J, Das R, Davidow L, Deakin JE, Fazzari MJ, Glass JL, Grabherr M, Greally JM, Gu W, Hore TA, Huttley GA, Kleber M, Jirtle RL, Koina E, Lee JT, Mahony S, Marra MA, Miller RD, Nicholls RD, Oda M, Papenfuss AT, Parra ZE, Pollock DD, Ray DA, Schein JE, Speed TP, Thompson K, VandeBerg JL, Wade CM, Walker JA, Waters PD, Webber C, Weidman JR, Xie X, Zody MC, Marshall Graves JA, Ponting CP, Breen M, Samollow PB, Lander ES, Lindblad-Toh K Broad Institute Genome Sequencing Platform and Broad Institute Whole Genome Assembly Team. Genome of the marsupial Monodelphis domestica reveals innovation in non-coding sequences. Nature. 2007;447:167–177. doi: 10.1038/nature05805. [DOI] [PubMed] [Google Scholar]
- Millan JL, Driscoll CE, Goldberg E. Epitopes of human testis-specific lactate dehydrogenase deduced from a cDNA sequence. Proc. Natl. Acad. Sci. USA. 1987;84:5311–5315. doi: 10.1073/pnas.84.15.5311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouse Sequencing Consortium. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- Odet F, Duan C, Willis WD, Goulding EH, Kung A, Eddy EM, Goldberg E. Expression of the gene for lactate dehydrogenase C (Ldhc) is required for male fertility. Biol. Reprod. 2008;79:26–34. doi: 10.1095/biolreprod.108.068353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ota T, Suzuki Y, Nishikawa T, Otsuki T, Sugiyama T, Irie R, Wakamatsu A, Hayashi K, Sato H, Nagai K, Kimura K, Makita H, Sekine M, Obayashi M, Nishi T, Shibahara T, Tanaka T, Ishii S, Yamamoto J, Saito K, Kawai Y, Isono Y, Nakamura Y, Nagahari K, Murakami K, Yasuda T, Iwayanagi T, Wagatsuma M, Shiratori A, Sudo H, Hosoiri T, Kaku Y, Kodaira H, Kondo H, Sugawara M, Takahashi M, Kanda K, Yokoi T, Furuya T, Kikkawa E, Omura Y, Abe K, Kamihara K, Katsuta N, Sato K, Tanikawa M, Yamazaki M, Ninomiya K, Ishibashi T, Yamashita H, Murakawa K, Fujimori K, Tanai H, Kimata M, Watanabe M, Hiraoka S, Chiba Y, Ishida S, Ono Y, Takiguchi S, Watanabe S, Yosida M, Hotuta T, Kusano J, Kanehori K, Takahashi-Fujii A, Hara H, Tanase TO, Nomura Y, Togiya S, Komai F, Hara R, Takeuchi K, Arita M, Imose N, Musashino K, Yuuki H, Oshima A, Sasaki N, Aotsuka S, Yoshikawa Y, Matsunawa H, Ichihara T, Shiohata N, Sano S, Moriya S, Momiyama H, Satoh N, Takami S, Terashima Y, Suzuki O, Nakagawa S, Senoh A, Mizoguchi H, Goto Y, Shimizu F, Wakebe H, Hishigaki H, Watanabe T, Sugiyama A, Takemoto M, Kawakami B, Yamazaki M, Watanabe K, Kumagai A, Itakura S, Fukuzumi Y, Fujimori Y, Komiyama M, Tashiro H, Tanigami A, Fujiwara T, Ono T, Yamada K, Fujii Y, Ozaki K, Hirao M, Ohmori Y, Kawabata A, Hikiji T, Kobatake N, Inagaki H, Ikema Y, Okamoto S, Okitani R, Kawakami T, Noguchi S, Itoh T, Shigeta K, Senba T, Matsumura K, Nakajima Y, Mizuno T, Morinaga M, Sasaki M, Togashi T, Oyama M, Hata H, Watanabe M, Komatsu T, Mizushima-Sugano J, Satoh T, Shirai Y, Takahashi Y, Nakagawa K, Okumura K, Nagase T, Nomura N, Kikuchi H, Masuho Y, Yamashita R, Nakai K, Yada T, Nakamura Y, Ohara O, Isogai T, Sugano S. Complete sequencing and characterization of 21,243 full-length human cDNAs. Nature Genetics. 2004;36:40–45. doi: 10.1038/ng1285. [DOI] [PubMed] [Google Scholar]
- Platypus Genome Sequencing Consortium. Genome analysis of the platypus reveals unique signatures of evolution. Nature. 2008;453:175–183. doi: 10.1038/nature06936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quattro JM, Woods HA, Powers DA. Sequence analysis of teleost retina-specific lactate dehydrogenase C: evolutionary implications for the vertebrate lactate dehydrogenase gene family. Proc. Natl Acad. Sci. USA. 1993;90:242–246. doi: 10.1073/pnas.90.1.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read JA, Winter VJ, Eszes CM, Sessions RB, Brady RL. Structural basis for altered activity of M- and H- isozyme forms of human lactate dehydrogenase. Protein Sci. 2001;43:175–185. doi: 10.1002/1097-0134(20010501)43:2<175::aid-prot1029>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- Schwede T, Kopp J, Guex N, Peitsch MC. SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res. 2003;31:3381–3385. doi: 10.1093/nar/gkg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeno T, Li SS-L. Structure of the human lactate dehydrogenase B gene. Biochem. J. 1989a;257:921–924. doi: 10.1042/bj2570921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeno T, Li SS-L. Human testicular lactate dehydrogenase gene is interrupted by six introns at positions homologous to those of LDH-A (muscle) and LDH-B (heart) genes. Biochem. Biophys. Res. Commun. 1989b;159:579–583. doi: 10.1016/0006-291x(89)90033-8. [DOI] [PubMed] [Google Scholar]
- Tsujibo H, Tiano HF, Li SS. Nucleotide sequences of the cDNA and an intronless pseudogene for human lactate dehydrogenase-A isozyme. Eur. J. Biochem. 1985;147:9–15. doi: 10.1111/j.1432-1033.1985.tb08711.x. [DOI] [PubMed] [Google Scholar]
- Wang H, Zhou Z, Lu L, Xu Z, Sha J. Cloning and characterization of a novel intronless lactate dehydrogenase gene from adult testis. Int. J. Mol. Med. 2005;15:949–953. [PubMed] [Google Scholar]
- Zinkham WH, Isensee H, Renwick JH. Linkage of lactate dehydrogenase B and C loci in pigeons. Science. 1969;164:185–187. doi: 10.1126/science.164.3876.185. [DOI] [PubMed] [Google Scholar]