Abstract
The scaffold protein Ste5 is required to properly direct signaling through the yeast mating pathway to the mitogen-activated protein kinase (MAPK), Fus3. Scaffolds are thought to function by tethering kinase and substrate in proximity. We find, however, that the previously identified Fus3-binding site on Ste5 is not required for signaling, suggesting an alternative mechanism controls Fus3’s activation by the MAPKK Ste7. Reconstituting MAPK signaling in vitro, we find that Fus3 is an intrinsically poor substrate for Ste7, although the related filamentation MAPK, Kss1, is an excellent substrate. We identify and structurally characterize a novel domain in Ste5 that catalytically unlocks Fus3 for phosphorylation by Ste7. This domain selectively increases the kcat of Ste7➔Fus3 phosphorylation by 5000-fold but has no effect on Ste7➔Kss1 phosphorylation. The dual requirement for both Ste7 and this Ste5 domain in Fus3 activation explains why Fus3 is selectively activated by the mating pathway, and not by other pathways that also utilize Ste7.
INTRODUCTION
Living cells receive vast amounts of environmental information, and a central question is how the cell’s system of signal transduction proteins is able to specifically process this information. This problem is particularly acute given that many closely related molecules (e.g. kinases, phosphatases, etc.) are involved in diverse, functionally distinct signaling pathways. An emerging paradigm is that, in many cases, signaling pathways are organized by scaffold proteins. Scaffolds are proteins that interact with multiple members of a pathway and are thought to function as “wiring” elements that by tethering pathway components into complexes and localizing them to specific sites in the cell, direct the flow of signaling information. Scaffolds are proposed to both enhance interactions between the correct signaling proteins and to insulate them from interactions with competing proteins (Bhattacharyya et al., 2006a; Bhattacharyya et al., 2006b; Burack et al., 2002; Burack and Shaw, 2000).
One of the first identified examples of a signaling scaffold is the Ste5 protein from Saccharomyces cerevisiae, which plays an essential role in signal transmission through the yeast mating pathway. When yeast are stimulated by mating pheromone from the opposite mating type, signal is transmitted from the mating receptor (Ste2) via a heterotrimeric G-protein (Gpa1, Ste4 and Ste18) to a mitogen-activated protein (MAP) kinase cascade. MAP kinase cascades are composed of three kinases that successively phosphorylate and activate one another: signal passes from a MAP kinase kinase kinase (MAPKKK) to a MAP kinase kinase (MAPKK) and finally to a MAP kinase (MAPK). In the mating pathway, signal is transmitted from the MAPKKK Ste11 to the MAPKK Ste7 to the MAPK Fus3. The Ste5 scaffold, although it has no catalytic domains (eg. kinase domains), is required for the mating response. Ste5 was initially identified as a scaffold protein because, by yeast two-hybrid assays, it was shown to have binding sites for all three MAPK cascade members (Ste11, Ste7, and Fus3) (Choi et al., 1994) and the Gβ protein, Ste4 (Whiteway et al., 1995). Interaction with Ste4 localizes the Ste5 complex to the membrane upon stimulation, allowing Ste11 to be activated by a membrane-localized (PAK) kinase, Ste20. Additionally, interaction of Ste5 with the kinases in the cascade is thought to promote their successive phosphorylation.
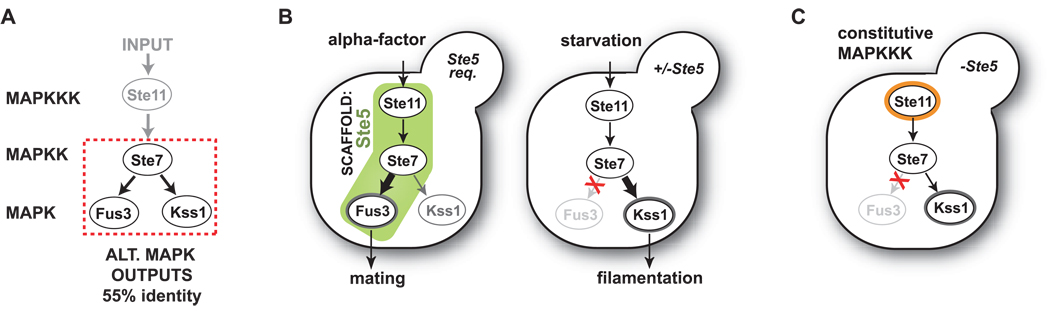
The need for robust mechanisms for controlling signaling specificity is particularly important for the mating pathway because of the potential for cross-signaling with other related MAPK pathways that use overlapping signaling components. For example, the filamentous growth pathway, which is activated by nitrogen starvation, requires kinases shared with the mating pathway: the MAPKKK Ste11 and the MAPKK Ste7 (although it does not require the scaffold Ste5). During the mating respose, signaling to Ste7 is primarily transmitted to the MAPK Fus3, while in the filamentation pathway signaling is transmitted to the MAPK Kss1. Here we focus on the critical question of how activated Ste7 chooses between the two MAP kinases, Fus3 and Kss1, which are 55% identical (Fig. 1A). Why does Ste7 that is activated by pheromone stimulation phosphorylate Fus3, whereas Ste7 that is activated by nitrogen starvation phosphorylate only Kss1? What is the role of the Ste5 scaffold in this specificity choice?
Figure 1. Ste5 scaffold protein is required for mating pathway signaling.
(A) The MAPKKK Ste11 and MAPKK Ste7 function in both the mating and filamentation pathways in yeast. Ste7 must select the appropriate MAPK to phosphorylate in response to input (Fus3 for α-factor, and Kss1 in response starvation). (B) During mating, stimulation with α-factor leads primarily to phosphorylation of Fus3. This reaction requires the scaffold protein Ste5. Starvation input specifically induces the filamentation response through phosphorylation of Kss1. The Ste5 scaffold is not required for filamentation. (C) Expression of a constitutively active allele of MAPKKK Ste11 in a strain lacking Ste5 results only in Kss1 phosphorylation (both Kss1 and Fus3 phosphorylation are observed in strains with Ste5), further indicating that the Ste5 scaffold is required, in vivo, for Ste11➔Ste7➔Fus3 signaling (Flatauer et al., 2005).
Despite the importance of Ste5 as a canonical example of a scaffold protein, little is understood about the biochemical mechanisms that scaffolds use to regulate MAPK signaling specificity. The simplest model for how a scaffold might promote phosphorylation of one substrate versus another is through tethering – by increasing the proximity and effective concentration of components in the scaffold complex. Tethering, appears to be important for certain key aspects of Ste5 function: mutation of the binding sites for the Ste11 and Ste7 kinases disrupts signal transmission, while re-recruitment of these proteins to the Ste5 complex via heterologous engineered protein-protein interactions or covalent fusion can partially rescue signaling (Harris et al., 2001; Park et al., 2003).
The mechanism by which Ste5 directs signaling from the MAPKK Ste7 to the MAPK Fus3, however, is far less clear. Is the scaffold needed to colocalize these kinases or does it play some other role? Previous work identified and characterized a binding site for Fus3 within Ste5. This ~30 amino acid peptide (288–316) binds Fus3 with an affinity of 1µM, and it stimulates partial Fus3 autophosphorylation (it promotes one of two phosphorylation events required for Fus3 activation) (Bhattacharyya et al., 2006a). Surprisingly, however, mutation of this Fus3 binding site does not block mating but actually increases mating output (as measured by transcription), suggesting that this site plays more of a tuning role, modulating signaling dynamics (Bhattacharyya et al., 2006a). Nonetheless, the scaffold as a whole is still absolutely required for signaling to Fus3. Thus, it appears that there may be another site in Ste5 that controls Fus3 activation and that the scaffold may be playing a more active or catalytic role in controlling signal transmission to this MAP kinase.
Here we have purified components of the mating and filamentous growth MAP kinase pathways (Ste7, Fus3, Kss1, and Ste5) in order to understand the role of scaffolds in specifying in MAPKK➔MAPK signal transmission. We find that Fus3 is intrinsically a poor substrate for activated Ste7, while Kss1 is intrinsically a very good substrate. A ~200 residue segment of Ste5, however, is sufficient to permit Ste7 phosphorylation of Fus3 but has no effect on Kss1 phosphorylation. This Ste5 fragment is distinct from the previously identified Fus3 binding site, and crystallographic studies show that it is an independently folding domain which we refer to as Ste5-ms (minimal scaffold). The Ste5-ms domain binds tightly to Ste7, but only very weakly to Fus3. However, mutational and kinetic studies show that the Ste5-ms fragment can catalytically unlock the Fus3 MAPK so that it is now a good substrate for Ste7. This domain specifically increases the kcat for the Ste7➔ Fus3 reaction by ~5000-fold, while it has no effect on the kcat or KM of the Ste7➔ Kss1 reaction. Fus3 appears to have evolved a structure that is “locked” to prevent stray activation by isolated forms of Ste7 (generated by non-mating inputs). Phosphorylation of Fus3 occurs only in the combined presence of Ste7 and Ste5, and this mechanism explains why Fus3 is only activated by mating input.
RESULTS
Fus3 is an intrinsically poor substrate for Ste7 that requires Ste5 as a co-activator
The MAPKK, Ste7, is used in two distinct yeast MAPK pathways, the mating and filamentous growth pathways. When stimulated by α-factor (pheromone input for the mating pathway), Ste7 primarily activates the mating-specific MAPK Fus3. However, when stimulated by starvation (input for the filamentation or haploid invasive growth pathway), Ste7 activates Kss1. How Ste7 makes the appropriate input-dependent substrate choice between Fus3 and Kss1 (Fig. 1A) is a challenging question, as the two alternative MAP kinases are very closely related (55% identity; >70% similarity, Supp. Fig. 8A). Previous genetic work indicates that the Ste5 scaffold is required to direct signal from a constitutively active MAPKKK Ste11 through Ste7 to the mating MAPK Fus3 (Fig. 1C) (Flatauer et al., 2005). Could Ste5 be playing a direct role in the selective activation of Fus3 by Ste7? To investigate the biochemical requirements for Ste7➔MAPK specificity, we purified key components and reconstituted this pathway step in vitro.
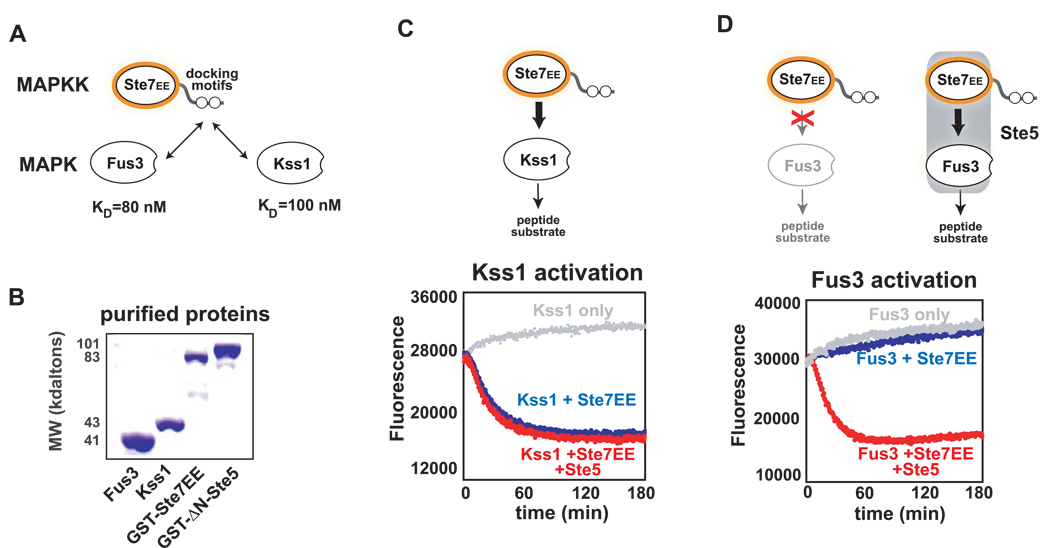
Previous studies have shown that in addition to any scaffold (Ste5) contributions, Ste7➔Fus3 phosphorylation requires direct docking interactions between the two proteins. Ste7 has two MAPK docking motifs on its N-terminus. These are ~10 residue peptide motifs (consensus motif: [RK][RK]X(4–6)LxL) that are found to mediate functional interactions between MAPKs and a variety of their regulators and substrates (Remenyi et al., 2006). At least one of these docking motifs is required for phosphorylation of either Fus3 or Kss1 by Ste7 (Bhattacharyya et al., 2006a; Remenyi et al., 2005). However, the docking sites in Ste7 cannot be sufficient to distinguish between Fus3 and Kss1, since they bind to both Fus3 and Kss1 with roughly equal affinity (stronger site KD ~ 100nM) (Remenyi et al., 2005).
We first investigated whether Ste7 could activate the MAPKs Fus3 and Kss1 in vitro. We expressed and purified the following recombinant proteins: Fus3, Kss1, a constitutively active form of Ste7 (Ste7EE – bearing S359E and T363E phosphomimic mutations in the Ste7 activation loop) (Maleri et al., 2004), and ΔN-Ste5 (Ste5, with an 279 residue N-terminal deletion, which makes the protein soluble and biochemically tractable) (Fig. 2B). The ability of Ste7EE to activate Fus3 or Kss1 was measured using a fluorescence-based MAPK assay (Trulight kinase assay, Calbiochem -- see Supp Fig. 2). We found that Ste7EE rapidly activated Kss1, either in the presence or absence of purified Ste5 (all components at 50nM) (Fig. 2C). In contrast, Ste7EE cannot activate Fus3, demonstrating that Fus3 is an intrinsically weak substrate for Ste7 (Fig. 2D). If, however, the Ste5 scaffold is added, Ste7 rapidly phosphorylates Fus3 at a rate comparable to Kss1 (Fig. 2D). These results indicate that Fus3 is inherently a poor substrate for Ste7, but that Ste5 can serve as a co-activator to permit efficient Ste7➔Fus3 phosphorylation.
Figure 2. Fus3 is intrinsically a poor substrate for Ste7, unless the Ste5 scaffold is present.
(A) Fus3 and Kss1 both bind tightly to docking motifs (D-motifs) on Ste7 (KD ~100nM for each MAPK). (B) Coomassie stained gel showing purified components of the mating and filamentation MAPK pathways. (C & D) Activation of Kss1 and Fus3 by Ste7EE in vitro measured using the Trulight kinase assay - in which phosphorylation of a MAPK-specific labeled peptide substrate results in a decrease in fluorescence over time (the peptide quenches signal of a sensor bead coated with fluorescent polymers) (See Supp. Fig. 2A–C). 50nM of each protein was used in these assays. Ste7EE rapidly activates Kss1, and addition of the Ste5 scaffold has no impact on the reaction. (D) Fus3 cannot be activated by Ste7EE, unless ΔN-Ste5 is added. These results demonstrate that Fus3 is intrinsically a very poor substrate for Ste7, and that Ste5 is a required co-activator in Ste7➔Fus3 phosphorylation
A novel domain in Ste5 is required for Fus3 phosphorylation by Ste7
How does the Ste5 scaffold permit Ste7➔Fus3 phosphorylation? The simplest model is that a protein scaffold like Ste5 acts as a tethering or co-localization device that enhances the interaction of proteins that interact poorly on their own (Fig. 3A). Consistent with a tethering model, mutagenesis of Ste5 and a prior yeast-two hybrid study have identified binding sites for both Ste7 and Fus3 within the Ste5 scaffold (Choi et al., 1994; Inouye et al., 1997a) (Fig. 3B). Conversely, two other results argue strongly that Ste5 is not acting as a simple tether to promote Ste7➔Fus3 phosphorylation. First, as described above, Fus3 can already bind tightly to Ste7 without the scaffold due to the MAPK docking motifs found at the N-terminus of Ste7 (Bardwell et al., 1996; Remenyi et al., 2005). Second, we show here that the previously identified Fus3 binding site in Ste5 (residues 288–316 in Ste5) is not required to promote the Ste7➔Fus3 reaction. A variant of Ste5, in which this site is mutated so that it no longer binds Fus3 (Ste5-ND) (Bhattacharyya et al., 2006a), is indistinguishable from the wild-type protein in its ability to promote the Ste7➔Fus3 reaction in vitro (Fig. 3B, C). In contrast when the MAPK docking sites in Ste7 are mutated, Ste7 cannot phosphorylate either Fus3 or Kss1, both in the presence or absence of Ste5 (Fig. 3E). Furthermore, when the previously identified Fus3 binding domain in Ste5 is mutated in vivo, mating output upon alpha-factor stimulation actually increases (Bhattacharyya et al., 2006a). Together, these results are consistent with a model in which this previously characterized Fus3 binding motif does not play a role in promoting the main flow of signaling information from the MAPKK Ste7 to the MAPK Fus3, but rather plays a modulatory role in tuning the quantitative and dynamic output of the pathway.
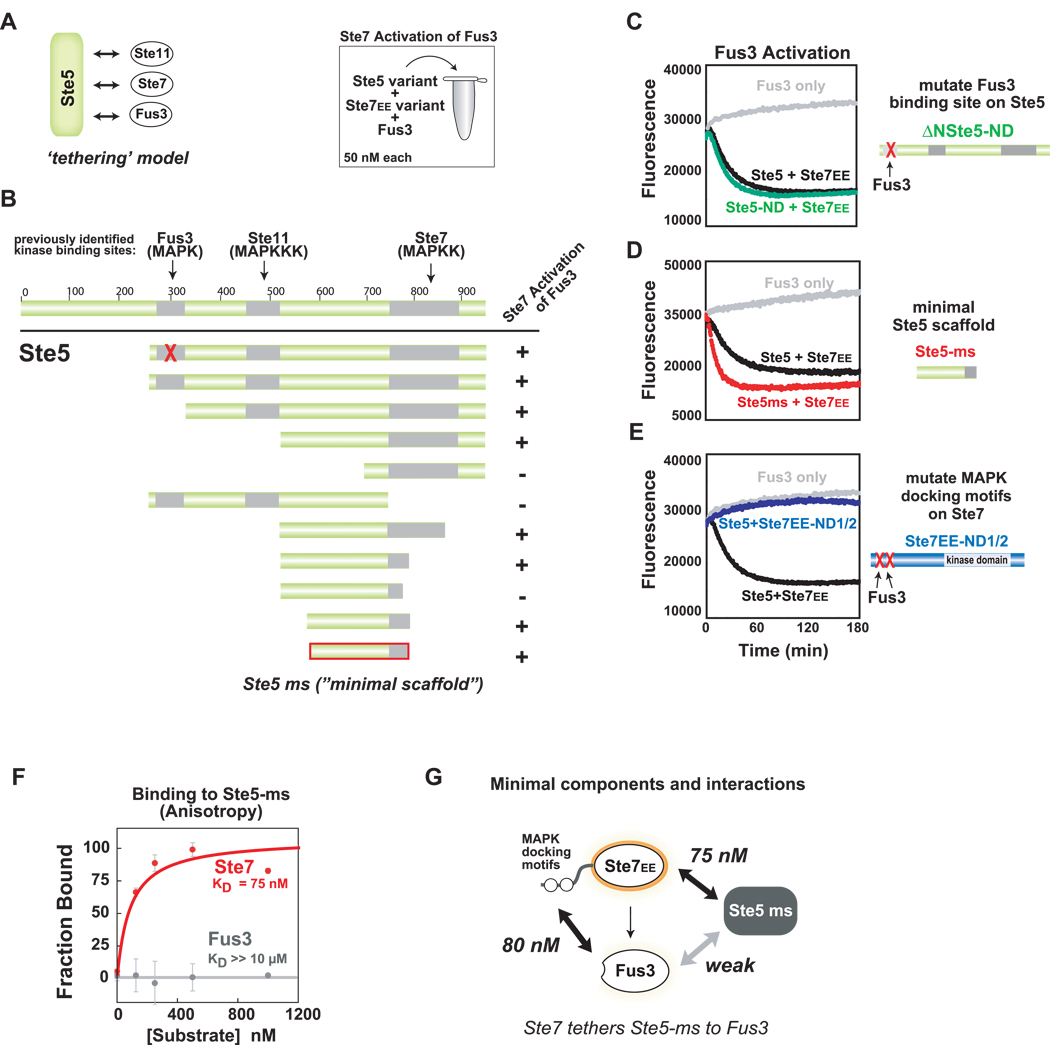
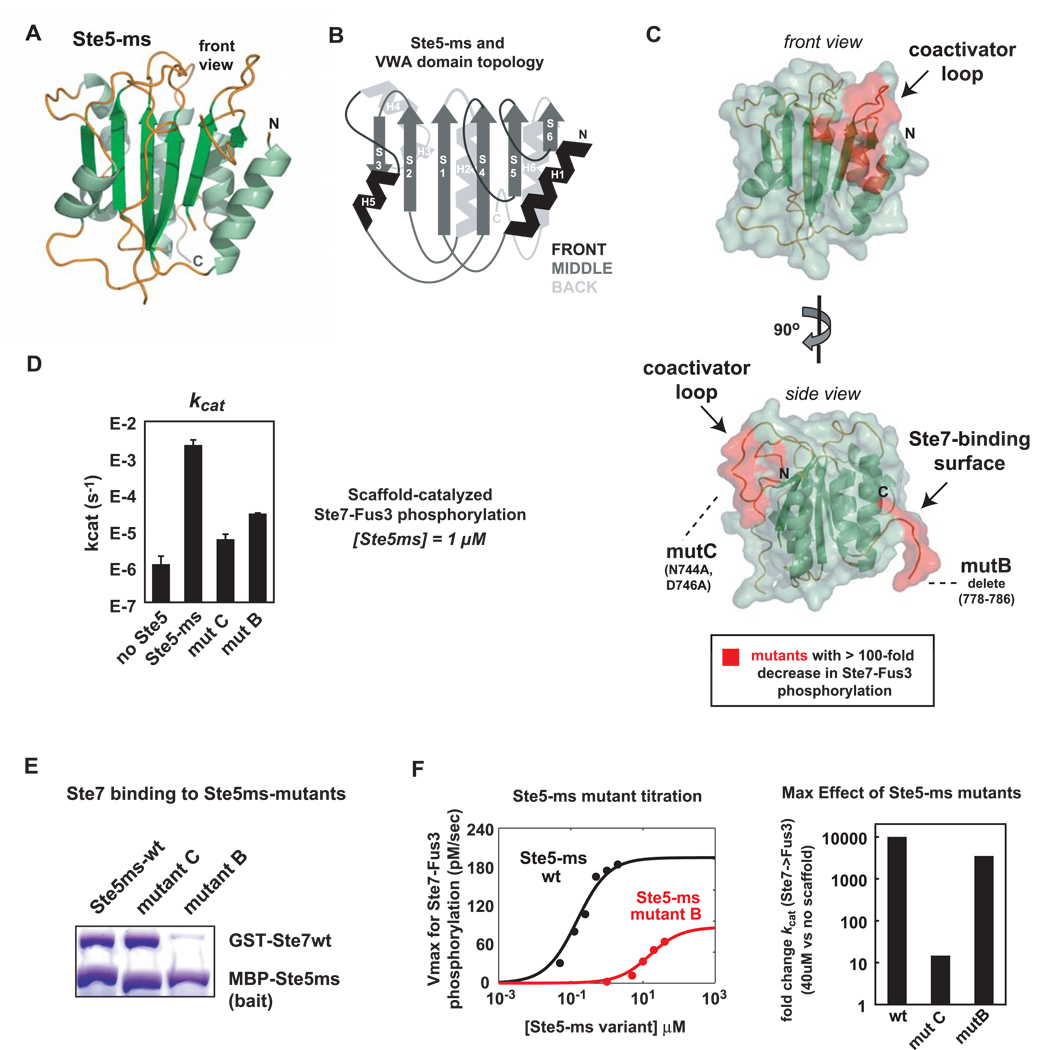
Figure 3. Ste5 contains a novel domain required for Ste7➔Fus3 phosphorylation.
(A) Ste5 is a large protein (917aa) that contains previously identified binding sites for the mating pathway kinases. Canonical tethering model proposes that Ste5 co-localizes three kinases in the mating pathway (Ste11, Ste7, Fus3) to promote signaling. (B) Deletion mapping identifies minimal region of Ste5 required for Ste7EE➔Fus3 phosphorylation in vitro. As in Figure 2, Trulight assay was used to measure Fus3 activation by Ste7EE. Amino acids 593–786 of Ste5 define the ‘minimal scaffold’ domain (Ste5-ms) sufficient to promote Ste7➔Fus3 phosphorylation. (C) Confirmation that the Fus3-binding region (KD = 1 µM) in Ste5 is not required for phosphorylation of Fus3 by Ste7EE. ΔN-Ste5-ND (green curve) is a variant of ΔN-Ste5 (black curve) bearing a mutation in the Fus3 binding region that disrupts interaction with Fus3. For panels C-E all reaction components are at 50 nM. (D) Ste5-ms domain is as active at the larger scaffold protein (ΔN-Ste5). (E) MAPK docking motifs on Ste7EE (KD ~ 100nM) are necessary for Fus3 activation. Mutation of these sites disrupt Ste7➔ Fus3 phosphorylation, even in the presence of Ste5 (purple curve). (F) Ste5-ms binds to Ste7 but not to Fus3. Interactions were measured with fluorescence polarization (anisotropy) using 5nM of fluoroscein-labeled Ste5-ms. (G) Minimal interactions necessary for formation of the Ste5-Ste7-Fus3 signaling complex.
The finding that the Fus3 binding domain in Ste5 is not required for mating signaling in vitro led us to postulate that there might be a different region of Ste5 that promotes Ste7➔Fus3 phosphorylation. Therefore, we performed deletion analysis to search for the minimal region of Ste5 that was capable of permitting Ste7➔Fus3 phosphorylation (Fig. 3B). We identified a ~200 residue fragment of Ste5 (593–786) that was sufficient for promoting Fus3 phosphorylation. As will be discussed later, structural analysis revealed that this fragment forms a unique, independently folded domain. This domain was at least as active as a larger fragment of Ste5 (ΔN-Ste5) in promoting Ste7➔Fus3 phosphorylation (Fig. 3D) and we refer to the domain as the Ste5 minimal scaffold (Ste5-ms). The Ste5-ms domain lacks the previously identified Fus3 and Ste11-binding regions but contains part of the previously mapped Ste7-binding region (Inouye et al., 1997a).
In fluorescence polarization binding studies, we found that the Ste5-ms domain binds tightly to Ste7 (KD = 75nM) (Fig. 3F) but does not detectably bind to Fus3 (Fig. 3F). The lack of a strong Fus3 binding site in the Ste5-ms fragment argues against a mechanism in which this fragment is acting as a passive tether, simply increasing the effective concentration of Ste7 and Fus3. Although colocalization of the two proteins does appear to be necessary, it is the direct docking interaction between the MAPKK Ste7 and MAPK Fus3 that plays this role (Fig. 3G). Tethering of the two proteins (Ste7 and Fus3) together, however, does not seem to be sufficient for Fus3 activation. Thus the Ste5-ms domain must play a distinct functional role in promoting phosphorylation.
Ste5-ms selectively improves kcat for Fus3 but not other substrates
To understand precisely how the Ste5-ms domain contributes to Fus3 phosphorylation we performed quantitative kinetic analyses (Fig. 4). We measured the kcat and KM of Fus3 and Kss1 phosphorylation by Ste7EE both in the presence and absence of the Ste5-ms fragment (scaffold concentration 1 µM). To simplify the kinetic analysis, we used a variant of Ste7 with a single docking site (mutant Ste7EE-ND2 has the second, weaker docking motif removed). This Ste7EE variant has the same kcat as Ste7EE with both docking motifs, and behaves similarly in other assays both in vivo (Bhattacharyya et al., 2006a) and in vitro (data not shown) (Remenyi et al., 2005). As a substrate, we used a catalytically-dead allele of the MAPK Fus3 (K42R) in order to eliminate background autophosphorylation that is observed for the wild-type protein.
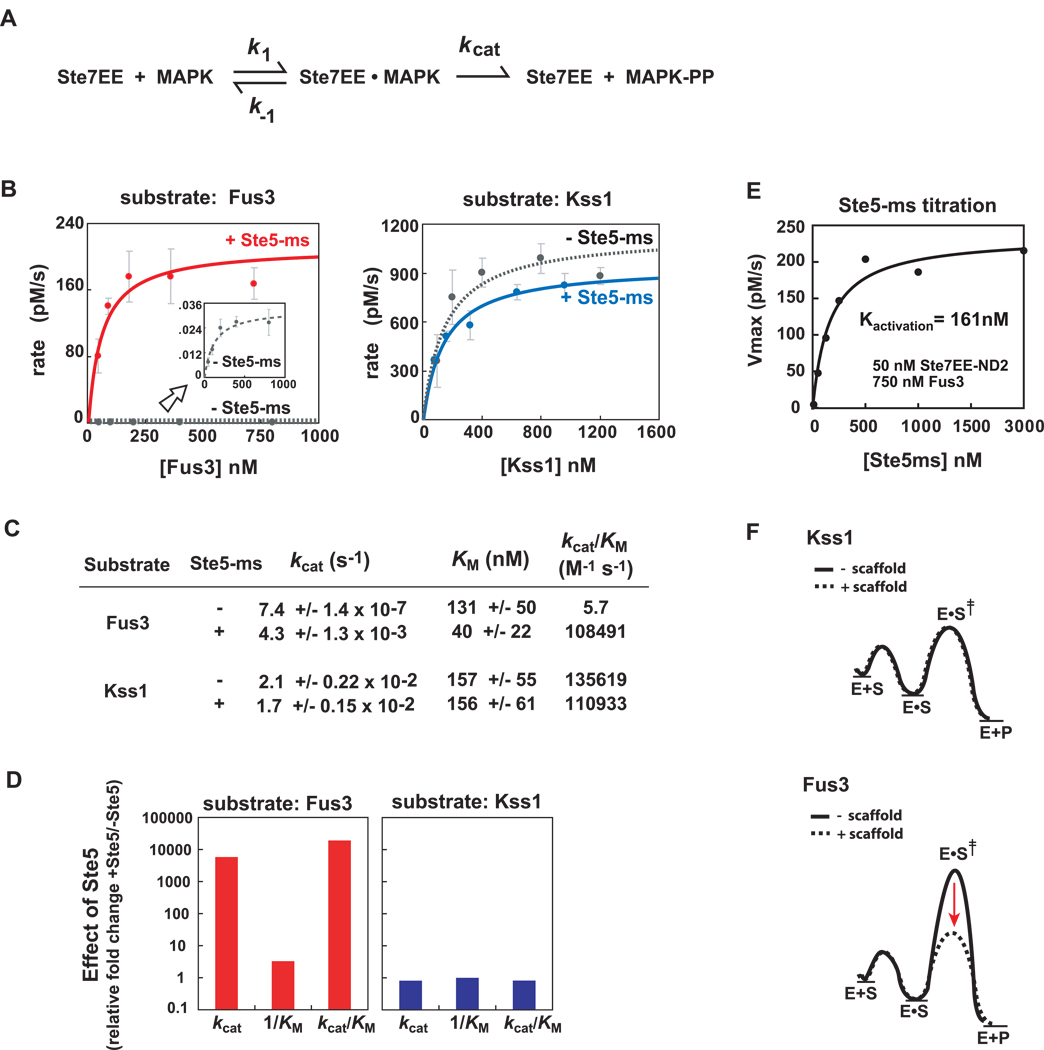
Figure 4. Ste5-ms domain selectively improves kcat (not KM) for the substrate, Fus3.
(A) Simple kinetic scheme for Ste7➔MAPK phosphorylation. Ste7EE enzyme converts substrate (MAPK) into doubly-phosphorylated product (MAPK-pp). Fus3 and Kss1 phosphorylation by Ste7EE was quantified using in vitro western blots with an anti-phospho p44/42 MAPK antibody (See Supp Fig. 1A–D and Supp Fig. 4). (B) Michaelis-Menten plots show Fus3 phosphorylation requires Ste5-ms, Kss1 phosphorylation does not. Ste7EE-ND2 (which contains only one MAPK docking motif, KD ~ 100nM), and Fus3-K42R (which is catalytically dead) were used to simplify the analyses. Kinase reactions contain 50nM Ste7EE-ND2, and a saturating concentration (1000nM, where appropriate) of Ste5-ms (see panel 4E). Fus3 activation by Ste7EE-ND2, in the absence Ste5-ms, is very slow but can be measured (inset graph). (C) Ste5-ms enhances the kcat of Ste7➔Fus3 phosphorylation by ~ 5000-fold, with negligible effect on KM. Ste5-ms has little or no effect on the kcat or KM of Kss1 phosphorylation. Overall specificity (kcat /KM) of Ste7 for Fus3 and Kss1 is comparable (~105 M−1 s−1). (D) Effect of Ste5 on Ste7➔MAPK phosphorylation reaction parameters, plotted as the fold-change in kcat, 1/KM, and kcatK/M for Fus3 and Kss1 activation by Ste7EE-ND2. Major effect of Ste5 is enhancement of the kcat for Fus3 phosphorylation. (E) Determination of the concentration of Ste5-ms required to drive Ste7➔Fus3 phosphorylation. 50nM Ste7EE was used along with a saturating amount of Fus3, (750nM, based on Fig. 4B). Rate of Fus3 activation reaches half-maximum at ~ 161nM Ste5-ms (+/− 60nM), which we infer is an apparent dissociation constant for the Ste7/Ste5-ms interaction. 1000 nM Ste5-ms, used in experiments described in panel 4B, represents a saturating concentration. (F) Reaction free energy diagram illustrating how Ste5-ms selectively lowers the energy of the transition state for Fus3 phosphorylation (dotted line).
These experiments show that the Ste5-ms domain enhances the kcat of Fus3 phosphorylation by Ste7EE by ~5000-fold, with little effect on the KM (Fig. 4C), further contradicting a potential tethering role for the Ste5-ms domain (which would be expected to lower the KM). This effect on kcat is highly substrate specific - the Ste5-ms domain has essentially no impact, positive or negative, on the kcat or KM of Kss1 phosphorylation by Ste7 (Fig. 4C, 4D). By varying the concentration of Ste5-ms in a reaction containing 50nM Ste7EE-ND2 and saturating (750nM) Fus3 we determined that the concentration of Ste5-ms required to maximally exert its effects is less than 1 µM. This titration experiment gives a midpoint of activation (Kactivation, an estimation of Ste7/Ste5-ms dissociation) of 161nM. This number is roughly the same as the KD for the Ste7-Ste5-ms interaction measured by anisotropy (75 nM), consistent with a model in which the Ste7/Ste5-ms complex is the catalytically competent complex.
A simple model for how the Ste5-ms domain kinetically modulates the Fus3 phosphorylation reaction is shown in the reaction coordinate free energy diagrams (Fig. 4F). Ste7EE is able to phosphorylate Kss1 efficiently in the presence or absence of Ste5 because it has a low transition state energy (E•S ‡). In contrast, Ste7EE is unable to phosphorylate Fus3, because it has a much higher transition state energy – Fus3 is an intrinsically poor substrate. The Ste5-ms scaffold domain is able to lower the energy of Fus3’s transition state, resulting in a higher kcat, thereby converting a very poor substrate into a good substrate, comparable to Kss1. Thus, the Ste5-ms domain is essentially serving as a substrate specific co-catalyst for Ste7➔Fus3 phosphorylation –a role that is conceptually similar to that of a cyclin which acts as a co-catalyst for the cyclindependent kinase (Cdk).
Ste5-ms is a folded domain with distinct surfaces for communicating with Ste7 and Fus3
To understand how the Ste5-ms domain might act as a substrate-specific co-catalyst, we determined the structure of the domain. We obtained crystals of the Ste5-ms fragment and solved the structure to 1.6 Å resolution (Fig. 5A, Supp. Fig. 5A, Supp. Table 2). This fragment adopts a well-ordered, independently folded structural domain. While primary sequence analysis (BLAST) failed to identify proteins clearly related to the Ste5-ms domain (outside of yeasts closely related to S. cerevisiae), the structural homology program DALI (Holm and Sander, 1996) showed that this domain shares the same fold as the von-Willebrand Type-A (VWA) domain found in extracellular matrix proteins and integrin receptors (Fig. 5B, Supp. Fig. 5B). Although many of known VWA domain proteins are extracellular, the most ancient VWA domains conserved across all eukaryotes appear to be intracellular proteins involved in diverse multiprotein assemblies (Whittaker and Hynes, 2002).
Figure 5. Ste5-ms is a folded domain with distinct surfaces important for kinase-binding and catalysis.
(A) Crystal structure of the Ste5 ms domain (1.6 Å resolution; data collection and refinement statistics can be found in Supp. Table 2). Structural figures were made using Pymol (DeLano, 2002). (B) Structural alignment using DALI illustrates the Ste5-ms domain is homologous to the von-Willebrand Type-A (VWA) domain. Cartoon of VWA domain fold and topology. (C) Ste5-ms has two distinct surfaces critical for Fus3 phosphorylation by Ste7 (identified by surface mutant scan of Ste5-ms for mutations with > 100-fold decrease in activity - see Supp Fig. 6 for full list of mutants used in the scanning experiment). One interface, the ‘coactivator loop’ (745–756) is critical for catalyzing Ste7➔Fus3 phosphorylation (phenotypes are represented by mutant ‘C’, N744A/D746A), and another interface is necessary for Ste7-binding (represented by mutant ‘B’, deletion of 778–786). (D) kcat of Ste7➔Fus3 phosphorylation reduced 100-fold for Ste5-ms mutant B and reduced nearly 1000-fold for mutant C. These mutants have no effect on the KM of Ste7-Fus3 phosphorylation (data not shown). Ste5-ms variants present at 1 µM, a concentration that saturates binding to Ste7 for Ste5-ms wild-type. (E) Pull-down assays show Ste5-ms mutant B is defective in binding to Ste7; mutant ‘C’ maintains Ste7 binding. Ste5-ms mutants were expressed as fusions to maltose binding protein (MBP) as a pull-down affinity tag. (F) Catalysis of Ste7➔Fus3 reaction by Ste5-ms mutant B, but not mutant C, can be restored by adding much higher concentrations of the mutant scaffold domain. Vmax for Ste7➔Fus3 reaction, measured using 50nM Ste7EE and 750nM Fus3. Point of half-max activation (Kact) gives apparent dissociation constants of Ste5-ms variants for Ste7. As expected, wild-type Ste5-ms has a Kactiv of 150nM, while Mutant ‘B’ had greatly diminished Kact = 15,500 nM, consistent with a defect in Ste7 binding. At high enough concentrations, mutant B can promote signaling to near wild-type levels. Ste5-ms mutant C shows a Kact close to wild-type (71nM) ( Supp Fig. 7B), but its Vmax at saturating concentrations is 1000-fold lower than wildtype (bar graph to right). This behavior is consistent with a defect in the catalytic step of Ste7➔Fus3 phosphorylation.
Using the Ste5-ms structure as a guide, we performed extensive mutagenesis of the ms domain surface to try to identify regions of the protein that are critical for catalysis (Supp. Fig. 6). Twenty one mutant proteins were generated, each containing a block of 2–3 mutant residues clustered together on the protein surface. Of these mutant proteins, six out of 21 showed greatly diminished (>100-fold decrease) ability to promote Ste7-to-Fus3 phosphorylation (Supp. Fig. 6B). We then screened these mutants for their effect on both binding to Ste7 and on the kcat of Fus3 phosphorylation (Supp. Fig. 6B, Supp Fig. 7A)
Five of the six mutations to the Ste5-ms domain that significantly disrupt activity cluster on two structurally and functionally distinct interfaces (Fig. 5C). The first interface contains four sets of mutations that selectively block catalysis without disrupting binding to Ste7; these mutations significantly reduce the kcat for Ste7EE-ND2➔Fus3 phosphorylation, but do not alter Ste7/Ste5-ms interaction (representative mutant ‘C’ is shown in Fig. 5D, Fig. 5E). This region is composed of a semi-disordered loop (residues 745–756) which we have named the ‘coactivator loop.’ We postulate that this loop plays a role in lowering the barrier of the Ste7➔Fus3 phosphorylation reaction, perhaps through transient interactions with Fus3.
A second interface, near the C-terminus of the Ste5-ms domain, appears to be involved in direct binding to Ste7. This interface consists of a negatively charged segment (DEHDDDDEEDN, residues 776–786). Mutant ‘B’ is a variant of the Ste5-ms domain in which the nine most Cterminal residues (778–786) have been deleted. This mutant is catalytically impaired, most likely because, as shown in pulldown assays, it has greatly reduced binding to Ste7 (Fig. 5D, 5E). In summary, there appear to be two functionally distinct surfaces on the Ste5-ms domain that are critical for its function in promoting Ste7➔ Fus3 phosphorylation: one that is responsible for association with Ste7, and a distinct surface that is responsible for Fus3-specific catalysis.
A prediction of this model is that the mutations that selectively reduce the affinity of the Ste5-Ste7 interaction should be able to rescue the Ste7➔ Fus3 reaction if added at much higher concentrations (kcat values shown in Fig. 4D were only measured at a concentration of 1 µM Ste5-ms). As predicted, a Ste5-ms protein bearing mutation B, which selectively disrupts Ste7 binding, has a kcat for the Ste7➔Fus3 reaction that is comparable to that of the wild-type protein, but only when added at ~100-fold higher concentrations (Fig. 5F). In contrast, addition of increasing amounts of a Ste5-ms protein bearing mutation C (a “coactivator” mutation) plateaus at a kcat that is 1000-fold lower than observed with wild-type Ste5-ms (does not result in increased kcat) (Supp. Fig. 7B).
The importance of these two regions within the Ste5-ms domain is also highlighted by alignment of homologs of Ste5 from other fungal species. The most conserved region of these Ste5 scaffold homologs corresponds to the Ste5-ms domains (Supp. Fig. 3B), and especially both within the coactivator loop (745–756) and across the previously defined Ste7 binding region (Supp Fig. 3C).
Ste5-ms domain catalytically unlocks Fus3 for phosphorylation by Ste7
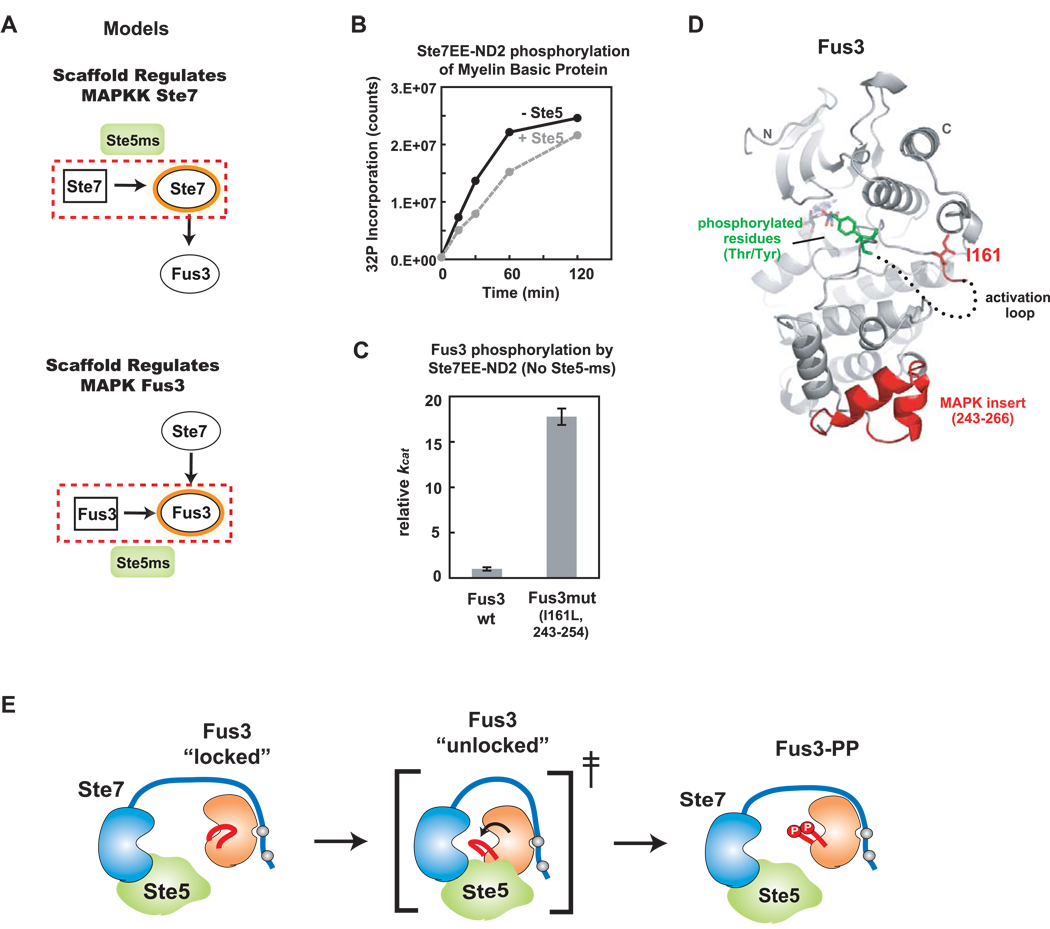
There are two distinct models for how the Ste5-ms domain promotes Ste7➔ Fus3 phosphorylation. One model is that the Ste5-ms primarily acts as an activator of Ste7 to enhance its overall catalytic activity (Fig. 6A – top), much as a cyclin activates a CDK. A second competing model is that Ste5-ms primarily acts to convert Fus3 from a poor Ste7 substrate to a good Ste7 substrate (Fig. 6A – bottom). A prediction of the first model is that addition of Ste5 to Ste7 will enhance its overall kinase activity toward any substrate. To test this model, we compared rates of phosphorylation of a general substrate, myelin basic protein, by Ste7EE, in the presence and absence of the Ste5-ms domain (Fig. 6B). The rates are indistinguishable, indicating that the Ste5-ms domain is not a general activator of Ste7. In addition, as described above, the addition of Ste5-ms has no effect on the Ste7➔Kss1 reaction (Fig. 4B, 4C). These two findings strongly disfavor a model where the Ste5-ms upregulates the general kinase activity of Ste7 to enhance Fus3 phosphorylation.
Figure 6. Ste5-ms catalytically unlocks Fus3 for phosphorylation by Ste7.
(A) Two potential models for how Ste5-ms enhances Ste7➔Fus3 phosphorylation. One model proposes that Ste5-ms primarily acts on Ste7; Ste7 is a poor enzyme that requires Ste5-ms binding to increase its activity (top). An alterative model hypothesizes that Ste5-ms acts primarily on Fus3 - converting it from a poor substrate to a good one (bottom). (B) Ste5-ms has no effect on overall catalytic activity of Ste7EE as tested against the general kinase substrate, Myelin Basic Protein (MBP) using a 32P kinase assay. (C) To identify elements in Fus3 that make it a poor substrate compared to Kss1, we made mutations in Fus3 that make it more similar in sequence to Kss1 (Supp. Fig. 8A,B). These mutants were tested for their ability to be phosphorylated by Ste7EE in the absence of Ste5 (Supp. Fig 8C–D). A combined mutation of I161L with replacement of the 243–254 ‘MAPK insertion loop’ (with the same region from Kss1) created a Fus3 mutant with a 20-fold increase in kcat compared to wild-type (reaction contains 50nM Ste7EE-ND2, 750nM Fus3 variant, no scaffold). (D) Crystal structure of Fus3 (Remenyi et al., 2005), showing positions of critical mutations in red (I161L, and MAPK insert 243–254). The activation loop (shown as dotted line; not fully visible in the crystal structure) sits between these two regions. Residues that become phosphorylated (T180 and Y182) shown in green. (E) A model for Ste5-ms action: Fus3’s activation loop normally adopts a “locked” conformation, but Ste5-ms interaction with Fus3 transiently (and only in the presence of Ste7) stabilizes a transition state in which Fus3’s activation loop is accessible to Ste7.
These results point toward the alternative model in which the Ste5-ms exerts its effect on the substrate – it selectively improves Fus3 as a substrate for Ste7. This model suggests that a key difference between the closely related, competing MAPK’s Kss1 and Fus3, is that Kss1 is already primed to be a good substrate for Ste7, but that Fus3 is, by itself, locked in a state that makes it a poor substrate. If this is true, then we reasoned it might be possible to make mutations in Fus3 that “unlock” it, making it more like Kss1 which can serve as a scaffold-independent substrate for Ste7. We made ten sets of mutations in Fus3 that make the sequence more like Kss1, based on sequence regions that diverge between Fus3 and Kss1. We tested the ability of activated Ste7 (Ste7EE-ND2) to phosphorylate these chimeric mutants in the absence of Ste5 (Supp Fig. 8A–D). We found that a mutation of residue I161L combined with replacement of residues 243–254 (a region known as the ‘MAPK insertion loop’) with the comparable insert from Kss1 resulted in a Fus3 variant that had an approximately 20-fold increase in kcat compared to Fus3 wild-type, in the absence of scaffold (Fig. 6C, Supp. Fig. 8D). Consistent with this result, previous studies showed that a I161L mutant in Fus3 could partially complement the loss of Ste5 scaffold in mating pathway activation in vivo (Brill et al., 1994). We mapped the location of mutations that “unlock” Fus3 onto its crystal structure (Remenyi et al., 2005), which shows that these residues lie near the Fus3 activation loop (Fig. 6D).
These data suggest a model for how Fus3 phosphorylation may be regulated by the Ste5 scaffold. We postulate that Fus3’s activation loop normally exists in a locked state so that it cannot be easily phosphorylated by Ste7. However, when the scaffold is present and bound to Ste7, the Ste5-ms domain may stabilize a transition-state conformation of Fus3’s activation loop that is accessible to Ste7 (Fig. 6E). The precise mechanism of how the activation loop structure and dynamics are altered remains to be elucidated.
DISCUSSION
Assisted catalysis and tethering: complementary mechanisms by which the Ste5 scaffold directs specificity of MAPK signaling
Scaffold proteins have emerged as important elements in determining the wiring of cell signaling pathways. The simplest model for how scaffolds direct signaling specificity is through tethering: co-recruiting components to the same site. In the case of the yeast mating MAPK scaffold Ste5, there is ample evidence that tethering plays a central role in its function: Ste5 interacts with the Gβ protein Ste4, the MAPKKK Ste11, and the MAPKK Ste7, and disruption of these interactions is sufficient to destroy proper signaling. Moreover, the effects of these mutations can be overcome by re-recruiting the missing components to the complex via heterologous interactions or protein fusions (Harris et al., 2001; Park et al., 2003). Nonetheless, it has been far less clear if Fus3 activation in the mating pathway is directed by Ste5 through a tethering mechanism, because disruption of the previously mapped Fus3 interaction site on Ste5 does not impair the mating response (Bhattacharyya et al., 2006a).
Here we show that the Ste5 scaffold protein plays a far more active, co-catalytic role in directing Ste7➔Fus3 signaling. A specific domain in Ste5, which we have named the minimal scaffold, or ‘ms’ domain, is a necessary co-factor for the Ste7➔Fus3 phosphorylation reaction: Fus3 is an extremely poor substrate in the absence of this Ste5 domain, although Ste7 is a perfectly competent enzyme. Conceptually, this domain of the scaffold is a required co-factor, much like a cyclin is a required co-factor for CDK. However, the Ste5-ms domain appears to act in a detailed manner that is quite unique: kinase accessory factors like a cyclin generally act by either globally increasing the kcat for kinase activity (usually by allosterically repositioning key catalytic residues) or by decreasing the KM for specific substrates via additional substrate recognition sites (Loog and Morgan, 2005; Pavletich, 1999). In this case, the Ste5 scaffold improves the kcat of the phosphorylation reaction, but in a manner that is only specific for one substrate, Fus3. The KM of Ste7➔Fus3 phosphorylation is likely dictated by the strength of MAPK docking interactions (data not shown).
A catalytic role for the Ste5 scaffold helps to explain several paradoxes concerning organizing factors like scaffolds that were presumed to function solely by a tethering mechanism. First, if a tethering scaffold is present at a higher concentration than its components, it might cause inhibition of pathway, by segregating individual components into different complexes. Second, if a tethering scaffold uses increased binding energy to shunt signaling specificity towards one substrate, then it may be more difficult to release this component. This issue is critical for a MAPK like Fus3, which must dissociate from the scaffold and enter the nucleus to exert many of its downstream effects. FRAP studies show that Fus3 rapidly dissociates from the Ste5 complex, more so than other pathway components (van Drogen and Peter, 2002; van Drogen et al., 2001). These two issues, however, are mitigated by a mechanism in which the scaffold plays a direct catalytic role. Inhibitory segregation would not be observed if the Ste7-Ste5 complex is the only unit that is able to activate Fus3 (Ste7 or Ste5 cannot activate Fus3 individually). In addition, the lack of a strong direct Ste5-ms/Fus3 interaction in the Fus3 activation step may allow reasonably rapid dissociation of active Fus3 from the complex (Maeder et al., 2007).
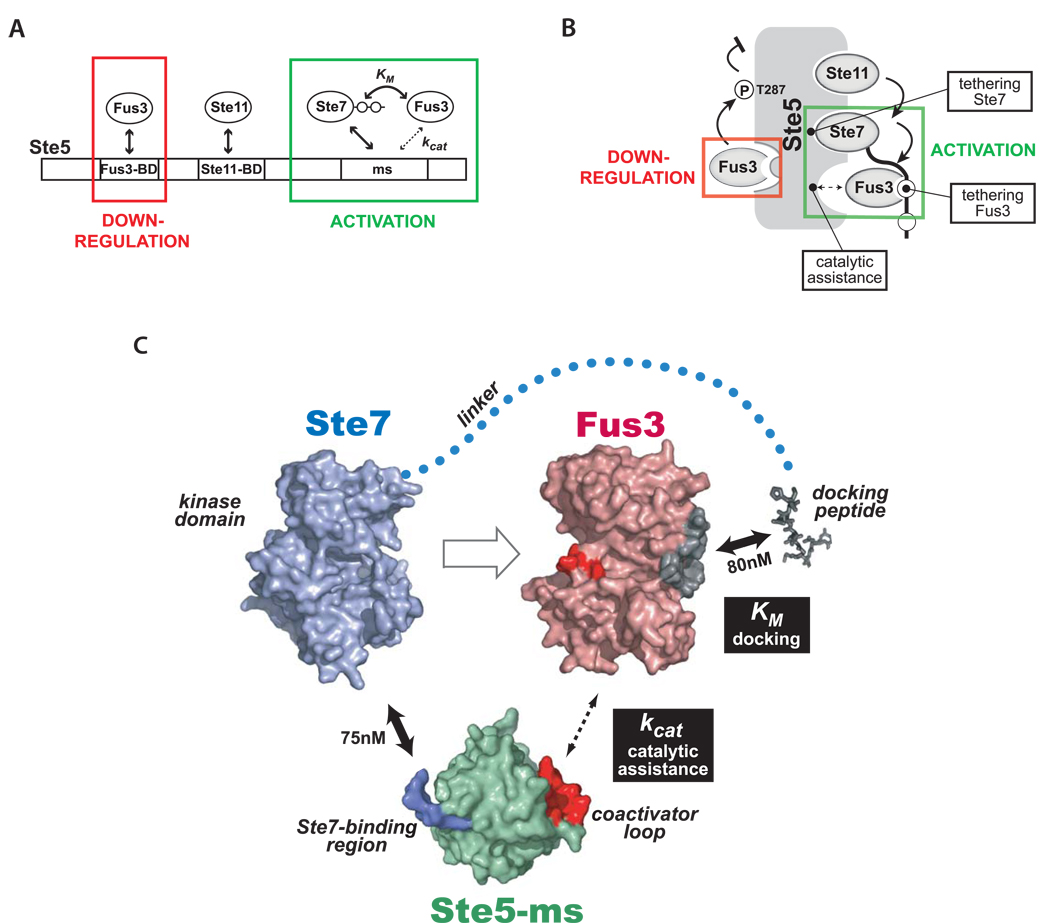
A revised model for how Ste5 coordinates the mating MAPK pathway
While Ste5 still acts as a central organizer of the mating MAPK pathway, our new findings force us to update the model of how Ste5 directs the flow of information (Figure 7). Most significantly, an updated model must include a role for the Ste5-ms domain in co-catalyzing Fus3 phosphorylation by Ste7. It is also now clear that there are both activating and downregulatory interaction sites for Fus3 on the scaffold (Figure 7A, Figure 7B). Activation of the mating response requires recruitment of Fus3 to Ste7 docking motifs, and a transient, catalytic interaction with Ste5-ms. Conversely, downregulation of pathway output is mediated in part by recruitment of Fus3 to its strong binding site on Ste5.
Figure 7. A new model for how the Ste5 scaffold controls information flow in the mating MAPK pathway.
(A) Ste5 has upregulatory (activating) and downregulatory interactions with Fus3. The strong, previously identified Fus3 binding site on Ste5 (Fus3-BD, KD = 1 µM) is not required for Ste7➔Fus3 phosphorylation, but rather is important for tuning down pathway output in vivo. Interactions that promote Fus3 phosphorylation involve the Ste5-ms domain (in cooperation with Ste7). (B) Cartoon summarizing various activities of Ste5. The Ste5-mediated complex has several critical tethering interactions (Ste5-Ste1 1, Ste5-Ste7, and Ste7-Fus3) essential for linear propagation of the mating pathway signal. In addition, Ste5-ms domain is an essential co-factor promoting catalysis of the Ste7➔Fus3 phosphorylation reaction. (C) Detailed model of minimal interactions in the mating scaffold complex required for Ste7➔Fus3 phosphorylation. Ste7 binds strongly to both Ste5-ms domain (via surface on Ste5-ms colored blue) and Fus3 (docking motifs on Ste7 bind to docking groove on Fus3 - colored gray), thereby tethering two proteins that normally interact only very weakly. Ste5-ms contains a coactivator loop (red surface) which promotes Fus3’s phosphorylation by Ste7. Fus3’s activation loop is colored red. Interaction affinities, where know, are indicated. Interactions that modulate kcat and KM of Fus3 phosphorylation by Ste7 are indicated by black boxes. Models for Fus3 (PDB code 2B9F) and Ste5-ms (this study) are derived from crystal structures. Ste7’s kinase domain was modeled from the structure of a homologous mammalian MAPKK (MKK7) using the threading program Phyre (Bennett-Lovsey et al., 2008).
An updated model of the mating pathway is summarized below. Binding of α-factor to its receptor (Ste2) leads to dissociation of the GB protein (Ste4) from the GA subunit (Gpa1). Activated Ste4, which is membrane tethered, binds to Ste5, recruiting it to the membrane, allowing the membrane-localized PAK kinase, Ste20, to phosphorylate and activate the MAPKKK Ste11 (bound to Ste5). Phosphorylated Ste11 then activates scaffold-associated MAPKK Ste7. We now understand that Ste7, only when phosphorylated and bound to the Ste5-ms domain can activate Fus3, since both Ste7 and the Ste5-ms domain required to work together catalytically to promote Ste7➔Fus3 phosphorylation. In support of this model, a single point mutation (E756G) in the coactivator loop of Ste5-ms (that impairs Ste5-ms function in vitro) destroys the ability of Ste7 to activate Fus3 during the mating response, but has no effect on Ste7➔Kss1 phosphorylation, in vivo (Schwartz and Madhani, 2006). In our model, Fus3 recruitment to the scaffold complex in not unimportant, but is carried out via a docking interaction with Ste7. These docking motifs are one of several absolutely required elements for Ste7➔ Fus3 phosphorylation. After activation, Fus3 dissociates from the Ste5 complex to enter the nucleus, where it can exert its downstream effects.
While there is a strong (KD = 1µM) binding site for Fus3 on Ste5 (residues 288–316), this site does not appear to play a significant role in directing the main forward flow of signaling information down the MAPKKK➔MAPKK➔MAPK cascade. Rather, this site downregulates mating signaling – through feedback phosphorylation of Ste5 by Fus3 – and thereby tunes the amplitude and dynamics of pathway output (Bhattacharyya et al., 2006a). Other studies have also suggested a role for this regulatory Fus3-binding domain in tuning the precise input/output behavior of the pathway: mutation of this domain leads to misregulation of mating projection formation, and improper decision making between budding, shmooing, and elongated growth cell fates (Hao et al., 2008; Maeder et al., 2007).
Evolution of new pathways: How the Ste5 scaffold may have facilitated the functional divergence of the Fus3 and Kss1 MAPKs
New signaling pathways are thought to emerge through duplication of signaling components, followed by their functional divergence. This mechanism of evolution raises issues of specificity – when components are duplicated, how is improper crosstalk avoided, given that they will interact with the same upstream and downstream partners? Based on their similarity, it seems likely that Fus3 and Kss1 originated from just this type of duplication event. Although a simple tethering scaffold protein can contribute to distinguishing the partners of such close homologs, it seems unlikely that a shift in relative affinities would be sufficient to completely prevent misactivation by the wrong upstream pathway. In this case, it seems particularly important that activated Ste7 that results from starvation input (filamentous growth pathway) does not lead to launching of the costly mating response.
To avoid misactivation, it appears that Fus3 has evolved a safety catch mechanism that distinguishes it from Kss1. We postulate a model in which the activation loop of Fus3 is “locked,” making it a poor substrate for Ste7 alone. However, this lock can be kinetically opened by the Ste5-ms domain. Thus, as discussed previously by Flatauer et al., Fus3 can only be phosphorylated by Ste7 that is activated and bound within the Ste5 complex. Active Ste7 associated with Ste5 is likely to only arise via activation by mating input. While it is formally possible that Ste7 activated by starvation input could subsequently bind to Ste5, there is evidence that Ste5 may not be competent for Fus3 activation in unstimulated cells. Ste5 translocates to the membrane upon alpha-factor stimulation (not by filamentation input) and it has been hypothesized that this translocation promotes a conformational change in Ste5 that is important for mating pathway activation (Flatauer et al., 2005; Inouye et al., 1997b; Sette et al., 2000). In support of this, a number of mutations or fusions to Ste5 that enhance membrane localization lead to increased mating signaling (Winters et al., 2005). It is possible that Ste7 only binds to the scaffold, or that Ste5-ms is only accessible for Ste7-Fus3 catalysis, when the Ste5 scaffold is in the proper conformation at the membrane.
Phylogenetic analysis of close fungal species supports this general duplication-divergence model involving scaffold co-catalysis (Supp. Fig. 9). Within the subphylum Saccharomycotina, the majority of genomes contain two homologs of the mammalian ERK MAPK (the subfamily encompassing Fus3 and Kss1), consistent with a duplication of this gene prior to this branchpoint. All ten fungi that contain a Ste5 sequence homolog fall within this subphylum and have a Fus3/Kss1 (ERK) duplication. This data is consistent with a model in which Ste5 was one solution for promoting the functional divergence of the duplicated MAP kinases, Fus3 and Kss1. Other mechanisms to diverge these kinases have also evolved: for example misactivation of the filamentous response by mating input is blocked by Fus3-induced degradation of the filamentous growth transcription factor, Tec1 (Bao et al., 2004; Chou et al., 2004). Interestingly, Tec1 is present only in the fungi that also have Ste5 (Supp. Fig. 9). Species within Saccharomycotina that lack Ste5 and Tec1 presumably have alternative mechanisms to promote functional divergence, perhaps yet undiscovered scaffolds. It will be exciting to see if other pathway scaffold proteins, including those involved in mammalian MAPK signaling (e.g. JIP, KSR, etc.) utilize a kind of direct catalytic assistance to promote specific kinase-substrate reactions, and whether these are associated with other evolutionary duplication-divergence branchpoints.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
For expanded methods, see Supplemental information. Fus3, Ste5 scaffold truncations, and Ste5-ms variants were expressed in Rosetta (DE3) pLysS E.coli cells. Ste7 variants and Kss1 were expressed in Spodoptera frugiperda (SF9) cells. Purification was carried out as described previously (Remenyi et al., 2005), with some modifications (see Supplemental).
In vitro Kinase Assays
(A) Trulight Kinase Assay
The Ste7-to-MAPK phosphorylation reactions were measured in a continuous, high-throughput fashion using the Trulight Superquenching Kinase Assay (Kit #539710, EMD Biosciences) in 96-well plates on a SpectraMax Gemini XS fluorescence plate-reader (Molecular Devices). Kinases and scaffold were added at 50nM concentration unless written otherwise. Trulight assay kit includes proprietary sensor beads coated with a fluorescent polymer, a MAPK-specific peptide (LVEPLTPSGEAPNQK) labeled with a Lissamine Rhodamine B quencher, and Assay Buffer. Kinase activity (phosphorylation of the peptide) is monitored as a loss in fluorescence over time. For more details on the Trulight Assay, see Supp. Fig. 2D–F.
(B) Quantitative anti-phospho MAPK Western blots
Quantitative in vitro western blots – used to monitor accumulation of pTyr/pThr on the activation loop of either Fus3 or Kss1 - were carried out using a primary anti-phospho p44/42 MAPK antibody (Cell Signal Technology, #9101) which recognizes both Fus3 and Kss1 equally (Supp. Fig. 1D), and a secondary IRDye 800CW Goat Anti-Rabbit IgG antibody (Licor, #926–32211). Kinase reactions contained 50nM enzyme (GST-Ste7EE or GST-Ste7EE-ND2) and 1 µM Ste5 scaffold, with varying concentration of substrate (either Fus3 or Kss1), unless otherwise noted. Standard kinase assay buffer included 100nM NaCl, 25mM Tris pH 8, 0.05% NP-40 and 2mM TCEP. Note, we varied the concentration of enzyme (10nM and 250nM Ste7EE-ND2) to test if this could drastically altered the kcat and KM values for Fus3 phosphorylation – it did not. Blots were visualized using the 800nm channel on Licor Odyssey Imaging System and quantified using Odyssey 2.1 software (see Supp. Fig. 4 for further details). Rate plots for Fus3 and Kss1 phosphorylation were fit to the Michaelis-Menten equation (V = k2[E][S]/(KM+[S])), using nonlinear least-squares method in Matlab. The KM and kcat values were calculated as the average of fitting the Michaelis-Menten equation to three separate curves (from three separate experiments), and errors reported as standard deviation. Kact plots were fit to a simple binding equation (y = a1 + a2(([X]/KD)/(1+([X]/KD)))), also in Matlab.
(C) Radioactive Kinase Assay
Phosphorylation of the general kinase substrate, Myelin Basic Protein (Sigma), by GST-Ste7EE was monitored by the rate of incorporation of 32P using autoradiography. Assay conditions included: 0.5 µM GST-Ste7EE, 2.5 µM Ste5-ms (where present), and standard kinase assay buffer (listed above) plus 10 µM 32P-ATP, 500 µM cold ATP, and 1mM MgCl2.
Protein Binding Assays and Structure Determination
For detailed methods on protein binding assays and structure determination see Supplemental information.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by fellowships from Genentech (MCG) and Achievement Rewards for College Scientists (MCG), and the Leukemia and Lymphoma Society (AR). It was also supported by grants from the NIH (WAL) and the Packard Foundation (WAL), The Wellcome Trust (AR) and a Marie Curie Reintegration Grant (AR). Thanks to P. Egea for help with structure determination, and to G. Narlikar, A. Chau, N. Helman, J. Zalatan for assistance and advice. Also thanks to J. Weissman, D. Morgan, C. Bashor, S Peisajovich, A. Watters, E. McCullagh, and H. Ramage, and members of the Lim Lab for thoughtful discussions. WAL would like to dedicate this paper to the memory of Jeremy R. Knowles who taught me the value of the free energy diagram.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Bao MZ, Schwartz MA, Cantin GT, Yates JR, 3rd, Madhani HD. Pheromone-dependent destruction of the Tec1 transcription factor is required for MAP kinase signaling specificity in yeast. Cell. 2004;119:991–1000. doi: 10.1016/j.cell.2004.11.052. [DOI] [PubMed] [Google Scholar]
- Bardwell L, Cook JG, Chang EC, Cairns BR, Thorner J. Signaling in the yeast pheromone response pathway: specific and high-affinity interaction of the mitogen-activated protein (MAP) kinases Kss1 and Fus3 with the upstream MAP kinase kinase Ste7. Mol Cell Biol. 1996;16:3637–3650. doi: 10.1128/mcb.16.7.3637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett-Lovsey RM, Herbert AD, Sternberg MJ, Kelley LA. Exploring the extremes of sequence/structure space with ensemble fold recognition in the program Phyre. Proteins. 2008;70:611–625. doi: 10.1002/prot.21688. [DOI] [PubMed] [Google Scholar]
- Bhattacharyya RP, Remenyi A, Good MC, Bashor CJ, Falick AM, Lim WA. The Ste5 scaffold allosterically modulates signaling output of the yeast mating pathway. Science. 2006a;311:822–826. doi: 10.1126/science.1120941. [DOI] [PubMed] [Google Scholar]
- Bhattacharyya RP, Remenyi A, Yeh BJ, Lim WA. Domains, Motifs, and Scaffolds: The Role of Modular Interactions in the Evolution and Wiring of Cell Signaling Circuits. Annu Rev Biochem. 2006b doi: 10.1146/annurev.biochem.75.103004.142710. [DOI] [PubMed] [Google Scholar]
- Brill JA, Elion EA, Fink GR. A role for autophosphorylation revealed by activated alleles of FUS3, the yeast MAP kinase homolog. Mol Biol Cell. 1994;5:297–312. doi: 10.1091/mbc.5.3.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burack WR, Cheng AM, Shaw AS. Scaffolds, adaptors and linkers of TCR signaling: theory and practice. Curr Opin Immunol. 2002;14:312–316. doi: 10.1016/s0952-7915(02)00347-3. [DOI] [PubMed] [Google Scholar]
- Burack WR, Shaw AS. Signal transduction: hanging on a scaffold. Curr Opin Cell Biol. 2000;12:211–216. doi: 10.1016/s0955-0674(99)00078-2. [DOI] [PubMed] [Google Scholar]
- Choi KY, Satterberg B, Lyons DM, Elion EA. Ste5 tethers multiple protein kinases in the MAP kinase cascade required for mating in S. cerevisiae. Cell. 1994;78:499–512. doi: 10.1016/0092-8674(94)90427-8. [DOI] [PubMed] [Google Scholar]
- Chou S, Huang L, Liu H. Fus3-regulated Tec1 degradation through SCFCdc4 determines MAPK signaling specificity during mating in yeast. Cell. 2004;119:981–990. doi: 10.1016/j.cell.2004.11.053. [DOI] [PubMed] [Google Scholar]
- DeLano WL. The Pymol Molecular Graphics System. San Carlos, CA: DeLano Scientific; 2002. [Google Scholar]
- Flatauer LJ, Zadeh SF, Bardwell L. Mitogen-activated protein kinases with distinct requirements for Ste5 scaffolding influence signaling specificity in Saccharomyces cerevisiae. Mol Cell Biol. 2005;25:1793–1803. doi: 10.1128/MCB.25.5.1793-1803.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao N, Nayak S, Behar M, Shanks RH, Nagiec MJ, Errede B, Hasty J, Elston TC, Dohlman HG. Regulation of cell signaling dynamics by the protein kinase-scaffold Ste5. Mol Cell. 2008;30:649–656. doi: 10.1016/j.molcel.2008.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris K, Lamson RE, Nelson B, Hughes TR, Marton MJ, Roberts CJ, Boone C, Pryciak PM. Role of scaffolds in MAP kinase pathway specificity revealed by custom design of pathway-dedicated signaling proteins. Curr Biol. 2001;11:1815–1824. [PubMed] [Google Scholar]
- Holm L, Sander C. Mapping the protein universe. Science. 1996;273:595–603. doi: 10.1126/science.273.5275.595. [DOI] [PubMed] [Google Scholar]
- Inouye C, Dhillon N, Durfee T, Zambryski PC, Thorner J. Mutational analysis of STE5 in the yeast Saccharomyces cerevisiae: application of a differential interaction trap assay for examining protein-protein interactions. Genetics. 1997a;147:479–492. doi: 10.1093/genetics/147.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inouye C, Dhillon N, Thorner J. Ste5 RING-H2 domain: role in Ste4-promoted oligomerization for yeast pheromone signaling. Science. 1997b;278:103–106. doi: 10.1126/science.278.5335.103. [DOI] [PubMed] [Google Scholar]
- Loog M, Morgan DO. Cyclin specificity in the phosphorylation of cyclin-dependent kinase substrates. Nature. 2005;434:104–108. doi: 10.1038/nature03329. [DOI] [PubMed] [Google Scholar]
- Maeder CI, Hink MA, Kinkhabwala A, Mayr R, Bastiaens PI, Knop M. Spatial regulation of Fus3 MAP kinase activity through a reaction-diffusion mechanism in yeast pheromone signalling. Nat Cell Biol. 2007;9:1319–1326. doi: 10.1038/ncb1652. [DOI] [PubMed] [Google Scholar]
- Maleri S, Ge Q, Hackett EA, Wang Y, Dohlman HG, Errede B. Persistent activation by constitutive Ste7 promotes Kss1-mediated invasive growth but fails to support Fus3-dependent mating in yeast. Mol Cell Biol. 2004;24:9221–9238. doi: 10.1128/MCB.24.20.9221-9238.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SH, Zarrinpar A, Lim WA. Rewiring MAP kinase pathways using alternative scaffold assembly mechanisms. Science. 2003;299:1061–1064. doi: 10.1126/science.1076979. [DOI] [PubMed] [Google Scholar]
- Pavletich NP. Mechanisms of cyclin-dependent kinase regulation: structures of Cdks, their cyclin activators, and Cip and INK4 inhibitors. J Mol Biol. 1999;287:821–828. doi: 10.1006/jmbi.1999.2640. [DOI] [PubMed] [Google Scholar]
- Remenyi A, Good MC, Bhattacharyya RP, Lim WA. The role of docking interactions in mediating signaling input, output, and discrimination in the yeast MAPK network. Mol Cell. 2005;20:951–962. doi: 10.1016/j.molcel.2005.10.030. [DOI] [PubMed] [Google Scholar]
- Remenyi A, Good MC, Lim WA. Docking interactions in protein kinase and phosphatase networks. Curr Opin Struct Biol. 2006;16:676–685. doi: 10.1016/j.sbi.2006.10.008. [DOI] [PubMed] [Google Scholar]
- Robinson FL, Whitehurst AW, Raman M, Cobb MH. Identification of novel point mutations in ERK2 that selectively disrupt binding to MEK1. J Biol Chem. 2002;277:14844–14852. doi: 10.1074/jbc.M107776200. [DOI] [PubMed] [Google Scholar]
- Schwartz MA, Madhani HD. Control of MAPK signaling specificity by a conserved residue in the MEK-binding domain of the yeast scaffold protein Ste5. Curr Genet. 2006;49:351–363. doi: 10.1007/s00294-006-0061-6. [DOI] [PubMed] [Google Scholar]
- Sette C, Inouye CJ, Stroschein SL, Iaquinta PJ, Thorner J. Mutational analysis suggests that activation of the yeast pheromone response mitogen-activated protein kinase pathway involves conformational changes in the Ste5 scaffold protein. Mol Biol Cell. 2000;11:4033–4049. doi: 10.1091/mbc.11.11.4033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Drogen F, Peter M. MAP kinase cascades: scaffolding signal specificity. Curr Biol. 2002;12:R53–R55. doi: 10.1016/s0960-9822(01)00672-8. [DOI] [PubMed] [Google Scholar]
- van Drogen F, Stucke VM, Jorritsma G, Peter M. MAP kinase dynamics in response to pheromones in budding yeast. Nat Cell Biol. 2001;3:1051–1059. doi: 10.1038/ncb1201-1051. [DOI] [PubMed] [Google Scholar]
- Whiteway MS, Wu C, Leeuw T, Clark K, Fourest-Lieuvin A, Thomas DY, Leberer E. Association of the yeast pheromone response G protein beta gamma subunits with the MAP kinase scaffold Ste5p. Science. 1995;269:1572–1575. doi: 10.1126/science.7667635. [DOI] [PubMed] [Google Scholar]
- Whittaker CA, Hynes RO. Distribution and evolution of von Willebrand/integrin A domains: widely dispersed domains with roles in cell adhesion and elsewhere. Mol Biol Cell. 2002;13:3369–3387. doi: 10.1091/mbc.E02-05-0259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winters MJ, Lamson RE, Nakanishi H, Neiman AM, Pryciak PM. A membrane binding domain in the ste5 scaffold synergizes with gbetagamma binding to control localization and signaling in pheromone response. Mol Cell. 2005;20:21–32. doi: 10.1016/j.molcel.2005.08.020. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.