Cohesin not only links sister chromatids but also inhibits the transcriptional machinery’s interaction with and movement along chromatin1–6. In contrast, replication forks must traverse such cohesin-associated roadblocks to duplicate the entire genome in S phase. How this transpires is unknown. Through single-molecule analysis, we demonstrate that the replication factor C (RFC)-Ctf18 clamp loader1, 7 controls the velocity, spacing, and restart activity of replication forks and is required for robust acetylation of cohesin’s Smc3 subunit and sister chromatid cohesion. Unexpectedly, we discovered that cohesin acetylation itself is a central determinant of fork processivity, as cells lacking the Eco1-related acetyltransferases Esco1 or Esco28–10 (including those derived from Roberts Syndrome patients, in whom ESCO2 is biallelically mutated11) or expressing a non-acetylatable form of Smc3 also harbor slow-moving replication forks. This defect was a consequence of cohesin’s hyperstable interaction with two regulatory cofactors, Wapl and Pds5a12, 13; removal of either cofactor allowed forks to progress rapidly without Esco1, Esco2, or RFCCtf18. Our results reveal a novel mechanism for clamp loader-dependent fork progression, mediated by the post-translational modification and structural remodeling of the cohesin ring. Loss of this regulatory mechanism leads to the spontaneous accrual of DNA damage and may contribute to the abnormalities of the Roberts Syndrome cohesinopathy.
The ring-shaped cohesin complex not only catenates sister DNAs but also acts as a structural barrier to other chromatin-based transactions. For example, in budding and fission yeast cohesin slides to the 3’ ends of convergently arrayed genes and prevents readthrough transcription by RNA polymerase II2, 14. In mammals, cohesin accumulates at insulators and boundary elements and blocks activation of adjacent promoters4, 5. These sites occur about once per 25 kb in the human and mouse genomes, an interval severalfold smaller than the average somatic replicon15, and are occupied by cohesin throughout the cell cycle4, 5. Cohesin can also bind regions between insulators in a distributive manner and pair sister chromatids through these sites5. Hence, nearly all forks must replicate through at least one cohesin-associated region during S phase. In addition to mediating genome duplication, fork passage is thought to lead to entrapment of nascent DNA strands within the cohesin ring, thereby generating functional cohesion between sister chromatids1. Cohesion establishment also requires acetylation of cohesin’s Smc3 subunit by Eco1-related acetyltransferases8–10, but how this modification affects cohesin’s interaction with the replication fork remains unclear.
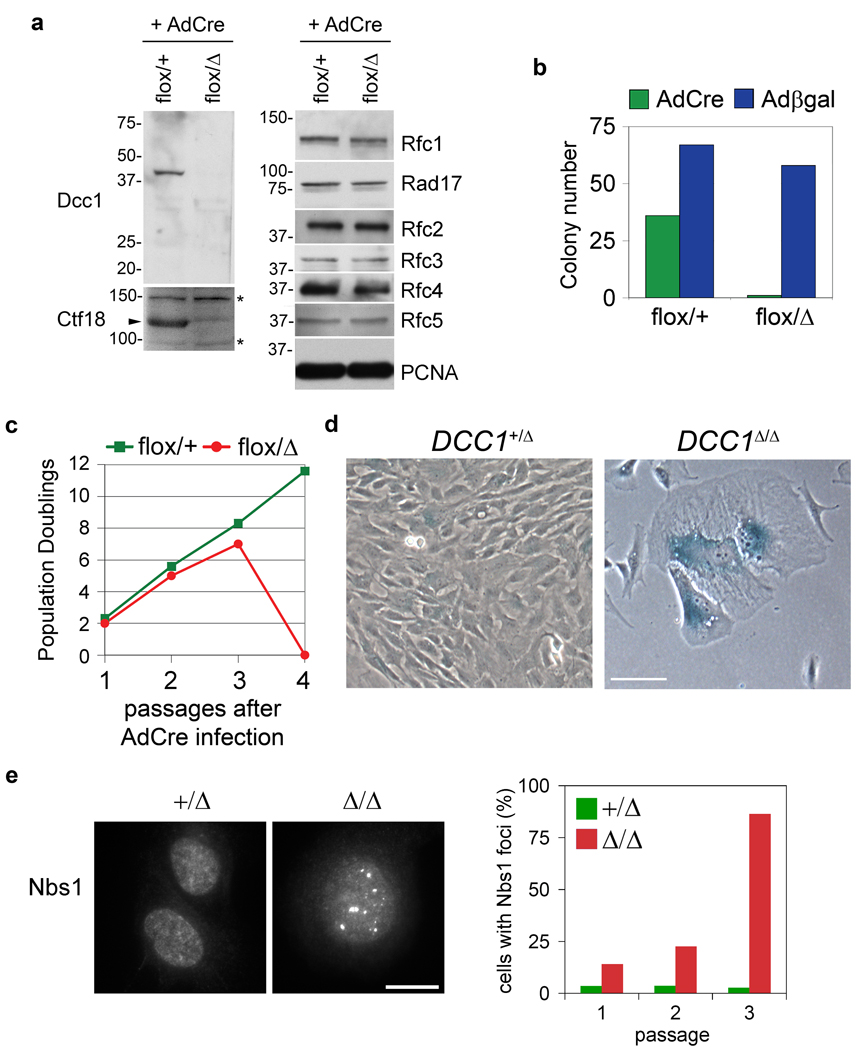
In addition to Eco1, cohesion establishment is aided by the replication factor C-Ctf18-Ctf8-Dcc1 (hereafter RFCCtf18) complex, which loads the polymerase processivity clamp PCNA onto replication forks in vitro and in vivo1, 7. Nevertheless, physical assays of replication fork dynamics have not uncovered this enzyme’s contribution during a normal S phase, nor is it known whether RFCCtf18 is important for Smc3 acetylation. To address these questions, human retinal pigment epithelial cells were subjected to two rounds of adeno-associated virus (AAV)-mediated homologous recombination, such that exon 2 of the DCC1 locus was either flanked by loxP sites or deleted outright (Supplementary Fig. 1). To inactivate RFCCtf18, DCC1flox/Δ cells were infected with an adenovirus expressing Cre recombinase (AdCre). The resulting DCC1Δ/Δ cells lacked detectable Dcc1 protein and had markedly reduced levels of Ctf18, but normal amounts of all other RFC subunits (Fig. 1a). Whereas RFCCtf18-deficient yeast grow robustly, DCC1Δ/Δ clones could not be isolated via limiting dilution (Fig. 1b), indicating that this complex is essential in mammals. To understand this requirement, DCC1flox/Δ cells were infected with AdCre en masse and monitored during serial passage. Within 7 divisions, DCC1Δ/Δ cells ceased to proliferate and exhibited hallmarks of premature senescence (increased size, flattened morphology, and expression of senescence-associated β-galactosidase; Fig. 1c,d). Senescence is commonly triggered by genotoxic stresses that activate the DNA damage response16; consistently, DCC1Δ/Δ cells accumulated high levels of Nbs1 foci during cell cycle exit (Fig. 1e). Unlike RFCRad17-deficient cells17, RFCCtf18-deficient cells activated Chk1 and imposed the G2/M checkpoint in response to genotoxic stress (Supplementary Fig. 2).
Figure 1. The RFCCtf18 complex is required to prevent accumulation of endogenous DNA damage and terminal senescence.
a, Homozygous deletion of DCC1 selectively inactivates RFCCtf18. b, Cells were infected with AdCre or Adβgal and scored for colony formation. All DCC1flox/Δ -derived clones retained the parental genotype. c, Cells were serially passaged to monitor proliferation. Senescent DCC1Δ/Δ cells at passage 3 failed to attach and survive after replating for passage 4, yielding a terminal PD value of zero. d, DCC1-null cells undergo senescence. Scale bar, 50 µm. e, DCC1Δ/Δ cells accumulate Nbs1-positive DNA damage foci. Scale bar, 10 µm.
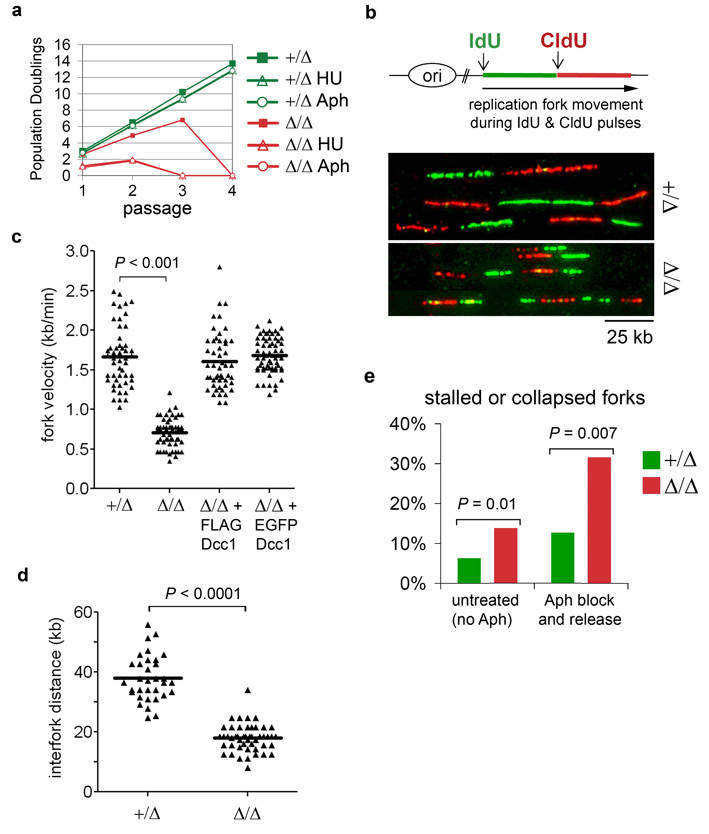
Unstable replication forks are a prime source of DNA damage in both prokaryotes and eukaryotes18. As an initial test of RFCCtf18’s contribution to fork stability, replication forks in newly generated DCC1+/Δ and DCC1Δ/Δ cells were transiently stalled with hydroxyurea (which depletes nucleotide pools) or aphidicolin (which inhibits replicative polymerases). RFCCtf18-deficient cells were hypersensitive to both replication inhibitors, as evidenced by their accelerated transit into senescence (Fig. 2a). To obtain a more precise view of replication dynamics, single-molecule analyses were performed on extended DNA fibers. Cells were sequentially pulse-labeled with the halogenated nucleosides iododeoxyuridine (IdU) and chlorodeoxyuridine (CldU), after which genomic DNA was gently extracted and stretched on glass slides, stained with IdU- and CldU-specific antibodies, and visualized microscopically (Fig. 2b). Unlike other assays for DNA synthesis, this technique affords specific and quantitative information on origin firing, replication fork movement, and fork recovery throughout the genome, based on examination of large numbers of individually labeled forks. Three effects of RFCCtf18 deficiency were evident in these assays. First, the velocity of individual forks decreased by more than 50% (0.7 ± 0.2 kb/min versus 1.7 ± 0.4 kb/min in DCC1+/Δ cells (mean ± s.d.); Fig. 2c), indicating a highly penetrant defect in replisome progression. Second, the interval between forks decreased significantly (from 38 ± 7.6 kb to 18 ± 4.5 kb; Fig. 2d), consistent with firing of dormant origins that usually are replicated passively19. Third, replication forks appeared to arrest or collapse at an elevated rate, as reflected by an excess of tracks labeled exclusively with IdU (Fig. 2e). As a further test, we measured the efficiency of fork recovery from an aphidicolin block, imposed between the IdU and CldU pulses. Over 30% of forks failed to restart under these conditions (Fig. 2e), thus explaining the sensitivity of DCC1Δ/Δ cells to replication inhibitors.
Figure 2. RFCCtf18 controls global replication dynamics and promotes fork reactivation after genotoxic stress.
a, Cells were transiently exposed to hydroxyurea (HU) or aphidicolin (Aph) and monitored for growth during subsequent passages. b, Single-molecule analysis of replication fork dynamics in DCC1+/Δ and DCC1Δ/Δ cells. Fork velocities (c) and distances between forks (d) were measured from 50 tracks per sample. e, Cells were either labeled with IdU and CldU as above (left columns), or treated with 10 µM aphidicolin for 2 hours after the IdU pulse, then washed and pulsed with CldU (right columns).
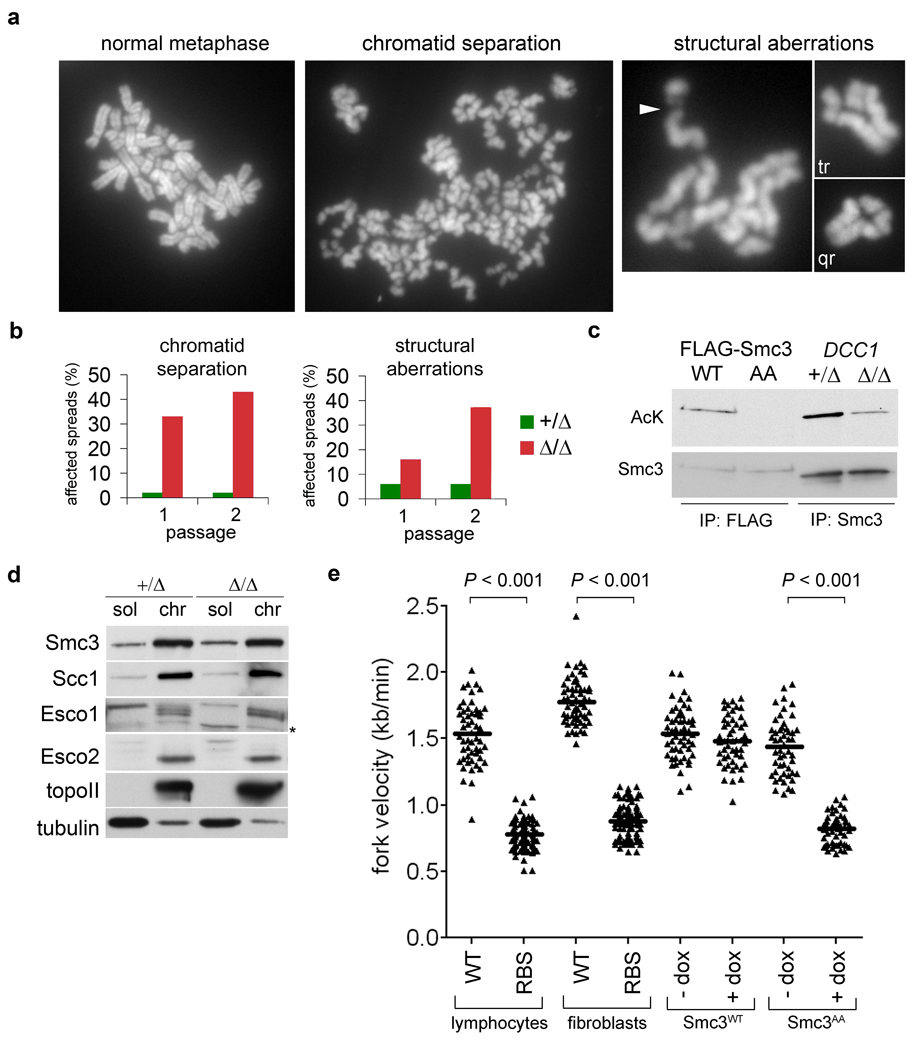
The role of the RFCCtf18 complex in cohesion establishment was also examined. As senescing cells are excluded from metaphase spreads, we abrogated this p53- and Rb-mediated response via expression of human papillomavirus E6 and E720. Bypassing senescence did not alter the requirement for RFCCtf18 in sustaining high fork velocity, origin dormancy, and replication restart (Supplementary Fig. 3), but did reveal precocious chromatid separation in over 40% of DCC1Δ/Δ cells, as well as structural aberrations (Fig. 3a,b). A similar mixture of cohesion loss and chromosome fragility occurs in cells expressing non-acetylatable Smc3 (ref. 8), suggesting that this modification may depend on RFCCtf18. To test this idea, cohesin was immunopurified and analyzed by quantitative immunoblotting with antibodies specific for acetylated lysine. Smc3 acetylation was reduced by 70% in DCC1Δ/Δ cells (Fig. 3c), despite proper targeting of cohesin and the Eco1-related acetyltransferases Esco1 and Esco2 to chromatin (Fig. 3d). Thus, RFCCtf18 promotes cohesion at a later step than the Cdc7 kinase and other pre-replicative complex components, which are required for cohesin’s deposition onto interphase chromosomes in Xenopus21. These data are compatible with a model wherein Esco1 and Esco2 are stimulated to acetylate cohesin through physical interactions with RFCCtf18-loaded PCNA1, 22.
Figure 3. Smc3 acetylation and replication fork progression are interdependent.
a and b, Karyotypic analysis of E6- and E7-expressing DCC1+/Δ and DCC1Δ/Δ cells. Chromatid gaps (arrowhead), triradial (tr), and quadriradial (qr) chromosomes are highlighted. c, Cells in (a) were synchronized in S phase with thymidine. Chromatin-associated Smc3 was immunopurified and blotted for acetylated lysine or total Smc3. Wildtype (WT) and non-acetylatable (AA) Smc3 variants8 were included as controls. d, Soluble and chromatin fractions from S-phase cells were analyzed by immunoblotting. e, Fork velocities were determined in cells from normal donors and RBS patients, and in cells expressing Smc3WT or Smc3AA from a doxycycline-inducible promoter.
Our results show that in the absence of RFCCtf18, replication fork processivity and Smc3 acetylation are both significantly diminished. We next asked whether these processes occur independently downstream of RFCCtf18 or are themselves linked. To this end, we analyzed replication dynamics in cells derived from patients with Roberts Syndrome/SC phocomelia (RBS), a severe hereditary disorder in which ESCO2 is biallelically mutated, resulting in weakened cohesion between sister centromeres11. Fork velocities were markedly reduced in cells from two unrelated RBS patients (Fig. 3e) and accompanied by spontaneous accumulation of DNA damage (Supplementary Fig. 4). Fork slowing was also observed in HeLa cells depleted of Esco2 or Esco1 via RNA interference (RNAi; see Fig. 4e). To test the contribution of cohesin acetylation more specifically, we analyzed replication forks in cells expressing wildtype (Smc3WT) or non-acetylatable Smc3 (Smc3AA) from a tetracycline-regulated promoter. Expression of Smc3AA alone reduced fork speeds from 1.5 ± 0.2 kb/min to 0.8 ± 0.1 kb/min (Fig. 3e), indicating that cohesin acetylation per se is required for processive DNA synthesis. In contrast, the expression of cohesin-regulated genes was insensitive to these manipulations (Supplementary Fig. 5 and Supplementary Table 1). We conclude that the acetylated form of cohesin specifically regulates replication fork progression, but is not required for transcriptional insulation.
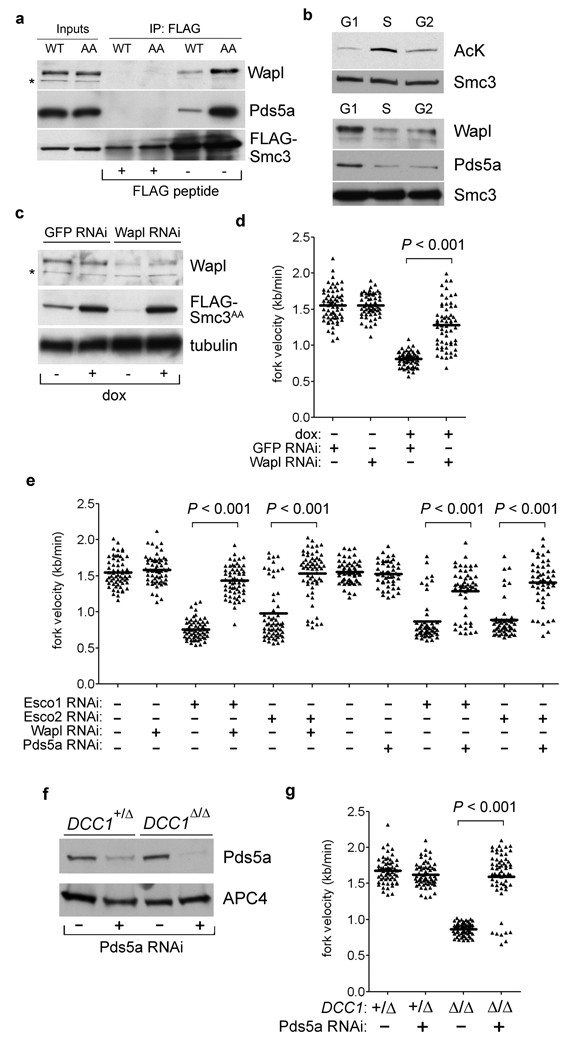
Figure 4. Processive fork movement requires acetylation-mediated dissociation of Pds5a and Wapl.
a, Smc3 variants were immunopurified and examined for Wapl and Pds5a binding. b, Cells were synchronized by serum withdrawal (G1 phase) or thymidine block and release (S and G2). Acetylation of chromatin-bound Smc3 (top) and association with Wapl and Pds5a (bottom) were determined as in Fig. 3c and 4a. c and d, Wapl depletion rescues fork progression in Smc3AA-expressing cells. e, Downregulating Wapl or Pds5a reverses fork slowing in Esco1- and Esco2-depleted cells. f and g, Pds5a depletion rescues fork progression in the absence of RFCCtf18.
How Smc3 acetylation affects cohesin biochemically is not well understood, but genetic evidence indicates that this modification acts in opposition to Rad61 (also called Wapl) and Pds5, which together bind cohesin and regulate its association with chromosomes10, 12, 13, 23–26. Strikingly, non-acetylatable human Smc3 copurified with increased amounts of both Wapl and Pds5a (Fig. 4a and Supplementary Fig. 6). Furthermore, this interaction was negatively correlated with Smc3 acetylation in synchronized cells (Fig. 4b and Supplementary Fig. 7). To determine if the excess stability of this interaction inhibits fork movement, RNAi was used to deplete Wapl and Pds5a in cells expressing Smc3AA (Fig. 4c,d) or concomitantly depleted of Esco1 or Esco2 (Fig. 4e and Supplementary Fig. 8). Strikingly, downregulating Wapl or Pds5a rescued fork progression in all instances but had no effect on its own, arguing that ring opening by the Wapl/Pds5a complex is not generally required for replication forks to move through cohesin-binding sites. Pds5a depletion also allowed forks in DCC1-null cells to move at normal rates (Fig. 4f–g). We conclude that the RFCCtf18 complex speeds chromosomal replication mainly by stimulating the acetylation-dependent remodeling of cohesin rings, rather than by activating DNA polymerases directly, at least in the presence of the replicative RFC.
Our analysis provides new and unexpected insights into the relationship between DNA replication and cohesion establishment. Previous studies did not detect an S phase delay in yeast ctf18 or eco1 mutants, leading to the conclusion that chromosomes are replicated in a similar fashion whether or not cohesion is established between nascent sister DNAs. In contrast, by applying sensitive single-molecule assays, we find that replication dynamics in human cells are highly attuned to the acetylation of Smc3 and possibly other substrates by Esco1 and Esco2. Notably, the fork’s response to deficiencies in cohesin acetylation is indistinguishable from the consequences of physically blocking the fork by other routes (e.g., nucleotide depletion, polymerase inhibition, or DNA alkylation), suggesting that a primary role of this modification is to switch cohesin from a configuration that obstructs the fork to one that permits its advancement. Furthermore, our genetic and biochemical data imply that this switch involves cohesin’s regulatory cofactors Pds5a and Wapl, rather than an intrinsic biophysical property of acetylated cohesin itself. To account for these observations, we propose that Wapl and Pds5a prevent the replication fork from sliding through the ring and/or interfere with cohesin’s stable embrace of the nascent sister DNAs (Supplementary Fig. 9). The possibility of an occlusive conformation is already suggested by Pds5’s bivalent interactions with opposite sides of the ring27, as well as flexion of the Smc1 and Smc3 coiled coils28. To neutralize this, the fork-associated RFCCtf18 complexes stimulate Esco1 and Esco2 to acetylate Smc3, causing the Wapl/Pds5a complex to dissociate. Cohesin could then adopt an open conformation that allows the fork to advance while stably trapping the nascent sister DNAs within the ring. This model would also explain the “anti-establishment” activities ascribed to Wapl and Pds5a10, 23, 25, 26, since hindrance of fork passage through the cohesin ring would not only slow chain elongation but also interfere with entrapment of sister chromatids. In addition to generating cohesion, entrapment may contribute positively to fork processivity by facilitating sister chromatid-dependent pathways of replication restart29.
Weakened centromere cohesion is a diagnostic hallmark of RBS11. However, we show here that this disorder is also associated with widespread slowing of the replication fork, presumably due to unopposed action of the Wapl/Pds5a complex on cohesin. Whether this chronic alteration in replication dynamics contributes to the abnormalities in RBS patients is unclear. However, DNA replication in general is known to be epigenetically coordinated with development. Whereas active origins are closely spaced during the initial cell cycles after fertilization, their density falls more than 10-fold during embryogenesis, resulting in a dramatic expansion of replicon size15. By impeding fork progression and reactivating dormant origins, defects in cohesion establishment may disrupt this epigenetic switch, causing replication stress and premature senescence or apoptosis of progenitor cells during development. Further work with in vivo models of RBS will be needed to test this hypothesis. Reducing fork velocity also results in the packaging of chromosomal DNAs into smaller than normal chromatin loops19. This effect has been suggested to involve changes in the replication-dependent modification of factors associated with matrix attachment regions and boundary elements, a proposal compatible with the known properties of cohesin4, 5. Its acetylation may therefore be required not only to produce cohesion locally, but also to shape mammalian genomes more globally.
Methods Summary
To mutate the DCC1 locus, hTERT-RPE1 cells were infected with AAV vectors with homology to sequences flanking exon 2. Clones with correct gene replacements were identified by PCR and Southern blotting. Purified adenoviruses expressing Cre recombinase or β-galactosidase were obtained from Baylor College of Medicine Vector Development Laboratory and applied at a multiplicity of infection (MOI) of 200. Senescence bypass was accomplished through retroviral expression of human papillomavirus E6 and E7 oncoproteins. RNAi was initiated by transfection of synthetic siRNA duplexes via Oligofectamine (Invitrogen), followed by analysis of protein depletion, replication fork movement, and sister chromatid cohesion 48 to 72 hours later. Techniques for preparing and analyzing IdU- and CldU-labeled DNA fibers were adapted from the procedures of Jackson and Pombo30. Images were acquired on a TE2000 microscope fitted with a 100x/1.4NA oil objective and analyzed using the NIS-Elements software package (Nikon Instruments).
Full methods and any associated references are available in the online version of the paper at www.nature.com/nature.
Supplementary Material
Acknowledgements
We thank D. Galloway, J. Hurwitz, J. Petrini, H. Nakao, and H. Zou for reagents, and S. Keeney, K. Marians, and J. Petrini for discussions and reading of the manuscript. We thank A. Viale, M. Hassimi, and the MSKCC Genomics Core Laboratory for assistance with microarray experiments. This work was supported by a grant from the National Institutes of Health and a Pew Scholar in the Biochemical Sciences award to P.V.J.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature. A figure summarizing the main result of this paper is also included as SI.
Author Contributions. M.-E.T. and P.V.J. designed experiments, M.-E.T., R.S., and S.R. performed experiments, J.Q. contributed reagents, M.-E.T., R.S., S.R., and P.V.J. analyzed the data, and M.-E.T. and P.V.J. wrote the paper.
Author Information. Reprints and permissions information is available at www.nature.com/reprints.
References
- 1.Lengronne A, et al. Establishment of sister chromatid cohesion at the S. cerevisiae replication fork. Mol Cell. 2006;23:787–799. doi: 10.1016/j.molcel.2006.08.018. [DOI] [PubMed] [Google Scholar]
- 2.Gullerova M, Proudfoot NJ. Cohesin complex promotes transcriptional termination between convergent genes in S. pombe. Cell. 2008;132:983–995. doi: 10.1016/j.cell.2008.02.040. [DOI] [PubMed] [Google Scholar]
- 3.Rubio ED, et al. CTCF physically links cohesin to chromatin. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:8309–8314. doi: 10.1073/pnas.0801273105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Parelho V, et al. Cohesins functionally associate with CTCF on mammalian chromosome arms. Cell. 2008;132:422–433. doi: 10.1016/j.cell.2008.01.011. [DOI] [PubMed] [Google Scholar]
- 5.Wendt KS, et al. Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature. 2008;451:796–801. doi: 10.1038/nature06634. [DOI] [PubMed] [Google Scholar]
- 6.Stedman W, et al. Cohesins localize with CTCF at the KSHV latency control region and at cellular c-myc and H19/Igf2 insulators. Embo J. 2008;27:654–666. doi: 10.1038/emboj.2008.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bermudez VP, et al. The alternative Ctf18-Dcc1-Ctf8-replication factor C complex required for sister chromatid cohesion loads proliferating cell nuclear antigen onto DNA. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:10237–10242. doi: 10.1073/pnas.1434308100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang J, et al. Acetylation of Smc3 by Eco1 is required for S phase sister chromatid cohesion in both human and yeast. Mol Cell. 2008;31:143–151. doi: 10.1016/j.molcel.2008.06.006. [DOI] [PubMed] [Google Scholar]
- 9.Unal E, et al. A molecular determinant for the establishment of sister chromatid cohesion. Science. 2008;321:566–569. doi: 10.1126/science.1157880. [DOI] [PubMed] [Google Scholar]
- 10.Ben-Shahar TR, et al. Eco1-dependent cohesin acetylation during establishment of sister chromatid cohesion. Science. 2008;321:563–566. doi: 10.1126/science.1157774. [DOI] [PubMed] [Google Scholar]
- 11.Vega H, et al. Roberts syndrome is caused by mutations in ESCO2, a human homolog of yeast ECO1 that is essential for the establishment of sister chromatid cohesion. Nat Genet. 2005;37:468–470. doi: 10.1038/ng1548. [DOI] [PubMed] [Google Scholar]
- 12.Gandhi R, Gillespie PJ, Hirano T. Human Wapl is a cohesin-binding protein that promotes sister-chromatid resolution in mitotic prophase. Curr Biol. 2006;16:2406–2417. doi: 10.1016/j.cub.2006.10.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kueng S, et al. Wapl controls the dynamic association of cohesin with chromatin. Cell. 2006;127:955–967. doi: 10.1016/j.cell.2006.09.040. [DOI] [PubMed] [Google Scholar]
- 14.Lengronne A, et al. Cohesin relocation from sites of chromosomal loading to places of convergent transcription. Nature. 2004;430:573–578. doi: 10.1038/nature02742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Walter J, Newport JW. Regulation of replicon size in Xenopus egg extracts. Science. 1997;275:993–995. doi: 10.1126/science.275.5302.993. [DOI] [PubMed] [Google Scholar]
- 16.Deng Y, Chan SS, Chang S. Telomere dysfunction and tumour suppression: the senescence connection. Nat Rev Cancer. 2008;8:450–458. doi: 10.1038/nrc2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang X, et al. Rad17 phosphorylation is required for claspin recruitment and Chk1 activation in response to replication stress. Mol Cell. 2006;23:331–341. doi: 10.1016/j.molcel.2006.06.022. [DOI] [PubMed] [Google Scholar]
- 18.Heller RC, Marians KJ. Replisome assembly and the direct restart of stalled replication forks. Nat Rev Mol Cell Biol. 2006;7:932–943. doi: 10.1038/nrm2058. [DOI] [PubMed] [Google Scholar]
- 19.Courbet S, et al. Replication fork movement sets chromatin loop size and origin choice in mammalian cells. Nature. 2008;455:557–560. doi: 10.1038/nature07233. [DOI] [PubMed] [Google Scholar]
- 20.Shay JW, Pereira-Smith OM, Wright WE. A role for both RB and p53 in the regulation of human cellular senescence. Exp Cell Res. 1991;196:33–39. doi: 10.1016/0014-4827(91)90453-2. [DOI] [PubMed] [Google Scholar]
- 21.Takahashi TS, Basu A, Bermudez V, Hurwitz J, Walter JC. Cdc7-Drf1 kinase links chromosome cohesion to the initiation of DNA replication in Xenopus egg extracts. Genes & development. 2008;22:1894–1905. doi: 10.1101/gad.1683308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moldovan GL, Pfander B, Jentsch S. PCNA controls establishment of sister chromatid cohesion during S phase. Mol Cell. 2006;23:723–732. doi: 10.1016/j.molcel.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 23.Tanaka K, Hao Z, Kai M, Okayama H. Establishment and maintenance of sister chromatid cohesion in fission yeast by a unique mechanism. Embo J. 2001;20:5779–5790. doi: 10.1093/emboj/20.20.5779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Losada A, Yokochi T, Hirano T. Functional contribution of Pds5 to cohesinmediated cohesion in human cells and Xenopus egg extracts. J Cell Sci. 2005;118:2133–2141. doi: 10.1242/jcs.02355. [DOI] [PubMed] [Google Scholar]
- 25.Rowland BD, et al. Building sister chromatid cohesion: Smc3 acetylation counteracts an antiestablishment activity. Mol Cell. 2009;33:763–774. doi: 10.1016/j.molcel.2009.02.028. [DOI] [PubMed] [Google Scholar]
- 26.Sutani T, Kawaguchi T, Kanno R, Itoh T, Shirahige K. Budding Yeast Wpl1(Rad61)-Pds5 Complex Counteracts Sister Chromatid Cohesion-Establishing Reaction. Curr Biol. 2009;19:492–497. doi: 10.1016/j.cub.2009.01.062. [DOI] [PubMed] [Google Scholar]
- 27.McIntyre J, et al. In vivo analysis of cohesin architecture using FRET in the budding yeast Saccharomyces cerevisiae. Embo J. 2007;26:3783–3793. doi: 10.1038/sj.emboj.7601793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sakai A, Hizume K, Sutani T, Takeyasu K, Yanagida M. Condensin but not cohesin SMC heterodimer induces DNA reannealing through protein-protein assembly. Embo J. 2003;22:2764–2775. doi: 10.1093/emboj/cdg247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Branzei D, Foiani M. Regulation of DNA repair throughout the cell cycle. Nat Rev Mol Cell Biol. 2008;9:297–308. doi: 10.1038/nrm2351. [DOI] [PubMed] [Google Scholar]
- 30.Jackson DA, Pombo A. Replicon clusters are stable units of chromosome structure: evidence that nuclear organization contributes to the efficient activation and propagation of S phase in human cells. The Journal of cell biology. 1998;140:1285–1295. doi: 10.1083/jcb.140.6.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.