Abstract
Genes are not simply turned on or off, but instead their expression is fine-tuned to meet the needs of a cell. How genes are modulated so precisely is not well understood. The glucocorticoid receptor (GR) regulates target genes by associating with specific DNA binding sites, the sequences of which differ between genes. Traditionally, these binding sites have been viewed only as docking sites. Using structural, biochemical, and cell-based assays, we show that GR binding sequences, differing by as little as a single base pair, differentially affect GR conformation and regulatory activity. We therefore propose that DNA is a sequence-specific allosteric ligand of GR that tailors the activity of the receptor toward specific target genes.
Allosteric mechanisms have evolved to modulate protein function. Allostery can enable a protein to integrate and respond to multiple signals. For example, nuclear hormone receptors, such as the glucocorticoid receptor (GR), use hormones as allosteric effectors of their transcriptional regulatory activity (1); additional inputs, such as phosphorylation, also affect GR function (2).
Upon hormone binding, GR associates with high affinity to genomic GR binding sequences (GBSs), typically imperfect palindromic, hexameric half sites separated by 3–base pair (bp) spacers (3). Within these 15-bp GBSs, five positions are nearly invariant, whereas the remainder can be altered with little effect on GR binding (4) and vary substantially among all functional GBSs (5). In contrast, certain GBSs linked to target genes are highly conserved across species (5). Similarly, single nucleotide differences in NF-κB binding sequences determine cofactor specificity for NF-κB dimers (6). Thus, the precise nucleotide sequence of a regulatory factor binding site, in addition to guiding the factor to specific genomic loci, may also specify the mode of transcriptional regulation.
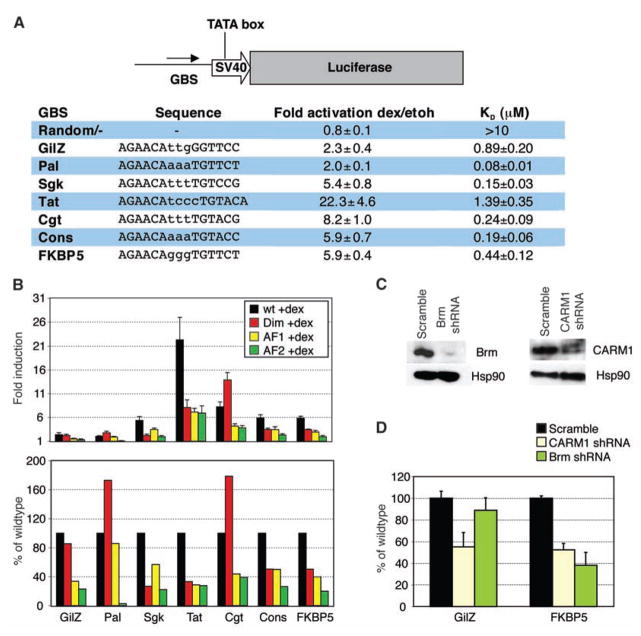
To examine GBS effects on GR activity, we constructed luciferase reporters, each containing a single 15-bp core GBS (Fig. 1A). The GBSs, differing by as little as 1 bp, were derived from endogenous target genes, matched the GR consensus motif (5), or were reportedly responsive (7). The reporters displayed comparable basal activities (fig. S1A), whereas induction by dexamethasone (dex), a synthetic glucocorticoid, varied from ~twofold (Pal, GilZ) to ~20-fold (TAT) (Fig. 1A).
Fig. 1.
GBSs differentially direct GR activity. (A) GBSs were cloned upstream of a minimal SV40 promoter driving luciferase. Transcriptional activities and binding affinities (humanGR-DBD 380 to 540) for each GBSs ± SEM are shown [number of independent experiments (n) ≥ 3]. KD, dissociation constant. (B) GBS- specific patterns of domain utilization. GBS reporters respond differentially to mutations in Dim (red, A477T), AF1 (yellow, E219K/F220L/W234R), and AF2 (green, E773R) domains. Fold induction by dex ± SEM (top) and percent induction by mutant GR relative to wild type (bottom) are shown (n ≥ 3). (C) Immunoblots demonstrating short hairpin–mediated RNA (shRNA) knock-down of Brm and CARM1. (D) GBS inductions after CARM1 or Brm knock-down, relative to scrambled shRNA ± SEM, are shown (n = 3).
However, gel shift assays revealed no correlation between in vitro GBS affinities and in vivo transcriptional activities (Fig. 1A and fig. S1C) for a GR fragment with DNA binding affinities comparable to that of full-length GR (8, 9). For example, TAT was 2- to 10-fold more active than the other GBSs but bound comparably to those with lower activities, whereas GBSs with similar transcriptional activities (Pal, GilZ) displayed different affinities. Moreover, merely reversing the orientation of asymmetric GBSs relative to the transcription start site, which presumably does not alter their affinities, altered their regulatory activities (fig. S2). These data suggest that GBS half sites confer unique function to the associated monomer.
Next, we tested the effects of mutating each of three GR surfaces implicated in transcriptional activation: (i) activation function 1 (AF1), (ii) AF2, and (iii) the dimerization region (Dim) (10). Wild-type GR or point mutants that abrogate each activity were cotransfected with GBS reporter plasmids into U2OS cells, which lack endogenous GR expression. Similar to endogenous target genes (10), we found a GBS-specific usage of GR surfaces (Fig. 1B). For example, GBSs that differed by 1 bp (Cgt, Sgk) differed in their dependence on the Dim domain, and GBSs with identical half sites but different spacer sequences (FKBP5, Pal) used all three surfaces differently. Pal and GilZ used different patterns of GR surfaces to arrive at their final indistinguishable activities. Furthermore, for a ~1-kb genomic fragment that recapitulated the domain utilization of endogenous GilZ [see supporting online material (SOM) text] (10, 11), changing only GBS sequences changed domain utilization, suggesting that GBSs direct GR activity, even in the context of large composite elements found at endogenous genes (fig. S3).
To assess GBS specific actions of GR cofactors, we knocked down expression of SWI/SNF subunit Brahma (Brm) (12) and coactivator-associated arginine methyltransferase 1 (CARM1) (13). Knock-down of CARM1 reduced activation for each GBS tested (Fig. 1, C and D), whereas Brm knock-down had effects ranging from only ~10% reduction on the GilZ GBS to ~60% at FKBP5 (Fig. 1, C and D). Thus, the influence of GBS sequences on GR, in turn, alters the composition or function of cognate regulatory complexes.
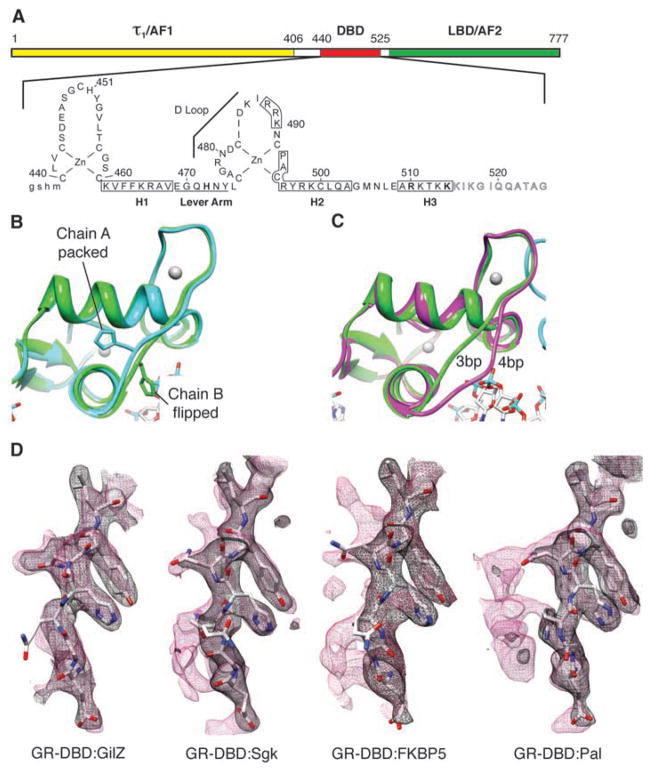
To test the structural basis for these sequence specific changes, we examined the GR–DNA binding domain (GR-DBD) (Fig. 2A) in complex with various GBSs by x-ray crystallography. Each GBS was bound asymmetrically by a GR-DBD dimer, with crystal packing contacts between dimerization domain residues on both monomers and a surface of one monomer, denoted here as chain A (fig. S4). By convention, we place chain A in contact with the conserved AGAACA half site and chain B in contact with the variable half site. The chain A crystal packing surface included parts of the DNA recognition helix, the Dim domain, and the “lever arm” (residues 469 to 474) (9, 14) loop connecting these two motifs (Fig. 2B). In contrast to a described structure (a palindrome with a non-canonical 4-bp spacer) (15), the 3-bp spacer allowed both recognition helices to make specific major groove contacts (figs. S4 and S5). The contact made by R466 (16) was invariant, whereas V462, and more profoundly K461, were GBS-specific (fig. S5). However, the lack of fixed DNA orientation (17) precluded definitive attribution of side chain contacts to individual bases.
Fig. 2.
DNA sequence-mediated structural differences in GR-DBD. (A) Domain structure of GR. τ1, tau1. (B) Overlay of chains A and B from GR-DBD:Pal complex shows packed and flipped conformations. (C) Overlay of chain B from GR-DBD complexed with 4-bp spacer (15) (magenta) and 3-bp spacer GBS (green). (D) Composite omit maps of GR-DBD complexed with different GBSs (GilZ, FKBP5, Sgk, and Pal) under the same conditions. Lever arm peptide is shown with 2Fo-Fc (black mesh) and composite omit map (red mesh) overlaid.
We also observed a C-terminal helix (H3; residues 509 to 515) that, as with progesterone receptor (PR) (18), made a nonspecific minor groove backbone contact 3 bp upstream of the GBS motif mediated by R510 (fig. S6). Distinct from PR, H3 of GR laid across the minor groove, presenting five lysines following R510 as potential sources of interaction. The R510A mutation reduced GR-DBD affinity for DNA (~threefold) and K514A (~twofold), whereas transcriptional activation was unaffected by the K514A mutation and increased by R510A, reaffirming that regulatory activity is not determined solely by affinity (fig. S6).
A comparison of 13 GR-DBD:GBS structures revealed that they were virtually superimposable, except for the lever arm. In every complex, H472 within the lever arm adopted one of two distinct orientations (Fig. 2B and fig. S7, A and B). In chain A of each structure, H472 packed identically in the core of the protein fold; in chain B, H472 was flipped out, and the lever arm conformation was more heterogeneous, particularly between complexes with a different spacer length (Fig. 2C). The chain B lever arm of four different GR-DBD:GBS complexes crystallized under the same conditions refined to a similar predominant conformation, albeit with higher B factors, indicating less well-defined structure. Composite omit maps (for an explanation, see SOM text) revealed additional electron density specifically at the lever arm, indicating discrete alternate conformations (Fig. 2D). The densities were distinct for each complex, suggesting that DNA sequence directs distinct changes in the lever arm.
The conformation of the lever arm appeared to be influenced by DNA backbone contacts at one or both ends. At one end, Y474 formed a weak hydrogen bond to the DNA backbone at GilZ but not the other GBSs (fig. S8). At the other end, E469 contacted the DNA backbone through K465. GilZ displayed the strongest interactions at these two sites and no alternate conformations, whereas for the other GBSs, the anchoring was weaker and exhibited alternate conformations (Fig. 2D). Thus, the conformation of the lever arm is specified by DNA topology, which, in turn, is determined by sequence. Crystallization of GR-DBD:GilZ complexes under a range of conditions revealed substantial lever arm heterogeneity, whereas GR-DBD:Sgk complexes were invariant under the crystallization conditions tested (fig. S7, C and D). Together, these structures reveal that the lever arm is conformationally sensitive, responding to small environmental shifts, including DNA sequence.
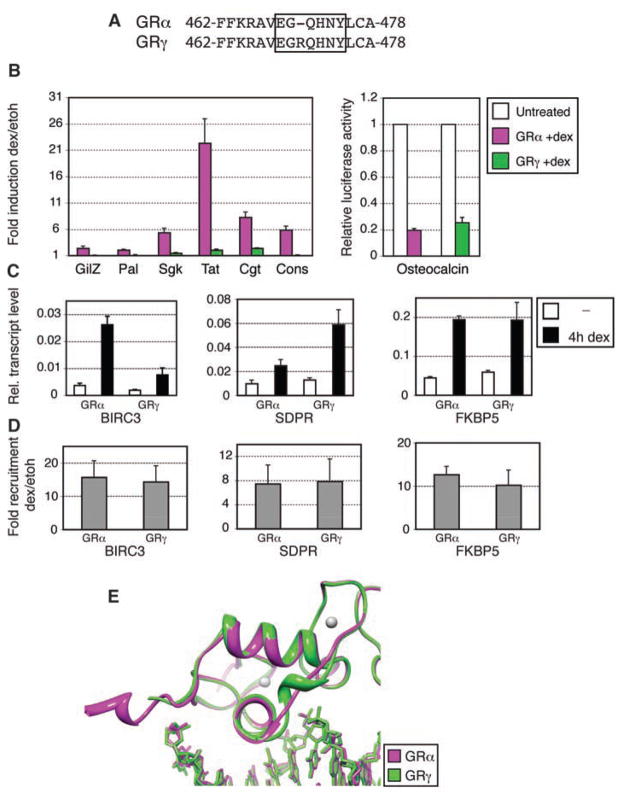
To assess the role of the lever arm in modulating GR activity, we studied GRγ, a splice variant that inserts an arginine in the lever arm (Fig. 3A) (19). Consistent with the initial description of GRγ (19), reporter assays revealed reduced activity for each GBS (Fig. 3B). Relative to the predominant GRα isoform, GRγ displayed normal DNA binding affinity (fig. S9) and near-normal repression of osteocalcin (20), suggesting that the lever arm selectively affects transcriptional activation. We then tested the role of the lever arm on activity at endogenous genes by comparing U2OS cells stably expressing GRα (21) or GRγ. The two isoforms were generally similar, but transcriptional regulation was distinct at a subset of target genes (Fig. 3C and fig. S10). Transcriptional activation of FKBP5 by GRγ was equivalent to GRα, whereas SDPR was elevated and BIRC3 was decreased by GRγ. Chromatin immunoprecipitation showed comparable GRα and GRγ occupancy (Fig. 3D) at these genes (5), indicating that the lever arm modulates events subsequent to GR:GBS binding to produce gene-specific, probably GBS-specific, regulation.
Fig. 3.
Activities and structure of GRγ. (A) GRγ amino acid sequence, showing Arg insertion in the lever arm. (B) U2OS cells were cotransfected with GRα or GRγ, together with GBS reporters (left) or with an osteocalcin reporter (right). Fold induction (left) and luciferase activity relative to untreated cells (right) ± SEM are shown (n = 3). (C) Regulation of endogenous target genes in U2OS cells stably expressing GRα or GRγ, measured by quantitative real-time fluorescence polymerase chain reaction. (D) Chromatin immunoprecipitation of GR at GBSs of isoform-specific target genes; GR recruitment upon dex treatment ± SEM is shown (n = 3). (E) Overlay of structures for GRα:FKBP5 and GRγ:FKBP5 complexes.
We crystallized GRγ-DBD:GBS complexes and found that the structures were similar to those with GRα, except within the lever arm (Fig. 3E). Base-specific and DNA backbone contacts were maintained (fig. S11), reflecting the similar affinities of GRα and GRγ. Thus, the conformational differences in the lever arm have functional consequences.
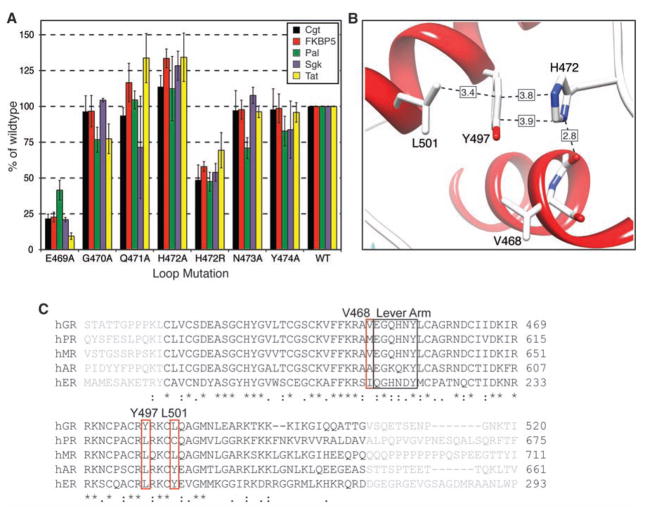
Mutational analysis of the lever arm revealed that E469 was an important residue for mediating activation (Fig. 4A) and that the H472A mutation produced increased activity, whereas H472R impaired GR activation. Several lever arm mutations had GBS-specific effects (Fig. 4A): N473A selectively reduced activation at Pal, whereas G470A reduced activation at Pal and Tat. Thus, different lever arm conformations may produce multiple functionally distinct interaction surfaces.
Fig. 4.
Receptor activity is modulated by lever arm residues. (A) H472 is critical for tuning activity. Effects of mutating lever arm residues were assayed using GBS reporters; activities are plotted as percentage of wild type ± SEM (n ≥ 3). (B) H472 resides in the DBD pocket formed by the carbonyl adjacent to V468, Y497, and L501. (C) Human DBD sequence alignments reveal variation at V468, Y497, and L501.
In chain A, the packing of H472 occurred through interactions with residues within the DBD core that are GR-specific within the nuclear hormone receptor family (Fig. 4C): Y497, L501, and the carbonyl adjacent to V468 (Fig. 4B). GRα complexes with 16- and 18-bp GBSs displayed different crystal packing, yet all exhibited the packed chain A and flipped out chain B (fig. S12). Disruption of chain A packing by a Y497L mutation (22) or by the R insertion in GRγ affected receptor activity in certain contexts, suggesting that packing of GRα chain A confers functional consequences quite distinct from those seen if both chains are flipped, as in GRγ and perhaps in other nuclear hormone receptors.
Protein functions have evolved commonly to be modulated by cellular signals. Based on our results, we propose that DNA sequences serve as one such signal, functioning as allosteric ligands that direct the activity of GR and probably other transcriptional regulators (23). Evidence for transduction of structural changes from DNA to other nuclear receptor domains has been described: DNA binding by GR induces secondary structure in its AF1 domain (24), and the estrogen receptor (ER) AF2 domain interacts with different cofactor peptides when ER is bound to different sequences (25, 26). We propose that conformational changes in the lever arm amplify signals at the reading head and transmit them to other domains. Our studies with the GR-DBD are an important first step establishing that distinct binding site sequences induce subtle structural differences that are propagated and functionally amplified in the context of the full-length receptor. Studies of other transcriptional regulatory factors imply that interpretation of sequence may be a general property of DNA binding proteins (6, 23, 27).
Supplementary Material
Footnotes
References and Notes
- 1.Nagy L, Schwabe JW. Trends Biochem Sci. 2004;29:317. doi: 10.1016/j.tibs.2004.04.006. [DOI] [PubMed] [Google Scholar]
- 2.Ismaili N, Garabedian MJ. Ann NY Acad Sci. 2004;1024:86. doi: 10.1196/annals.1321.007. [DOI] [PubMed] [Google Scholar]
- 3.Strahle U, Klock G, Schutz G. Proc Natl Acad Sci USA. 1987;84:7871. doi: 10.1073/pnas.84.22.7871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.La Baer J, Yamamoto KR. J Mol Biol. 1994;239:664. doi: 10.1006/jmbi.1994.1405. [DOI] [PubMed] [Google Scholar]
- 5.So AY, Chaivorapol C, Bolton EC, Li H, Yamamoto KR. PLoS Genet. 2007;3:e94. doi: 10.1371/journal.pgen.0030094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Leung TH, Hoffmann A, Baltimore D. Cell. 2004;118:453. doi: 10.1016/j.cell.2004.08.007. [DOI] [PubMed] [Google Scholar]
- 7.Iniguez-Lluhi JA, Pearce D. Mol Cell Biol. 2000;20:6040. doi: 10.1128/mcb.20.16.6040-6050.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Godowski PJ, Rusconi S, Miesfeld R, Yamamoto KR. Nature. 1987;325:365. doi: 10.1038/325365a0. [DOI] [PubMed] [Google Scholar]
- 9.Amino acid numbering refers to rat GR. We conducted affinity assays with the use of human GR-DBD 380 to 540 (rat 401 to 558).
- 10.Rogatsky I, et al. Proc Natl Acad Sci USA. 2003;100:13845. doi: 10.1073/pnas.2336092100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang JC, et al. Proc Natl Acad Sci USA. 2004;101:15603. doi: 10.1073/pnas.0407008101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Muchardt C, Yaniv M. EMBO J. 1993;12:4279. doi: 10.1002/j.1460-2075.1993.tb06112.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ma H, et al. Curr Biol. 2001;11:1981. doi: 10.1016/s0960-9822(01)00600-5. [DOI] [PubMed] [Google Scholar]
- 14.van Tilborg MA, et al. J Mol Biol. 2000;301:947. doi: 10.1006/jmbi.2000.4001. [DOI] [PubMed] [Google Scholar]
- 15.Luisi BF, et al. Nature. 1991;352:497. doi: 10.1038/352497a0. [DOI] [PubMed] [Google Scholar]
- 16.Single-letter abbreviations for the amino acid residues are as follows: A, Ala; C, Cys; D, Asp; E, Glu; F, Phe; G, Gly; H, His; I, Ile; K, Lys; L, Leu; M, Met; N, Asn; P, Pro; Q, Gln; R, Arg; S, Ser; T, Thr; V, Val; W, Trp; and Y, Tyr.
- 17.Materials and methods are available as supporting material on Science Online.
- 18.Roemer SC, et al. Mol Endocrinol. 2006;20:3042. doi: 10.1210/me.2005-0511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kasai Y. FEBS Lett. 1990;274:99. doi: 10.1016/0014-5793(90)81339-p. [DOI] [PubMed] [Google Scholar]
- 20.Meyer T, Gustafsson JA, Carlstedt-Duke J. DNA Cell Biol. 1997;16:919. doi: 10.1089/dna.1997.16.919. [DOI] [PubMed] [Google Scholar]
- 21.Rogatsky I, Trowbridge JM, Garabedian MJ. Mol Cell Biol. 1997;17:3181. doi: 10.1128/mcb.17.6.3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heck S, et al. EMBO J. 1994;13:4087. doi: 10.1002/j.1460-2075.1994.tb06726.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lefstin JA, Yamamoto KR. Nature. 1998;392:885. doi: 10.1038/31860. [DOI] [PubMed] [Google Scholar]
- 24.Kumar R, et al. J Biol Chem. 1999;274:24737. doi: 10.1074/jbc.274.35.24737. [DOI] [PubMed] [Google Scholar]
- 25.Heery DM, Kalkhoven E, Hoare S, Parker MG. Nature. 1997;387:733. doi: 10.1038/42750. [DOI] [PubMed] [Google Scholar]
- 26.Hall JM, McDonnell DP, Korach KS. Mol Endocrinol. 2002;16:469. doi: 10.1210/mend.16.3.0814. [DOI] [PubMed] [Google Scholar]
- 27.Scully KM, et al. Science. 2000;290:1127. doi: 10.1126/science.290.5494.1127. [DOI] [PubMed] [Google Scholar]
- 28.We thank E.C. Bolton, A. Kroch, J. J. Miranda, and A. Frankel for reviewing the manuscript. Work by M.A.P. was performed under NIH grant GM08537 and by M.A.P. and S.H.M. as fellows of the Leukemia and Lymphoma Society. We received research support from NIH grants (to K.R.Y., D.L.B., and L.C.). Coordinates and structure factors have been deposited in the Protein Data Bank with accession codes 3FYL, 3G6P, 3G6Q, 3G6R, 3G6T, 3G6U, 3G8U, 3G97, 3G8X, 3G99, 3G9I, 3G9J, 3G9M, 3G9O, and 3G9P. K.R.Y. is a paid consultant with Merck and Company.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.