Abstract
We characterized the resonance energy transfer interactions for conjugates consisting of QD donors self-assembled with three distinct fluorescent protein acceptors: two monomeric fluorescent proteins, the dsRed derivative mCherry or yellow fluorescent protein and the multi-chromophore b-phycoerythrin light harvesting complex. Using steady-state and time-resolved fluorescence, we showed that nonradiative transfer of excitation energy in these conjugates can be described within the Förster dipole-dipole formalism, with transfer efficiencies that vary with the degree of spectral overlap, donor-acceptor separation distance and the number of acceptors per QD. Comparison between the quenching data and simulation of the conjugate structures indicated that while energy transfer to monomeric proteins was identical to what was measured for QD-dye pairs, interactions with b-phycoerythrin were more complex. For the latter, the overall transfer efficiency results from the cumulative contribution of individual channels between the central QD and the chromophores distributed throughout the protein structure. Due to the biocompatible nature of fluorescent proteins, these QD-assemblies may have great potential for use in intracellular imaging and sensing.
Keywords: Quantum dot, self-assembly, fluorescent protein, fluorescence, FRET, Förster, resonance energy transfer
Introduction
Luminescent semiconductor quantum dots (QDs) have generated an intense interest for use as fluorescent probes in biological labeling and imaging.1-6 They are larger in size than their molecular scale dye counterparts (typical radii of 2.5 - 6 nm) which implies that multiple biological molecules (i.e. proteins, peptides or DNA) can be simultaneously conjugated to an individual QD without losing their biological functions; this can also provide added advantages such as higher local avidity. There has also been a growing interest in creating hybrid QD-protein conjugates for use in applications where the proteins provide biospecific functionality while the QD allows fluorescent tracking. These include developing assemblies with catalytic activity, as probes to detect specific protein targets in vitro, as vehicles to understand issues associated with drug delivery, and as conjugates that enable long-term in vivo tracking and sensing.3,5-9 Developing QDs as platforms for immobilizing proteins and peptides with control over the number and orientation of these receptors will be key to implementing many of these applications. Fluorescent proteins provide an ideal model system for developing and testing such QD-bioconjugates as they can allow concurrent monitoring of nanoparticle-biomolecule interactions. They can also permit the use of fluorescence resonance energy transfer (FRET) interactions to gain information on conjugate structure and valence (i.e, the number of proteins attached to the nanocrystal) and the ability to monitor the fate of such hybrid assemblies inside live cells.
A few preliminary studies characterizing FRET between QDs and fluorescent proteins or testing their mechanism of intracellular delivery have already been reported.10-12 Dennis and Bao examined energy transfer in assemblies of micelle-encapsulated (commercially available) CdSe-ZnS QDs self-assembled with mOrange, Tdtomato and mCherry.10 They demonstrated that the energy transfer efficiency strongly depended on both the degree of spectral overlap and conjugate valence. In another elegant demonstration, Woggon and Niemeyer assembled hybrid nanoparticle-protein/DNA conjugates consisting of QDs simultaneously coupled to fluorescent proteins, oligonucleotides and dyes, and characterized the resulting two- and three-chromophore interactions. They showed that within the three-chromophore system, where a relay fluorophore plays the dual roles of an intermediary acceptor and a donor, the range of energy transfer interactions can be extended beyond the usual 10-100Å window.11,13 We recently demonstrated that the additional functionalization of similar QD-fluorescent protein conjugates with small cell penetrating peptides could allow their efficient intracellular delivery which presents the opportunity for investigating and tracking these assemblies in vivo.12,14
Here, we extend the previous findings and demonstrate that QDs can function as effective self-assembly vehicles for a variety of structurally and photophysically diverse fluorescent proteins. CdSe-ZnS core-shell QDs were self-assembled with either the monomeric yellow fluorescent (YFP) or mCherry proteins via metal-affinity coordination, and the multi-chromophore b-phycoerythrin (abbreviated as b-PE) coupled via biotin-Streptavidin interactions. We used steady-state and time-resolved fluorescence measurements to monitor the energy transfer interactions in these conjugates. Our results showed that the transfer efficiencies for these pairs depend on the degree of spectral overlap between QD emission and protein absorption, the center-to-center separation distance and conjugate valence for monomer protein. For b-PE, multichannel FRET interactions with the individual chromophores in the protein structure had to be taken into account in order to properly interpret the efficiency data. Furthermore, we show that energy transfer data can overall be interpreted within the Förster dipole-dipole formalism. Information on conjugate conformation derived from energy transfer data together with QD structure and the protein crystallographic coordinates provided insight into the architectures of these hybrid assemblies.
Materials and Methods
Quantum Dots
We used four sets of CdSe-ZnS core-shell QDs with emission maxima centered at ~510, 520, 540 and 550 nm. Nanocrystals were synthesized using high temperature reaction of organometallic precursors in hot coordinating solvents.15-18 They were made hydrophilic by exchanging the native capping shell with either neat dihydrolipoic acid (DHLA) or a mixture of DHLA-PEG600 and biotin-terminated DHLA-PEG400 (9:1 ratio DHLA-PEG600:DHLA-PEG400-biotin). DHLA-PEG400,600 designate dihydrolipoic acid appended with polyethylene glycol segments of MW = 400 or 600.14,19 The chemical structures of the ligands are provided in the Supporting Information.
Fluorescent Proteins and Self-Assembly of Quantum Dot Bioconjugates
YFP and mCherry are monomeric proteins (MW ~27 kD) derived from the jellyfish Aequorea and the tetrameric mRFP red fluorescent protein, respectively; they originate from the Tsien laboratory.20-22 Both proteins have similar tertiary β-barrel structures formed from arranging 11 β-strands into a hollow cylinder with a centrally located chromophore. The active chromophore in each arises from the rearrangement of several key residues during the maturation process, similar to the p-hydroxybenzylideneimidazolinone chromophore in GFP which originates from modification of the Ser-Tyr-Gly residues at position 65-67.20-22 b-PE is a member of the chromophore bearing phycobiliproteins found in the macromolecular light-harvesting phycobilisome complexes of red algae and cyanobacteria.23,24 b-PE is photophysically and structurally different from YFP and mCherry. These multi-subunit protein complexes have a large molecular weight (MW ~240 kD, ~10 times larger than YFP or mCherry monomers) with ~34 covalently attached bilins (open chain tetrapyrrole chromophores) per functional moiety; this results in an extinction coefficient ~100 times larger that of monomeric fluorescent proteins. This fluorescent protein has been extensively utilized as a sensitive and biocompatible fluorescent probe.23
The yellow fluorescent protein was originally obtained on plasmid pRSET B (Invitrogen, Carlsbad, CA) within a gene encoding a blue fluorescent protein - glucose binding protein - YFP FRET-based glucose sensor.25 The final YFP gene encoding a His6 sequence after the start ATG, Asp-Gln, an Xpress Epitope (Asp-Leu-Tyr-Asp-Asp-Asp-Asp-Lys) and another His6 sequence at the start of the YFP coding sequence was constructed, expressed and purified over Ni-NTA Resin (Qiagen, Valencia, CA) as described in references12,26. The mCherry expression plasmid and usage license was obtained from Clontech (Mountain View, CA). The protein, inserted between BamH1 and EcoR1 of the multi-cloning site on plasmid pRSET B (Invitrogen, Carlsbad, CA), was expressed under the T7 promoter with a N-terminal linker of 35 residues that encodes a His6 sequence after the start methionine, a T7 gene 10 leader sequence, an Xpress Epitope and an enterokinase domain. The plasmid was transformed into Rosetta 2 (DE3) cells (Novagen, San Diego, CA) and starter cultures were grown under 2% glucose suppression, then cells were induced with 1 mM isopropyl-beta-D-thiogalactopyranoside overnight at 30°C. Mature mCherry was purified over Ni-NTA Resin (Qiagen, Valencia, CA). Maltose binding protein (MBP) was prepared as described in reference27. One-to-one b-phycoerythrin-Streptavidin conjugates (1 mg/mL, abbreviated as b-PE) were obtained from Invitrogen.
Self-Assembly of Quantum Dot Bioconjugates
We utilized two self-assembly approaches to form our QD-protein bioconjugates, namely metal-affinity coordination directly between CdSe-ZnS nanocrystals and a polyhistidine (His)n sequence appended onto the protein termini or biotin-avidin coupling. YFP and mCherry were engineered to express N-termini bearing (His)6 sequences and these were separated from their respective proteins by spacer segments to allow access to the QD surface. Streptavidin-modified b-PE was coupled to QDs functionalized with DHLA-PEG400-biotin. Both conjugation approaches provided tight coupling between QDs and protein, with reported dissociation constants KD of ~1-10 × 10-9 M for QD-His6 binding and ~10-15 M for biotin-avidin, respectively.28,29
His-appended YFP was self-assembled on DHLA QDs together with His5-appended MBP (mixed surface conjugates) at a constant total valence of 12 proteins per QD, while varying the fraction of YFP in the conjugate. This configuration allows us to maintain a given number of proteins per nanocrystal and removes issues associated with small changes in QD PL observed when assembled varying number of His-appended proteins onto DHLA-capped nanocrystals.27 The indicated molar ratios of YFP and MBP were added to 30 picomoles of 510 nm DHLA QDs in 100 μL 10 mM Tris-Cl pH 8 (0.3 μM QD) and allowed to self-assemble for ~30 mins at room temperature. A similar method was used for assembling mCherry onto 550 nm DHLA QDs. b-PE-Streptavidin was coupled to either 520 nm or 540 nm-emitting DHLA-PEG-biotin QDs by incubating the reagents in phosphate buffered saline (137 mM NaCl, 10 mM Phosphate, 2.7 mM KCl, pH 7.4, PBS) overnight at 4°C. Selected QD-fluorescent protein bioconjugates were also separated and characterized on agarose gels buffered with 1xTBE (0.89M Tris, 0.89M boric acid and 0.02M EDTA, pH 8.3) which provided an additional means of verifying conjugate formation. Representative gel images are provided in the Supporting Information.
FRET Data Collection and Analysis
Steady-state fluorescent spectra from solutions of QD-protein bioconjugates and proteins alone (controls) were collected on a Tecan Safire Dual Monochromator Multifunction Microtiter Plate Reader (Tecan, Research Triangle Park, NC) using 325 nm excitation. Following subtraction of the direct excitation contribution to the acceptor from the control solutions, the composite spectra were deconvoluted to separate the QDs and fluorescent protein signals. This allowed us to account for the loss of QD PL together with the increase in the protein signal due to energy transfer. The steady-state experiments were complemented (only for QD-mCherry assemblies) with fluorescence lifetime measurements performed using a time-correlated single-photon counting system with a temporal resolution of 50 ps. A brief description of the system can be found in the Supporting Information. The QD and mCherry fluorescence intensities were integrated over narrow windows centered at 537 nm and 620 nm, respectively, in order to isolate signals that are representative of QD and mCherry emissions. Fluorescence decay traces of the QD signal with time (collected for the indicated protein-to-QD ratio, n) were fit to a three-exponential function:
| (1) |
where t is time, Ai is weighting parameter associated with each decay time. An average amplitude-weighted lifetime defined as:
| (2) |
was extracted from the fit using FluoFit (Picoquant, Berlin Germany). Average energy transfer efficiency E was extracted from either the steady-state or time-resolved fluorescence data for each set of QD-conjugates using the expressions:
| (3) |
where FD and FDA are, respectively, the fluorescence intensities of the donor alone and donor in the presence of acceptor(s); similarly, τD and τDA designate the QD excited-state lifetimes without and with the protein acceptor.13 The metal-His driven self-assembly (applied to YFP and mCherry) provides conjugates with a centro-symmetric distribution of acceptors around the QD, with a constant average center-to-center separation distance r.27,30 If analyzed within the Förster dipole-dipole formalism, the energy transfer efficiency can be fit to the expression:27
| (4) |
where n is the average number of fluorescent proteins per QD, and R0 is the Förster separation distance corresponding to 50% energy transfer efficiency;13 R0 is expressed as:
| (5) |
where nD is the refractive index of the medium, QD is the QD PL quantum yield, I designates the integral of the spectral overlap function, J(λ), defined as:
| (6) |
with εA(λ) being the acceptor extinction coefficient spectrum and PLD-corr is the normalized donor emission spectrum. A value of κp2 = 2/3 for the dipole orientation factor was used for our QD-protein assemblies in Eq. 5.27,31 For conjugates having small numbers of acceptors (n < 5), heterogeneity in the conjugate valence was taken into account using the Poisson distribution function, p(N,n), when fitting the efficiency data:32
| (7) |
where n designates the exact numbers of acceptors (valence) for conjugates with a nominal average valence of N. We have previously shown that the valence of QD-protein bioconjugates formed, for example, via metal-affinity self-assembly follows a statistical distribution that is well described by the Poisson function (see reference 32 for additional details).
Results and Discussion
Spectral Overlap of the Quantum Dot-Fluorescent Protein Pairs
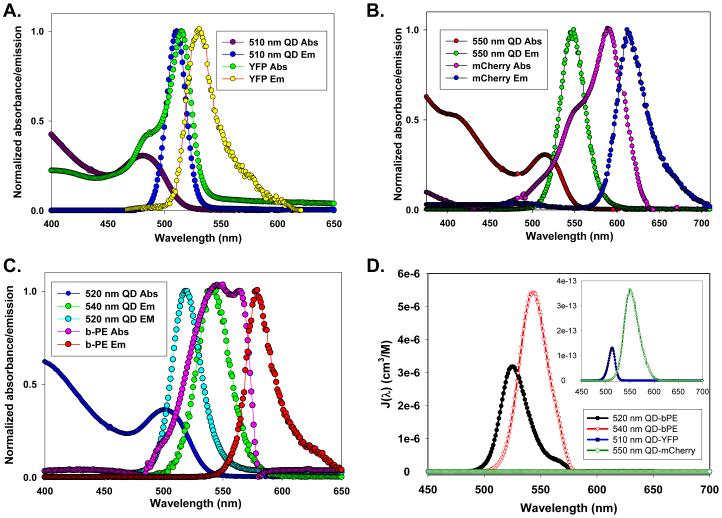
Figure 1 shows the absorption and emission spectra for the various QD-fluorescent pairs used together with the corresponding spectral overlap function J(λ). Table I presents the relevant QDs and fluorescent protein photophysical characteristics, the acceptor extinction coefficients at the peak absorption together with the corresponding R0 values for the various QD-protein pairs. Data indicate that R0 varies from one pair to another, depending on the degree of spectral overlap and the acceptor extinction coefficient spectrum. As previously demonstrated for QD-dye pairs, R0 and thus the rate of energy transfer can potentially be optimized by choosing a QD donor that offers the best spectral overlap with a particular fluorescent protein acceptor.27 For example, by switching from 520 nm to 540 nm-emitting QDs, R0 was increased from 44 Å to 53 Å for the b-PE acceptor, which constitutes a substantial extension of the distance range allowing efficient FRET interactions. Similarly, two QD samples with distinct emissions were used with YFP or mCherry to optimize the spectral overlap for each pair. We should emphasize that for QD-b-PE pairs, the overlap integral I and thus R0 were estimated using the extinction coefficient of an individual bilin chromophore (~71,000 M-1cm-1) and not the extinction coefficient spectrum for the full protein; this is reasonable since all chromophores in the protein are identical. This assumption is more appropriate than treating the whole protein as a monomeric chromophore acceptor (as was done for YFP and mCherry above), since the bilins are located at different separation distances and thus interact with the QD via distinct energy transfer channels (and rates). YFP and mCherry in comparison each presents a single chromophore located at the center of the β-barrel and can be treated as point acceptors.
Figure 1. Photophysical characteristics of the fluorophores.
Normalized absorption and PL spectra of the 510 nm QD-YFP pair (A), 550 nm QD-mCherry pair (B), and 520 nm QD-b-PE and 540 nm QD-b-PE pairs (C). Spectral overlap functions for the four pairs are shown in panel (D). The inset shows a close up of the QD-YFP and QD-mCherry overlap functions.
Table I. Selected characteristics of the quantum dot - fluorescent protein pairs used in this study.
| Quantum Dot Donor | Fluorescent Protein Acceptor | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| QD λmax emission |
Quantum yield |
Conjugation strategy |
Protein acceptor |
λmax absorption |
λmax emission |
Extinction coeff. (M-1cm-1) |
Quantum yield |
R0 | r |
| 510 nm | 12 % | Metal-affinity | YFP | 516 nm | 529 nm | 20,200 | 60 % | 39Å | 62 Å |
| 550 nm | 20 % | Metal-affinity | mCherry | 587 nm | 610 nm | 71,000 | 22 % | 49Å | 56 Å |
| 520 nm | 20 % | Biotin/Streptavidin | b-PE | 545,565 nm | 575 nm | 2,410,000 | 98 % | 44Å* | --- |
| 540 nm | 19 % | Biotin/Streptavidin | b-PE | 545,565 nm | 575 nm | 2,410,000 | 98 % | 53Å* | --- |
YFP - yellow fluorescent protein, b-PE - b-phycoerythrin
R0 calculated for interactions with a single bilin chromophore Standard deviations associated with the experimental r values were ~ 5-7%.
Steady-state and Time-resolved Fluorescence
Steady-state fluorescence data
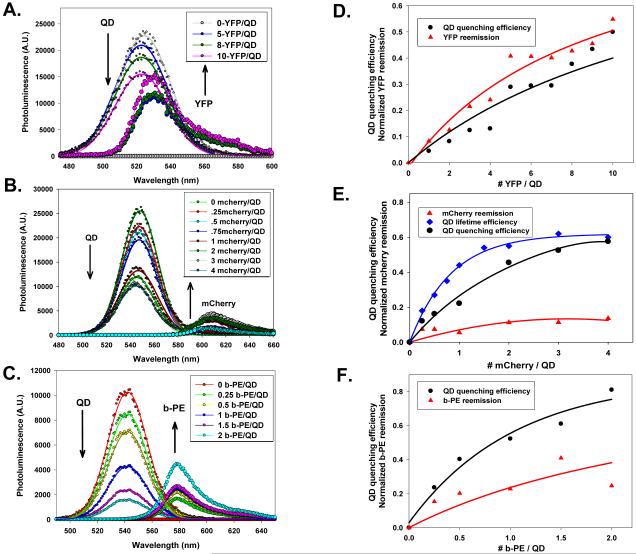
Figures 2A-C show the deconvoluted PL emission spectra for an increasing number of proteins per QD for the three sets of conjugates. Samples were excited at 325 nm, which coincides with the absorption minima for all three proteins, in order to reduce the direct excitation of the acceptor. This contribution (determined from protein only control samples), though small, was carefully subtracted prior to deconvolution. Data clearly show that for all three sets of conjugates, there is a consistent and progressive loss of the QD PL accompanied with a gradual increase in the protein emission, as the number of acceptors per QD-conjugate increases. The corresponding QD PL quenching efficiencies derived from the above spectra (following deconvolution of the raw spectra for every QD-FP pair) along with the sensitized-acceptor reemission as a function of conjugate valence are shown in Figure 2D-F. The acceptor reemission varied from one set to the other and did not always directly trace the QD PL loss, which can be attributed to differences in protein fluorescent yields and their sensitivity to the surrounding environment. Even though the general trend for the QD PL loss and acceptor gain are comparable, there are substantial differences in the exact values, with the most pronounced quenching efficiencies measured for the 540 nm QD-b-PE pair and smallest measured for the 510 nm QD-YFP pair. For example at n = 2, ~ 80% PL loss was measured for the 540 nm QD-b-PE conjugates compared to less than 10% quenching for the 510 nm QD-YFP pair. This is due to a combination of differences in the spectral overlap and conjugate architecture. Similarly, the higher quenching efficiency measured for the 550 nm QD-mCherry pair compared to the 510 nm QD-YFP can also be primarily attributed to a larger spectral overlap integral (or higher R0, see Table I).
Figure 2. Fluorescence spectra and quenching efficiencies.
(A-C) Representative composite and deconvoluted spectra showing individual PL contributions as a function of the protein-to-QD ratio for: 510 nm QD-YFP, 550 nm QD-mCherry, and 540 nm-QD-b-PE assemblies. (D-E) Plots of the corresponding quenching efficiencies along with the acceptor-sensitized emission. In panel (E), the energy transfer efficiency extracted from changes in the QD excited state lifetime are also shown for the 550 nm QD-mCherry pair. Data are corrected for the direct excitation of the b-PE. QD emission profiles were fit with a Gaussian function.
Time-resolved fluorescence data
We focused on the 550 nm QD-mCherry pair as a representative and easier to analyze system. The other two conjugates either did not allow easy separation of the donor and acceptor signals because of a strong overlap of their emissions, or explored a limited range of conjugate valence in order to reduce the direct excitation contribution of the acceptor (case of 540 nm QD-b-PE pair). Figures 3A,B show plots of QDs and mCherry intensity versus time for several protein-to-QD ratios immediately following the laser pulse. We verified that at the excitation wavelength and conjugate concentrations used, the direct excitation contribution to the protein signal was essentially negligible. Figure 3C shows the normalized intensity-time traces at a few representative ratios along with the traces collected for the control samples (QDs and protein only). For each mCherry:QD ratio the QD trace shown in Figure 3A was analyzed and an average exciton lifetime extracted using Equation 2 (see Table II). There is a ratio-dependent reduction (shortening) of the average QD lifetime with increasing conjugate valence for n ≤ 4, before saturation at higher values. The energy transfer efficiencies vs. n extracted from the lifetime data (using Equation 3) are in a reasonable agreement with those derived from steady-state PL (Figure 2E). Efficiencies were consistently higher than those extracted from steady-state measurements, though. This may be attributed to the non-exponential decay traces and the complex method used to extract the exciton lifetime (multi-exponential fit) combined with a small leakage of the protein signal into the QD detection channel. Regardless, the trend clearly follows the ensemble fluorescence data.
Figure 3. Time-resolved fluorescence data for 550 nm QD-mCherry conjugates.
(A) Plot of 550 nm QD PL intensity (monitored at 537 nm) versus time for several mCherry-to-QD ratios. (B) Plot of mCherry intensity (monitored at 620 nm) versus time for several mCherry-to-QD ratios. (C) Superimposed plots of normalized QD and mCherry intensity vs. time for selected ratios.
Table II. Donor-acceptor excited state lifetimes for 550 nmDHLA QDs assembled with increasing mCherry protein. Amplitude weighted average lifetimes (τAv) are given in nanoseconds.
| Ratio of mCherry/QD | QD donor1 | mCherry acceptor2 | mCherry acceptor rise time3 |
|---|---|---|---|
| 0 | 8.51 (QD only) | 1.55 (mcherry alone) | -------- |
| 0.25 | 6.98 | 4.32 | 0.80 |
| 0.5 | 6.19 | 4.10 | 0.75 |
| 0.75 | 5.51 | 3.69 | 0.77 |
| 1 | 4.74 | 3.44 | 0.74 |
| 1.5 | 3.89 | 3.44 | 0.70 |
| 2 | 3.86 | 3.41 | 0.64 |
| 3 | 3.22 | 3.00 | 0.69 |
| 4 | 3.37 | 3.02 | 0.67 |
| 6 | 3.51 | 3.30 | 0.60 |
| 8 | 3.47 | 3.06 | 0.67 |
| 10 | 3.67 | 3.12 | 0.63 |
| 15 | 3.89 | 3.13 | 0.56 |
Fitted with an average of 3 or 4 lifetime components.
Fitted with an average of 2 or 3 lifetime components.
Extracted from the data shown in Figure 3B. QD and mCherry lifetimes monitored at 537 nm and 620 nm, respectively. Standard deviations were < 5% for all values.
In comparison, the mCherry intensity-time traces show a more complex behavior, with a clear rise of the signal immediately after the excitation pulse, followed by a more progressive decay at longer time. In addition, we observed acceleration of the signal rise with increasing ratio n (Table II). This behavior is typical of a FRET acceptor intensity-time trace, where rapid transfer of excitation energy from an excited donor to a ground state acceptor manifests in a rapid increase of the acceptor signal with time;13 it also reflects the simultaneous and cumulative energy transfer to several acceptors. Decay behavior at longer times is dominated by the characteristics of the QD donor. This is not unexpected since the natural decay time of the acceptor is short (1.5 ns), and thus signal at longer times will ultimately follow the tail of the QD excitation. Additional details showing analysis of the mCherry fluorescence rise and decay dynamics is presented in the Supporting Information.
Overall the steady-state and time-resolved fluorescence data confirm that the signature observed for these pairs is due to energy transfer between QDs and fluorescent proteins, and that the process is essentially driven by dipole-dipole coupling and can be treated within the Förster formalism. This manifests in the progressive increase in the donor (QD) PL quenching with increasing protein-to-QD ratio. It also manifests in a ratio-dependent shortening of the exciton lifetime of the QD donor. Analysis of the QD PL quenching data within the framework of the Förster dipole-dipole formalism taking into account conjugate heterogeneity where necessary (using Eq. 7) for the two QD-monomeric fluorescent protein assemblies, YFP and mCherry, provided estimates for the separation distances r (see Table I). The value for the QD-YFP pair might be subject to a slightly larger uncertainty given the difficulties associated with deconvolution of the spectra shown in Figure 2A. Nevertheless, the r values for these two pairs are close, which is consistent with the overall structure of these QD-protein conjugates. We should emphasize that separation distance measured for the present QD-mCherry assemblies are comparable to those reported for the same protein assembled via metal-His affinity onto commercial carboxyl-functionalized EviTag QDs.10 In that report, the QD PL quenching similarly reached saturation at ~4 proteins/QD. The much higher rate of quenching measured for QD-b-PE conjugates can be primarily attributed to the nature of the protein. (Similar data were collected from 520 nm-QD-b-PE conjugates.) Treating the QD-b-PE as a single donor - single acceptor (simple pair) is not appropriate for these conjugates, given the characteristics of the protein. With the large number of individual chromophores distributed across the protein structure, the QD-b-PE assembly is essentially equivalent to a system consisting of one donor interacting with an array of multiple acceptors distributed at varying separation distances (see below).
Modeling of QD-Protein Structures
Using the dimensions of the nanocrystals, the FRET derived center-to-center separation distances, and aided by the crystallographic structures of the proteins, we modeled the conformation of our QD-protein conjugates. To do this we combine information on: 1) the QD-ligand structure, 2) how the proteins are expected to self-assemble onto the QD surface, 3) separation distance r for each system, and 4) the protein structure as determined by x-ray crystallography. This approach of correlating the model structure with the fluorescence data has allowed us to gain additional insights into the overall conformation of the conjugates and their potential as sensing assemblies.30,33 The protein structures were obtained from the Protein Data Bank (www.rcsb.org/pdb) and include entries 1B8D for b-PE, 1F0B for YFP, 1RXJ for Streptavidin and 2H5Q for mCherry. QD radii were extracted from x-ray scattering and TEM measurements.15,34 The conformations of DHLA, DHLA-PEG600 and DHLA-PEG400-biotin ligand bound to the nanocrystal were modeled using Chem3D and the MM2 module (CambridgeSoft, www.cambridgesoft.com). The hybrid conjugates were assembled from their components using MidasPlus (www.cgl.ucsf.edu) where torsion angles were adjusted to match the measured separation distances.35,36 Images were rendered in MidasPlus and assembled from individual composite sections (required only for the larger b-PE complex) in Photoshop (Adobe, San Jose CA). Modeling the conformational structures of our QD-protein conjugates has inherent uncertainties that result from errors in the separation distance measurements, the availability of the crystal structures in the PDB and the assumptions made in the comparison.
Figure 4A shows a representative structure for the 510 nm-QD-YFP conjugates; a DHLA-capped QD with one YFP is shown for simplicity. The core-shell QD is represented by a sphere of ~25 Å radius and the DHLA solubilizing layer by ~10-11 Å crimson crown.15,27 The YFP β-barrel structure has dimensions of circa 41 × 28 Å (length × barrel diameter). The YFP chromophore (magenta) was placed ~62 Å from the QD center (distance determined from FRET). In assembling a model structure of the QD-protein conjugate, we neglected the contributions of the His tag, since polyhistidine is proven to interact directly with the QD surface.28 We also did not include the 23-residue linker between the terminal (His)6 and the YFP in the simulated structure, because there are no coordinates available for it in the PBD; the lack of structural data despite multiple occurrences of homologous sequences in the PDB indicate that this region is not well ordered and likely has many conformations in the solid state.28,30,33 This analysis suggests that allowing for some rotational freedom around the cylinder axis (while self-assembled via the histidine tag on the QD), the optimal configuration of the protein consists of the β-barrel aligned with its base facing the QD surface. This analysis also suggests that up to ~15-YFP can be self-assembled around each 510 nm QD, in agreement with valences anticipated for similar size proteins.37 To compensate for the rather weak spectral overlap (r/R0 ~1.5), one could use an enhanced YFP variant such as ‘Venus’ which has a higher extinction coefficient; this could improve the measured FRET efficiency for such conjugates.22,38,39
Figure 4. Model structures for QD-YFP and QD-mCherry conjugates.
(A) 510 nm QD-YFP structure. QD with radius of ~28 Å (blue) is surrounded by a DHLA shell of 10 Å (crimson). The YFP is attached to the QD via a double His6 sequence; only the N-terminal His6 is shown in green. The YFP residue attached to the double (His)6 linker is shown in red. The central YFP chromophore is shown in magenta. The yellow line corresponds to the 62 Å experimental separation distance. (B) Rendition of a 550 nm QD-mCherry structure using 56 Å experimental separation distance (yellow), with the protein chromophore shown in red. The mCherry residue attached to the (His)6-linker sequence is shown in green, but the 35 amino acid linker and terminal His6 sequence are not shown.
The 56 Å average separation distance measured for QD-mCherry conjugates is slightly smaller than that for the QD-YFP pair, even though the two proteins have nearly identical molecular weights and structures; 550 nm QDs have a radius of ~30 Å. mCherry is expressed with a slightly longer 35 amino acid N-terminal linker between the His6 tag and the protein. A search in the PDB found more than 25 identical linker sequences present in various crystallized protein structures as it is a commonly expressed N-terminal portion of the pRSET B plasmid but similar to the YFP linker above, no coordinates for this particular region were available. We thus assumed that it had a flexible conformation and its contribution allows for additional freedom of lateral ‘rotation’ of the mCherry while assembled onto the QD. This may also explain the slightly smaller r value extracted for this conjugate. For this pair, an estimated ~15-18 mCherry proteins can be immobilized on the QDs.
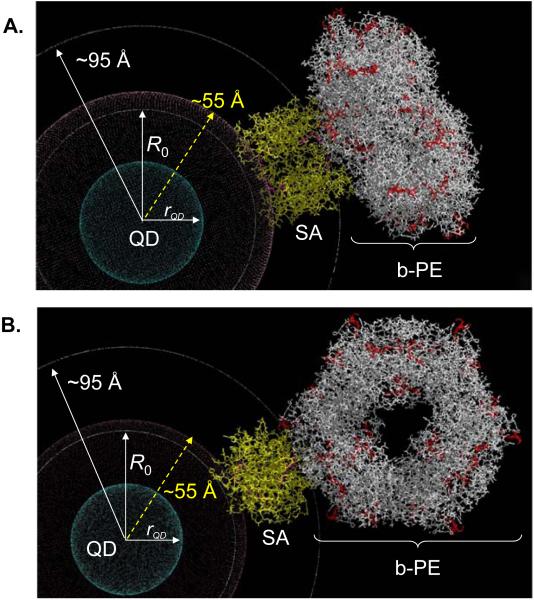
The QD-b-PE conjugate has a starkly different structure from that of the QD-monomeric-protein conjugates. b-PE consists of α,β paired monomers associated into a symmetrical hexameric ring structure (α/β)6 with a diameter of 110 Å (~2x that of the QD) and a thickness of 60 Å formed around a central channel with a diameter of 35-45 Å.40,41 The supplied b-PE is covalently linked to a single Streptavidin (on average), but the SA-b-PE conjugate structure is heterogeneous since the coupling chemistry used does not allow for a unique attachment site. Figure 5 shows models of 2 representative configurations of b-PE bound to the QDs via a Streptavidin bridge. The central QD is surrounded by a crimson sphere of ~25 Å (i.e., ~55 Å overall radius) representing the DHLA-PEG/DHLA-PEG-Biotin solubilizing and functionalizing layer; this energy minimized value accounts for ligand flexibility in solution. The Streptavidin tetramer is shown in yellow, with the biotin-binding sites highlighted in dark purple. Interpenetration between the Streptavidin and the ligands is meant to account for the fact that the biotin binding pocket is deep inside the protein. The b-PE is shown in white with the location of the ~34 individual chromophores in red. The two configurations shown in Figure 5A,B correspond to the hexamer ‘disk’ either laying with its face parallel to the QD surface, or facing away from the QD surface, respectively. Other conformers are also possible (for example, Streptavidin attached in the central b-PE pocket), and these results represent a range of possible conformations rather than a set of fixed acceptor sites. The concentric white circle closest to the QD (in both panels) corresponds to the R0 value of ~53Å. The 2nd outer white concentric circle is a visual distance marker set at ~95 Å from the QD center and corresponds to the overall QD-Streptavidin radius; it is the closest point of b-PE approach to the QD. In these representations, the range of distances sampled by the chromophores was set between 100 Å and 210 Å. To take into account the multi-chromophore nature of b-PE, we analyzed the quenching efficiency for the QD-b-PE pair using a modified expression of Equation 4, where the efficiency is now a sum of the individual FRET channels between the QD and all the chromophores distributed across the protein structure:
| (8) |
where n is the number of proteins around the central QD and ri designates the separation distance between the nanocrystal center and chromophore i in the protein (1 ≤ i ≤ 34). We assumed that the chromophores were optically identical and their positions were invariant by rotation around the axis perpendicular to the b-PE tore. The position/coordinates of each chromophore with respect to the QD could then be described by two parameters: QD center-to-b-PE center distance, RQD:b-PE, and the angle between the b-PE axis and the QD:b-PE center-to-center axis, θQD:b-PE (see schematic in Figure 6A and Supporting Information). Using the b-PE crystallographic structure, we developed a geometrical expression for the distances ri for each chromophore i in the overall protein structure, as a function of RQD:b-PE and θQD:b-PE. Replacing each ri by this geometrical expression in Equation 8, we obtained a theoretical FRET efficiency that depended only on RQD:b-PE and θQD:b-PE. Using this expression and the experimental FRET efficiency obtained from Figure 2C,F, we calculated the separation distance RQD:b-PE corresponding to different protein orientations (i.e., varying θQD:b-PE between 0 and 90 degrees). We found that these fitted parameters corresponded to separation distances RQD:b-PE ranging from ~102 Å, when the b-PE ‘disk’ structure lays parallel to the QD surface (closest separation) to a distance of ~125 Å when it is perpendicular to the QD (Figure 6B). For the 540 nm emitting QDs, a slightly larger distance of ~2.5-3 Å was measured concomitant with the larger size
Figure 5. Models of the QD-b-PE conjugate structure/conformation.
(A) b-PE is parallel to the QD surface (θQD:b-PE = 0°), and (B) b-PE fully extended away from the QD (θQD:b-PE = 90°). Central QD with radius of ~29 Å (rQD) shown in blue is surrounded by a crimson shell of ~25 Å thickness representing the DHLA-PEG-biotin. The intermediary Streptavidin (SA) protein is shown in yellow with biotin-binding sites highlighted in purple. The b-PE ring structure is shown in white with the multiple chromophores highlighted in red. The inner concentric white circle corresponds to the R0 (53 Å) for the 540 nm-QD-b-PE assembly. The 2nd outer white circle is a visual distance marker set at ~95 Å from the QD center and represents the closest approach of the b-PE to the QD.
Figure 6. Correlations between b-PE structure and FRET data.
(A) Schematic representation of possible conformations. θQD:b-PE is the angle between the QD-b-PE center-to-center axis and the axis perpendicular to the b-PE structure. RQD:b-PE is the QD-b-PE center to center separation distance. (B) QD-b-PE center-to-center distances estimated from FRET efficiencies and plotted versus RQD:b-PE (correspond to 0° ≤ θQD:b-PE ≤ 90°) for 540 nm (circles) and 520 nm (triangles) QDs. (C) Schematic mapping of the FRET efficiency for the QD-b-PE conjugate showing the effects of distance and distribution of the chromophores in the protein. Two representative b-PE conformations (dotted) along with the region explored by all possible conformations sampled by the protein on the nanocrystal (dashed). The color intensities indicate the FRET efficiencies between the QD and individual dyes in the b-PE (see bar on the right).
The structure shown in Figures 5 and 6 suggests that all the chromophores are essentially located at a distance exceeding 2 × R0 (~106 Å). Thus individually, the rate of FRET experienced by each chromophore when it interacts with the QD center is very small (<5%). The schematics in Figure 6C show the range of FRET efficiencies expected between the QD and bilin choromophores. Yet a large FRET efficiency (~0.6 at a valence of 1) was measured for this QD-protein pair. This is a direct reflection of the unique property of the multi-chromophore b-PE protein acceptor used. The measured FRET efficiency results from multiple FRET channels between the central nanocrystal and the chromophores distributed within the protein structure, which cumulatively results in high measured quenching efficiencies. We should also emphasize that FRET experiments using commercially available Streptavidin-QDs (inherently large in size), required the use of a large number of dye-acceptors per QD to achieve sizable energy transfer efficiencies.42 Multi-chromophore proteins such as b-PE could provide an easy alternative to the design and assembly of targeted QD biosensors based on FRET interactions.
Conclusions
We have characterized the energy transfer interactions between luminescent QDs and several fluorescent proteins brought in close proximity via two self-assembly approaches, metal-affinity coordination and biotin-avidin coupling. This study combines aspects of two structurally and photo-physically different fluorescent probes that have attracted an increasing interest in the past decade, inorganic QDs and engineered fluorescent proteins. We used very dissimilar fluorescent protein acceptors: two monomeric (single chromophore) proteins, YFP and mCherry, and a large molecular weight multichromophore protein, b-PE. Steady-state and time-resolved fluorescence measurements proved that energy transfer in these systems is fully consistent with the Förster dipole-dipole interaction formalism, as demonstrated for QD-dye pairs.27 Analysis also confirmed that the transfer efficiency could be easily controlled and tuned by varying the degree of spectral overlap and the number of proteins assembled on the QD surface.27,30 Furthermore, our study demonstrated that when using the b-PE multi-chromophore protein as acceptor, limitations due to the rather large separation distances could be circumvented, as high energy transfer efficiencies were measured for overall separation distances exceeding 100 Å even at low valences. This property results from the cumulative interactions of all the individual chromophores arrayed within the protein structure with the same QD donor. This is a rather remarkable finding as it opens up the possibility of further advantage of QD donors to probe biological interactions and processes that occur over larger distances than those initially considered as the boundaries for FRET (10 - 100 Å).13 It also opens up new opportunities in biological sensing and tracking where one can take advantage of the growing range of available fluorescent proteins combined with their natural biocompatibility.22,43,44
Supplementary Material
Acknowledgements
The authors acknowledge the CB Directorate/Physical S&T Division (DTRA), ONR, NRL and the NRL-NSI for financial support. K.B. and D.F. acknowledge ASEE and NRC fellowships through NRL, respectively. GB and AMD acknowledge support by the National Heart Lung and Blood Institute of the NIH as a Program of Excellence in Nanotechnology (HL80711).
Footnotes
Supporting Information Available. Ligand structures, agarose gel electrophoresis and further discussion of the lifetime collection and modeling. This information is available free of charge via the Internet at www.acs.org.
References
- 1.Lidke DS, Nagy P, Heintzmann R, Arndt-Jovin DJ, Post JN, Grecco HE, Jares-Erijman EA, Jovin TM. Quantum dot ligands provide new insights into erbB/HER receptor-mediated signal transduction. Nat. Biotechnol. 2004;22:198–203. doi: 10.1038/nbt929. [DOI] [PubMed] [Google Scholar]
- 2.Delehanty JB, Mattoussi H, Medintz IL. Delivering quantum dots into cells: strategies, progress and remaining issues. Anal. Bioanal. Chem. 2009;393(4):1091–1105. doi: 10.1007/s00216-008-2410-4. [DOI] [PubMed] [Google Scholar]
- 3.Medintz I, Uyeda H, Goldman E, Mattoussi H. Quantum dot bioconjugates for imaging, labeling and sensing. Nat. Mater. 2005;4:435–446. doi: 10.1038/nmat1390. [DOI] [PubMed] [Google Scholar]
- 4.Michalet X, Pinaud FF, Bentolila LA, Tsay JM, Doose S, Li JJ, Sundaresan G, Wu AM, Gambhir SS, Weiss S. Quantum dots for live cells, in vivo imaging, and diagnostics. Science. 2005;307:538–544. doi: 10.1126/science.1104274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alivisatos AP, Gu W, Larabell CA. Quantum dots as cellular probes. Annu. Rev. Biomed. Eng. 2005;7:55–76. doi: 10.1146/annurev.bioeng.7.060804.100432. [DOI] [PubMed] [Google Scholar]
- 6.Klostranec JM, Chan WCW. Quantum Dots in Biological and Biomedical Research: Recent Progress and Present Challenges. Adv. Mater. 2006;18:1953–1964. [Google Scholar]
- 7.Luccardini C, Yakovlev A, Gaillard S, van’t Hoff M, Alberola AP, Mallet J-M, Parak WJ, Feltz A, Oheim M. Getting across the plasma membrane and beyond: Intracellular uses of colloidal semiconductor nanocrystals. J. Biomed. Biotechnol. 2007 doi: 10.1155/2007/68963. Article ID 68963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Emerich DF, Thanos CG. The pinpoint promise of nanoparticle-based drug delivery and molecular diagnosis. Biomol. Eng. 2006;23:171–184. doi: 10.1016/j.bioeng.2006.05.026. [DOI] [PubMed] [Google Scholar]
- 9.Hild WA, Breunig M, Goepferich A. Quantum dots - Nano-sized probes for the exploration of cellular and intracellular targeting. Eur. J. Pharmaceutics and Biopharmaceutics. 2008;68:153–68. doi: 10.1016/j.ejpb.2007.06.009. [DOI] [PubMed] [Google Scholar]
- 10.Dennis AM, Bao G. Quantum dot-fluorescent protein pairs as novel fluorescence resonance energy transfer probes. Nano Lett. 2008;8:1439–1445. doi: 10.1021/nl080358+. [DOI] [PubMed] [Google Scholar]
- 11.Lu H, Schops O, Woggon U, Niemeyer CM. Self-assembled donor comprising quantum dots and fluorescent proteins for long-range fluorescence resonance energy transfer. J. Am. Chem. Soc. 2008;130:4815–4827. doi: 10.1021/ja078243f. [DOI] [PubMed] [Google Scholar]
- 12.Medintz IL, Pons T, Delehanty JB, Susumu K, Brunel FM, Dawson PE, Mattoussi H. Intracellular delivery of quantum dot-protein cargos mediated by cell penetrating peptides. Bioconj. Chem. 2008;19:1785–1795. doi: 10.1021/bc800089r. [DOI] [PubMed] [Google Scholar]
- 13.Lakowicz JR. Principles of Fluorescence Spectroscopy. Third Edition Springer; New York: 2006. [Google Scholar]
- 14.Susumu K, Uyeda HT, Medintz IL, Pons T, Delehanty JB, Mattoussi H. Enhancing the stability and biological functionalities of quantum dots via compact multifunctional ligands. J. Am. Chem. Soc. 2007;129:13987–96. doi: 10.1021/ja0749744. [DOI] [PubMed] [Google Scholar]
- 15.Dabbousi BO, Rodriguez-Viejo J, Mikulec FV, Heine JR, Mattoussi H, Ober R, Jensen KF, Bawendi MG. (CdSe)ZnS Core-Shell Quantum Dots: Synthesis and Optical and Structural Characterization of a Size Series of Highly Luminescent Materials. J. Phys. Chem. B. 1997;101:9463–9475. [Google Scholar]
- 16.Hines MA, Guyot-Sionnest P. Synthesis and characterization of strongly luminescing ZnS-Capped CdSe nanocrystals. J. Phys. Chem. 1996;100:468–471. [Google Scholar]
- 17.Peng ZA, Peng X. Formation of high-quality CdTe, CdSe, and CdS nanocrystals using CdO as precursor. J. Am. Chem. Soc. 2001;123:183–184. doi: 10.1021/ja003633m. [DOI] [PubMed] [Google Scholar]
- 18.Clapp AR, Goldman ER, Mattoussi H. Capping of CdSe-ZnS quantum dots with DHLA and subsequent conjugation with proteins. Nat. Protocols. 2006;1:1258–66. doi: 10.1038/nprot.2006.184. [DOI] [PubMed] [Google Scholar]
- 19.Mei BC, Susumu K, Medintz IL, Mattoussi H. Polyethylene glycol-based bidentate ligands to enhance quantum dot and gold nanoparticle stability in biological media. Nat. Protocols. 2009;4:412–423. doi: 10.1038/nprot.2008.243. [DOI] [PubMed] [Google Scholar]
- 20.Tsien RY. Building and breeding molecules to spy on cells and tumors. Febs Letters. 2005;579:927–932. doi: 10.1016/j.febslet.2004.11.025. [DOI] [PubMed] [Google Scholar]
- 21.Tsien RY. The green fluorescent protein. Annu. Rev. Biochem. 1998;67:509–544. doi: 10.1146/annurev.biochem.67.1.509. [DOI] [PubMed] [Google Scholar]
- 22.Shaner NC, Campbell RE, Steinbach PA, Giepmans BNG, Palmer AE, Tsien RY. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp red fluorescent protein. Nat. Biotechnol. 2004;22:1567–1572. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- 23.Glazer AN. Phycobiliproteins - a Family of Valuable, Widely Used Fluorophores. J. Appl. Phycology. 1994;6:105–112. [Google Scholar]
- 24.Glazer AN, Clark JH. Phycobilisomes - Macromolecular Structure and Energy-Flow Dynamics. Biophys. J. 1986;49:115–116. doi: 10.1016/S0006-3495(86)83616-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Deuschle K, Okumoto S, Fehr M, Looger LL, Kozhukh L, Frommer WB. Construction and optimization of a family of genetically encoded metabolite sensors by semirational protein engineering. Protein Science. 2005;14:2304–2314. doi: 10.1110/ps.051508105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fehr M, Frommer WB, Lalonde S. Visualization of maltose uptake in living yeast cells by fluorescent nanosensors. Proceed. Nat. Acad. Sci. (USA) 2002;99:9846–9851. doi: 10.1073/pnas.142089199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Clapp AR, Medintz IL, Mauro JM, Fisher BR, Bawendi MG, Mattoussi H. Fluorescence resonance energy transfer between quantum dot donors and dye-labeled protein acceptors. J. Am. Chem. Soc. 2004;126:301–310. doi: 10.1021/ja037088b. [DOI] [PubMed] [Google Scholar]
- 28.Sapsford KE, Pons T, Medintz IL, Higashiya S, Brunel FM, Dawson PE, Mattoussi H. Kinetics of metal-affinity driven self-assembly between proteins or peptides and CdSe-ZnS quantum dots. J. Phys. Chem. C. 2007;111:11528–11538. [Google Scholar]
- 29.Hermanson GT. Bioconjugate Techniques. Second Edition ed. Academic Press; San Diego: 2008. [Google Scholar]
- 30.Medintz IL, Clapp AR, Brunel FM, Tiefenbrunn T, Uyeda HT, Chang EL, Deschamps JR, Dawson PE, Mattoussi H. Proteolytic activity monitored by fluorescence resonance energy transfer through quantum-dot-peptide conjugates. Nat. Mater. 2006;5:581–589. doi: 10.1038/nmat1676. [DOI] [PubMed] [Google Scholar]
- 31.Medintz IL, Mattoussi H. Quantum dot-based resonance energy transfer and its growing application in biology. Phys. Chem. Chem. Phys. 2009;11:17–45. doi: 10.1039/b813919a. [DOI] [PubMed] [Google Scholar]
- 32.Pons T, Medintz IL, Wang X, English DS, Mattoussi H. Solution-phase single quantum dot fluorescence resonant energy transfer sensing. J. Am. Chem. Soc. 2006;128:15324–15331. doi: 10.1021/ja0657253. [DOI] [PubMed] [Google Scholar]
- 33.Boeneman K, Mei B, Dennis A, Bao G, Deschamps JR, Mattoussi H, Medintz IL. Sensing Caspase 3 activity with quantum dot-fluorescent protein assemblies. J. Am. Chem. Soc. 2009;131:3828–3829. doi: 10.1021/ja809721j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mattoussi H, Cumming AW, Murray CB, Bawendi MG, Ober R. Properties of CdSe nanocrystal dispersions in the dilute regime: Structure and interparticle interactions. Phys. Rev. B. 1998;58:7850–7863. [Google Scholar]
- 35.Huang CC, Pettersen EF, Klein TE, Ferrin TE, Langridge R. Conic - a Fast Renderer for Space-Filling Molecules with Shadows. J. Molecular Graphics. 1991;9:230. doi: 10.1016/0263-7855(91)80016-s. &. [DOI] [PubMed] [Google Scholar]
- 36.Ferrin TE, Huang CC, Jarvis LE, Langridge R. The Midas Display System. J. Molecular Graphics. 1988;6:13. &. [Google Scholar]
- 37.Mattoussi H, Mauro JM, Goldman ER, Anderson GP, Sundar VC, Mikulec FV, Bawendi MG. Self-Assembly of CdSe-ZnS Quantum Dot Bioconjugates Using an Engineered Recombinant Protein. J. Am. Chem. Soc. 2000;122:12142–12150. [Google Scholar]
- 38.Miyawaki A, Sawano A, Kogure T. Lighting up cells: labelling proteins with fluorophores. Nat. Cell Biol. 2003:S1–S7. [PubMed] [Google Scholar]
- 39.Shaner NC, Steinbach PA, Tsien RY. A guide to choosing fluorescent proteins. Nat. Methods. 2005;2:905–909. doi: 10.1038/nmeth819. [DOI] [PubMed] [Google Scholar]
- 40.Ficner R, Huber R. Refined crystal structure of phycoerythrin from Porphyridium cruentum at 0.23-nm resolution and localization of the γ subunit. Eur. J. Biochem. 1993;218:103–106. doi: 10.1111/j.1432-1033.1993.tb18356.x. [DOI] [PubMed] [Google Scholar]
- 41.Glazer AN. Light Harvesting by Phycobilisomes. Ann. Rev. Biophys. Biophys. Chem. 1985;14:47–77. doi: 10.1146/annurev.bb.14.060185.000403. [DOI] [PubMed] [Google Scholar]
- 42.Levy M, Cater SF, Ellington AD. Quantum-dot aptamer beacons for the detection of proteins. Chembiochem. 2005;6:2163–2166. doi: 10.1002/cbic.200500218. [DOI] [PubMed] [Google Scholar]
- 43.Caruthers SD, Wickline SA, Lanza GM. Nanotechnological applications in medicine. Curr. Opinion Biotechnol. 2007;18:26–30. doi: 10.1016/j.copbio.2007.01.006. [DOI] [PubMed] [Google Scholar]
- 44.Nahar M, Dutta T, Murugesan S, Asthana A, Mishra D, Rajkumar V, Tare M, Saraf S, Jain NK. Functional polymeric nanoparticles: An efficient and promising tool for active delivery of bioactives. Critical Reviews in Therapeutic Drug Carrier Systems. 2006;23:259–318. doi: 10.1615/critrevtherdrugcarriersyst.v23.i4.10. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.