Abstract
Background
The extent to which immunologic and clinical biomarkers influence HIV outcomes remains incompletely characterized, particularly for non-B subtypes. Based on data supporting in vitro HIV protein-specific CD8 T-lymphocyte responses as correlates of immune control in cross-sectional studies, we assessed the relationship of these responses, along with established HIV biomarkers, with rates of CD4 decline in subtype-C infection.
Methods
Bi-variate and multivariate mixed effects models were used to assess the relationship of baseline CD4, plasma viral load (pVL), HLA class I alleles, and HIV protein-specific CD8 responses with rate of CD4 decline in a longitudinal population-based cohort of 300 therapy-naïve, chronically infected adults with baseline CD4>200 cells/mm3 and pVL>500 copies/ml, over a median 25 months follow-up.
Results
In bi-variate analyses, baseline CD4, pVL and possession of a protective HLA allele correlated significantly with rate of CD4 decline. No relationship was observed between HIV protein-specific CD8 responses and CD4 decline. Results from multivariate models, incorporating baseline CD4 (201–350 and >350), pVL (≤100,000 and >100,000), HLA (protective vs. not), yielded the ability to discriminate CD4 declines over a 10-fold range: the highest rate of decline was observed among individuals with CD4>350, pVL>100,000 with no protective HLA alleles (−59 cells/mm3/year), while the slowest decline was observed in individuals with CD4 201–350, pVL≤100,000 and a protective allele (−6 cells/mm3/year).
Conclusions
The combination of plasma viral load and HLA class I type, but not in vitro HIV protein-specific CD8 responses, differentiates rates of CD4 decline in chronic subtype-C infection better than either marker alone.
Keywords: HIV subtype C, disease progression, CD4 decline, HLA class I, HIV-specific CTL responses
Introduction
Rates of CD4 decline vary widely among HIV-1 infected individuals, however the factors influencing these differences remain incompletely characterized. Determinants of HIV outcomes include plasma viral load (pVL), CD4 cell count [1], and to a lesser extent Human Leukocyte Antigen (HLA) class I profile [2–4] and other human genetic polymorphisms [5]. However, less than half of population variation in rates of CD4 decline can be explained by differences in pVL [1, 6], and only a minor portion of pVL set point variation is explained by expression of HLA-B*57 and other key polymorphisms in the Major Histocompatibility Complex (MHC) region [7, 8]. Investigation of additional correlates of CD4 decline is clearly necessary. Moreover, given the global HIV subtype distribution, there is a critical need to expand outcomes research in non-B subtypes.
Cross-sectional studies have reported associations between in vitro HIV protein-specific CD8 T-lymphocyte responses and clinical disease markers in untreated chronic infection. In particular, detection of Gag (most notably p24)-specific responses have been associated with lower pVL and/or higher CD4 counts [9–15], while detection of responses to Envelope and/or Accessory proteins have been associated with higher pVL [10, 11]. Although these observations suggest that CD8 responses against certain viral targets may be more beneficial than others, the cross-sectional nature of these studies precludes inference of cause and effect. Smaller observational studies have reported links between Gag-specific responses and slow or nonprogressive infection [16–19], however the relationship between CD8 responses and HIV outcomes has rarely been investigated in a population-based setting [20], and thus remains unclear. We therefore sought to evaluate whether in vitro HIV protein-specific CD8 T-cell responses, along with pVL and HLA class I types, correlate with rates of CD4 decline in chronic untreated subtype C infection.
Methods
Patient Selection
The study group comprised antiretroviral naïve, chronically subtype C-infected adults from the Sinikithemba cohort based at McCord Hospital, Durban, South Africa, enrolled between August 2003 and June 2006 [10, 21]. Sociodemographic/pVL/CD4 data were collected at baseline. Follow-up CD4 and pVL measurements were performed at 3- and 6-month intervals, respectively. Of the 449 original participants, 68 were excluded due to lack of CD8 T-cell response data. A further 7 were excluded due to missing age, 1 due to pediatric infection and 3 due to missing baseline clinical data. As our objective was to characterize typical rates of CD4 decline in chronic infection, spontaneous HIV controllers (pVL<500;N=10), and individuals with advanced disease eligible for antiretroviral treatment (baseline CD4<200;N=60) were excluded, yielding a final N=300. Informed consent was obtained from all patients and approval was obtained from the appropriate Institutional Review boards.
Laboratory Methods
In vitro CD8 T-cell responses were measured by interferon-gamma (IFN-γ) Enzyme-Linked Immunosorbent Spot (ELISpot) assays using a set of 410 18-mer overlapping peptides spanning the HIV-1 subtype C proteome [10]. Initially, responses were assessed to pools of 11 or 12 peptides, after which responses to individual peptides were confirmed independently. Negative controls were performed in quadruplicate. Responses >100 spot-forming cells [SFC]/106 cells after subtraction of average background were considered positive. Responses were classified by protein: Gag, Pol, Env, and Accessory/Regulatory (“Acc/Reg”, comprising Tat, Rev, Vpr, Vpu, Vif and Nef) [10]. In the primary analysis, individuals were classified as “non-responders” or “responders” if they exhibited positive responses to 0 or ≥1 peptides within each protein, respectively.
High-resolution HLA class I typing was performed using molecular methods [21]. In the primary analysis, “protective” alleles were defined as B*57, B*5801, and B*4201, as these alleles are significantly associated with lower pVL in subtype C infection after correction for HLA linkage disequilibrium [21].
Statistical Analysis
Bi-variate and multivariate mixed effects models were constructed to assess the relationship between immunologic and clinical biomarkers (independent variables) and rate of CD4 decline (dependent variable). To enhance transparency of interpretation, all primary analyses featured biomarkers as binary variables: baseline CD4 count (201–350 vs. >350 cells/mm3), pVL (mean of all measurements during untreated follow-up; ≤100,000 vs. >100,000 copies/ml), HLA class I (possession of a protective allele vs. not), and HIV protein-specific CD8 T-cell activity (responders vs. nonresponders). Secondary analyses employing alternative binary classifiers and treating biomarkers as continuous variables were also performed. Biomarkers exhibiting associations with CD4 cell decline at p<0.1 in bi-variate analyses were advanced in the multivariate model. Individuals were censored at their last visit up to April 15, 2008. Individuals initiating HAART (N=33) were censored on their treatment initiation date. P-values <0.05 were considered statistically significant. Analyses were performed using SAS 9.1.
Results
Baseline Characteristics
The cohort was stratified by baseline CD4 (201–350 vs. >350 cells/mm3) (Table 1). Age, gender and HLA distributions did not differ significantly between these strata. No significant differences in the frequency of CD8 responses to different HIV proteins were observed, however there was a trend towards more frequent p24Gag responses in individuals in the upper compared to the lower CD4 stratum (p=0.08). Individuals in the upper CD4 stratum had lower pVL and longer follow-up times compared to those in the lower CD4 stratum (p<0.0001).
Table 1.
Baseline Characteristics of the study group
| Overall | Baseline CD4 stratum |
p | ||
|---|---|---|---|---|
| (N=300) | 201–350 (N=114) |
>350 (N=186) |
||
| Age (median, IQR) | 31 (27 – 37) | 32 (27 – 38) | 31 (26 – 36) | 0.25 |
|
| ||||
| Gender (Female) (%) | 245 (82) | 90 (79) | 155 (83) | 0.34 |
|
| ||||
| pVL >100,000 copies/ml (%) | 107 (36) | 57 (50) | 50 (27) | <0.0001 |
|
| ||||
| CTL responders (%) | ||||
| p24Gag | 232 (77) | 82 (72) | 150 (81) | 0.08 |
| Pol | 244 (81) | 98 (86) | 146 (78) | 0.11 |
| Env | 127 (42) | 52 (46) | 75 (40) | 0.37 |
| Acc/Reg | 263 (88) | 97 (85) | 166 (89) | 0.29 |
|
| ||||
| Protective HLA allele (%) | 116 (39) | 49 (43) | 67 (36) | 0.23 |
|
| ||||
| CD4 measurements/subject | 8 (4 – 11) | 7 (3 – 9) | 9 (6 – 12) | <0.0001 |
|
| ||||
| Median Follow-up months (IQR) | 25 (15 – 41) |
21 (10 – 28) |
27 (20 – 45) |
<0.0001 |
IQR=Interquartile range
Correlation between biomarkers and CD4 decline: Bi-variate analysis
In bi-variate analyses, individuals with baseline CD4>350 exhibited two times higher rates of CD4 decline compared to individuals with baseline CD4 201–350 cells/mm3 (Table 2). Similarly, rates of CD4 decline among individuals with pVL>100,000 were nearly twice as high as those with pVL≤100,000 copies HIV RNA/ml. Expressed as a continuous variable, every 100-cell increment in baseline CD4 resulted in loss of an additional 9.1 cells/mm3 per year (standard error 0.89 cells/mm3 per year; p<0.0001). Similarly, every log10 increment in pVL resulted in loss of an additional 11.0 cells/mm3 per year (standard error 2.37 cells/mm3 per year; p<0.0001). A modest but significantly lower rate of CD4 decline was observed in individuals possessing at least one subtype-C-specific protective HLA allele (B*57, B*5801 or B*4201) compared to individuals not possessing a protective allele (Table 2).
Table 2.
Clinical/immunogenetic biomarkers and rates of CD4 decline: Bi-Variate analyses.
| Biomarker | Group | Frequency (%) | CD4 cell decline (cells/mm3 per year) | p-value |
|---|---|---|---|---|
| Primary Analysis: biomarkers as binary variables | ||||
| Baseline CD4 | >350 | 186 (62) | −39.9 | <0.0001 |
| 201–350 | 114 (38) | −19.3 | ||
| pVL | >100,000 | 107 (36) | −51.8 | <0.0001 |
| ≤100,000 | 193 (64) | −28.1 | ||
| HLA | Non-protective | 184 (61) | −40 | 0.0002 |
| protective | 116 (39) | −27.2 | ||
The data, however, provided insufficient statistical evidence supporting an association between in vitro HIV protein-specific CD8 responses and rates of CD4 decline when the cohort was divided into “nonresponders” or “responders” to each individual HIV protein (Table 3). Moreover, the magnitude of the differences in CD4 decline between responders vs. nonresponders revealed no trends suggestive of clinical importance.
Table 3. HIV protein-specific CD8 responses and rates of CD4 decline: Bi-Variate anlayses.
In the primary analysis (where individuals are classified as “nonresponder” and “responder” if they responded to 0 or ≥1 peptide within the indicated protein), results are expressed in terms of absolute rates of CD4 decline (cells/mm3 per year) for each category. In the secondary analysis (where biomarkers are treated as continuous variables), results are expressed in terms of the difference in rate of CD4 decline (cells/mm3 per year) per every indicated increment in the biomarker. For example, every per-unit increment in the absolute breadth of Gag p24 responses is associated with a difference in rate of CD4 decline of +0.44 cells/mm3 per year.
| Biomarker | Group | Frequency (%) | CD4 cell decline (cells/mm3 per year) | p-value |
|---|---|---|---|---|
| Primary Analysis: responder vs. nonresponder | ||||
| p24Gag | nonresponder | 68 (23) | −33.3 | 0.77 |
| responder | 232 (77) | −34.7 | ||
| Pol | nonresponder | 56 (19) | −36.4 | 0.65 |
| responder | 244 (81) | −34.1 | ||
| Env | nonresponder | 173 (58) | −36.6 | 0.18 |
| responder | 127 (42) | −32.1 | ||
| Acc/Reg | nonresponder | 37 (12) | −35.4 | 0.84 |
| responder | 263 (88) | −34.3 | ||
| Secondary Analyses: CD8 responses as continuous variables | ||||
|---|---|---|---|---|
| Difference in rate of CD4 decline cells/mm3 per year, per indicated increment in biomarker (SE*) | p-value | |||
| Absolute Breadth (per unit increment) | ||||
| p24Gag | 0.44 (1.0) | 0.65 | ||
| Pol | 0.98 (0.8) | 0.24 | ||
| Env | 0.98 (1.8) | 0.58 | ||
| Acc/Reg | 0.42 (0.8) | 0.61 | ||
| Relative Breadth (per 10% increment) | ||||
| p24Gag | −0.01 (0.83) | 0.99 | ||
| Pol | 1.0 (0.9) | 0.26 | ||
| Env | 2.2 (1.6) | 0.2 | ||
| Acc/Reg | 0.2 (1.0) | 0.84 | ||
| Magnitude (per increment of 100 SFC/million) | ||||
| p24Gag | 0.2 (0.1) | 0.05 | ||
| Pol | 0.1 (0.1) | 0.09 | ||
| Env | 0.3 (0.2) | 0.29 | ||
| Acc/Reg | 0.1 (0.1) | 0.15 | ||
SE= standard error
Secondary analyses featuring alternative binary classifiers of absolute breadth (≤1 vs. ≥2 responses), relative breadth (proportion of total responses directed against each HIV protein; <25% vs. ≥25%) and relative magnitude (proportion of the total magnitude of responses against each HIV protein; <25% vs. ≥25%) also failed to reveal significant correlations with CD4 decline (not shown). Similarly, treatment of absolute breadth, relative breadth and magnitude as continuous variables also failed to reveal significant correlations with CD4 decline (Table 3), with the exception of a weak association between higher-magnitude p24Gag responses and slower CD4 decline. The magnitude of this association, however, was extremely modest (every 100 SFC/million increment was associated with a difference in the rate of CD4 decline of +0.2 cells/mm3 per year) and did not remain significant after consideration for multiple tests. Note that evaluation of responses to the whole Gag (as opposed to p24 only) yielded comparable results in all analyses (not shown).
Based on data suggesting that the ability to target specific functionally-constrained regions in Gag may be associated with protective effects mediated by the expression of specific HLA alleles [22, 23], we investigated whether CD8 responses to these specific regions may be associated with rate of CD4 decline in our cohort. We thus defined “functionally-constrained” peptides as described in [22], meaning those including one or more of the following Gag residues: 146, 147, 177, 182, 186, 242, 247, 302, 310, 456.
In bi-variate analyses, we observed no statistically significant nor clinically relevant differences in rates of CD4 decline between individuals who did not respond to any functionally-constrained Gag region (N=112; −36.1 cells/yr) vs. those who responded to at least one (N=188, −33.9 cells/yr, p=0.56). Similarly, restricting this analysis to individuals expressing at least one protective HLA allele failed to yield significant differences in rates of CD4 decline between responders and nonresponders to these regions (p=0.42, not shown). Finally, no significant relationship between the absolute breadth of proteome-wide CD8 responses and CD4 decline was observed (p>0.29, not shown).
Based on the lack of evidence supporting a clear relationship between HIV protein-specific CD8 responses and CD4 decline in bi-variate analyses, CD8 responses were not advanced to the multivariate model.
Correlation between biomarkers and CD4 decline: Multivariate analysis
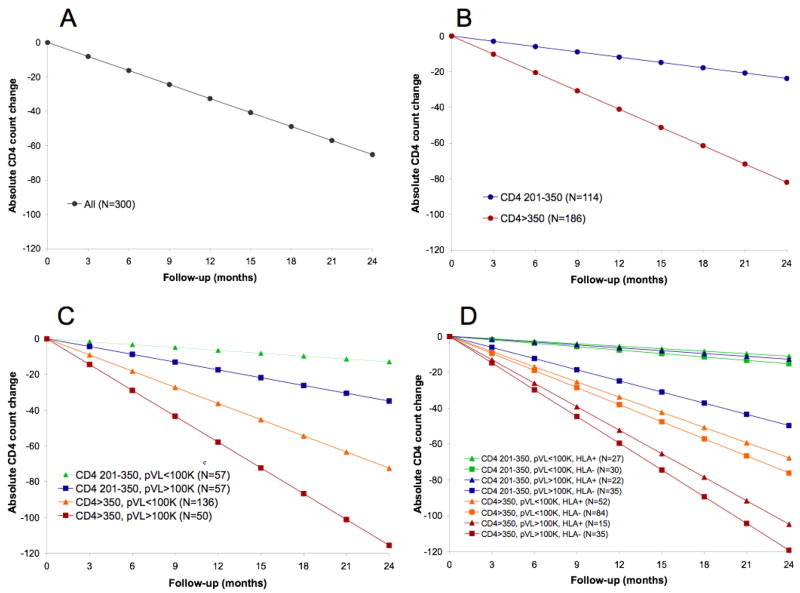
In multivariate analyses including baseline CD4, pVL and HLA as binary variables as per the primary analysis, the overall mean rate of CD4 decline was −32.6 cells/year (Figure, panel A). In a subsequent model, stratification by baseline CD4 yielded mean declines of −41.0 vs. −11.9 cells/yr in individuals with baseline CD4>350 vs. 201–350 (p<0.0001; panel B). Further stratification by pVL revealed faster CD4 decline in individuals with pVL>100,000 compared to pVL≤100,000 in both CD4 strata: mean absolute rates of decline were −57.8 vs. −36.2 cells/year for baseline CD4>350, and −17.4 vs. −6.5 cells/year for baseline CD4 201–350 (overall p<0.0001, panel C). In pairwise comparisons designating the CD4 201–350/pVL≤100,000 as the reference group, all differences in rates of CD4 decline were statistically significant at p<0.05.
Figure. Absolute rates of CD4 decline, stratified by baseline CD4, plasma viral load and HLA class I alleles.
Results of multivariate, sequentially-built mixed effects models show mean rates of CD4 decline in the overall cohort (panel A); stratified by baseline CD4 count (panel B); stratified by baseline CD4 and pVL (panel C); and stratified by baseline CD4, pVL and the posession of a protective HLA allele (panel D). Individuals in different subgroups within the same stratum can be distinguished by the use of different shades of the same color; for example, individuals with baseline CD4 201–350 and >350 are represented by blue/green and red/orange lines, respectively, in figures C and D.
Addition of HLA to the model further discriminated rates of decline in all groups (p<0.0001, panel D), although the magnitude of separation for the CD4 201–350/pVL≤100,000 group was relatively minor. In the CD4>350 stratum, the fastest decline was observed in patients with pVL>100,000 with no protective alleles (−59.6 cells/yr), while the slowest decline was observed in patients with pVL≤100,000 and a protective allele (−33.9 cells/yr). In the CD4 201–350 stratum, corresponding rates of CD4 decline were −24.8 and −5.5 cells/yr, respectively. In pairwise comparisons designating CD4 201–350/pVL≤100,000/HLA+ as the reference group, all differences except the CD4 201–350/pVL≤100,000/HLA− and CD4 201–350/pVL>100,000/HLA+ groups were statistically significant (p<0.01).
Sensitivity analyses
Several post-hoc sensitivity analyses were performed. Firstly, data were re-analyzed in terms of relative CD4 decline (% decline from baseline). In bi-variate analyses, pVL and HLA, but not baseline CD4 count or CD8 T-cell responses, correlated significantly with % CD4 decline/year (not shown). In multivariate model “A” adjusting for baseline CD4, pVL and HLA, the overall relative rate of CD4 decline in the cohort was 6.4% cells/year. Stratification by baseline CD4 revealed declines of 7.2% and 4.2% in CD4>350 vs. 201–350, respectively, a difference which was statistically significant (model B, p<0.0001). Addition of pVL revealed greater relative CD4 declines among individuals with pVL>100,000 vs. ≤100,000: 10.5% vs. 6.3% cells/year in the CD4>350 stratum, compared to 6.0% vs. 2.4% cells/year in the CD4 201–350 stratum. Again, model C was significant at p<0.0001, with all pairwise comparisons statistically significant using CD4 201–350/pVL<100,000 as a reference group (p<0.0001). However, adding HLA to this model further discriminated rates of CD4 decline for the CD4 201–350/pVL>100,000 group only: while previously 6.0%/year in model C, rates were now 8.2% vs. 2.4% cells/year in individuals lacking or having a protective allele, respectively (model D, overall p<0.0001).
Second, based on data from subtype C cohorts in Zambia [24] and South Africa ([25], and P. Goulder unpublished data) we expanded the definition of “protective” alleles to include B*3910, B*8101, and B*13. Possession of a protective allele (N=134; 45%) was associated with slower CD4 decline (bi-variate p=0.0002). Consistent with the primary analysis, sequential addition of CD4, pVL, and HLA to multivariate models A–D resulted in incremental ability to discriminate rates of CD4 decline in all resulting groups (p<0.0001).
Third, to examine potential biases due to informative censoring, we restricted the analysis to the first 7 clinic visits only (18 months) in individuals with a minimum of 7 visits. Of the original 300 patients, 193 (64.3%) met these criteria (N=136 and 57 with baseline CD4 >350 and 201–350, respectively). In multivariate analyses adjusting for baseline CD4, pVL and HLA, higher absolute rates of CD4 decline were observed among individuals with baseline CD4>350 (−22.7 cells/yr) compared to CD4 201–350 (−10.1 cells/yr), (model B, p=0.0032). Further stratification by pVL in model C revealed faster CD4 decline among those with pVL>100,000 compared to ≤100,000 regardless of baseline CD4: rates were −30.1, −20.8, −15.5 and −6.2 cells/yr, respectively, for individuals with CD4>350/pVL>100,000 (N=30), CD4>350/pVL≤100,000 (N=106), CD4 201–350/pVL>100,000 (N=23), CD4 201–350/pVL≤100,000 (N=34), overall p=0.005. However, further stratification by HLA in model D did not yield significantly improved ability to discriminate rates of CD4 decline, possibly due to diminished statistical power in this subanalysis.
Finally, a mixed effects model treating baseline CD4 and pVL as continuous variables and expressing CD4 decline in absolute terms was performed using a backwards selection procedure. In this analysis, expression of nonprotective HLA alleles was associated with the loss of an additional 6.6 cells/mm3 per year compared to expression of a protective allele (p=0.04). Moreover, the interaction between log10 pVL and baseline CD4 was significantly associated with differences in rate of CD4 decline. Specifically, every 100-unit increase in the product of log10 pVL*baseline CD4 resulted in the loss of an additional 3.2 cells/mm3 per year (Standard Error 1.2 cells/mm3 per year, p=0.002).
Discussion
Elucidating the relationship between HIV-specific CD8 T-cell responses and longitudinal outcomes has important implications for the design of vaccines and immunotherapeutic strategies aimed at stimulating these responses. Indeed, consistent correlations between Gag-specific CD8 responses and favorable clinical profiles in cross-sectional studies of chronic infection [9, 11, 12], including an expanded version of the present cohort [10] suggest that strategies aimed at stimulating Gag-specific responses may be beneficial. Cross-sectional studies, however, do not address whether Gag-specific responses in chronic infection are predictive of subsequent disease outcomes - a critical question addressed by few population-based longitudinal studies. In the present study, we observed neither statistically significant nor clinically relevant evidence of an association between either the magnitude or frequency (absolute or relative) of in vitro CD8 T-cell responses to any HIV protein, and subsequent rate (either absolute or relative) of subsequent CD4 decline in chronic untreated subtype C infection. A subanalysis investigating the relationship between CD8 responses to key functionally-constrained regions in Gag also failed to reveal any significant association with rates of CD4 decline.
These results contrast with a smaller subtype B study of 31 individuals reporting slower CD4 decline among individuals with ≥50% (vs. <50%) of CD8 responses directed at Gag [19]. In the current study, grouping by these criteria yielded too few patients with ≥50% Gag responses to robustly verify these findings. Note that, on average, the clade B cohort not only had more dominant Gag responses but also had higher CD4 counts and lower pVL [19] compared to the present cohort, possibly indicating earlier disease stage (note that infection dates were unknown for both cohorts). If the predictive ability of Gag-specific responses differs throughout the disease course [26], this could help reconcile these findings. Likely due to smaller sample size, the clade B study did not adjust for baseline CD4, pVL and HLA, which may also affect interpretation.
Our finding that protective HLA allele expression, but not CD8 T-cell responses, is associated with CD4 decline is somewhat surprising given that the most likely mechanism of HLA-mediated control of HIV is through CD8 T-cell responses. A possible explanation for this finding is that in vitro CD8 responses measured during relatively advanced infection may no longer be relevant to certain disease outcomes, even though these responses may have contributed to HIV containment in earlier disease stages. This may be due in part to the decline in immune function over the disease course, or to the accumulation of escape mutations in the autologous virus that may compromise the ability to detect responses to the corresponding consensus peptide. Indeed, the use of consensus (vs. autologous) peptides may underestimate response rates by up to 30% [27]. Note that our findings do not exclude the possibility that Gag-specific responses measured earlier in infection represent important and clinically relevant predictors of outcome. Indeed, there is some evidence supporting the relevance of acute/early CD8 responses to disease outcomes [28–31], although this remains controversial [32–34].
In this chronically-infected cohort, mean rates of CD4 decline were calculated based on initial CD4 count, pVL and HLA type. Analysis of these biomarkers in combination discriminated rates of CD4 decline better than each individually; rates of CD4 decline ranged from −6 to −60 cells/mm3 per year depending on the combination of biomarkers expressed. Of interest, within the CD4 201–350 stratum, comparable rates of CD4 decline were observed for pVL≤100,000 (regardless of HLA) and pVL>100,000 with a protective HLA, whereas those with pVL≤100,000 and no protective HLA had significantly faster CD4 decline, suggesting that having either lower pVL or a protective allele may be beneficial at low CD4. We suggest that population-level rates of CD4 decline could be applied in the clinical setting, for example, to estimate the time until antiretroviral treatment may be required in patients presenting with chronic infection.
Some limitations deserve mention. First, the restriction of the study to individuals with CD4>200 cells/mm3 with unknown infection dates raises the possibility of a survivor bias. To address this, we stratified the cohort by baseline CD4 and performed sensitivity analyses reporting CD4 decline on a relative (%) scale and “informative censoring” analyses restricting the two CD4 strata to equal follow-up, both of which yielded results consistent with the original findings. The IFN-γ ELISpot assay used to measure CD8 responses also has some limitations [35], including its use of supraphysiological levels of synthetic HIV peptides that bypass the normal intracellular antigen processing pathways, its use of consensus peptides which may not reflect immune responses to the autologous virus [27], and the fact that CD8 function is measured indirectly (by quantification of a secreted cytokine [27, 35, 36]). Despite these limitations, this assay is widely used to assess vaccine-induced and naturally occurring CD8 responses [37–39]. Finally, due to the measurement of CD8 responses at a single timepoint, we were unable to investigate the correlation between long-term stability of CD8 responses and CD4 decline [17].
In conclusion, we observed neither statistically significant nor clinically important correlations between in vitro HIV-specific CD8 T-cell responses and rates of CD4 decline in chronic subtype C infection. The combination of pVL and HLA class I type, however, predict CD4 decline significantly better than either of these biomarkers alone.
Acknowledgments
The authors wish to thank Sisters Kesia Ngwenya, Thandi Cele, Thandi Sikhakane and Nokuthula Lutuli for their invaluable support as clinic staff; Nolwandle Mngquandaniso, Dhanwanthie Ramduth, Nonhlanhla Nene and Nompumelelo Gumbi for expert technical assistance; Wendy Mphatswe and the management of McCord hospital for her support of the Sinikithemba cohort. Finally, the authors gratefully acknowledge the individuals who participated in the Sinikithemba study, without whom this research would not have been possible.
Funding Statement: ZLB is supported by a post-doctoral fellowship from the Canadian Institutes of Health Research (CIHR). TN holds the South African Department of Science and Technology/National Research Foundation Research Chair in Systems Biology of HIV/AIDS. This research was supported in part by funds from the Bill and Melinda Gates Foundation, the International AIDS Vaccine Initiative (IAVI), the South African AIDS Vaccine Initiative (SAAVI) and the Center for AIDS Research (CFAR) at Harvard University. It was also supported in part by NIH grant # R01-AI067073 and NIH contract # N01-AI-15422 (BDW), NIH grant R01-AI46995 (PG) and funds from the Wellcome Trust (PG). This project has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under Contract No. HHSN261200800001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government. This research was supported in part by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research. Sources of support were not involved in the design and conduct of the study, nor were they involved in the collection, analysis, and interpretation of the data, or in the preparation, review, or approval of the manuscript.
Footnotes
Brief summary of article’s main points: Combining plasma viral load and HLA class I allele information, but not in vitro HIV-specific CD8 responses, allows the differentiation of rates of CD4 decline in a large longitudinal cohort of chronically-subtype C infected individuals
Conflict of Interest statement: The authors declare no conflict of interest.
References
- 1.Mellors JW, Munoz A, Giorgi JV, et al. Plasma viral load and CD4+ lymphocytes as prognostic markers of HIV-1 infection. Ann Intern Med. 1997 Jun 15;126(12):946–54. doi: 10.7326/0003-4819-126-12-199706150-00003. [DOI] [PubMed] [Google Scholar]
- 2.Kaslow RA, Carrington M, Apple R, et al. Influence of combinations of human major histocompatibility complex genes on the course of HIV-1 infection. Nat Med. 1996 Apr;2(4):405–11. doi: 10.1038/nm0496-405. [DOI] [PubMed] [Google Scholar]
- 3.Gao X, Nelson GW, Karacki P, et al. Effect of a single amino acid change in MHC class I molecules on the rate of progression to AIDS. N Engl J Med. 2001 May 31;344(22):1668–75. doi: 10.1056/NEJM200105313442203. [DOI] [PubMed] [Google Scholar]
- 4.Carrington M, O’Brien SJ. The influence of HLA genotype on AIDS. Annu Rev Med. 2003;54:535–51. doi: 10.1146/annurev.med.54.101601.152346. [DOI] [PubMed] [Google Scholar]
- 5.Dean M, Carrington M, Winkler C, et al. Genetic restriction of HIV-1 infection and progression to AIDS by a deletion allele of the CKR5 structural gene. Hemophilia Growth and Development Study, Multicenter AIDS Cohort Study, Multicenter Hemophilia Cohort Study, San Francisco City Cohort, ALIVE Study. Science. 1996 Sep 27;273(5283):1856–62. doi: 10.1126/science.273.5283.1856. [DOI] [PubMed] [Google Scholar]
- 6.Rodriguez B, Sethi AK, Cheruvu VK, et al. Predictive value of plasma HIV RNA level on rate of CD4 T-cell decline in untreated HIV infection. JAMA. 2006 Sep 27;296(12):1498–506. doi: 10.1001/jama.296.12.1498. [DOI] [PubMed] [Google Scholar]
- 7.Fellay J, Shianna KV, Ge D, et al. A whole-genome association study of major determinants for host control of HIV-1. Science. 2007 Aug 17;317(5840):944–7. doi: 10.1126/science.1143767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Limou S, Le Clerc S, Coulonges C, et al. Genomewide Association Study of an AIDS-Nonprogression Cohort Emphasizes the Role Played by HLA Genes (ANRS Genomewide Association Study 02) J Infect Dis. 2009 Feb 1;199(3):419–26. doi: 10.1086/596067. [DOI] [PubMed] [Google Scholar]
- 9.Edwards BH, Bansal A, Sabbaj S, Bakari J, Mulligan MJ, Goepfert PA. Magnitude of functional CD8+ T-cell responses to the gag protein of human immunodeficiency virus type 1 correlates inversely with viral load in plasma. J Virol. 2002 Mar;76(5):2298–305. doi: 10.1128/jvi.76.5.2298-2305.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kiepiela P, Ngumbela K, Thobakgale C, et al. CD8+ T-cell responses to different HIV proteins have discordant associations with viral load. Nat Med. 2007 Jan;13(1):46–53. doi: 10.1038/nm1520. [DOI] [PubMed] [Google Scholar]
- 11.Novitsky V, Gilbert P, Peter T, et al. Association between virus-specific T-cell responses and plasma viral load in human immunodeficiency virus type 1 subtype C infection. J Virol. 2003 Jan;77(2):882–90. doi: 10.1128/JVI.77.2.882-890.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zuniga R, Lucchetti A, Galvan P, et al. Relative dominance of Gag p24-specific cytotoxic T lymphocytes is associated with human immunodeficiency virus control. J Virol. 2006 Mar;80(6):3122–5. doi: 10.1128/JVI.80.6.3122-3125.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Masemola A, Mashishi T, Khoury G, et al. Hierarchical targeting of subtype C human immunodeficiency virus type 1 proteins by CD8+ T cells: correlation with viral load. J Virol. 2004 Apr;78(7):3233–43. doi: 10.1128/JVI.78.7.3233-3243.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Geldmacher C, Currier JR, Herrmann E, et al. CD8 T-cell recognition of multiple epitopes within specific Gag regions is associated with maintenance of a low steady-state viremia in human immunodeficiency virus type 1-seropositive patients. J Virol. 2007 Mar;81(5):2440–8. doi: 10.1128/JVI.01847-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tsegaye A, Ran L, Wolday D, et al. HIV-1 Subtype C gag-specific T-cell responses in relation to human leukocyte antigens in a diverse population of HIV-infected Ethiopians. J Acquir Immune Defic Syndr. 2007 Aug 1;45(4):389–400. doi: 10.1097/QAI.0b013e318059beaa. [DOI] [PubMed] [Google Scholar]
- 16.Klein MR, van Baalen CA, Holwerda AM, et al. Kinetics of Gag-specific cytotoxic T lymphocyte responses during the clinical course of HIV-1 infection: a longitudinal analysis of rapid progressors and long-term asymptomatics. J Exp Med. 1995 Apr 1;181(4):1365–72. doi: 10.1084/jem.181.4.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tsegaye A, Ran L, Wolday D, et al. Stable pattern of HIV-1 subtype C Gag-specific T-cell responses coincides with slow rate of CD4 T-cell decline in HIV-infected Ethiopians. AIDS. 2007 Jan 30;21(3):369–72. doi: 10.1097/QAD.0b013e32801222e3. [DOI] [PubMed] [Google Scholar]
- 18.Riviere Y, McChesney MB, Porrot F, et al. Gag-specific cytotoxic responses to HIV type 1 are associated with a decreased risk of progression to AIDS-related complex or AIDS. AIDS Res Hum Retroviruses. 1995 Aug;11(8):903–7. doi: 10.1089/aid.1995.11.903. [DOI] [PubMed] [Google Scholar]
- 19.Peretz Y, Tsoukas CM, Bernard NF. HIV Gag-specific immune responses predict the rate of CD4 decline. AIDS. 2008 Jun 19;22(10):1222–4. doi: 10.1097/QAD.0b013e3283021a76. [DOI] [PubMed] [Google Scholar]
- 20.Chouquet C, Autran B, Gomard E, et al. Correlation between breadth of memory HIV-specific cytotoxic T cells, viral load and disease progression in HIV infection. AIDS. 2002 Dec 6;16(18):2399–407. doi: 10.1097/00002030-200212060-00004. [DOI] [PubMed] [Google Scholar]
- 21.Kiepiela P, Leslie AJ, Honeyborne I, et al. Dominant influence of HLA-B in mediating the potential co-evolution of HIV and HLA. Nature. 2004 Dec 9;432(7018):769–75. doi: 10.1038/nature03113. [DOI] [PubMed] [Google Scholar]
- 22.Matthews PC, Prendergast A, Leslie A, et al. Central Role of Reverting Mutations in Hla Associations with Hiv Viral Setpoint. J Virol. 2008 Jul 2; doi: 10.1128/JVI.00580-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Streeck H, Lichterfeld M, Alter G, et al. Recognition of a defined region within p24 gag by CD8+ T cells during primary human immunodeficiency virus type 1 infection in individuals expressing protective HLA class I alleles. J Virol. 2007 Jul;81(14):7725–31. doi: 10.1128/JVI.00708-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tang J, Tang S, Lobashevsky E, et al. Favorable and unfavorable HLA class I alleles and haplotypes in Zambians predominantly infected with clade C human immunodeficiency virus type 1. J Virol. 2002 Aug;76(16):8276–84. doi: 10.1128/JVI.76.16.8276-8284.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Honeyborne I, Prendergast A, Pereyra F, et al. Control of human immunodeficiency virus type 1 is associated with HLA-B*13 and targeting of multiple gag-specific CD8+ T-cell epitopes. J Virol. 2007 Apr;81(7):3667–72. doi: 10.1128/JVI.02689-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jiao Y, Xie J, Li T, et al. Correlation between gag-specific CD8 T-cell responses, viral load, and CD4 count in HIV-1 infection is dependent on disease status. J Acquir Immune Defic Syndr. 2006 Jul;42(3):263–8. doi: 10.1097/01.qai.0000221692.00091.a2. [DOI] [PubMed] [Google Scholar]
- 27.Altfeld M, Addo MM, Shankarappa R, et al. Enhanced detection of human immunodeficiency virus type 1-specific T-cell responses to highly variable regions by using peptides based on autologous virus sequences. J Virol. 2003 Jul;77(13):7330–40. doi: 10.1128/JVI.77.13.7330-7340.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Altfeld M, Kalife ET, Qi Y, et al. HLA Alleles Associated with Delayed Progression to AIDS Contribute Strongly to the Initial CD8(+) T Cell Response against HIV-1. PLoS Med. 2006 Oct;3(10):e403. doi: 10.1371/journal.pmed.0030403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Musey L, Hughes J, Schacker T, Shea T, Corey L, McElrath MJ. Cytotoxic-T-cell responses, viral load, and disease progression in early human immunodeficiency virus type 1 infection. N Engl J Med. 1997 Oct 30;337(18):1267–74. doi: 10.1056/NEJM199710303371803. [DOI] [PubMed] [Google Scholar]
- 30.Patke DS, Langan SJ, Carruth LM, et al. Association of Gag-specific T lymphocyte responses during the early phase of human immunodeficiency virus type 1 infection and lower virus load set point. J Infect Dis. 2002 Oct 15;186(8):1177–80. doi: 10.1086/343811. [DOI] [PubMed] [Google Scholar]
- 31.Ndongala ML, Peretz Y, Boulet S, et al. HIV Gag p24 specific responses secreting IFN-gamma and/or IL-2 in treatment-naive individuals in acute infection early disease (AIED) are associated with low viral load. Clin Immunol. 2009 Jan 7; doi: 10.1016/j.clim.2008.11.006. [DOI] [PubMed] [Google Scholar]
- 32.Schellens IM, Borghans JA, Jansen CA, et al. Abundance of early functional HIV-specific CD8+ T cells does not predict AIDS-free survival time. PLoS ONE. 2008;3(7):e2745. doi: 10.1371/journal.pone.0002745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cao J, McNevin J, Holte S, Fink L, Corey L, McElrath MJ. Comprehensive analysis of human immunodeficiency virus type 1 (HIV-1)-specific gamma interferon-secreting CD8+ T cells in primary HIV-1 infection. J Virol. 2003 Jun;77(12):6867–78. doi: 10.1128/JVI.77.12.6867-6878.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gray CM, Mlotshwa M, Riou C, et al. Human immunodeficiency virus-specific gamma interferon enzyme-linked immunospot assay responses targeting specific regions of the proteome during primary subtype C infection are poor predictors of the course of viremia and set point. J Virol. 2009 Jan;83(1):470–8. doi: 10.1128/JVI.01678-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang OO. Will we be able to ‘spot’ an effective HIV-1 vaccine? Trends Immunol. 2003 Feb;24(2):67–72. doi: 10.1016/s1471-4906(02)00034-0. [DOI] [PubMed] [Google Scholar]
- 36.Jamieson BD, Yang OO, Hultin L, et al. Epitope escape mutation and decay of human immunodeficiency virus type 1-specific CTL responses. J Immunol. 2003 Nov 15;171(10):5372–9. doi: 10.4049/jimmunol.171.10.5372. [DOI] [PubMed] [Google Scholar]
- 37.Russell ND, Hudgens MG, Ha R, Havenar-Daughton C, McElrath MJ. Moving to human immunodeficiency virus type 1 vaccine efficacy trials: defining T cell responses as potential correlates of immunity. J Infect Dis. 2003 Jan 15;187(2):226–42. doi: 10.1086/367702. [DOI] [PubMed] [Google Scholar]
- 38.Yang OO. Aiming for successful vaccine-induced HIV-1-specific cytotoxic T lymphocytes. AIDS. 2008 Jan 30;22(3):325–31. doi: 10.1097/QAD.0b013e3282f29491. [DOI] [PubMed] [Google Scholar]
- 39.McElrath MJ, De Rosa SC, Moodie Z, et al. HIV-1 vaccine-induced immunity in the test-of-concept Step Study: a case-cohort analysis. Lancet. 2008 Nov 29;372(9653):1894–905. doi: 10.1016/S0140-6736(08)61592-5. [DOI] [PMC free article] [PubMed] [Google Scholar]