Abstract
Colorectal cancer arises from the progressive accumulation of mutations and epigenetic alterations in colon epithelial cells. Such alterations often deregulate signaling pathways that affect the formation of colon cancer, such as the Wnt, RAS-MAPK and TGF-β pathways. The tumor promoting effects of mutations in genes, such as APC, have been demonstrated in cancer cell lines and in mouse models of intestinal cancer; however, the biological effects of most epigenetic events identified in colorectal cancer remain unknown. Consequently, we assessed whether the aberrant methylation of TSP1, the gene for thrombospondin 1, a regulator of TGF-β ligand activation, is an epigenetic mechanism for inhibiting the TGF-β signaling pathway. We found methylated TSP1 occurs in colon cancer cell lines (33%), colon adenomas (14%) and colon adenocarcinomas (21%). In primary colorectal cancers, loss of TSP1 expression correlated with impaired TGF-β signaling as indicated by decreased Smad2 phosphorylation and nuclear localization. Furthermore, methylation-induced silencing of TSP1 expression reduced the concentration of secreted active TGF-β1 and attenuated TGF-β signaling. Reversal of TSP1 methylation resulted in increased TSP1 mediated activation of the latent LAP:TGF-β complex and subsequent TGF-β receptor activation. Our results demonstrate that the aberrant methylation of TSP1 has biological consequences and provide evidence that the aberrant methylation of TSP1 is a novel epigenetic mechanism for suppressing TGF-β signaling in colorectal cancer.
Keywords: Thrombospondin, transforming growth factor β, methylation, colorectal cancer, epigenetics
Introduction
Colorectal cancer is the third most common cancer in the United States. In 2008, approximately 154,000 new cases of colorectal cancer will be diagnosed and more than 52,000 people are predicted to die from the disease 1. Colon cancer arises from the accumulation of DNA alterations in colon epithelial cells, which mediate the initiation and progression of this cancer. Mutations in genes such as KRAS, TP53 and members of the transforming growth factor (TGF-β) signaling pathway play a pathogenic role in the polyp-carcinoma sequence 2–4.
TGF-β is a pluripotent cytokine that has a multitude of effects on epithelial cells including the inhibition of proliferation, the induction of apoptosis and the stimulation of differentiation. Inactivating mutations in central members of the TGF-β signaling pathway, TGFBR2 and SMAD4, have been identified in approximately half of colon cancers 5–8. The biological effects of TGF-β in the colon and the identification of inactivating mutations in TGFBR2 and SMAD4 in colon cancers have demonstrated that the TGF-β signaling pathway can act as a colon cancer tumor suppressor pathway.
The TGF-β signaling pathway consists of the TGF-β ligand; the heteromeric TGF-β receptor composed of the type I and type II TGF-β receptors (TGFBR1 and TGFBR2); and the post-receptor signaling proteins, including SMAD2, 3, and 4. TGF-β1 is the most commonly expressed TGF-β ligand in adult epithelial tissues and is secreted in a latent form, which is noncovalently associated with the latency-associated peptide (LAP). The N-terminal region of LAP is the domain that interacts with TGF-β 1 and mediates the latency of the complex 9. The dissociation of TGF-β:LAP is induced in vivo by plasmin or cathepsin as well as by thrombospondin 1 (TSP1) and is a required step for the activation of secreted TGF-β10, 11. The activation of TGF-β1 is one of the primary mechanisms for regulating the activity of the TGF-β signaling pathway 9. Once TGF-β1 is activated, it can bind to and activate the TGF-β receptor, composed of TGFBR1 and TGFBR2, inducing post-receptor signaling pathways 12, 13. The biological events induced by TGF-β, such as apoptosis or inhibition of proliferation, then mediate the tumor suppressor effects of this signaling pathway in the colon 5.
The common occurrence of mutations in genes that encode proteins that operate in individual signaling pathways, such as the Wnt signaling pathway or Ras-Raf signaling pathway, has led to the concept that these mutations contribute to cancer formation through the deregulation of these tumor suppressor signaling pathways14. More recently, genes in signaling pathways implicated in cancer formation have also been shown to be targets of aberrant CpG island DNA methylation, which represses gene expression. For example, silencing of SFRP2 by methylation can promote the Wnt signaling pathway in colorectal cancer and is an alternate and complementary mechanism to APC mutation for activating this pathway 15. In addition to SFRP2, other aberrantly methylated genes are found commonly in colon adenomas and colon adenocarcinomas, and it has been suggested that they also promote tumor formation through signal pathway deregulation 16. TSP1 is one of the genes that has been found to be aberrantly methylated in some colorectal cancers as well as in neuroblastomas, and gastric cancers. It has been suggested methylated TSP1 may promote tumorigenesis through its effects on angiogenesis 17–19. We hypothesized that in light of TSP1’s known role in mediating TGF-β1 activation that the aberrant methylation of TSP1 may promote tumor formation through inhibiting the TGF-β signaling pathway. We now present results that demonstrate the aberrant methylation of TSP1 is a novel epigenetic mechanism for inhibiting the TGF-β signaling pathway in colorectal cancer.
Material and methods
Cell lines
SW-48, RKO, LoVo, V411, Moser, V241, FET and Raji cells were used in these studies and were cultured in DMEM+10% FBS (Gibco/Invitrogen, Carlsbad, California). The SW-48 cell line was obtained from ATCC (CCL-231, Manassas, VA). The RKO and LoVo cell lines were provided by Dr. Sanford Markowitz (Case Western Reserve University, Cleveland, OH). The human Burkitt lymphoma cell line Raji was kindly provided by Jennifer Pietenpol (Vanderbilt University, Nashville, TN). The Moser and FET cells were a generous gift from Michael Brattain (Roswell Park Cancer Institute, Buffalo, NY).
With regards to the treatment with 5′-Aza 2′ deoxycytidine (5Aza) treatment, the cells were grown to 70% confluence and then treated with 1 μM of 5Aza (Sigma, A3656) or vehicle only (DMSO) overnight. After an overnight incubation, the media was replaced with serum free DMEM. The cells were grown for an additional 48, 96 and 144 hours. The cells and conditioned media were harvested at these time points. The peptide LSKL was obtained from Genemade Synthesis, Inc, (San Francisco, CA).
Plasmids
The pBLAST49-hTSP1 expression vector was obtained from InvivoGen, (San Diego, California,). The pRC-CMV-TGFBR2 expression vector was provided by Sanford Markowitz (Ireland Cancer Center, Case Western Reserve University, Cleveland, OH). The p3TP-Lux vector was kindly provided by Joan Massague (Memorial Sloan Kettering Cancer Center, New York, New York). The pRL-TK vector was purchased from Promega (Madison, WI).
Tissue samples
Colon adenomas and adenocarcinomas were collected from patients treated at Vanderbilt University Medical Center, the Department of Veterans Affairs Tennessee Valley Health Care System, and Meharry Medical Center (Nashville, TN) and at University Hospitals of Cleveland (Cleveland, OH) following protocols approved by the Institutional Review Board of each institution. All tissue samples were formalin-fixed and paraffin-embedded tissue blocks obtained from the pathology archives and were collected randomly based on tissue availability. The cases included 42 colon adenomas, 22 primary colon adenocarcinomas, and 15 samples of normal colon mucosa obtained from colon resection specimens from patients who had diverticular disease or had colon cancer. In addition, two tissue microarrays (TMA) derived from 38 patients’ resections of metastatic colon cancer resections to the liver were constructed. Four 1.5 mm cores from each lesion of routinely processed formalin-fixed paraffin embedded adenocarcinoma were used. The TMAs were constructed using a Beecher Instruments Manual Tissue Arrayer 1.
DNA extraction and sodium bisulfite treatment
DNA extraction from the cell lines and tissue samples was performed as previously published. DNazol (Invitrogen) was used to extract DNA from the cell lines 2. The DNA was modified with sodium bisulfite for use in methylation specific PCR (MSP) assays as previously described 20.
Methylation Specific PCR (MSP)
MSP primers were designed to amplify the methylated and unmethylated alleles for TSP1 and methylation-specific PCR assay conditions were determined so that specific reaction products were obtained from each respective set of primers. Each PCR reaction mix consisted of a total volume of 20 μl composed of PCR buffer (Qiagen, Valencia, CA), 200 pM deoxynucleotide triphosphate mix (Applied Biosystems, Foster City, CA), 500 nM of each primer (Sigma Genosys, The Woodlands, TX), 1 unit of HotStar Taq DNA polymerase (Qiagen), and bisulfite-modified DNA. The primers sequences and reaction conditions have been previously published 21. All the samples were subjected to at least two independent rounds of sodium bisulfite treatment and MSP assays. Control samples from cells with known methylated and unmethylated TSP1 were included in each MSP assay to confirm the technical success of the assays. The MSP products were subjected to horizontal gel electrophoresis in a 1.5% agarose gel, stained with ethidium bromide, and visualized with UV transillumination using an Eagle Eye Imaging system (Stratagene, La Jolla, CA).
Quantitative RT-PCR
RNA was extracted from the cell lines using TRI reagent (Molecular Research Center, Cincinnati, OH) following the manufacturer’s recommended protocol. One microgram of total RNA from each sample was reverse transcribed using oligo d(T) priming and Superscript-II reverse-transcriptase (Invitrogen).
TaqMan On-Demand primers and probes were used to determine the relative expression levels of TSP1 (Assay number Hs00170236_m1, Applied Biosystems, Foster City, CA) and 18S in all samples (Assay number Hs99999901_s1, Applied Biosystems,). 18S RNA was used as an internal control. The reactions were run in triplicate in the ABI Prism 7700 detection system (Applied Biosystems) and results were analyzed with SDS 2.1 software.
Immunostaining
For TSP1 immunostaining, tissue sections were deparaffinized, rehydrated, and endogenous peroxidase activity was blocked with H2O2 treatment using standard methods 22. Antigen unmasking was achieved by incubation in boiling sodium citrate buffer pH 6.0 (Biogenex, San Ramon, CA) for 15 minutes. Non-specific binding was blocked by treating the sections with 1.5% horse normal serum for 10 minutes, and the immunostaining was performed with the mouse monoclonal antibody anti-TSP1 (ab2 Lab Vision, Fremont, CA) diluted 1:2000 incubated at 4°C overnight. The Vectastain ABC Kit (PK-4002, Vector labs, Burlingame, CA) was used to obtain the final stain. The tissue sections were then coverslipped and analyzed with an Eclipse 80i compound microscope (Nikon). Photomicrographs were obtained using a CCD camera (Micropublisher, Coolpix 3.3). The evaluation of positive TSP1 expression in tumors was performed using a semiquantitative scoring of the percentage of positive tumor cells. TSP1 expression was scored positive when >40% of the tissue analyzed was immunoreactive, which is similar to a scoring system that has been used by Zlobec et al, who demonstrated that the scoring system is reproducible among pathologists 23.
siRNA transfection
1–2 ×105 FET cells were cultured in a 24 well plate in complete media and then transfected 24 hours after the cells were seeded. The media was replaced by serum free media and the cells were transfected with the siRNA for TSP1 (cat # L-019743-00, Dharmacon) and control siRNA (cat # D-001810-03-05, Dharmacon), using Lipofectamine 2000 (cat # 11668-019, Invitrogene) following the manufacturer’s protocols. Forty-eight hour after the transfection, activated TGF-β1 or the TGF-β:LAP complex was added to the media. The media was collected 24 hours later. RNA from the cells was extracted at this time as well.
TGF-β1 ELISA
Media was collected from cells treated with 5′-aza-2′-deoxycytidine or transfected with the pBLAST49-hTSP1 expression vector described above. The concentration of TGF-β1 was measured with the TGF-β1 Quantikine ELISA assay (DB-100, R&D Systems, Minneapolis, MN) following the manufacturer’s protocol with the following modification. In order to measure the concentration of active and latent TGF-β, the media was divided. The first portion of the media was activated with hydrochloric acid and the remaining portion of the same media was assessed without any hydrochloric acid treatment. The ELISA assay results were measured using a VERSAmax microplate spectrophotometer (Molecular Devices, Sunnyvale, CA) set at 450 nm.
Formation of the TGF-β:LAP complex
TGF-β1 (2ng) (101-B1-010, R&D Systems) was added to recombinant human LAP (30ng) (246-LP, R&D Systems). The complex was allowed to form at room temperature for at least 2 hours before being added to the media.
3TP-Lux luciferase reporter assay
SW-48 cells were transfected with the pBLAST49-hTSP1 vector using FuGene (11814443001, Roche) following the manufacturer’s protocol or were treated with 5′-aza 2′ deoxycitidine in a six well plate as previously published 5.
Results
Loss of TSP1 expression and aberrantly methylated TSP1 is present in both colon adenomas and adenocarcinomas
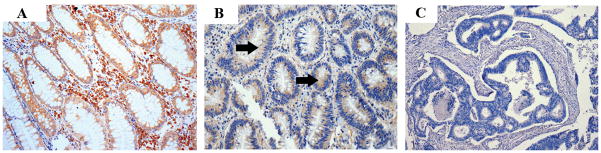
The overall frequency of TSP1 loss in colorectal cancer was assessed by immunostaining normal colon (N=14), colon adenomas (N=4), and colon adenocarcinomas (N=51). TSP1 immunoreactivity was observed in 86% (N=12/14) of normal colon mucosa, 50% (N=2/4) of colon adenomas, 33% of primary cancers (N=4/12), and 25% (N=10/39) of liver metastases, demonstrating that TSP1 loss is common in colon cancers (Figure 1). The absence of expression in two of the normal colon samples may reflect low levels of expression that are below the limits of detection of the immunostaining assay or may reflect a field defect as these samples were obtained from individuals with concurrent colon cancer. The significance of the lack of expression of TSP1 in the normal colon is not clear at this time.
Figure 1.
Results of immunostaining for TSP1 in colon tissue samples. Photomicrographs of representative samples are shown (magnification 40X and 100X). The expression of TSP1 in samples of normal colon (A), adenoma (B) and adenocarcinoma (C) is demonstrated by the presence of brown staining. TSP1 expression in adenocarcinomas is lower than in normal tissue and adenomas (C). The arrows indicate the areas of TSP1 immunoreactivity.
In light of the frequent loss of TSP1 expression in colon adenomas and adenocarcinomas, we next determined the frequency of aberrantly methylated TSP1 in colorectal cancer cell lines, primary colon adenomas and adenocarcinomas. The methylation status of TSP1 was assessed in a region that is 30 bp upstream from the transcription start site, which has been demonstrated to be involved with methylation-induced transcriptional repression 17, 21. We found aberrantly methylated TSP1 in 33% of colon cancer cell lines (N=3/9), 14% of colon adenomas (N=6/42) and 21% of adenocarcinomas (N=18/96) (Figure 2). (Note: A portion of the adenomas and cancers used in this study have been reported previously 21.) Importantly, no methylated TSP1 was detected in normal colon mucosa samples (N=10), providing evidence that TSP1 hypermethylation is a cancer-specific epigenetic event. In order to assess the correlation between TSP1 methylation and TSP1 expression, TSP1 immunostaining was performed in a subset of the cancer cases described above (N=19). We observed that none of the tumors that carried methylated TSP1 (N=5) expressed TSP1 in the neoplastic tissue and that 78% of tumors with unmethylated TSP1 (N=14) expressed TSP1. Of interest, some of the tumors with absent TSP1 expression in the tumor cells did show TSP1 expression in the adjacent stroma (Supplementary figure).
Figure 2.
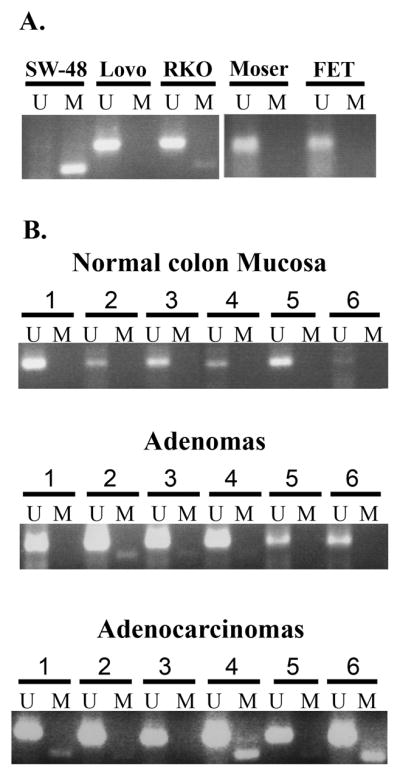
A. Representative TSP1 MSP assay results for cell lines. U, PCR product from the MSP assay using primers specific for the unmethylated allele; M, PCR product from the MSP assay using primers specific for the methylated allele. B. Representative results from the TSP1 MSP analysis of normal colon mucosa, adenomas and adenocarcinomas using primers for methylated (M) and unmethylated (U) alleles of TSP1. No methylated TSP1 was detected in any of the normal colon mucosa samples. Approximately 20% of colon neoplasms were found to have methylated TSP1. The unmethylated allele present in the tumors that were shown to have methylated TSP1 is presumably from intermixed normal tissue.
The aberrant methylation of TSP1 induces silencing of TSP1 expression
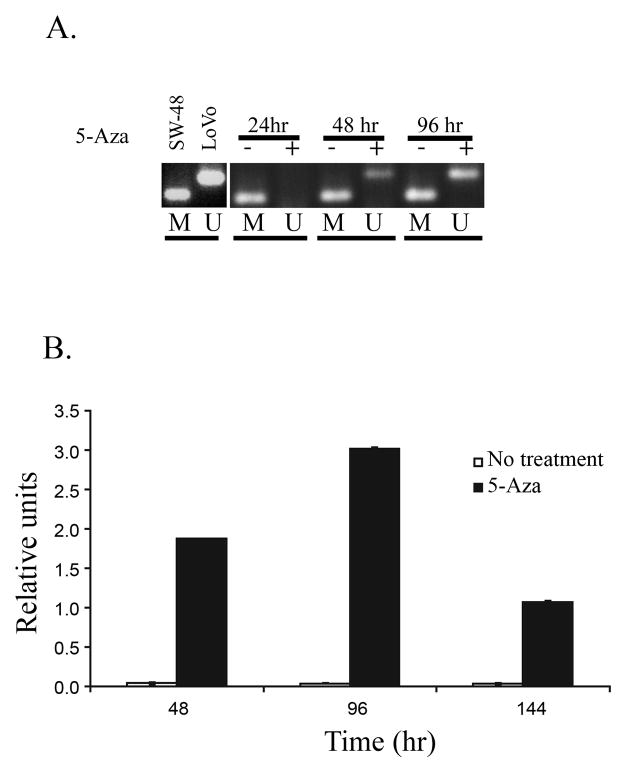
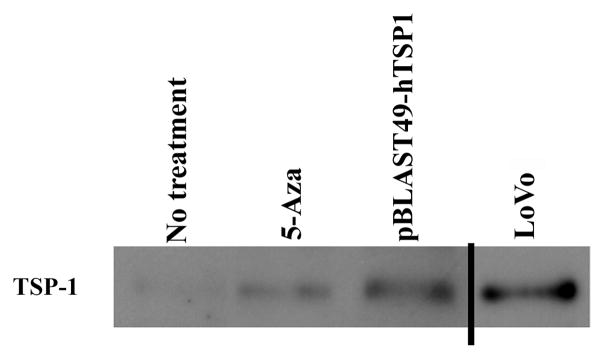
We next evaluated the effect of aberrant methylation of TSP1 on TSP1 mRNA expression and protein secretion. Treatment of SW-48 cells with 5′-Aza-2′deoxycytidine (5′-Aza) resulted in the demethylation of TSP1 and a 1.5–2 fold increase in expression of TSP1 that was detected after 96 hours of treatment (Figures 3A and 3B) (Student’s t-test; p=0.042). Moreover, secreted TSP1 was present in the conditioned media of SW-48 after treatment with 5′-Aza, consistent with the effects of 5′-Aza observed on mRNA expression and demonstrating increased levels of secreted TSP1 secondary to 5′-Aza induced demethylation of the gene (Figure 4). These results suggest the aberrant methylation of TSP1 suppresses TSP1 mRNA expression and TSP1 protein secretion in colorectal cancer.
Figure 3.
Re-expression of TSP1 in SW-48 cells after treatment with 5′-Aza-2′deoxycytidine (5′-Aza). A. TSP1 MSP results from cells that were treated for 24, 48 and 96 hours with 5′-Aza. The presence of unmethylated DNA can be seen after 48 and 96 hours of treatment. B. TSP1 mRNA levels assessed by quantitative RT-PCR for TSP1 expression, treatment with 5′-Aza resulted in a significant increase in the levels of TSP1 mRNA in SW-48 cells.
Figure 4.
TSP1 is secreted from SW-48 cells after treatment with 5′-Aza. Western blot analysis for TSP1 in conditioned media from SW-48 and LoVo colon cancer cells. Conditioned media was collected from SW-48 cells transfected with TSP1 or after treatment with 5′-Aza and analyzed. The amount of TSP1 observed in conditioned media from the LoVo colon cancer cell line, which has unmethylated TSP1, is similar to that from the SW-48 cells after TSP1 transfection or treatment with 5′-Aza.
TSP1 inactivation inhibits the activation of secreted TGF-β1
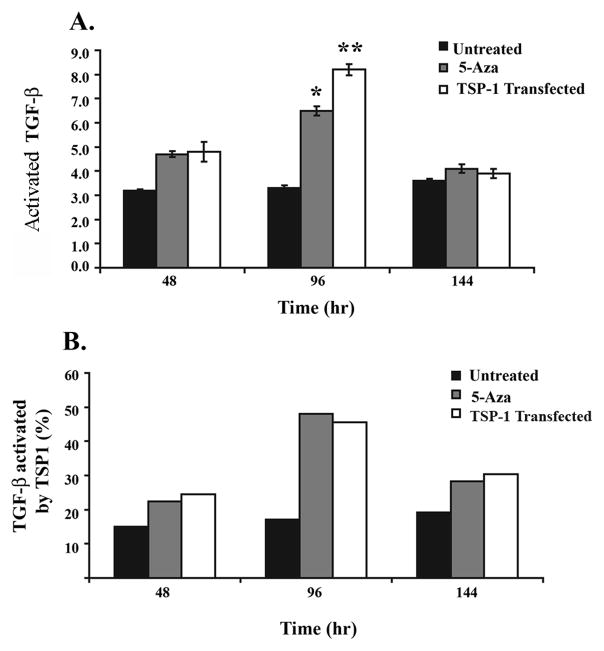
Upon providing evidence that the aberrant methylation of TSP1 inhibits its expression, we assessed how TGF-β1 activation is affected by the loss of TSP1 secretion. Demethylation of TSP1 with 5′-Aza resulted in a two fold increase in activated TGF-β1 at 96 hours. (Figure 5A) The concentration of total TGF-β1 did not change after treatment with 5′-Aza (data not shown), demonstrating that the effect of 5′-Aza on TGF-β1 activation was not simply a consequence of increased production of total TGF-β1 (Figure 5A).
Figure 5.
Increased secreted activated TGF-β1 is present in conditioned media from SW-48 cells after treatment with 5′-Aza or transfection with TSP1. Results from TGF-β1 ELISA for secreted active and total TGF-β1 in conditioned media from SW-48 after treatment with 5′-Aza or transfection with TSP1. A. Increased activated TGF-β1, compared to the basal levels, is present 48 and 96 hours after the initiation of treatment with 5′-Aza or after transfection with TSP1, with the most significant increase observed at 96 hours. B. The percentage of activated TGF-β1 is also increased at 48, 96, and 144 hours after initiation of treatment with 5′-Aza or transfection with TSP1. The percentage of activated TGF-β1 present in the treatment groups at 144 hours is similar to 48 hours, which is likely a result of loss of TSP1 expression that occurs after transient transfection with TSP1 or after the cessation of treatment with 5′-Aza. (** students t-test; p=0.035 and * students t-test; p=0.049.)
In order to provide evidence for the specificity of the 5′-Aza treatment on TGF-β1 activation through the induction of TSP1, we also assessed the effect of TSP1 reconstitution by gene transfection on TGF-β1 activation (Figure 5A). Interestingly, we observed similar effects on TGF-β1 activation in the SW-48 cells reconstituted with TSP1 compared to the 5′-Aza treatment, although the magnitude of the effect was greater at 96 hours in the TSP1 reconstituted SW-48 cells compared to the 5′-Aza treated cells, consistent with the increased amount of secreted TSP1 observed in SW-48 cells transfected with TSP1. These results suggest ~25% of the total activated TGF-β1 present in the media is activated by TSP1 (Figure 5B). Interestingly, 144 hours after treatment, the levels of activated TGF-β1 returned to the basal levels, which likely is a consequence of waning levels of TSP1 that occurs at 144 hours after 5′-Aza treatment or the transient transfection with TSP1 (Figure 3.) These results provide additional indirect evidence that the increased in TGF-β1 activation observed in this study is a consequence of TSP1 activity.
To further assess if TSP1 silencing can affect the activation of TGF-β1, TSP1 expression was inhibited using siRNA in the FET colon cancer cell line, which carries unmethylated TSP1 and expresses the gene. We observed decreased TSP1 expression and reduced activation of TGF-β1 in FET transfected cells with siRNA against TSP1 (Students t-test; p=0.0002).
Aberrant methylation of TSP1 can suppress TGF-β signaling by impairing the activation of TGF-β1
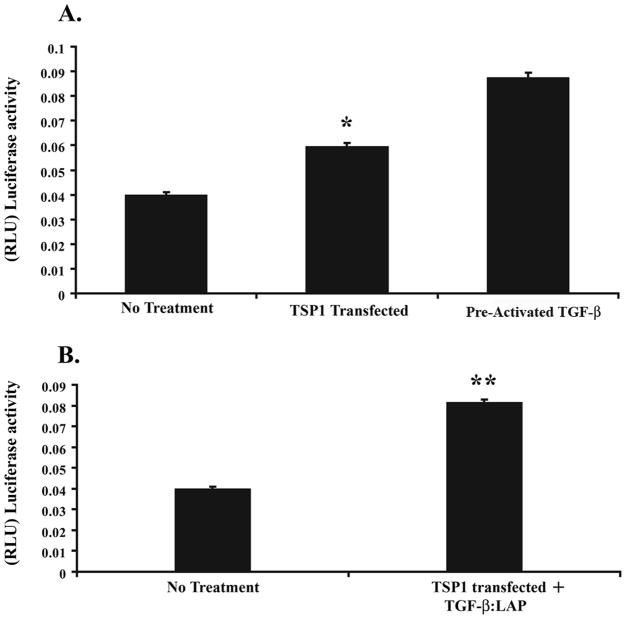
After demonstrating that TSP1 methylation can affect the concentration of secreted, activated TGF-β1 in colon cancer, we assessed the functional consequences of this effect on TGF-β mediated signaling. Using the 3TP-Lux reporter assay, we observed a ~ 30% increase in 3TP-Lux activity after TSP1 transfection (Figure 7A). As TSP1 is known to activate TGF-β1 by a specific mechanism, which is the disassociation of LAP from TGF-β1, we also assessed the effect of TSP1 reconstitution on activating pre-formed TGF-β1:LAP complexes and inducing TGF-β signaling. We observed increased 3TP-Lux activity at 96 hours after TSP1 reconstitution (Figure 7B). Additionally in cells in which the TGF-β1:LAP complex was added but that were not transfected with TSP1, no increase in 3TP-Lux reporter activity was observed. The 3TPLux activity in the “No treatment” group likely reflects the effects of autocrine TGF-β1 and activation of this autocrine TGF-β1 by non-TSP1 mediated mechanisms, such as plasmin. Similarly, 3TPLux activity is present in the “No treatment and TGF-β:LAP” group. This activation of TGF-β1 is presumably mediated by plasmin or another TSP1 independent mechanism. The “activated TGF-β” group was treated with acid-activated exogenous TGF-β and reveals the maximal 3TPLux response possible in the SW48 cells.
Figure 7.
TSP1 activates TGF-β present in the TGF-β1:LAP complex and induces Smad signaling. A. 3TPLux luciferase reporter activity in the SW-48 cell line 48 hours after transfection with TSP1. The 3TP-Lux reporter activity is increased approximately 1.2X in the TSP1 transfected SW-48 cells and 1.8X in the SW-48 cells treated with activated TGF-β (2ng/ml) (101-B1-010, R&D Systems) when compared with non-treated SW-48 cells (*p=0.0005, Student’s t-test analysis). B. 3TPLux luciferase reporter activity in the SW-48 cell line after addition of preformed TGF-β1:LAP and transfection with TSP1. The 3TP-Lux reporter activity is increased approximately 1.3X after 96 hours of treatment compared to the baseline level. (**p= 0.003; Student’s t-test analysis).
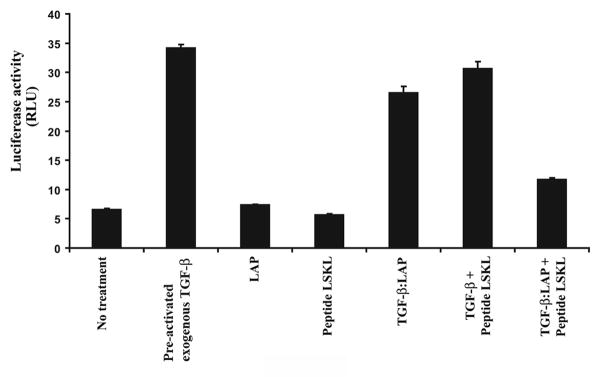
The region of TSP1 that is responsible for the activation of TGF-β1 is localized in the amino end of the TSR2 domain. This portion of the protein interacts with the LSKL sequence present in the LAP inducing the conformational change of LAP that results in the dissociation of TGF-β from the TGF-β1:LAP complex. In order to confirm that the effect we observed on TGF-β pathway activation was due to TSP1 activating the latent TGF-β:LAP complex and not through a nonspecific mechanism, we assessed the effect of an inhibitory peptide that contains the LSKL motif on TGF-β pathway activation. The LSKL peptide inhibits the ability of TSP1 to induce the dissociation of LAP from TGF-β. We observed that the LSKL peptide inhibited the activation of the TGF-β signaling pathway reporter 3TP-Lux in SW-48 cells transfected with TSP1 (Figure 8). Of note, we observed 3TP-Lux reporter activity in the no treatment group, which presumably reflects autocrine TGF-β1 that is being activated by TSP1 independent mechanisms, such as plasmin. The “TGF-β” group demonstrates the maximal amount of 3TP-Lux activity that can be induced by adding pre-activated TGF-s to the SW-48 cells. Comparison of the other treatment groups to the “TGF-β” group reveals that transfected TSP1 can induce substantial TGF-β activation and pathway activity in the SW-48 cells treated with a TGF-β:LAP complex.
Figure 8.
TGF-β pathway activation measured by 3TP-Lux reporter luciferase activity. The SW-48 cell line was transfected with 3TP-Lux and TSP1, and then treated with the agents noted along the X-axis. As expected, the addition of exogenous pre-activated TGF-β1 (101-B1-010, R&D Systems) induces the highest activity of the pathway and demonstrates the maximal amount of induction possible in the SW48 cells. When the complex TGF-β:LAP is added in presence of TSP1 (transfected cells), the pathway is activated. The inhibitory peptide LSKL attenuates the 3TP-Lux activity in the cells with and without the TGF-β:LAP complex. In light of the specificity of TSP1 on the activation of TGF-β1 through dissociating the TGF-β LAP complex, these results confirm that TSP1 is inducing the activation of the TGF-β pathway through effects on activation of TGF-β1.
TSP1 expression is associated with the nuclear localization of Smad2 in primary colorectal neoplasms
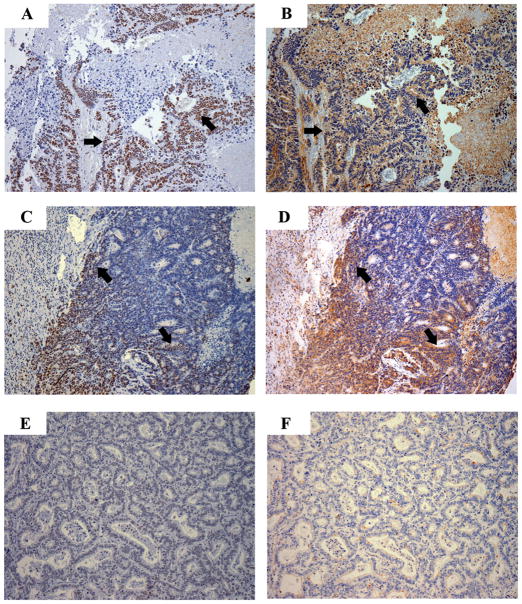
Our studies in the SW-48 and FET cell lines demonstrated in vitro that the absence of TSP1 inhibits TGF-β pathway activation. We next assessed the correlation between TSP1 expression and TGF-β pathway activation in primary colon cancer by determining the correlation between nuclear phosphorylated Smad2 and TSP1 in normal colon, colon adenomas, and colon adenocarcinomas. We found that TSP1 expression correlated with phosphorylated Smad2 in the majority of the cases (N=17/20) (Pearson rank test, p=0.032) (Figure 9). Of note, we also observed phosphorylated Smad2 in the absence of TSP1 expression in three tumors, which suggests alternate mechanisms can activate TGF-β1 in these tumors, such as plasmin or cathepsin. We also observed that in one of the cases, cells that expressed TSP1 did not have detectable phosphorylated Smad2. In this case, we presume that the TGF-β signaling pathway has been disrupted at the level of the receptor or post-receptor elements.
Figure 9.
Results of immunostaining for phosphorylated Smad2 (pSmad2) (A, C, and E) and TSP1 (B, D, and F) in primary colorectal cancers. A-B. Nuclear pSmad2 is present in most of the epithelial cells and TSP1 expression is present. C-D. Heterogeneity of expression of pSmad2 and TSP1 is present in this tumor. Co-localization of pSmad2 and TSP1 can be appreciated. E-F. No expression of pSmad2 or TSP1 is present. (Magnification: 100X).
In summary, we have demonstrated TSP1 expression is often reduced in colorectal neoplasms and the aberrant methylation of TSP1 is one mechanism responsible for the silencing of TSP1. We have further shown the loss of TSP1 attenuates the activation of secreted TGF-β and the SMAD signaling pathway in in vitro systems. Finally, our assessment of colon neoplasms is also consistent with an effect of aberrant methylation of TSP1 on the formation of primary colorectal cancers being the attenuation of TGF-β signaling in these tumors.
Discussion
TGF-β can inhibit cell proliferation, induce apoptosis, and induce the terminal differentiation of normal colon epithelial cells and colon adenoma cells 24, 25. The TGF-β pathway is inactivated or impaired in >50% of colon adenocarcinomas through a variety of mechanisms including the mutation of TGFBR2, SMAD4, and SMAD2, the overexpression of inhibitory Smad7, and the increased expression of Smad signaling repressors such as Ski or SnoN 3, 4, 22, 26–28. We now provide evidence that an epigenetic event, the aberrant methylation of TSP1, can impair TGF-β signaling in colorectal cancer, providing evidence for a tumor promoting biological effect of the methylation of TSP1 as well as for a novel mechanism for impairing TGF-β signaling in colorectal cancer.
The aberrant methylation of tumor suppressor genes has been shown to affect many genes in colon cancer, including MLH1, HLTF, CDKN2A, SLC5A8, and is believed to contribute to the clonal progression of the tumors since aberrant DNA methylation can inactivate the function of genes 18, 21, 29–31. Indeed, the aberrant methylation of SFRP2, TIMP3, HIC1, and ITGA4 have all been shown to have tumor promoting effects in cancer cell lines 15, 32–34. Nonetheless, it is also clear that some of the aberrantly methylated genes are hitchhiker or passenger events that do not influence the pathogenesis of the tumors 35, 36. Thus, the functional consequences that DNA hypermethylation and hypomethylation have on the behavior of the cancer cells remains to be determined for the majority of the genes that have been shown to be aberrantly methylated in cancer 33, 37–39
In this study, we have demonstrated that the aberrant methylation of TSP1 results in loss of TSP1 expression and in attenuation of TGF-β1 activation and in TGF-β signaling pathway activity. These results provide support that epigenetic alterations can play a pathogenic role in colon cancer formation by inhibiting TGF-β signaling. In approximately 75% of colon cancer cell lines, there is resistance to TGF-β mediated growth inhibition, and in many colorectal cancers, the TGF-β signaling pathway is inhibited by mutational inactivation of the TGF-β receptor or SMADs 5. However, in approximately 30–40% of colon cancers, the mechanism(s) resulting in impaired TGF-β signaling is not known. Some investigators have shown that epigenetic alterations can affect KM23, TGFBI (Betaig-h3 gene), and RUNX3, which are all genes involved with regulation of the TGF-β signaling pathway, but none of these studies have demonstrated that the aberrant methylation of these genes directly alters the TGF-β signaling pathway 40–42. We have provided evidence that TGF-β1 activation is impaired when TSP1 is aberrantly methylated resulting in the suppression of TGF-β receptor activation. These results provide the first evidence of which we are aware that an epigenetic event can attenuate TGF-β responsiveness in colon cancer. It is also possible that the aberrant methylation of TSP1 may promote angiogenesis through effects on VEGF or through effects on extracellular matrix remodeling, which was not investigated in this study 17, 43.
The demonstration of aberrant methylation and functional inactivation of TSP1 also provides support that TSP1 is a true tumor suppressor gene that is subject to epigenetic inactivation in cancer. Prior to the discovery of aberrant methylation of TSP1, there was no genetic or epigenetic evidence that supported the role of TSP1 as a bona fide tumor suppressor gene 43. Earlier studies have shown TSP1 is a downstream target of TP53 mutations and a regulator of VEGF, which suggests TSP1 is a tumor suppressor gene 44, 45. The identification of aberrantly methylated TSP1 in colorectal cancer, neuroblastomas, and gliomas demonstrates, that like CDKN2A, TSP1 is susceptible to epigenetic alterations even though it does not appear to be commonly mutated in cancer 17, 43, 46. It is not clear whether the stroma can be a source for TSP1 in the setting of a cancer that has silenced TSP1 expression. The immunostaining results we have obtained demonstrate a strong correlation between TSP1 methylation and lack of TSP1 expression as well as between TSP1 expression and the presence of phosphorylated Smad2. These results suggest that the tumor cells are a major source of TSP1 and that methylation of TSP1 substantially reduces TSP1 activity in the tumors.
In summary, we have identified a novel epigenetic mechanism for suppressing the TGF-β signaling pathway. We have provided evidence that the aberrant methylation of TSP1 impairs the activation of TGF-β1 in colon cancer cell lines and propose that the effect is similar in primary human colon cancer. Furthermore, these results provide evidence that the aberrant methylation of TSP1 is an epigenetic event that silences this tumor suppressor gene and results in biological events that would promote the development of colon cancer, suggesting that targeting therapy towards aberrant DNA methylation could inhibit cancer cells at least in part by inducing the TGF-β signaling pathway. Additional studies should help clarify the ultimate effects of the aberrant methylation of TSP1 and the effects of therapy directed towards this epigenetic event on colon cancer formation given the complex effects of TSP1 and TGF-β on cancer cells and the cancer-associated stroma.
Supplementary Material
Supplementary Figure. Tumor immunostaining showing differences in TSP1 expression. A. Immunostaining results from a tumor that has unmethylated TSP1. The expression of the protein is detected mainly in the stroma (blue arrow) and not in tumor cells (black arrow). B. TSP1 expression in a tumor in which TSP1 is methylated. Faint immunostaining is detected in this sample.
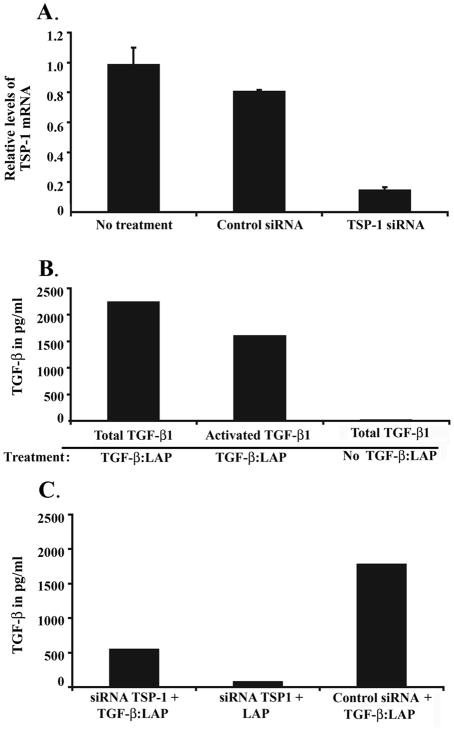
Figure 6.
A. Assessment of TSP1 mRNA levels by quantitative RT-PCR after transfection with siRNA against TSP1. TSP1 expression is reduced to approximately 15% of basal levels by siRNA. No effect of the control siRNA on TSP1 expression is observed. B. Assessment of total and activated TGF-βin FET cells treated with TGF-β:LAP. Activated TGF-β1 is detected in FET cells that were treated with the TGF-β:LAP complex, such activation represents ~50% of the total TGF-β present in the media. TGF-β1 activation was not detected in cells in which the TGF-β:LAP complex was not added. C. Assessment of TGF-β:LAP complex activation in FET cells after treatment with siRNA against TSP1. A substantial reduction in activated TGF-β is observed. No effect on TGF-β activation is observed in the FET cells treated with the control siRNA. These experiments were done in duplicate with a representative set of results shown.
Acknowledgments
Sources of Support: This work was supported by a Damon Runyon-Lilly Clinical Investigator Award from the Damon Runyon Cancer Research Fund, a VA Presidential Early Career Award for Scientists and Engineers from the Department of Veterans Affairs, and from a pilot project award from the Early Detection and Intervention Initiative (FHCRC Comprehensive Cancer Center Support Grant) (to WMG).
References
- 1.Jemal A, Siegel R, Ward E, Murray T, Xu J, Thun MJ. Cancer Statistics. CA Cancer J Clin. 2007;57(1):43–66. doi: 10.3322/canjclin.57.1.43. [DOI] [PubMed] [Google Scholar]
- 2.Grady W, Rajput A, Myeroff L, Liu D, Kwon K-H, Willis J, Markowitz S. Mutation of the type II transforming growth factor-β receptor is coincident with the transformation of human colon adenomas to malignant carcinomas. Cancer Research. 1998;58:3101–04. [PubMed] [Google Scholar]
- 3.Munoz NM, Upton M, Rojas A, Washington MK, Lin L, Chytil A, Sozmen EG, Madison BB, Pozzi A, Moon RT, Moses HL, Grady WM. Transforming growth factor beta receptor type II inactivation induces the malignant transformation of intestinal neoplasms initiated by Apc mutation. Cancer Research. 2006;66(20):9837–44. doi: 10.1158/0008-5472.CAN-06-0890. [DOI] [PubMed] [Google Scholar]
- 4.Grady WM, Markowitz SD. Genetic and epigenetic alterations in colon cancer. Annual Reviews Genomics and Human Genetics. 2002;3:101–28. doi: 10.1146/annurev.genom.3.022502.103043. [DOI] [PubMed] [Google Scholar]
- 5.Grady W, Myeroff L, Swinler S, Rajput A, Thiagalingam S, Lutterbaugh J, Neumann A, Brattain M, Chang J, Kim S-J, Kinzler K, Vogelstein B, et al. Mutational inactivation of transforming growth factor s receptor type II in microsatellite stable colon cancers. Cancer Research. 1999;59:320–24. [PubMed] [Google Scholar]
- 6.Liu B, Parsons R, Papadopoulos N, Nicolaides NC, Lynch HT, Watson P, Jass JR, Dunlop M, Wyllie A, Peltomaki P, de la Chapelle A, Hamilton SR, et al. Analysis of mismatch repair genes in hereditary non-polyposis colorectal cancer patients. Nat Med. 1996;2(2):169–74. doi: 10.1038/nm0296-169. [DOI] [PubMed] [Google Scholar]
- 7.Derynck R, Akhurst RJ, Balmain A. TGF-beta signaling in tumor suppression and cancer progression. Nat Genet. 2001;29(2):117–29. doi: 10.1038/ng1001-117. [DOI] [PubMed] [Google Scholar]
- 8.Ku JL, Park SH, Yoon KA, Shin YK, Kim KH, Choi JS, Kang HC, Kim IJ, Han IO, Park JG. Genetic alterations of the TGF-beta signaling pathway in colorectal cancer cell lines: a novel mutation in Smad3 associated with the inactivation of TGF-beta-induced transcriptional activation. Cancer Lett. 2007;247(2):283–92. doi: 10.1016/j.canlet.2006.05.008. [DOI] [PubMed] [Google Scholar]
- 9.Sha X, Yang L, Gentry LE. Identification and analysis of discrete functional domains in the pro region of pre-pro-transforming growth factor beta 1. J Cell Biol. 1991;114(4):827–39. doi: 10.1083/jcb.114.4.827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Crawford SE, Stellmach V, Murphy-Ullrich JE, Ribeiro SM, Lawler J, Hynes RO, Boivin GP, Bouck N. Thrombospondin-1 is a major activator of TGF-beta1 in vivo. Cell. 1998;93(7):1159–70. doi: 10.1016/s0092-8674(00)81460-9. [DOI] [PubMed] [Google Scholar]
- 11.Murphy-Ullrich JE, Poczatek M. Activation of latent TGF-beta by thrombospondin-1: mechanisms and physiology. Cytokine Growth Factor Rev. 2000;11(1–2):59–69. doi: 10.1016/s1359-6101(99)00029-5. [DOI] [PubMed] [Google Scholar]
- 12.Massague J. TGF-β signaling: receptors, transducers, and mad proteins. Cell. 1996;85:947–50. doi: 10.1016/s0092-8674(00)81296-9. [DOI] [PubMed] [Google Scholar]
- 13.Bierie B, Moses HL. TGF-beta and cancer. Cytokine Growth Factor Rev. 2006;17(1–2):29–40. doi: 10.1016/j.cytogfr.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 14.Chung D. The genetic basis of colorectal cancer:insights into critical pathways of tumorigenesis. Gastroenterology. 2000;119:854–65. doi: 10.1053/gast.2000.16507. [DOI] [PubMed] [Google Scholar]
- 15.Suzuki H, Watkins DN, Jair KW, Schuebel KE, Markowitz SD, Dong Chen W, Pretlow TP, Yang B, Akiyama Y, Van Engeland M, Toyota M, Tokino T, et al. Epigenetic inactivation of SFRP genes allows constitutive WNT signaling in colorectal cancer. Nat Genet. 2004;36(4):417–22. doi: 10.1038/ng1330. [DOI] [PubMed] [Google Scholar]
- 16.Baylin SB, Ohm JE. Epigenetic gene silencing in cancer - a mechanism for early oncogenic pathway addiction? Nat Rev Cancer. 2006;6(2):107–16. doi: 10.1038/nrc1799. [DOI] [PubMed] [Google Scholar]
- 17.Yang QW, Liu S, Tian Y, Salwen HR, Chlenski A, Weinstein J, Cohn SL. Methylation-associated silencing of the thrombospondin-1 gene in human neuroblastoma. Cancer Research. 2003;63(19):6299–310. [PubMed] [Google Scholar]
- 18.Lee S, Hwang KS, Lee HJ, Kim JS, Kang GH. Aberrant CpG island hypermethylation of multiple genes in colorectal neoplasia. Lab Invest. 2004;84(7):884–93. doi: 10.1038/labinvest.3700108. [DOI] [PubMed] [Google Scholar]
- 19.Kim HC, Kim JC, Roh SA, Yu CS, Yook JH, Oh ST, Kim BS, Park KC, Chang R. Aberrant CpG island methylation in early-onset sporadic gastric carcinoma. J Cancer Res Clin Oncol. 2005;131(11):733–40. doi: 10.1007/s00432-005-0017-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grady WM, Willis J, Guilford PJ, Dunbier AK, Toro TT, Lynch H, Wiesner G, Ferguson K, Eng C, Park JG, Kim SJ, Markowitz S. Methylation of the CDH1 promoter as the second genetic hit in hereditary diffuse gastric cancer. Nat Genet. 2000;26(1):16–7. doi: 10.1038/79120. [DOI] [PubMed] [Google Scholar]
- 21.Kim YH, Petko Z, Dzieciatkowski S, Lin L, Ghiassi M, Stain S, Chapman WC, Washington MK, Willis J, Markowitz SD, Grady WM. CpG island methylation of genes accumulates during the adenoma progression step of the multistep pathogenesis of colorectal cancer. Genes Chromosomes Cancer. 2006;45(8):781–9. doi: 10.1002/gcc.20341. [DOI] [PubMed] [Google Scholar]
- 22.Biswas S, Chytil A, Washington K, Romero-Gallo J, Gorska AE, Wirth PS, Gautam S, Moses HL, Grady WM. Transforming growth factor beta receptor type II inactivation promotes the establishment and progression of colon cancer. Cancer Research. 2004;64(14):4687–92. doi: 10.1158/0008-5472.CAN-03-3255. [DOI] [PubMed] [Google Scholar]
- 23.Zlobec I, Vuong T, Hayashi S, Haegert D, Tornillo L, Terracciano L, Lugli A, Jass J. A simple and reproducible scoring system for EGFR in colorectal cancer: application to prognosis and prediction of response to preoperative brachytherapy. Br J Cancer. 2007;96(5):793–800. doi: 10.1038/sj.bjc.6603619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang CY, Eshleman JR, Willson JK, Markowitz S. Both transforming growth factor-beta and substrate release are inducers of apoptosis in a human colon adenoma cell line. Cancer Res. 1995;55(21):5101–5. [PubMed] [Google Scholar]
- 25.Markowitz S, Myeroff L, Cooper M, Traicoff J, Kochera M, Lutterbaugh J, Swiriduk M, Willson J. A benign cultured colon adenoma bears three genetically altered colon cancer oncogenes, but progresses to tumorigenicity and transforming growth factor-beta independence without inactivating the p53 tumor suppressor gene. Journal of Clinical Investigation. 1994;93:1005–13. doi: 10.1172/JCI117048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wotton D, Massague J. Smad transcriptional corepressors in TGF beta family signaling. Curr Top Microbiol Immunol. 2001;254:145–64. [PubMed] [Google Scholar]
- 27.Kim BC, Mamura M, Choi KS, Calabretta B, Kim SJ. Transforming growth factor beta 1 induces apoptosis through cleavage of BAD in a Smad3-dependent mechanism in FaO hepatoma cells. Mol Cell Biol. 2002;22(5):1369–78. doi: 10.1128/mcb.22.5.1369-1378.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim SJ, Im YH, Markowitz SD, Bang YJ. Molecular mechanisms of inactivation of TGF-beta receptors during carcinogenesis. Cytokine Growth Factor Rev. 2000;11(1–2):159–68. doi: 10.1016/s1359-6101(99)00039-8. [DOI] [PubMed] [Google Scholar]
- 29.Bai AHC, Tongg JHM, To K-F, Chan MWY, Man EPS, Lo K-W, Lee JFY, Sung JJY, Leung WK. Promoter hypermethylation of tumor-related genes in the progression of colorectal neoplasia. International Journal of Cancer. 2004;112:846–53. doi: 10.1002/ijc.20485. [DOI] [PubMed] [Google Scholar]
- 30.Moinova HR, Chen WD, Shen L, Smiraglia D, Olechnowicz J, Ravi L, Kasturi L, Myeroff L, Plass C, Parsons R, Minna J, Willson JK, et al. HLTF gene silencing in human colon cancer. Proc Natl Acad Sci U S A. 2002;99(7):4562–7. doi: 10.1073/pnas.062459899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li H, Myeroff L, Smiraglia D, Romero MF, Pretlow TP, Kasturi L, Lutterbaugh J, Rerko RM, Casey G, Issa JP, Willis J, Willson JK, et al. SLC5A8, a sodium transporter, is a tumor suppressor gene silenced by methylation in human colon aberrant crypt foci and cancers. Proc Natl Acad Sci U S A. 2003;100(14):8412–7. doi: 10.1073/pnas.1430846100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Park J, Song SH, Kim TY, Choi MC, Jong HS, Kim TY, Lee JW, Kim NK, Kim WH, Bang YJ. Aberrant methylation of integrin alpha4 gene in human gastric cancer cells. Oncogene. 2004;23(19):3474–80. doi: 10.1038/sj.onc.1207470. [DOI] [PubMed] [Google Scholar]
- 33.Chen WD, Han ZJ, Skoletsky J, Olson J, Sah J, Myeroff L, Platzer P, Lu S, Dawson D, Willis J, Pretlow TP, Lutterbaugh J, et al. Detection in fecal DNA of colon cancer-specific methylation of the nonexpressed vimentin gene. J Natl Cancer Inst. 2005;97(15):1124–32. doi: 10.1093/jnci/dji204. [DOI] [PubMed] [Google Scholar]
- 34.Bachman KE, Herman JG, Corn PG, Merlo A, Costello JF, Cavenee WK, Baylin SB, Graff JR. Methylation-associated silencing of the tissue inhibitor of metalloproteinase-3 gene suggest a suppressor role in kidney, brain, and other human cancers. Cancer Research. 1999;59(4):798–802. [PubMed] [Google Scholar]
- 35.Ohm JE, Baylin SB. Stem cell chromatin patterns: an instructive mechanism for DNA hypermethylation? Cell Cycle. 2007;6(9):1040–3. doi: 10.4161/cc.6.9.4210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Widschwendter M, Fiegl H, Egle D, Mueller-Holzner E, Spizzo G, Marth C, Weisenberger DJ, Campan M, Young J, Jacobs I, Laird PW. Epigenetic stem cell signature in cancer. Nat Genet. 2007;39(2):157–8. doi: 10.1038/ng1941. [DOI] [PubMed] [Google Scholar]
- 37.Keshet I, Schlesinger Y, Farkash S, Rand E, Hecht M, Segal E, Pikarski E, Young RA, Niveleau A, Cedar H, Simon I. Evidence for an instructive mechanism of de novo methylation in cancer cells. Nat Genet. 2006;38(2):149–53. doi: 10.1038/ng1719. [DOI] [PubMed] [Google Scholar]
- 38.Baylin SB, Bestor TH. Altered methylation patterns in cancer cell genomes: cause or consequence. Cancer Cell. 2002;1:299–305. doi: 10.1016/s1535-6108(02)00061-2. [DOI] [PubMed] [Google Scholar]
- 39.Grem JL, Danenberg KD, Behan K, Parr A, Young L, Danenberg PV, Nguyen D, Drake J, Monks A, Allegra CJ. Thymidine kinase, thymidylate synthase, and dihydropyrimidine dehydrogenase profiles of cell lines of the National Cancer Institute’s Anticancer Drug Screen. Clin Cancer Res. 2001;7(4):999–1009. [PubMed] [Google Scholar]
- 40.Campbell IG, Phillips WA, Choong DY. Genetic and epigenetic analysis of the putative tumor suppressor km23 in primary ovarian, breast, and colorectal cancers. Clin Cancer Res. 2006;12(12):3713–5. doi: 10.1158/1078-0432.CCR-06-0800. [DOI] [PubMed] [Google Scholar]
- 41.Shao G, Berenguer J, Borczuk AC, Powell CA, Hei TK, Zhao Y. Epigenetic inactivation of Betaig- h3 gene in human cancer cells. Cancer Research. 2006;66(9):4566–73. doi: 10.1158/0008-5472.CAN-05-2130. [DOI] [PubMed] [Google Scholar]
- 42.Suzuki M, Shigematsu H, Shames DS, Sunaga N, Takahashi T, Shivapurkar N, Iizasa T, Frenkel EP, Minna JD, Fujisawa T, Gazdar AF. DNA methylation-associated inactivation of TGFbeta-related genes DRM/Gremlin, RUNX3, and HPP1 in human cancers. Br J Cancer. 2005;93(9):1029–37. doi: 10.1038/sj.bjc.6602837. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 43.Li Q, Ahuja N, Burger PC, Issa JP. Methylation and silencing of the Thrombospondin-1 promoter in human cancer. Oncogene. 1999;18(21):3284–9. doi: 10.1038/sj.onc.1202663. [DOI] [PubMed] [Google Scholar]
- 44.Dameron KM, Volpert OV, Tainsky MA, Bouck N. Control of angiogenesis in fibroblasts by p53 regulation of thrombospondin-1. Science. 1994;265(5178):1582–4. doi: 10.1126/science.7521539. [DOI] [PubMed] [Google Scholar]
- 45.Ren B, Yee KO, Lawler J, Khosravi-Far R. Regulation of tumor angiogenesis by thrombospondin-1. Biochim Biophys Acta. 2006;1765(2):178–88. doi: 10.1016/j.bbcan.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 46.Toyota M, Ho C, Ahuja N, Jair KW, Li Q, Ohe-Toyota M, Baylin SB, Issa JP. Identification of differentially methylated sequences in colorectal cancer by methylated CpG island amplification. Cancer Research. 1999;59(10):2307–12. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure. Tumor immunostaining showing differences in TSP1 expression. A. Immunostaining results from a tumor that has unmethylated TSP1. The expression of the protein is detected mainly in the stroma (blue arrow) and not in tumor cells (black arrow). B. TSP1 expression in a tumor in which TSP1 is methylated. Faint immunostaining is detected in this sample.