Abstract
Three closely related clones of leukemic lymphoid CEM cells were compared for their gene expression responses to the glucocorticoid dexamethasone (Dex). All three contained receptors for Dex, but only two responded by undergoing apoptosis. After a time of exposure to Dex that ended late in the interval preceding onset of apoptosis, gene microarray analyses were carried out. The results indicate that the expression of a limited, distinctive set of genes was altered in the two apoptosis-prone clones, not in the resistant clone. That clone showed altered expression of different sets of genes, suggesting that a molecular switch converted patterns of gene expression between the two phenotypes: apoptosis-prone and apoptosis-resistant. The results are consistent with the hypothesis that altered expression of a distinctive network of genes after glucocorticoid administration ultimately triggers apoptosis of leukemic lymphoid cells. The altered genes identified provide new foci for study of their role in cell death.
Keywords: Human lymphoid cells, CEM, Apoptosis, Glucocorticoid sensitivity/resistance, Microarray analysis, Novel genes, Gene expression profile, Data mining
For almost 60 years, it has been known that glucocorticoids are capable of lysing certain classes of immature lymphoid cells [1], and this knowledge has been applied widely in the treatment of lymphoid malignancies [2]. With the recognition of apoptosis as a form of suicidal cell death, it became clear that lymphoid cells also underwent this process when exposed to pharmacological concentrations of glucocorticoids [3,4]. Lymphoid cell apoptosis was shown to be absolutely dependent on glucocorticoid occupancy of the glucocorticoid receptor (GR), a specific intracellular glucocorticoid-activated transcription factor [5]. This implicated altered gene regulation in apoptosis, evidence for which came from experiments on thymocytes and malignant lymphoid cells. In both, general inhibitors of transcription and/or translation were used to show that blocking macromolecular synthesis also blocked various morphological and biochemical indicators of apoptosis [4,6 –10]. Timing was critical for these experiments, because the complete block of either transcription or translation would in itself be lethal after a time. Thus the paradigm was established that in these lymphoid apoptotic systems, alterations in gene regulation led to eventual apoptotic death. It is now clear that, in general, apoptosis occurs by the precise regulation of a multitude of genes, rather than one or a few genes [11].
The search for genes whose regulation is required for steroid-induced apoptosis of lymphoid cells has engrossed many laboratories for years. By knowledge of metabolic processes, biochemical pathways, regulatory systems, and cellular organelles, as well as by various forms of differential gene product screening, attempts have been made to discover the critically regulated genes and their mRNA and protein products [12–17]. This sustained effort has, in fact, produced several interesting and valuable leads. In thymocytes or malignantly transformed lymphoid cells, various landmarks in the processes leading to apoptosis have been established. Yet it is clear that these experiments, though based on the best biological intuitions or early screening techniques, have not completely defined the pathway. Until recently, the technologies available for relatively unbiased screening simply have not had the resolving power necessary to identify the necessary and sufficient genes involved. Now, with the advent of microarray technology, this long-sought goal is within reach. Several studies of lymphoid cell gene expression have now been carried out using this new technology, and a few have begun to examine the effects of glucocorticoids on gene expression. These studies, though exciting, have only begun to unravel the skein of complex gene regulations involved. We present here the initial results from the examination of glucocorticoid-induced changes in gene expression in a closely related set of three malignant lymphoid cell clones. Comparisons of genes regulated in the three clones have resulted in identification of a relatively small number of genes that correlate with cellular sensitivity to glucocorticoids’ evocation of apoptosis.
CEM cells are a line of lymphoblastic cells originally derived from a child with acute lymphoblastic leukemia [18]. Like all uncloned cell lines, the original CEM cells are a mixed population of clones with varied genotypes and phenotypes. Because of this, we have carried out all our studies on clones isolated from the original CEM line. Clones CEM-C7 and CEM-C1 provided our prototypical glucocorticoid-sensitive and -resistant phenotypes, respectively. Because of the well-known genetic and phenotypic drift of all types of cells growing continuously, and of eukaryotic tissue culture cells especially [19–21], we recently recloned each of the parental clones to repurify the original phenotypes. This established CEM-C7 subclone CEM-C7-14 as the prototypic GR-sensitive clone and CEM-C1 subclone CEM-C1-15 as the GR-resistant clone. In the process, we captured a clone from CEM-C1 that had reverted to sensitivity. In this paper, we describe and compare the changes in gene expression caused by exposure to the synthetic steroid dexamethasone (Dex) seen in the cells of these three clones—two sensitive and one resistant to glucocorticoid-induced apoptosis.
Sensitive CEM cells must be incubated in concentrations of steroid sufficient to occupy fully all GRs present in the cells for ≥24 h before the onset of biochemical and most morphological evidence of apoptosis can be measured [5]. The exception regarding morphology is classic pre-apoptotic cell shrinkage that begins as early as 6 h after steroid is added. Nevertheless, if glucocorticoid is removed or anti-glucocorticoid added at times up to ~24 h, all processes are reversible and the cells continue to grow without pause. Beyond 24 h, increasing numbers of cells become locked in the apoptotic processes, which removal of steroid does not reverse.
We therefore have chosen to identify the genes that have changed at a point just before the irreversible steps of true apoptosis begin. This tested our working hypothesis that the sensitive subclone CEM-C7-14 and the sensitive revertant CEM-C1-6 shared a set of altered gene products that would be distinctively different from any genes induced or suppressed in the resistant subclone, CEM-C1-15.
Results
Characteristics of CEM clones
Three clones of CEM cells were compared: two (C7-14 and C1-6) undergo apoptosis when exposed to 1 μM Dex; the third (C1-15) is completely resistant to the growth-inhibiting and apoptotic effects of the steroid. All three are recent subclones of the original C7 (sensitive) and C1 (resistant) clones, derived from the uncloned CEM cell line [22,23]. Subclones C7-14 and C1-15 were chosen for study because they re-established the pseudodiploid karyotypes and steroid response phenotypes of the original parental clones. They contain essentially equal levels of GRs. Sub-clone C1-6 represents a rare spontaneous revertant from C1 that has recovered an apoptotic response to Dex. C1-6 cells are hyperploid, with a mean chromosome number of 94. Some of the properties of the three clones are compared in Table 1 and Fig. 1.
Table 1.
Physiochemical characteristics of CEM clones
| CEM subclonesa |
Sensitive/resistantb | GR sites/cellc | Kdc | Doubling timed |
% Hyperploide |
Pseudodiploidf |
|---|---|---|---|---|---|---|
| C7-14 | Sens | 9,900 ± 2,600 | 12 ± 3.2 | 24.9 ± 0.0 | 0 | 47,XX,t(1;2)(p13;q21),del(9)(p13q13),+20 |
| C1-6 | Sens | 27,200 ± 3,200 | 7 ± 1.7 | 25.4 ± 2.9 | 84 | 94,XXXX,del(9)(p12),del(9)(p12),inv(9)(p13q13), inv(9)(p13q13),+20,+20 |
| C1-15 | Res | 10,352 ± 506 | 21 ± 10 | 21.9 ± 1.5 | 0 | 47,XX,del(9)(p12),inv(9)(p13q13),+20 |
Subclones were obtained by the soft agar method [24]
Resistant or sensitive to apoptosis in 10−6 M Dex
Average number of Dex binding sites/cell and Kd nM SD from 2– 4 multipoint Scatchard analyses
Average log phase population doubling time in hours ± SD, n = 3
Average percent hyperploidy in 25 chromosome spreads on each of 3 separate slides
Karyotyped by GopalRao V.N. Velagaleti, Director, Cytogenetics Laboratory, UTMB
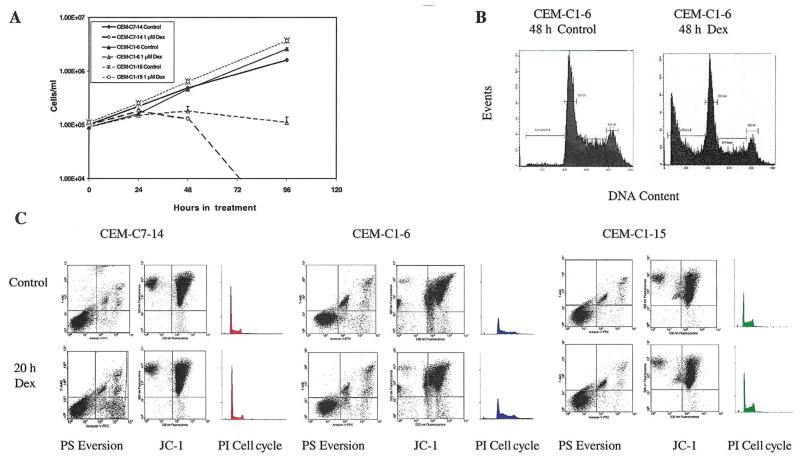
Fig. 1.
Characteristics of CEM clones. (A) Growth curves for CEM-C7-14, CEM-C1-6, and CEM-C1-15 cells treated with vehicle (Control) or 1 μM Dex. Cell viability was determined using Trypan Blue exclusion and a hemacytometer. (B) Propidium iodide (PI) analysis of CEM-C1-6 cells for apoptotic, sub-G1 levels of DNA after 48 h of vehicle (Control) or 1 μM Dex treatment. (C) Flow-cytometric analyses of CEM-C7-14, CEM- C1-6, and CEM-C1-15 cells treated with vehicle (Control) or 1 μM Dex for 20 h. Cells were evaluated using Annexin-V-FTC/7AA-D to assess phosphatidylserine (PS) membrane eversion (lower right quadrant = intact cells with everted PS), JC-1 to assess mitochondrial integrity (lower right quadrant = depolarized mitochondria), and PI staining of DNA for apoptosis and cell cycle analysis (subdiploid apoptotic population left of initial peak).
The onset of overt cell loss in the sensitive clones occurs after ~24 h (Fig. 1A). The C1-6 revertant-to-sensitive clone is less stable in phenotype than C7-14, and rapidly accumulates a subpopulation of resistant cells, indicated by its growth curve leveling off after 24 h, as a result of the relatively high rate of reaccumulation of resistant cells in the C1-6 population. As employed herein, however, the vast majority of the C1-6 cells underwent apoptosis after ≥48 h in Dex, as demonstrated by typical DNA breakdown to subdiploid levels by propidium iodide (PI) staining (Fig. 1B). Fig. 1C shows that in the two sensitive clones after 20 h in Dex, cell death had hardly begun according to several markers of apoptosis: phosphotidylserine (PS) eversion, mitochondrial membrane integrity, and PI staining of cellular DNA. No significant difference from control is seen in JC-1 stains for mitochondrial integrity or in the population with subdiploid DNA (PI, P = 0.5). The PS eversion of the lipid from the inner to the outer surface of the plasma membrane, an early event in apoptosis, was just beginning in one of the sensitive clones after 20 h. From these experiments and from previous knowledge of the timing of responses to steroids in CEM-C7 and CEM-C1 cells, we hypothesized that a complex sequence of changes in gene expression occurs in the sensitive lymphoid cells over a period of many hours, eventuating in one or more irreversible steps that triggers the acute apoptotic process. Clones C7-14 and C1-6 should display these gene expression changes; clone C1-15 should not.
To test the hypothesis, we analyzed gene expression profiles in the three clones by microarray analysis. Data were obtained on genes with altered expression after 20 h of continual exposure to Dex, a time late in the preliminary period leading to apoptosis. On three occasions over the course of 1 year, cells in mid-logarithmic growth at ~4 × 105 cells/ml were treated with ethanol vehicle or 10−6 M Dex. After 20 h, RNA was extracted from the cells and delivered to the UTMB Genomics Core Laboratory for target labeling, hybridization, and initial chip data analysis, using the Affymetrix HG_U95Av2 chip.
Validation of microarray data
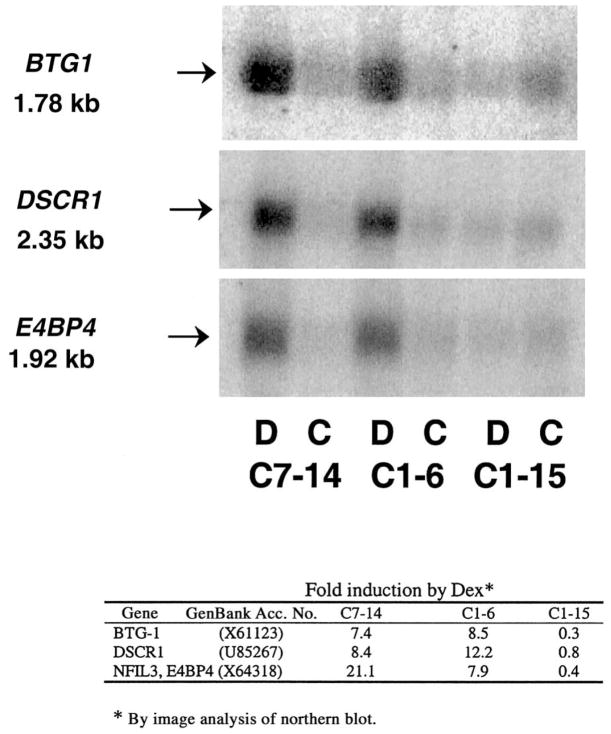
Several data validation experiments were conducted. Initially, the Genomics Core Laboratory independently tested the general reproducibility of gene expression displayed by the chips by analyzing a single RNA sample on three separate chips, not related to this study. The number of genes whose expression appeared to vary >2-fold was 12 of ~6,000 genes expressed, or ≤0.2%. In addition to this general validation, the data from our chips were checked in several ways. First, our microarray data confirmed the regulation of several genes that we had earlier identified by manual experiments in the sensitive parental C7 clone [5,25]. Thus, c-myc and ornithine decarboxylase were suppressed and GRα and c-jun were induced in the C7-14 subclone. Second, from the microarray data sets we arbitrarily selected three genes we had not studied before, that according to the chip data were induced in the sensitive cells. We tested for regulation of these three genes by northern blot analysis using actin as a normalizing control. The data (Fig. 2) clearly showed that there was induction of the mRNA of each by Dex in the sensitive clones but not in the resistant one. To our knowledge, none of these genes, B-cell translocation gene 1 (BTG1), Down syndrome candidate region 1 (DSCR1), or E4 promoter-binding protein 4 (E4BP4), had previously been associated with glucocorticoid-evoked lymphoid cell apoptosis. Finally, we noted that several of the genes detected as regulated were represented at two independent locations on the chips; hence, they provided internal independent controls that the observed change was not due to random causes. Together, these results suggested that the data from the gene chips reflected the behavior of gene expression in the cells.
Fig. 2.
Northern blots for genes BTG1, DSCR1, and E4BP4 in CEM-C7-14, CEM-C1-6, and CEM-C1-15 cells after treatment with vehicle (C) or 1 μM Dex (D) for 20 h. The table below depicts quantification of fold induction by densitometric analyses, normalized lane by lane to actin expression on the same membrane.
Cluster analyses and evaluation of randomness
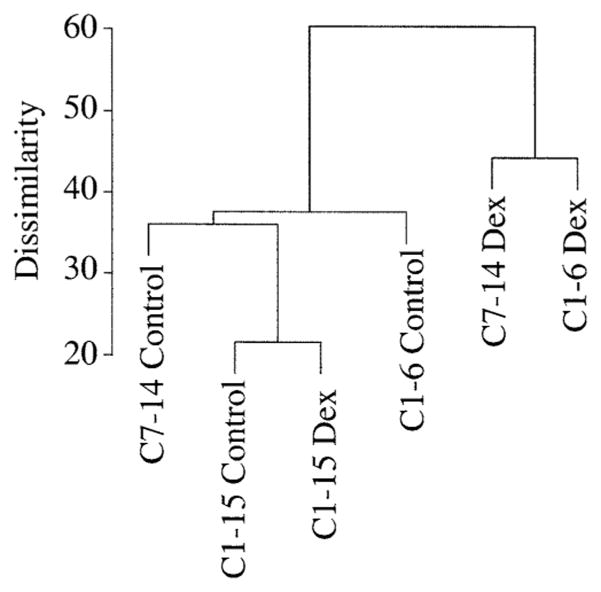
The patterns of basal gene versus altered expression were then compared by cluster analyses. Of the 12,626 genes on the Affymetrix HG_U95Av2 chip, approximately half were detected as expressed in the basal state. One-way ANOVA analysis of these at the 95% confidence limit revealed that in the basal state the resistant C1-15 clone and the Dex-sensitive revertant C1-6 clone clustered more closely than either did to the sensitive cone C7-14 (not shown). After Dex treatment, 751 genes were altered in expression at the 95% confidence limit in the two sensitive clones and not in the resistant clone. When clustering was carried out using only these altered genes, the sensitive revertant from C1 now shifted so as to cluster more closely with the inherently sensitive clone C7-14 and away from the resistant sister clone C1-15 (Fig. 3). This is consistent with our hypothesis. However, ANOVA-based statistical methods operate on the premise that the populations considered follow a Gaussian distribution. For fold-change data in cellular gene expression, this may not be so, because the apparent increases are often bunched in the very low-fold range. Several calculations were carried out to estimate the possibility that even at the 95% confidence limit there remained a considerable chance that some genes scored as altered could have been random events. In one such calculation, unpaired Student’s t-tests were conducted between the glucocorticoid-sensitive cell lines C1-6 and C7-14 (control plus Dex, eight chips) and the glucocorticoid-resistant cell line C1-15 (control plus Dex, four chips); (for technical reasons, data from only two of the three experiments could be used). The cumulative distribution of “fold changes” was calculated for the genes that were identified as “significantly different” in the t-tests (P < 0.05). “Fold changes” is defined as the ratio (≥1) of the mean of the two groups of samples to be compared, and the cumulative distribution of “fold changes” is the number of significant genes within specified limits. To estimate how many of the “significant” genes determined by the t-tests might have occurred solely by chance, we produced 20 permutated data sets (by shuffling the 12 data pieces for each gene randomly) and conducted t-tests for the permutated data sets. We than calculated the cumulative fold changes for the means of the results obtained from the 20 permutated data sets. The results indicated that in the original data, 1,697 genes were considered different at <2.5-fold and 238 genes were considered above 2.5-fold different. In the simulated data, 467 genes were found different <2.5-fold and 48.5 different above 2.5-fold. Thus, the ratios random/observed were 0.275 for genes <2.5-fold different and 0.204 for genes >2.5-fold different. Consequently, if one used data from only two experiments, a substantial fraction of “different” genes might be due to randomness, the more so at fold differences <2.5.
Fig. 3.
Cluster analysis of CEM-C7-14, CEM-C1-6, and CEM-C1-15 cells after treatment with vehicle (Control) or 1 μM Dex for 20 h. One-way ANOVA analysis at the 95% confidence limit was carried out on the genes that were altered in expression in the two sensitive clones (C7-14 and C1-6) and not in the resistant clone (C1-15).
Identification of a unique set of genes relevant to glucocorticoid-evoked apoptosis
Comparison of the expression of genes in the control, non-steroid-treated state showed that there are 73 genes expressed uniquely >2-fold higher in C1-15 cells, the resistant clone, than in either of the two sensitive clones. The two sensitive clones together basally expressed a different 37 genes >2-fold over C1-15 cells (data not shown). Examination of these genes showed no obvious explanation for the resistance of C1-15 cells or the sensitivity of the others.
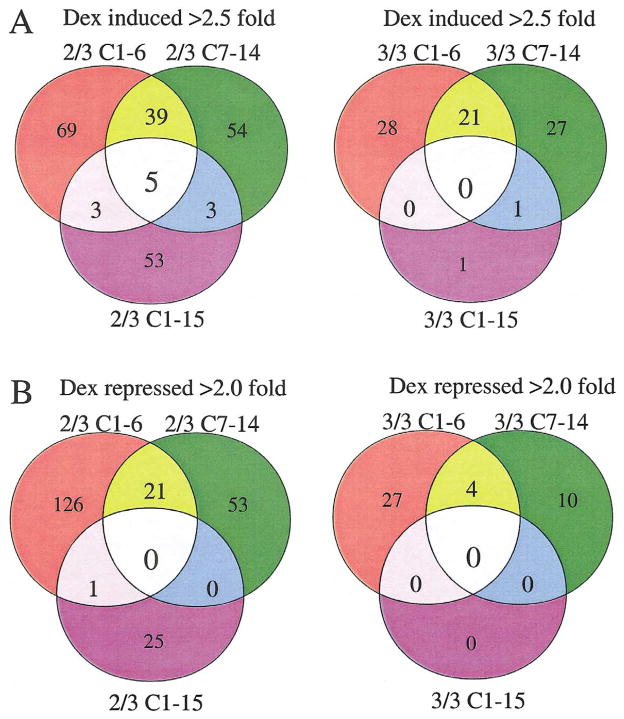
Working from the assumption that only genes reproducibly up- or downregulated in the sensitive clones could truly pertain to the cells’ eventual apoptosis, we carried out analyses using the following criteria. Only genes that were induced >2.5-fold or de-induced >2-fold were considered. These limits were chosen to eliminate most random changes, understanding that they may exclude some relevant genes whose fold change falls outside the cutoffs. We then required that each gene show this level of change in at least two of the three experiments. That is, to be included, an induced or repressed gene had to meet the cutoff criteria in at least two out of three data sets of each sensitive clone and not appear as regulated in the resistant clone more than once. When the arrays were evaluated at this level of stringency, a panel of 39 induced and 21 de-induced genes emerged that were distinctively regulated in the two sensitive clones. The pattern for induced gene changes according to these criteria is shown as a Venn diagram in Fig. 4A (left-hand side). Of the 39 induced genes, 7 were identified at two separate positions in the array. Each sensitive clone showed induction of a substantial additional number of genes unique to itself. The resistant clone C1-15 showed consistent induction of a similar number of uniquely induced genes, not regulated in the sensitive clones. A few inductions were shared between resistant clone C1-15 and one or the other of the sensitive clones (three each). Five genes frequently showed induction in all three clones. The right-hand side (Fig. 4A) shows the results when the criteria were tightened to require a fold change beyond the setpoints in each of the three experiments. By these more stringent criteria, a subset of 21 genes from the 39 was induced in every experiment in both C1-6 and C7-14 cells, but 3 of these had been induced over the limits in C1-15 cells twice. Therefore they were excluded from further consideration. Upon examination of the raw data for the remaining genes that had not met the limits in one of the three experiments, we found that there had been some induction in all cases. When all data were combined, these genes each showed on average increases of ≥2.5-fold. One gene was induced in both C7-14 and C1-15 cells but not in C1-6 cells, and one other gene was uniquely induced every time in C1-15 cells. No genes were always induced in all three clones.
Fig. 4.
Venn diagrams of CEM-C7-14, CEM-C1-6, and CEM-C1-15 genes induced >2.5-fold or repressed >2-fold after treatment with 1 μM Dex for 20 h. (A) Genes induced in at least two out of three experiments on left-hand side; genes induced in all three experiments on right-hand side. (B) Genes repressed in at least two out of three experiments on left-hand side; genes repressed in all three experiments on right-hand side.
The analogous Venn diagrams for genes de-induced >2-fold are displayed in Fig. 4B. When we required that the repression beyond the stated limit be seen in each of the three experiments, 4 genes were identified (Fig. 4B, right-hand side). An additional 27 genes were always repressed in clone C1-6, and a different set of 10 was repressed in clone C7-14. When we permitted one instance of the three to be less than the chosen cutoff (Fig. 4B, left-hand side), 21 genes were identified as repressed in both C1-6 and C7-14, and the numbers of genes repressed uniquely in each of the three clones increased considerably. Notably, in no case was a repressed gene found to be shared in both the sensitive and resistant clones. Table 2 lists the induced and de-induced genes for the sensitive clones. Many genes that emerged by this analysis show interesting groupings of properties. Others are as yet of unknown function. Clone C1-15, though resistant to Dex-evoked apoptosis, showed induction of 53 genes and repression of 25 genes in two of the three experiments (Figs. 4A and 4B, left-hand side), although these numbers dropped to 1 and 0, respectively, when we required the event in each of the three experiments (Figs. 4A and 4B, right-hand side). When combined, the data identify 78 candidate regulated genes unique to the C1-15 cells.
Table 2.
Glucocorticoid-regulated genes distinctive to CEM cells destined for apoptosis
| A Induced genes | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Probe i.d. | GenBank Acc. No. | Potential GREsa |
TF sites ± 100 ntb |
Namec | |||||
| Palindrome | 1/2 GRE | AP-1 | c/EBP | NFκB | Oct-1 | CREB | |||
| 37294_at | X61123 | 1 | 1 | 2 | BTG1 | ||||
| 41592_at | AB000734 | SOCS-1 | |||||||
| 36227_at, 1370_at | AF043129, M29696 | 1 | 1 | 2 | 2 | IL7R | |||
| 36591_at, 330_s_at | X06956, HG2259-HT2348 | TUBA1 | |||||||
| 36231_at | AC002073 | clone DJ51 5N1 | |||||||
| 37544_at | X64318 | E4BP4 | |||||||
| 41872_at | AF073308 | 1 | 1 | 1 | DFNA5 | ||||
| 35985_at | AB023137 | paralemmin 2 | |||||||
| 38717_at | AL050159 | 1 | 1 | 1 | 1 | DKFZp586A0522 | |||
| 32112_s_at, 32113_at | AI800499, U83115 | AIM1 | |||||||
| 32168_s_at | U85267 | DSCR1 | |||||||
| 995_g_at | X58288 | PTPRM | |||||||
| 41524_at, 656_at | L08488 | 1 | 1 | 1 | INPP1 | ||||
| 1102_s_at, 36690_at | M10901 | GRα | |||||||
| 38378_at | M37033 | MOX44 | |||||||
| 1427_g_at | D89077 | 1 | 1 | 2 | 1 | SLAP | |||
| 32542_at | AF063002 | SLIM1 | |||||||
| 735_s_at | HG2167-HT2237 | 2 | 4 | 1 | 2 | 2 | PKHt31 | ||
| 1461_at | M69043 | MAD-3, NFKBI | |||||||
| 31508_at | S73591 | 1 | 1 | 2 | 3 | 2 | VDUP1 | ||
| 32215_i_at, 32216_r_at | AB020685 | KIAA0878 | |||||||
| 31611_s_at | AF032457 | BIMEL | |||||||
| 35854_at | L14269 | 1 | 3 | 1 | SLC18A2 | ||||
| 35164_at | AF084481 | DFNA6 | |||||||
| 37112_at | AB002384 | DIFF40 | |||||||
| 38866_at | Z82206 | 2 | 1 | 11 | GRAP2 | ||||
| 32526_at | AA149644 | JAM3 | |||||||
| 1717_s_at | U45878 | 1 | 1 | 1 | 5 | BIRC3 | |||
| 36634_at | U72649 | 1 | 1 | 4 | 1 | 3 | BTG2 | ||
| 38799_at | AF068706 | AP1G2 | |||||||
| 37645_at | Z22576 | 1 | 2 | 1 | 2 | CD69 | |||
| 1814_at, 1815_g_at | D50683 | TGFBR2 | |||||||
| 390_at | X85740 | CMKBR4 | |||||||
| 38671_at | AB014520 | 1 | 1 | KIAA0620 | |||||
| 38661_at | X75315 | RNPC1 | |||||||
| 35917_at | W26631 | MAP1A | |||||||
| 33804_at | U43522 | 1 | 2 | 4 | 5 | PTK2B | |||
| 35763_at | AB011112 | 1 | 4 | 1 | KIAA0540 | ||||
| 32227_at | X17042 | 2 | 2 | 3 | 3 | 1 | PRG1 | ||
| B Repressed genes | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Probe i.d. | GenBank Acc. No. | Potential GREs |
TF sites ± 100 nt |
Name | |||||
| Palindrome | 1/2 GRE | AP-1 | c/EBP | NFκB | Oct-1 | CREB | |||
| 1077_at | M29474 | RAG1 | |||||||
| 37393_at | L19314 | HRY | |||||||
| 40455_at | AB020637 | 1 | 1 | 4 | KIAA0830 | ||||
| 1973_s_at, 37724_at | V00568 | 1 | 1 | 3 | 1 | c-myc | |||
| 40692_at | M99439 | 2 | 2 | 1 | 1 | TLE4 | |||
| 34805_at | AA195301 | 1 | 1 | 3 | 1 | 1 | 2 | MGC2574 | |
| 34517_at | X66435 | HMGCS1 | |||||||
| 38277_at | M29550 | 1 | 7 | 2 | 1 | PPP3CA | |||
| 38505_at | AL050151 | DKFZp586J0720 | |||||||
| 39235_at | AC003038 | 1 | 1 | 1 | 2 | 2 | 1 | DNA from cosmidR30923 | |
| 32118_at | AF076838 | 1 | 1 | 2 | RAD17 | ||||
| 34341_at | U00238 | 1 | 2 | 2 | GPAT | ||||
| 41861_at | AL050019 | DKFZp564C186 | |||||||
| 2061_at | L12002 | ITGA4 | |||||||
| 31838_at | U79274 | 1 | 3 | 1 | 1 | HSU79274 | |||
| 34998_at | AF059531 | PRMT3 | |||||||
| 41259_at | AI553745 | HSPC111 | |||||||
| 40729_s_at | Y14768 | LTB | |||||||
| 40020_at | AB011536 | CELSR3 | |||||||
| 35246_at | U18934 | 1 | 1 | TYRO3 | |||||
| 40982_at | AA926957 | 1 | 2 | FLJ10534 | |||||
Evaluations of GRE quality were based on criteria in Beato et al. [26].
Selected sites for transcription factors shown within 100 nucleotides of a GRE.
See GenBank or Celera databases for aliases.
Promoter analysis of glucocorticoid-regulated genes
We scanned the promoter/regulatory regions of the genes in Table 2A, for full palindromic glucocorticoid response elements (GREs) or hexameric “1/2 GREs.” The analysis identified 17 induced genes with one or more 1/2 GREs. Detailed examination of the sequences of these GREs showed 9 to be candidates for full, quasi-palindromic GREs; the remainder appeared to be 1/2 GREs. Sequences in the vicinity of the GREs often showed binding sites for CREB, c/EBP, NFκB, Oct1, and AP-1 transcription factors, with which the GR is known to be capable of interacting (Table 2).
The promoter/regulatory regions of the 21 genes in the sensitive cells whose mRNA products fell after glucocorticoid treatment revealed 11 that contained one or more 1/2 GREs (Table 2B). Of these, four appeared to be classic GRE palindromes. The specific “negative” GRE sequences described in two de-induced genes [27,28] were not found.
Discussion
The mechanism by which glucocorticoids cause the apoptosis of leukemic lymphoid (or normal) cells remains unknown. We subscribe to the general hypothesis that the activation of the actual apoptotic machinery requires a preceding period of gene regulation in the treated cell. We further hypothesized that during this period a reversible series of events takes place, and this group of events constitutes a network of regulatory interactions that culminate in one or more irreversible biochemical “triggers” being pulled, setting off apoptosis. In various systems of cultured, transformed lymphoid cells this preceding time period is rather long, on the order of 24 h.
We have taken advantage of this by studying a human leukemic lymphoid system in which a closely related set of three clones are compared at a time close to but before the irreversible apoptotic events are set in motion. The three clones all were derived from the CEM cell line and are recent subclones of a pair of clones isolated some time ago. C7 was the prototypical sensitive clone, and C1 the prototypical resistant clone. Among the C1 subclones, we serendipitously subcloned C1-6; one that had reverted to sensitivity. Therefore, our hypothesis predicts that C1-6 would show a set of Dex-regulated genes coincident with the critical set in C7, and that these would differ from what occurs in C1-15. We tested this prediction with gene array analysis. Considerable effort was spent in validating the data, and as given in Results, the validation tests strongly suggest that the data sets obtained were representations of the cellular responses. Over the course of 1 year, data from three experiments at the same time point after addition of Dex were obtained. This longitudinal approach strengthens the argument that the results found are consistent behaviors and not due to the idiosyncrasies of cellular activities at a particular point in time.
Although the Affymetrix gene chip employed does not represent the totality of genes expressed in human lymphoid cells, it is an excellent starting point, because the genes on this chip all represent identified full-length cDNAs. Cluster analysis based on control expression levels placed C1-6 closer to its sister clone C1-15 than to the inherently sensitive clone C7-14. These relationships change when one carries out cluster analysis based on the genes that are induced after adding Dex. Using that set only, C1-6 clusters more closely to C7-14 than to C1-15.
A number of genes were induced in response to the glucocorticoid in at least two of three experiments in the resistant clone C1-15; however, in this clone few genes were induced ≥2.5-fold in all three experiments. Initial examination of the induced genes in C1-15 again showed no obvious anti-apoptotic genes. The substantial number of genes found frequently induced or repressed in clone C1-15 indicates that the GR in these cells is capable of gene regulation, although the genes regulated differ from those in the apoptosis-sensitive clones. We have already shown that for a transfected, GR-driven gene in C1-15 cells, the level of induction is substantially enhanced by treatment with fors-kolin to activate the protein kinase A pathway [29]. It will be interesting to study the effects of such treatment on C1-15 endogenous genes.
Because cluster analysis of C7-14 and C1-6 after treatment with Dex showed that C1-6 became more closely clustered with C7-14, the other pro-apoptotic clone, we reasoned that the genes that led to apoptosis would have to be very consistent in their induction. By applying more stringent criteria than those used in the cluster analyses, we identified 39 genes that were induced ≥2.5-fold and 21 genes that are suppressed ≥2-fold in both of the sensitive clones. Obviously, using other criteria can produce additional genes in both the induced and repressed categories. Our chosen criteria, however, make these initial limited sets likely to be at least some of the genes relevant to the biological phenomenon of interest. By allowing the fold induction in one of three experiments to fall below the arbitrary limit, we took into account the possibility of one result being randomly aberrant. Of course, for a gene to be required for apoptosis, it would be expected to change in every experiment. In fact, the genes identified all did so (see Results). The variability cannot be attributed to GR mRNA fluctuations, because the chip data show that GR α and β mRNA levels essentially remained constant. It is intriguing that many of the genes identified encode proteins involved in regulatory pathways, plus pro- or anti-apoptotic functions. Additionally, among the glucocorticoid-induced genes are some contributing to structural integrity and membrane transport. Other induced genes include several that encode for cellular structural proteins.
Among the interesting genes induced at 20 h, one is Bim-EL, a pro-apoptotic member of the bcl2 family [30]. The late increase in such genes may precipitate apoptosis. Another gene specifically induced by Dex in glucocorticoid-sensitive cells is the DSCR1 gene [31]. DSCR1 encodes monocyte-enriched calcineurin interacting protein 1 (MCIP-1), which blocks calcineurin-dependent transcription [32]. Another target of glucocorticoid-mediated induction specific for glucocorticoid-sensitive cells is the gene encoding the adenovirus E4 promoter-binding protein-4 (E4BP4/NFIL3A), a transcriptional repressor in the bZIP family of transcription factors [33]. Also induced is inositol polyphosphate-1-phosphatase, an enzyme of the phosphatidylinositol signaling pathway that hydrolyzes the 1-phosphate from inositol triphosphate and inositol diphosphate. It has been implicated as an inhibitor of DNA synthesis [34]. Closer investigation of the Dex-mediated induction of the human receptor-like protein tyrosine phosphatase μ [35] and the protein tyrosine kinase interacting protein SOCS-1/TIP3 [36] (Table 2) may shed light on the role of protein tyrosine phosphorylation in glucocorticoid-evoked lympholysis. Growth-regulatory genes induced by Dex include BTG1, an antiproliferative gene that arrests cells in G0/G1 phase of the cell cycle and induces apoptosis in NIH3T3 cells [37]; Mad3, a c-Myc antagonist and hence pro-apoptotic in lymphoid cells [38]; and BIRC-3/IAP-1 (Table 2), which has been implicated as an anti-apoptotic factor [39]. This could represent a cellular attempt to activate compensatory anti-apoptotic mechanisms. Similar to a recently published report [40], our microarray analysis also demonstrated glucocorticoid-mediated induction of the interleukin-7 (IL-7) receptor α. In addition, Dex induces a number of gene products whose functions are either unknown or not directly apparent in glucocorticoid-mediated responses in lymphoid cells.
Among the repressed genes, we noted HRY, the human homolog of the Drosophila melanogaster gene hairy, which plays a key role in development and cell proliferation [41]. It contains a bHLH motif similar to c-myc, which also is repressed. Recombination activation gene-1 (RAG1) encodes a protein that regulates V(D)J recombination and immunoglobulin and T-cell diversity and may be active in immune cell survival [42].
As knowledge has grown regarding the ways by which glucocorticoids, through the GR, regulate gene transcription, it has become obvious that there are several mechanisms. These include GR homodimers binding directly to specific DNA response elements, GR-DNA interactions with heterologous transcription factors at composite DNA-binding sites, and strictly heterologous interactions in which the GR is tethered to DNA indirectly. Composite sites contain both a site for the heterologous factor and a 1/2 GRE. In heterologous protein-protein interactions, the GR may be tethered to DNA through the heterologous transcription factor or GR-factor interactions may prevent DNA binding.
The genes regulated in our experiments provided an opportunity to examine promoters to see what types of regulatory elements might be present in relevant genes. Each promoter/regulator region was examined up to −2.5 kb upstream from the transcription start site. In 11 of the promoters we identified a classic quasi-palindromic GRE; 5 of those contained an additional 1/2 GRE. We found 4 of the 11 in downregulated genes. Presumably in these, the negative regulation was due to a dominant repressive effect of other sites. In another 17 genes we noted at least one 1/2 GRE. In the vicinity of both types of GREs were found sites for other transcription factors known to interact with the GR, including those for Oct1, AP-1, c/EBP, CREB, and NFκB. The remainder of the induced genes showed no GRE in the 2.5-kb region examined. It may be that some of the induced genes are stimulated as secondary events as a consequence of changes in other regulatory gene products. Thus some of the changes seen could be indirect and not primary GR-mediated changes in transcription. For example, relief of repression due to downregulation of a repressor gene would cause upregulation of the repressed gene. With respect to the negatively regulated genes’ promoters, we found fewer GREs and neither of the negative GREs described for the pro-opiomelanocortin and prolactin genes.
Our microarray profiling generally correlates with published reports on known glucocorticoid-mediated effects on the transcription of individual genes. Our results confirm early observations that glucocorticoids modulate expression of a finite set of genes, rather than causing generalized transcriptional changes [12–17]. Only a few reports of microarray analyses of glucocorticoid-mediated gene regulation have been published: thus, detailed comparison of specific glucocorticoid-regulated genes so identified is difficult because of the differences in cells tested, the timing of Dex treatment, and/or the specific gene probes contained within the different arrays used. Glucocorticoid treatment of peripheral blood mononuclear cells showed an induction of cytokines, scavenger and Toll-like receptors, with mixed effects on apoptosis-related gene clusters [43]. In an attempt to segregate genes that cause growth arrest from those that cause apoptosis, Tonko et al. [44] compared glucocorticoid-regulated gene expression profiles in proliferating versus G1/G0-arrested CEM-C7-H2 cells, a subclone from the original glucocorticoid-sensitive CEM-C7 cells described here. Subsequently, the same laboratory obtained single-experiment data at early times after Dex addition to other CEM and to GR-containing Jurkat cells [45]. In both studies, single samples were taken at 2 or 3 and 8 h after the addition of Dex. Time-matched controls were not obtained. As the authors remarked, it is difficult to be certain which, among the genes possibly regulated in those studies, are random occurrences. Nevertheless, we note that several genes seen as regulated in our data also appear to be regulated in their more limited data set taken at earlier time points. In sensitive cells these include AIM1, DSCR1, and IL-7 receptor (IL7R) α (induced); c-myc, HRY, and RAG1 (repressed).
A death-associated protein kinase (DAP-kinase) family reported in the literature to participate in apoptotic cascades initiated by interferon-γ, tumor necrosis factor-α (TNFα), activated Fas, and detachment from extracellular matrix was evaluated [46]. Our arrays showed that there was basal expression of some family members but no regulation by Dex or differential expression in sensitive versus resistant clones.
In sum, our results support our initial hypothesis that a distinctive set of glucocorticoid-regulated genes would be found in cells destined to undergo apoptosis. Our data also suggest that the resistant clone C1-15 does have an active GR, but that a quite different gene set is regulated by it. We interpret this result as an indication that a regulatory switch has shifted the entire pattern of responses. The data here should provide a basis of comparison for glucocorticoid gene-regulatory effects in other apoptotically inclined lymphoid systems.
Materials and methods
Reagents
Dex and other reagent-grade chemicals were purchased from Sigma-Aldrich Chemical Company (St. Louis, MO, USA). The RNeasy total RNA isolation kit was from Qiagen (Santa Clara, CA, USA), T7-oligo (dT) promoter primer from Affymetrix (Santa Clara, CA, USA), and the BioArray high yield RNA transcription labeling kit from Enzo Biochem (New York, NY, USA). The following reagents were purchased from Invitrogen (Carlsbad, CA, USA): SuperScript II reverse transcriptase, dNTP mix, Escherichia coli DNA ligase, E. coli DNA polymerase I, RNase H, and T4 DNA polymerase. The Zeta Probe GT genomic blotting membrane for northern blots was from Bio-Rad (Richmond, CA, USA), the RediPrime II random primed DNA labeling system from Amersham-Pharmacia (Piscataway, NJ, USA), the α-[32P]dCTP from ICN (Irvine, CA, USA), and QuikHyb hybridization solution from Stratagene (La Jolla, CA, USA).
Cell culture
All CEM clones used in this study were cloned in 1996 in semisolid agarose medium in the absence of any selective pressure [47]. Thereafter, the cells were carefully maintained in logarithmic growth in Cellgro RPMI 1640 tissue culture medium (Mediatech, Herndon, VA, USA) supplemented with 5% FBS from Atlanta Biologicals (Norcross, GA, USA) at 37°C in a humidified 5% CO2, 95% air incubator.
RNA extraction
When cells had grown to a density of 4 × 105 cells/ml, they were treated with either ethanol vehicle (≤1% final concentration) or 1 μM Dex in vehicle for 20 h. Approximately 1 × 107 cells were harvested, washed once with chilled PBS, pH 7.4 (Cellgro) and resuspended in lysis buffer (RNeasy kit). The cell lysate was passed through a QIAshredder column (Qiagen) and processed for total RNA isolation as per the protocol provided (RNeasy kit). RNA samples were stored at −70°C in ethanol until used for either GeneChip analysis or northern hybridization. In each experiment an aliquot fraction of the culture was retained and followed for growth and apoptosis. The full apoptotic effect was seen in each experiment in the two sensitive clones, whereas the C1-15 clone was entirely resistant.
Target labeling and hybridization
First-strand cDNA synthesis was done using 10–25 μg of total RNA, a T7-(dT)24 oligomer (5′-GGCCAGTGAATTGTA ATACGACTCACTATAGGGAGGCGG-(dT)24-3′) and SuperScript II reverse transcriptase (Life Technologies, Baltimore, MD, USA). Second-strand synthesis converted the cDNA into a double-stranded DNA template, which was subjected to an in vitro transcription reaction using bacteriophage T7 RNA polymerase. The cRNA or target RNAs were labeled with biotin during the in vitro transcription reaction, then fragmented to a mean size of 200 bases to facilitate their hybridization to probe sequences of the HG_U95Av2 Affymetrix GeneChip Arrays. Each target RNA sample was initially hybridized to a test array that contained a set of probes representing genes commonly expressed in a majority of human cells, for example, actin, transferrin receptor, transcription factor ISGF-3, 18S RNA, 28S RNA, and Alu, to confirm the successful labeling of the target RNAs and prevent the use of degraded or nonrepresentative target RNA samples. Hybridization of GeneChip arrays was done at 45°C for 16 h in 0.1 M MES buffer, pH 6.6, 1 M NaCl, 0.02 M EDTA, and 0.01% Tween 20 detergent. Four prokaryotic genes (bioB, bioC, and bioD from the E. coli biotin synthesis pathway and cre the re-combinase gene from P bacteriophage) were added to the hybridization cocktail as internal controls. Arrays were washed using both nonstringent (SSPE-Tween 20 detergents, 25°C) and stringent (1 M NaCl, 50°C) conditions before staining with phycoerythrin-streptavidin (10 μg/ml final concentration). GeneChip arrays were scanned using a Gene Array Scanner (Hewlett Packard, Palo Alto, CA, USA) and analyzed using the Affymetrix GeneChip Suite 4.0 software.
Northern blotting
DNA fragments corresponding to the specified cDNA sequences were amplified from CEM-C7-14 cells treated with 1 μM Dex by RT-PCR using the following primers: BTG1 (361–751 bp, GenBank Accession No. X61123): sense, 5′-CCGTGTCCTTCATCTCCAAG-3′; antisense, 5′-CTGATTCGGCTGTCTACCAT-3′; DSCR1 (266– 659 bp, GenBank Accession No. U85267): sense, 5′-GGACAT-CACCTTTCAGTATT-3′; antisense, 5′-TTCCTCTTCT-TCCTCCTTCT-3′; and E4BP4 (273–1,587 bp, GenBank Accession No. X64318): sense, 5′-CAATGTGGACAA-GATGATGGTC-3′; antisense, 5′-AGCAGAGATTGGTT-GTGTGG-3′. The resulting PCR products were confirmed by sequencing and then were used as probes in northern hybridizations. A 245-bp (947–704 bp, GenBank Accession No. X00351) β-actin cDNA fragment from Ambion (Austin, TX, USA; catalog no. 7424) was labeled to serve as a probe for normalization between lanes. Total RNA (5 μg) from ethanol or 1 μM Dex-treated CEM-C7-14, CEM-C1-6, and CEM-C1-15 cells was separated on a 1.4% agarose gel under denaturing conditions (6% formaldehyde, 1× MOPS-EDTA buffer, pH 7.4, and 1 μg/ml ethidium bromide) for 16 h at 20 V. The resolved RNA was transferred on to ZetaProbe membrane by overnight capillary action, and the membrane was baked at 85°C in a vacuum oven for 45 min. For probe preparation, the appropriate gel-purified PCR products or DNA fragments were labeled with α[32P]dCTP using the Rediprime II random primed DNA labeling system (Amersham Biosciences). The membrane was prehybridized in QuikHyb solution (Stratagene) for 1 h at 60°C. Approximately 1 × 107 c.p.m. of the appropriate radiolabeled DNA probe was boiled with salmon sperm DNA (final concentration of 100 μg/ml), chilled on ice, and added to the QuikHyb prehybridization solution. Hybridization was carried out for 3 h at 60°C. The hybridized membrane was washed once at room temperature with 2 × SSC and 0.1% SDS (w/v) for 15 min, and then washed successively in 0.1% SSC (v/v) and 0.1% SDS for 30 min at 50, 55, and 57°C. After hybridization, membranes were stripped and reprobed for actin. Each membrane was scanned using the Molecular Dynamics Storm PhosphorImager (Sunnyvale, CA, USA) and the bands quantitated on an Alpha Innotech Imager (San Leandro, CA, USA).
Data analysis
Three independent experiments were done within a span of 1 year. In each, the ethanol- and Dex-treated samples from every cell clone were assessed. The raw data were obtained using Affymetrix software. Details regarding Affymetrix GeneChip design and the Affymetrix GeneChip Suite 4.0 algorithms can be obtained from the Affymetrix expression analysis technical manual or their website (http://www.affymetrix.com). Initial data processing was done using the Affymetrix GeneChip Suite 4.0, which uses absolute analysis algorithms to yield an average difference value for each of the 12,626 genes on the HG_U95Av2 array. Analyses were conducted with the software package GeneSpring (Silicon Genetics, Redwood City, CA, USA), version 4.2.1. Affymetrix 5.0 Pivotdata text files for control and treated chips were imported into GeneSpring and combined into single experiments for each replicate. For each individual experiment, control and treated chip were normalized together using the 50th percentile distribution of all genes.
Creating significant gene lists
To determine the genes regulated by Dex, the signals on chips receiving the products derived from Dex-treated cells were filtered to find the 5,000– 6,000 genes called “present,” and all other genes were dismissed for that particular experiment. All present genes were then compared to their counterparts on the control chips to find those that were induced at least 2.5-fold or suppressed at least 2.0-fold. These lists were then compared to elucidate those genes induced or suppressed in at least two out of the three experiments. Genes regulated by Dex in each of the three clones were compared with the results expressed by Venn diagrams and lists of specific genes. Differences in basal gene expression were studied using Significant Analysis of Microarrays software (Stanford University, Stanford, CA, USA). An unpaired Student’s t-test was used to determine genes that were significantly different for one group to another, keeping the false discovery rate below 1%.
DNA-binding site analysis
Using the program BLASTN, GenBank sequences for the genes identified in Table 2 were compared to the Celera Discovery System database to obtain their genomic sequences. From these, ~2.5 kb upstream of the presumptive transcription initiation site was saved for each gene. The sequence was then explored for possible transcription factor sites, using Match version 1.5 within the TRANSFAC Professional 6.2 database, which contains a library of mononucleotide weight matrices. Only high-quality matrices (predetermined on the TRANSFAC database) were used in our search. Matrices for GRE and other transcription factors that are known to bind GR (i.e., NFκB, CREB, AP-1, Oct1, and c/EBP) were employed to search the sequences using a user-defined 0.80 as the cutoff for core and matrix similarity.
Clustering
All replicate chips were merged into single experiments. Consolidated control and treated experiments were compared using a Welch-ANOVA to reveal any significant genes (P < 0.05). The significant genes list was used as a filter to cluster individual experiments using a variety of different algorithms.
Acknowledgments
The authors thank GopalRao V.M. Velagaletia and the staff of the Cytogenetics Laboratory at UTMB for karyotyping the clones used in this study. We also wish to recognize the contribution of Michelle Guigneaux of the DNA Recombinant Laboratory at UTMB for technical assistance in the microarray analyses.
References
- 1.Dougherty TF, White A. Functional alterations in lymphoid tissue induced by adrenal cortical secretion. Am J Anat. 1945;77:81–116. [Google Scholar]
- 2.Gaynon PS, Carrel AL. Glucocorticosteroid therapy in childhood acute lymphoblastic leukemia. Adv Exp Med Biol. 1999;457:593–605. doi: 10.1007/978-1-4615-4811-9_66. [DOI] [PubMed] [Google Scholar]
- 3.Blewitt RW, Abbott AC, Bird CC. Mode of cell death induced in human lymphoid cells by high and low doses of glucocorticoid. Br J Cancer. 1983;47:477–486. doi: 10.1038/bjc.1983.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wyllie AH, Morris RG, Smith AL, Dunlop D. Chromatin cleavage in apoptosis: association with condensed chromatin morphology and dependence on macromolecular synthesis. J Pathol. 1984;142:67–77. doi: 10.1002/path.1711420112. [DOI] [PubMed] [Google Scholar]
- 5.Thompson EB. Glucocorticoids and oxysterols in lymphoid apoptosis. In: O’Malley BW, editor. Hormones and Signaling. Vol. 1. Academic Press; San Diego, CA: 1998. pp. 1–40. [Google Scholar]
- 6.Compton MM, Haskil JS, Cidlowski JA. Analysis of glucocorticoid actions on rat thymocyte deoxyribonucleic acid by fluorescence-activated flow cytometry. Endocrinology. 1988;122:2158–2164. doi: 10.1210/endo-122-5-2158. [DOI] [PubMed] [Google Scholar]
- 7.Bansel N, Houle A, Melnykovych G. Apoptosis: mode of cell death induced in T-cell leukemia lines by dexamethasone and other agents. FASEB J. 1991;5:211–216. doi: 10.1096/fasebj.5.2.2004665. [DOI] [PubMed] [Google Scholar]
- 8.Wood AC, Waters CM, Garner A, Hickman JA. Changes in c-myc expression and the kinetics of dexamethasone-induced programmed cell death (apoptosis) in human lymphoid leukemic cells. Br J Cancer. 1994;69:663–669. doi: 10.1038/bjc.1994.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Perrin-Wolff M, Bertoglio J, Bressac B, Bohuon C, Pallardy M. Structure-activity relationships in glucocorticoid-induced apoptosis in T lymphocytes. Biochem Pharmacol. 1995;50:103–110. doi: 10.1016/0006-2952(94)00527-s. [DOI] [PubMed] [Google Scholar]
- 10.Ramdas J, Harmon JM. Glucocorticoid-induced apoptosis and regulation of NFkB activity in human leukemic T cells. Endocrinology. 1998;139:3813–3821. doi: 10.1210/endo.139.9.6180. [DOI] [PubMed] [Google Scholar]
- 11.Aravind L, Dixit VM, Koonin EV. Apoptotic molecular machinery: vastly increased complexity in vertebrates revealed by genome comparisons. Science. 2001;291:1279–1290. doi: 10.1126/science.291.5507.1279. [DOI] [PubMed] [Google Scholar]
- 12.Briehl MM, Flomerfelt FA, Wu XP, Miesfeld RL. Transcriptional analyses of steroid-regulated gene networks. Mol Endocrinol. 1990;4:287–294. doi: 10.1210/mend-4-2-287. [DOI] [PubMed] [Google Scholar]
- 13.Harrigan MT, Baughman G, Campbell NF, Bourgeois S. Isolation and characterization of glucocorticoid- and cyclic AMP-induced genes in T lymphocytes. Mol Cell Biol. 1989;9:3438–3446. doi: 10.1128/mcb.9.8.3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yuh YS, Thompson EB. Glucocorticoid effect on oncogene/growth gene expression in human T-lymphoblastic leukemic cell line CCRF-CEM: specific c-myc RNA suppression by dexamethasone. J Biol Chem. 1989;264:10904–10910. [PubMed] [Google Scholar]
- 15.Harrigan MT, Campbell NF, Bourgeois S. Identification of a gene induced by glucocorticoids in murine T-cells: a potential G protein-coupled receptor. Mol Endocrinol. 1991;5:1331–1338. doi: 10.1210/mend-5-9-1331. [DOI] [PubMed] [Google Scholar]
- 16.Baughman G, Harrigan MT, Campbell NF, Nurrish SJ, Bourgeois S. Genes newly identified as regulated by glucocorticoids in murine thymocytes. Mol Endocrinol. 1991;5:637–644. doi: 10.1210/mend-5-5-637. [DOI] [PubMed] [Google Scholar]
- 17.Chapman MS, et al. Isolation of differentially expressed sequence tags from steroid-responsive cells using mRNA differential display. Mol Cell Endocrinol. 1995;108:R1–R7. doi: 10.1016/0303-7207(95)03481-l. [DOI] [PubMed] [Google Scholar]
- 18.Foley GE, et al. Continuous culture of human lymphoblasts from peripheral blood of a child with acute leukemia. Cancer Res. 1965;8:522–529. doi: 10.1002/1097-0142(196504)18:4<522::aid-cncr2820180418>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 19.Howell AL, Richie ER. Phenotypic drift and clonal variation in differentiation antigen expression on AKR T-cell lymphoma lines grown in vitro. Nat Immun Cell Growth Regul. 1983;3:143–154. [PubMed] [Google Scholar]
- 20.Leglise MC, Dent GA, Ayscue LH, Ross DW. Leukemic cell maturation: a review. Blood Cells. 1988;13:319–337. [PubMed] [Google Scholar]
- 21.Leger I, Thomas M, Ronot X, Brugal G. Detection of chromosome 1 aberrations by fluorescent in situ hybridization (FISH) in the human breast cancer cell line MCF-7. Anal Cell Pathol. 1993;5:299–309. [PubMed] [Google Scholar]
- 22.Norman MR, Thompson EB. Characterization of a glucocorticoid-sensitive human lymphoid cell line. Cancer Res. 1977;37:3785–3791. [PubMed] [Google Scholar]
- 23.Harmon JM, Thompson EB. Isolation and characterization of dexamethasone-resistant mutants from human lymphoid cell line CEM-C7. Mol Cell Biol. 1981;1:512–521. doi: 10.1128/mcb.1.6.512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harmon JM, Thompson EB, Baione KA. Analysis of glucocorticoid-resistant human leukemic cells by somatic cell hybridization. Cancer Res. 1985;45:1587–1593. [PubMed] [Google Scholar]
- 25.Miller AL, Johnson BH, Medh RD, Townsend CM, Thompson EB. Glucocorticoids and polyamine inhibitors synergize to kill human leukemic CEM cells. Neoplasia. 2002;4:68–81. doi: 10.1038/sj.neo.7900208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Beato M, Chalepakis G, Schauer M, Slater EP. DNA regulatory elements for steroid hormones. J Steroid Biochem. 1989;32:737–748. doi: 10.1016/0022-4731(89)90521-9. [DOI] [PubMed] [Google Scholar]
- 27.Drouin J, et al. Glucocorticoid receptor binding to a specific DNA sequence is required for hormone-dependent repression of pro-opiomelanocortin gene transcription. Mol Cell Biol. 1989;9:5305–5314. doi: 10.1128/mcb.9.12.5305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Subramaniam N, Cairns W, Okret S. Studies on the mechanism of glucocorticoid-mediated repression from a negative glucocorticoid response element from the bovine prolactin gene. DNA Cell Biol. 1997;16:153–163. doi: 10.1089/dna.1997.16.153. [DOI] [PubMed] [Google Scholar]
- 29.Medh RD, Saeed MF, Johnson BH, Thompson EB. Resistance of human leukemic CEM-C1 cells is overcome by synergism between glucocorticoid and protein kinase A pathways: correlation with c-Myc suppression. Cancer Res. 1998;58:3684–3693. [PubMed] [Google Scholar]
- 30.O’Connor L, et al. Bim: a novel member of the Bcl-2 family that promotes apoptosis. EMBO J. 1998;17:384–395. doi: 10.1093/emboj/17.2.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fuentes JJ, et al. A new human gene from the Down syndrome critical region encodes a proline-rich protein highly expressed in fetal brain and heart. Hum Mol Genet. 1995;4:1935–1944. doi: 10.1093/hmg/4.10.1935. [DOI] [PubMed] [Google Scholar]
- 32.Rothermel B, et al. A protein encoded within the Down syndrome critical region is enriched in striated muscles and inhibits calcineurin signaling. J Biol Chem. 2000;275:8719–8725. doi: 10.1074/jbc.275.12.8719. [DOI] [PubMed] [Google Scholar]
- 33.Cowell IG, Skinner A, Hurst HC. Transcriptional repression by a novel member of the bZIP family of transcription factors. Mol Cell Biol. 1992;12:3070–3077. doi: 10.1128/mcb.12.7.3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Woodcock EA, et al. Inositol polyphosphate 1-phosphatase is a novel antihypertrophic factor. J Biol Chem. 2002;277:22734–22742. doi: 10.1074/jbc.M110405200. [DOI] [PubMed] [Google Scholar]
- 35.Gebbink MF, et al. Cloning, expression and chromosomal localization of a new putative receptor-like protein tyrosine phosphatase. FEBS Lett. 1991;290:123–130. doi: 10.1016/0014-5793(91)81241-y. [DOI] [PubMed] [Google Scholar]
- 36.Ohya K, et al. SOCS-1/JAB/SSI-1 can bind to and suppress Tec protein-tyrosine kinase. J Biol Chem. 1997;272:27178–27182. doi: 10.1074/jbc.272.43.27178. [DOI] [PubMed] [Google Scholar]
- 37.Matsuda S, Rouault J, Magaud J, Berthet C. In search of a function for the TIS21/PC3/BTG1/TOB family. FEBS Lett. 2001;497:67–72. doi: 10.1016/s0014-5793(01)02436-x. [DOI] [PubMed] [Google Scholar]
- 38.Haskill S, et al. Characterization of an immediate-early gene induced in adherent monocytes that encodes I κB-like activity. Cell. 1991;65:1281–1289. doi: 10.1016/0092-8674(91)90022-q. [DOI] [PubMed] [Google Scholar]
- 39.Liston P, et al. Suppression of apoptosis in mammalian cells by NAIP and a related family of IAP genes. Nature. 1996;379:349–353. doi: 10.1038/379349a0. [DOI] [PubMed] [Google Scholar]
- 40.Franchimont D, et al. Positive effects of glucocorticoids on T cell function by up-regulation of IL-7 receptor α. J Immunol. 2002;168:2212–2218. doi: 10.4049/jimmunol.168.5.2212. [DOI] [PubMed] [Google Scholar]
- 41.Feder JM, Li L, Jan LY, Jan YN. Genomic cloning and chromosomal localization of HRY, the human homolog to the Drosophila segmentation gene, hairy. Genomics. 1994;20:56–61. doi: 10.1006/geno.1994.1126. [DOI] [PubMed] [Google Scholar]
- 42.Alt FW, Rathbun G, Oltz E, Taccioli G, Shinkai Y. Function and control of recombination-activating gene activity. Ann NY Acad Sci. 1992;651:277–294. doi: 10.1111/j.1749-6632.1992.tb24626.x. [DOI] [PubMed] [Google Scholar]
- 43.Galon J, et al. Gene profiling reveals unknown enhancing and suppressive actions of glucocorticoids on immune cells. FASEB J. 2002;16:61–71. doi: 10.1096/fj.01-0245com. [DOI] [PubMed] [Google Scholar]
- 44.Tonko M, Ausserlechner MJ, Bernhard D, Helmberg A, Kofler R. Gene expression profiles of proliferating vs. G1/G0 arrested human leukemia cells suggest a mechanism for glucocorticoid-induced apoptosis. FASEB J. 2001;15:693–699. doi: 10.1096/fj.00-0327com. [DOI] [PubMed] [Google Scholar]
- 45.Obexer P, Certa U, Kofler R, Helmberg A. Expression profiling of glucocorticoid-treated T-ALL cell lines: rapid repression of multiple genes involved in RNA-, protein-, and nucleotide synthesis. Oncogene. 2001;20:4324–4336. doi: 10.1038/sj.onc.1204573. [DOI] [PubMed] [Google Scholar]
- 46.Shohat G, et al. The pro-apoptotic function of death-associated protein kinase is controlled by a unique inhibitory autophosphorylation-based mechanism. J Biol Chem. 2001;276:47460–47467. doi: 10.1074/jbc.M105133200. [DOI] [PubMed] [Google Scholar]
- 47.Juneja HS, Harvey WH, Brasher WK, Thompson EB. Successful in vitro purging of leukemic blasts from marrow by Cortivazol, a pyrazolosteroid: a preclinical study for autologous transplantation in acute lymphoblastic leukemia and non-Hodgkin’s lymphoma. Leukemia. 1995;9:1771–1778. [PubMed] [Google Scholar]