Abstract
Mutations in cdk5rap2 are linked to autosomal recessive primary microcephaly, and attention has been paid to its function at centrosomes. In this report, we demonstrate that CDK5RAP2 localizes to microtubules and concentrates at the distal tips in addition to centrosomal localization. CDK5RAP2 interacts directly with EB1, a prototypic member of microtubule plus-end tracking proteins, and contains the basic and Ser-rich motif responsible for EB1 binding. The EB1-binding motif is conserved in the CDK5RAP2 sequences of chimpanzee, bovine, and dog but not in those of rat and mouse, suggesting a function gained during the evolution of mammals. The mutation of the Ile/Leu-Pro dipeptide within the motif abolishes EB1 interaction and plus-end attachment. In agreement with the mutational analysis, suppression of EB1 expression inhibits microtubule tip-tracking of CDK5RAP2. We have also found that the CDK5RAP2–EB1 complex regulates microtubule dynamics and stability. CDK5RAP2 depletion by RNA interference impacts the dynamic behaviors of microtubules. The CDK5RAP2–EB1 complex induces microtubule bundling and acetylation when expressed in cell cultures and stimulates microtubule assembly and bundle formation in vitro. Collectively, these results show that CDK5RAP2 targets growing microtubule tips in association with EB1 to regulate microtubule dynamics.
INTRODUCTION
Microtubules (MTs) have highly dynamic and polarized tubular structures, which are essential in controlling a variety of cellular events. A group of diverse proteins binds to the plus ends of growing MTs and are therefore named plus-end tracking proteins (+TIPs). These proteins include EB1, the dynactin subunit p150glued, adenomatous polyposis coli protein (APC), cytoplasmic linker proteins (CLIPs), and CLIP-associating proteins (Carvalho et al., 2003; Galjart, 2005; Akhmanova and Steinmetz, 2008). Several mechanisms have been proposed for +TIPs to achieve their localization to growing microtubule (MT) tips. EB1, a prototypic +TIP, recognizes and accumulates at growing MT tips, perhaps via an autonomous mechanism (Tirnauer et al., 2002; Bieling et al., 2007, 2008). Other mechanisms known for plus-end tracking include “hitchhiking” through interaction with a +TIP such as EB1 and motor-driven plus-end–directed transport (Carvalho et al., 2003; Galjart, 2005; Akhmanova and Steinmetz, 2008). A +TIP may adopt several mechanisms for plus-end targeting. For example, APC moves along MTs in three distinct ways: direct association with MTs, hitchhiking on EB1, and kinesin-mediated transport (Askham et al., 2000; Mimori-Kiyosue et al., 2000b; Jimbo et al., 2002; Kita et al., 2006).
EB1 represents a highly conserved group of proteins that localize to cytoplasmic MTs (Vaughan, 2005; Akhmanova and Steinmetz, 2008). In mammalian cells, EB1 has two homologues: EB2 and EB3. Among them, EB1 and EB3 but not EB2 are enriched at growing MT tips. In addition, EB1 is ubiquitously expressed and best characterized, whereas EB3 is primarily expressed in neurons and muscle cells. The structure of EB1 comprises a calponin homology domain responsible for MT binding, a coiled-coil region responsible for its homodimerization, and a tail region for binding to most known +TIPs (Bu and Su, 2003; Hayashi and Ikura, 2003; Hayashi et al., 2005; Honnappa et al., 2005; Slep et al., 2005). At the plus ends, EB1 promotes MT polymerization and prevents MTs from pausing (Tirnauer et al., 2002; Rogers et al., 2002; Busch and Brunner, 2004). EB1 has intrinsic activity of promoting MT assembly, and such activity is tightly controlled by its tail region (Ligon et al., 2003; Hayashi et al., 2005). It is interesting that the autoinhibition can be relieved by binding APC or p150glued to the EB1 tail (Nakamura et al., 2001; Hayashi et al., 2005; Honnappa et al., 2005; Slep et al., 2005), implicating cooperation between EB1 and other +TIPs in the regulation of plus-end dynamics.
CDK5RAP2 (also known as Cep215) is a protein whose mutations associate with a neurogenic disorder, autosomal recessive primary microcephaly (Ching et al., 2000; Andersen et al., 2003; Bond et al., 2005). Recently, CDK5RAP2 has been characterized for its function at centrosomes. In somatic cells, CDK5RAP2 localizes to centrosomes throughout the cell cycle (Bond et al., 2005; Graser et al., 2007; Fong et al., 2008). It binds to the γ-tubulin ring complex and is required for the centrosomal assembly of γ-tubulin, playing a critical role in the MT-organizing function of centrosomes (Fong et al., 2008). CDK5RAP2 also has been shown to have an indirect function in centrosome cohesion (Graser et al., 2007). Therefore, CDK5RAP2 may play a multifunctional role at centrosomes.
In this report, we show that CDK5RAP2 tracks MT plus ends via association with EB1. CDK5RAP2 contains the basic and Ser-rich motif that has been found in several +TIPs and is responsible for EB1 interaction. It is interesting that the EB1-binding motif is conserved in the chimpanzee, bovine, and dog but not in the rat and mouse sequences of CDK5RAP2, pointing to a function gained during evolution. The suppression of CDK5RAP2 expression or the coexpression of CDK5RAP2 and EB1 impacts MTs. Furthermore, the CDK5RAP2–EB1 complex promotes MT assembly in vitro. Together, these results indicate that CDK5RAP2 forms a plus-end complex with EB1 to regulate MT dynamics.
MATERIALS AND METHODS
DNA Clones and Reagents
DNA plasmids were constructed with standard molecular cloning techniques and mutations were created by polymerase chain reaction (PCR) methods. The constructs were all verified by DNA sequencing. The cDNA of mouse CDK5RAP2 (GenBank accession AK129411) was obtained from the Kazusa DNA Research Institute (Chiba, Japan). The EB1-green fluorescent protein (GFP) plasmid (Mimori-Kiyosue et al., 2000a) was a gift from Dr. Yuko Mimori-Kiyosue (KAN Research Institute, Kobe, Japan) and the mCherry-α-tubulin plasmid (Shaner et al., 2004) was from Dr. Roger Y. Tsien (University of California, San Diego, La Jolla, CA). Small interfering RNA (siRNA) duplexes against human EB1 (UUGCCUUGAAGAAAGUGAA; Louie et al., 2004) and CDK5RAP2 (UGGAAGAUCUCCUAACUAA; Fong et al., 2008) were used as reported previously. The siRNA-resistant construct of CDK5RAP2 was prepared by introducing four silent substitutions into the targeted site (i.e., the resulting sequence is TGGAGGAACTGCTAACGAA).
The antibody recognizing a carboxy-terminal region of CDK5RAP2 was described previously (Fong et al., 2008). The following antibodies were purchased: anti-acetylated-α-tubulin, anti-FLAG (monoclonal M2 and polyclonal) from Sigma-Aldrich (St. Louis, MO), anti-α-tubulin from Calbiochem (San Diego, CA), anti-His6 (H-15) and anti-GFP from Santa Cruz Biotechnology (Santa Cruz, CA), and anti-EB1 and anti-active caspase-3 from BD Biosciences (San Jose, CA). A rabbit anti-pericentrin antibody (polyclonal M8) was obtained from Dr. Stephen J. Doxsey (University of Massachusetts, Amherst, MA). The monoclonal antibody GT335 was from Dr. Carsten Janke (Centre de Recherches de Biochimie Macromoléculaire, Montpellier, France).
Cell Culture, Transfection, and Stable Line Generation
Human embryonic kidney (HEK) 293T, HeLa, and U2OS were cultured in DMEM plus 10% fetal bovine serum (Invitrogen, Carlsbad, CA). DNA plasmids and siRNA duplexes were transfected into cells using Lipofectamine Plus and Lipofectamine 2000 (Invitrogen), respectively. To generate stable transfectants of yellow fluorescent protein (YFP)-α-tubulin (Clontech, Mountain View, CA), EB1-GFP, and GFP-CDK5RAP2, cells were selected with 2 mg/ml G418 (Invitrogen) 24 h after transfection. Resistant clones were picked and cultured. Expression of the transfected genes was examined by immunoblotting.
Immunofluorescence Imaging
Cells grown on coverglasses were fixed with 4% paraformaldehyde in phosphate-buffered saline (PBS) at room temperature for 20 min. Methanol fixation also was used to evaluate the centrosomal localization of CDK5RAP2. Images were acquired by confocal laser scanning microscopy (LSM510 META; Carl Zeiss Microimaging, Thornwood, NY) or by wide-field microscopy using an inverted microscope (Eclipse TE2000; Nikon, Tokyo, Japan) equipped with the camera SPOT RT1200 (Diagnostic Instruments, Sterling Heights, MI). Nuclear DNA was labeled with 1 μM Hoechst 33528 (Sigma-Aldrich). Antigen blocking was performed by incubating the CDK5RAP2 antibody with antigen protein in excess before immunostaining.
Time-lapse Microscopy
Cells grown on 35-mm glass-bottomed dishes were changed to the phenol red-free medium before being imaged on the Nikon microscope equipped with an incubator to maintain the culture conditions (e.g., 37°C and 5% CO2). Live-cell imaging was performed using an EMCCD camera (SPOT BOOST BT2100; Diagnostic Instruments). TIF image stacks were exported as MOV files using MetaMorph (Molecular Devices, Sunnyvale, CA). The play-back rate is 5 frames/s. To determine the dynamic behaviors of MTs, data were collected by tracking MT ends using the “track points” function of MetaMorph and were transferred to Excel (Microsoft, Redmond, WA) to plot the life history (Rusan et al., 2001). The phases of growth, shrinkage, and pause were then identified, and the time spent in each phase was determined. To derive rescue frequency, the number of transitions from shrinkage to growth or pause was divided by the duration of shrinkage. To calculate catastrophe frequency, the number of transitions from growth to shrinkage and pause to shrinkage was divided by the duration of growth and pause.
To determine the amount of CDK5RAP2 at MT plus ends, the fluorescence intensity was measured from a rectangle covering the entire tip area (8 × 30 pixels, width × height). The intensity was also measured from a cytoplasmic area of same size. After background subtraction, the intensity ratios of MT tips to cytoplasm were determined and presented as a histogram.
Preparation of Recombinant Proteins
Escherichia coli BL21(DE3) was used to express glutathione transferase (GST) fusion and His6-tagged proteins from pGEX and pET21b constructs, respectively. The expressed proteins were purified by binding His6 to Ni2+-nitrilotriacetic acid resin (QIAGEN, Valencia, CA) or GST to glutathione (GSH)-Sepharose (GE Healthcare, Chalfont St. Giles, Buckinghamshire, United Kingdom) as described in previous reports (Lim et al., 2004; Fu et al., 2006).
Immunoprecipitation and In Vitro Protein Binding
Cell extracts were prepared in the buffer of 25 mM Tris-HCl, pH 7.4, 0.5% NP-40, 100 mM NaCl, 5 mM MgCl2, 5 mM NaF, 20 mM β-glycerophosphate, 1 mM dithiothreitol, 1 mM EDTA, and the Compete Protease Inhibitor Cocktail (Roche Diagnostics, Indianapolis, IN). After the extracts were clarified by centrifugation (13,000 × g for 15 min), proteins were immunoprecipitated using anti-FLAG M2-coupled beads (Sigma-Aldrich) or otherwise antibodies coupled to protein A/G-Agarose (Invitrogen) as indicated. Immunoprecipitated products were detected by immunoblotting. To test protein binding in vitro, the CDK5RAP2 fragment 926-1208 and its L938A/P939A mutant prepared as recombinant GST proteins were incubated at 4°C with His6-EB1 in the buffer described above containing 2 mg/ml bovine serum albumin. In a control, GST was used instead of the CDK5RAP2 fragment. After the incubation, the GST proteins were bound to GSH-Sepharose and retrieved for SDS-polyacrylamide gel electrophoresis (PAGE) and immunoblotting.
MT Assembly
The light scattering assay was carried out with 18 μM tubulin purified from porcine brain and free of MT-associated proteins as described in a previous report (Hou et al., 2007). To test MT-elongating activity, MT seeds, prepared by shearing Taxol-stabilized MTs with a 26-gauge needle, were included in assembly assays. MT assembly also was examined by polymerization of rhodamine-labeled (Cytoskeleton, Denver, CO) and unlabeled tubulin (1:20) for epifluorescence microscopy (He et al., 2008).
RESULTS
CDK5RAP2 Localizes to MTs and Concentrates at the Distal Tips
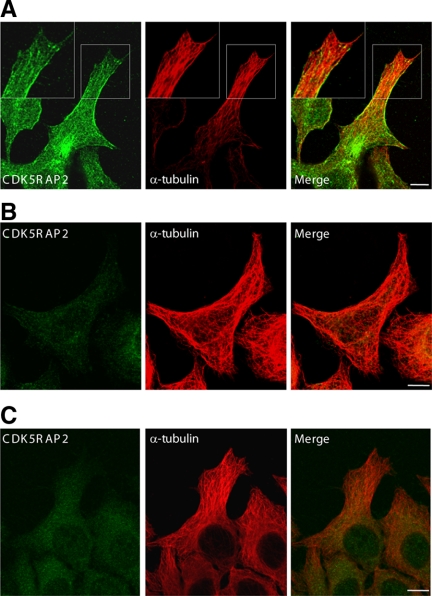
In a previous report, we showed CDK5RAP2 association with MTs in an MT-pelleting assay (Fong et al., 2008). We extended this observation and further examined the cytoplasmic distribution of CDK5RAP2. Cell cultures were fixed in paraformaldehyde and then examined by immunofluorescence confocal microscopy. CDK5RAP2 staining displayed diffuse and punctuate patterns in the cytoplasm, in addition to the centrosomal localization (Figure 1A). In the cell lamellae, CDK5RAP2 signals showed filamentous structures coaligned with MTs (Figure 1A). The filamentous patterns were most evident near the extending edge of cell protrusions, where CDK5RAP2 labeling was concentrated at the MT distal tips (Figure 1A). The staining of CDK5RAP2 to MTs and the distal tips are specific, because the filamentous patterns were eliminated upon CDK5RAP2 depletion by using RNA interference (RNAi) or by preincubation of the antibody with antigen (Figure 1, B and C). The CDK5RAP2 tracking of growing MT tips also was demonstrated by time-lapse microscopy (see below).
Figure 1.
CDK5RAP2 localization to MTs and the distal tips. (A) HeLa cells were stained with anti-CDK5RAP2 and anti-α-tubulin antibodies for confocal microscopy. The boxed areas are enlarged in insets. (B) HeLa cells depleted of CDK5RAP2 by RNAi were immunostained as described in A. (C) The CDK5RAP2 antibody was blocked with antigen protein and then subjected to immunostaining of HeLa cells. Bars, 10 μm.
CDK5RAP2 Is an EB1-binding Protein
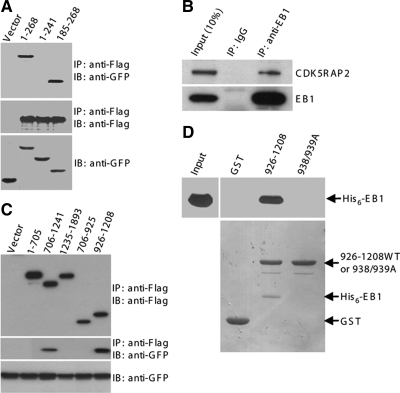
Because EB1 is a core component of protein complexes at MT plus ends, we tested the potential interaction between CDK5RAP2 and EB1. HEK293T was transiently transfected with CDK5RAP2 and EB1-GFP or the vector for coimmunoprecipitation. Immunoblots revealed the presence of EB1-GFP but not GFP by itself in the immunoprecipitate of ectopically expressed CDK5RAP2 (Figure 2A). We further probed the interaction using EB1 fragments. The tail fragment EB1(185-268) was readily detected to coimmunoprecipitate with CDK5RAP2 (Figure 2A). In contrast, EB1(1-241), the construct lacking the tail, did not show any binding activity (Figure 2A). Therefore, CDK5RAP2 binds to the EB1 tail, which is similar to several +TIPs such as APC and p150glued (Askham et al., 2002). We next performed coimmunoprecipitation of the endogenous proteins. Immunoprecipitation of endogenous EB1 specifically coprecipitated endogenous CDK5RAP2 (Figure 2B), confirming the interaction under physiological concentrations.
Figure 2.
CDK5RAP2 binds to EB1. (A) HEK293T extracts coexpressing Flag-CDK5RAP2 and either GFP or GFP-tagged EB1 proteins were subjected to anti-FLAG immunoprecipitation (IP). The immunoprecipitates (50%) and the cell extracts (5%) were analyzed by immunoblotting (IB). EB1 constructs used were the full-length protein (i.e., 1–268) and two fragments. (B) Endogenous EB1 was immunoprecipitated from HEK293T extracts to detect coimmunoprecipitation of endogenous CDK5RAP2. The immunoprecipitates (100%) and the cell extract (10%) were analyzed. (C) CDK5RAP2 fragments (FLAG-tagged) were coexpressed with EB1-GFP. The anti-FLAG immunoprecipitates (50%) and the cell extracts (5%) were probed on immunoblots. (D) In a GST pull-down assay, His6-EB1 was tested for binding to GST or the CDK5RAP2 fragment in fusion with GST. 938/939A, 926-1208(L938A/P939A). The pull-downs (20%) and the His6-EB1 input (10%) were resolved by SDS-PAGE and transferred to membranes. Top, anti-His6 immunoblots. Bottom, membrane stained with Ponseau S.
To delineate the EB1-binding domain, CDK5RAP2 constructs expressing various regions were used to test EB1 binding by coimmunoprecipitation. The fragment 926-1208 but not the other regions were found to be responsible for EB1 interaction (Figure 2C). The coimmunoprecipitation experiments described above revealed the association of CDK5RAP2 with EB1 as well as the binding regions but did not show whether the binding is direct or indirect. To address this, we prepared recombinant proteins of 926-1208 and EB1 by bacterial expression for testing the binding in vitro. We also prepared the L938A/P939A mutant of 926-1208, which lacks EB1-binding activity (see below). EB1 readily bound to 926-1208 but not to the L938A/P939A mutant or the tag of the recombinant protein (Figure 2D), indicating direct association of EB1 with the middle region of CDK5RAP2.
CDK5RAP2 Contains the Basic and Ser-rich Motif
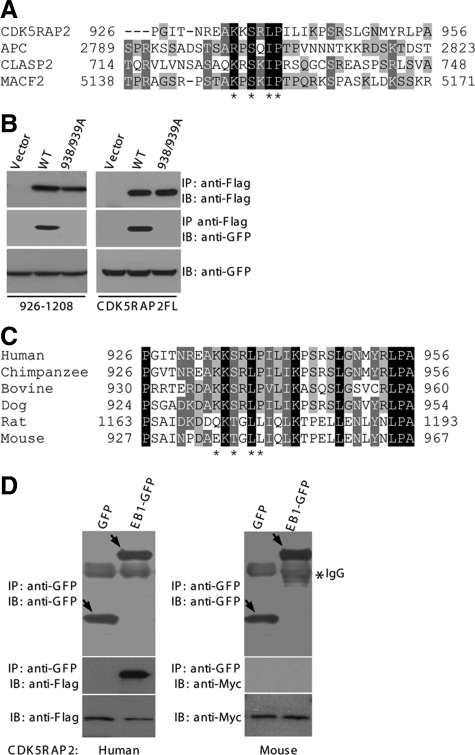
The EB1-binding sites of APC and several other +TIPs have been defined as a small region rich in Ser and basic residues and containing conserved motifs (Galjart, 2005; Honnappa et al., 2005; Mimori-Kiyosue et al., 2005; Slep et al., 2005). Scanning the EB1-binding region of CDK5RAP2 revealed a short segment, 926–956, exhibiting features of the EB1-binding domains found in the above-mentioned +TIPs (Figure 3A). Within the EB1-binding domain of APC, Ile2805-Pro2806 is critically involved in the interaction with EB1 (Honnappa et al., 2005). This dipeptide motif is conservatively substituted in CDK5RAP2 as Leu-Pro (i.e., Leu938-Pro939; Figure 3A). We double mutated Leu938-Pro939 to Ala and tested the mutational effect on EB1 binding. The binding assays showed that the mutation abolished the EB1-binding activity of 926-1208 as well as that of the full-length protein (Figure 3B). Together, CDK5RAP2 harbors a basic and Ser-rich sequence for EB1 interaction.
Figure 3.
CDK5RAP2 contains the basic and Ser-rich motif. (A) Alignment of CDK5RAP2(926-956) with several known basic and Ser-rich sequences. Asterisks denote residues conserved in the motif. (B) HEK293T was double transfected with FLAG-tagged CDK5RAP2 and EB1-GFP for anti-FLAG immunoprecipitation. The immunoprecipitates (50%) and the cell extracts (5%) were analyzed on immunoblots. The CDK5RAP2 constructs used were the full-length (CDK5RAP2FL) and the 926-1208 fragment. WT, wild type; 938/939A, L938A/P939A mutant. (C) Sequence alignment of CDK5RAP2 from different mammalian species. GenBank accession numbers are as follows: chimpanzee, NP_001035901; bovine, XP_584826; dog, XP_855524; rat, XP_575844; and mouse, NP_666102. (D) HEK293T extracts coexpressing EB1 and CDK5RAP2 were subjected to anti-GFP immunoprecipitation. The immunoprecipitates (100%) and the cell extracts (5%) were analyzed. CDK5RAP2 constructs are the human protein (FLAG-tagged) and the mouse counterpart (Myc-tagged).
In a search of GenBank databases, we found putative CDK5RAP2 sequences from several lower mammalian species, including mouse, rat, dog, bovine, and chimpanzee. Across this range of species, CDK5RAP2 shows a high degree of sequence homology ranging from 70 to 99%. We then examined EB1-binding sequences. The segment 926–956 of human CDK5RAP2 shares significant overall homologies with the corresponding regions from these sequences found in lower mammals (Figure 3C). The EB1-binding motif predicted from the alignment of the basic and Ser-rich sequences is well conserved in the sequences from chimpanzee, bovine, and dog but not in the murine sequences (Figure 3C). In particular, two residues including Pro of the critical Ile/Leu-Pro motif are substituted in the rat and mouse sequences. We expressed mouse CDK5RAP2 for testing in an EB1-binding assay. Immunoprecipitation of EB1 failed to pull down mouse CDK5RAP2, whereas the immunoprecipitation specifically coprecipitated the human protein in a control assay (Figure 3D). In addition, the mouse protein did not show plus-end tracking property when expressed, even though it localized to the centrosomes (data not shown). These analyses indicate that the MT tip-binding function of CDK5RAP2 is not conserved in the rat and mouse proteins.
CDK5RAP2 Tracks Growing MT Tips in an EB1-dependent Manner
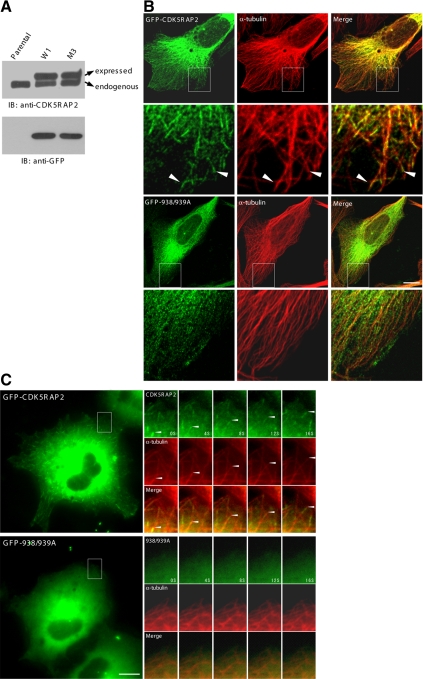
We proceeded to investigate CDK5RAP2 association with MT plus ends and whether the plus-end association requires EB1 binding. To this end, U2OS sublines were generated to stably express CDK5RAP2 or the EB1-binding–deficient mutant L938A/P939A. The proteins were expressed in fusion with GFP at the amino terminus. Several stable transfectants were selected with the proteins expressed at levels close to the endogenous amount. The results were shown below using the clones W1 and M3, which expressed the CDK5RAP2 wild type and the mutant, respectively. In these clones, the amount of ectopically expressed CDK5RAP2 was ∼1.4-fold of the endogenous level (Figure 4A). All experiments were repeated with at least two other selected clones of similar expression levels.
Figure 4.
CDK5RAP2 targets to MT distal ends in a manner dependant on EB1 interaction. (A) Stable U2OS lines were analyzed on immunoblots (IBs). Parental, parental U2OS; W1, a subline expressing GFP-CDK5RAP2; M3, a subline expressing the L938A/P939A mutant (GFP-938/939A). (B) The stable lines were immunostained for MTs (anti-α-tubulin). (C) Time-lapse microscopy was performed on the stable cells. mCherry-α-tubulin was transiently expressed at low levels. Time series of the boxed areas are enlarged. Bars, 10 μm.
The distribution patterns of CDK5RAP2 stably expressed in U2OS cells were first examined by immunofluorescence microscopy. The wild-type protein decorated along the shaft of MTs and concentrated at the distal end regions in addition to centrosomal localization (Figure 4B). The L938A/P939A mutant displayed a diffuse pattern without obvious localization to MTs and the distal tips despite its retained centrosomal localization (Figure 4B). To conduct time-lapse microscopy, mCherry-α-tubulin was expressed at low levels to allow imaging of MTs. In live cells, CDK5RAP2 wild type displayed highly dynamic comet-like fluorescence patterns that highlighted the growing MT tips and moved toward the cell periphery (Figure 4C and Supplemental Movie 1), demonstrating the plus-end tracking behavior. We then proceeded to analyze the stable line of the L938A/P939A mutant. The mutant protein did not show MT plus-end tracking behavior (Figure 4C and Supplemental Movie 2). Together, the analyses on the wild-type and mutant proteins indicate that the association of EB1 targets CDK5RAP2 to MTs and to the distal ends.
We tested whether the plus-end tracking of CDK5RAP2 depends on MT assembly/disassembly dynamics. The stable cells of GFP-CDK5RAP2 were treated with low-dosage nocodazole (0.1 μM), which perturbs MT dynamics but does not disrupt the array organization (Vasquez et al., 1997). The treatment quickly eliminated the dynamic signals of CDK5RAP2 (Supplemental Movie 3), which is similar to the effect observed for EB1 (Mimori-Kiyosue et al., 2000a). These data revealed the attachment of CDK5RAP2 with dynamic MT plus ends.
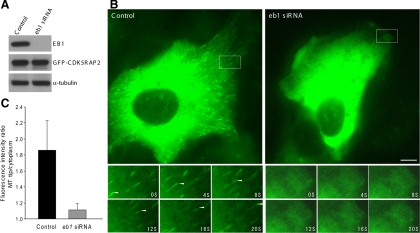
Using the stable cells of GFP-CDK5RAP2, we also examined CDK5RAP2 dynamic movement under the condition of EB1 depletion. The transfection of an eb1-targeting siRNA effectively reduced the EB1 level (by >90%) but did not affect the expression of CDK5RAP2 and tubulin (Figure 5A). In cells transfected with eb1 siRNA, GFP-CDK5RAP2 showed little MT localization and plus-end tracking (Figure 5, B and C, and Supplemental Movie 4). To assess more precisely the plus-end tracking activities, we determined the ratio of GFP-CDK5RAP2 intensities between MT tips and cytoplasm. The suppression of EB1 expression dramatically reduced CDK5RAP2 at the plus ends to the levels close to that of the cytoplasm (Figure 5C). This result supports the principal role of EB1 in CDK5RAP2 attachment to the plus ends.
Figure 5.
Depletion of EB1 inhibits CDK5RAP2 tracking MT plus ends. (A) The stable line of GFP-CDK5RAP2 (clone W1) was transfected with an eb1-targeting siRNA or the control. The protein levels of EB1, GFP-CDK5RAP2, and α-tubulin were detected. (B) Live cells of W1 were imaged for GFP-CDK5RAP2. Below are time series of the boxed areas. Bar, 10 μm. (C) Histogram shows the fluorescence intensity ratios of growing MT tips to cytoplasm. Eight control and twelve EB1-depleted cells were analyzed; 10 MTs were chosen from each cell for analysis. Statistical analysis was performed using Student's unpaired two tails t test (p < 0.001).
The CDK5RAP2–EB1 Complex Regulates MT Dynamics
To probe CDK5RAP2 function in the control of MT dynamics, we performed RNAi-mediated CDK5RAP2 depletion and imaged MTs in live cells. The suppression of CDK5RAP2 expression did not significantly affect EB1 tracking of MT plus ends. We derived the parameters of MT dynamic instability from the time-lapse microscopic data of interphase cells expressing YFP-α-tubulin. The dynamic properties measured from control cells (Table 1) were similar to those reported previously (Rusan et al., 2001). MTs in cells depleted of CDK5RAP2 were less dynamic than those in the control cells. The most notable effects of the depletion were shortened growth and prolonged pause of MTs (Table 1). CDK5RAP2 seems to have growth-promoting and pause-suppressing effects on MTs.
Table 1.
MT dynamic properties
| Control(30 MTs/7cells) | CDK5RAP2si(40 MTs/7cells) | CDK5RAP2si+ 938/939A(30 MTs/5cells) | |
|---|---|---|---|
| Growth rate (μm/min) | 12.0 ± 3.3 | 12.1 ± 4.2 | 12.3 ± 5.8 |
| Shrinkage rate (μm/min) | 18.4 ± 12.6 | 17.9 ± 10.8 | 16.8 ± 11.2 |
| Rescue frequency (s−1) | 0.335 ± 0.146 | 0.301 ± 0.147 | 0.360 ± 0.169 |
| Catastrophe frequency (s−1) | 0.077 ± 0.039 | 0.067 ± 0.023 | 0.073 ± 0.032 |
| Time in growth (%) | 25.6 | 17.8* | 20.3* |
| Time in shrinkage (%) | 18.2 | 16.1 | 16.3 |
| Time in pause (%) | 56.2 | 66.1* | 63.4* |
U2OS cells stably expressing YFP-α-tubulin were transfected with the control or cdk5rap2-targeting siRNA (CDK5RAP2si). The L938A/P939A mutant construct (938/939A) is resistant to the siRNA duplex used and expresses the mCherry-tagged protein. Live cells were imaged 72 h after transfection for 90 s at a 2-s interval. MT tips near the cell periphery were chosen to plot the MT life histories. Statistical analysis was performed using Student's unpaired two-tailed t test for comparison between two groups. Presented are mean values ± SD for growth and shrinkage rates and rescue and catastrophe frequencies. Asterisks indicate p < 0.05 compared with the control data.
Because CDK5RAP2 is required for γ-tubulin assembly onto centrosomes, CDK5RAP2 depletion delocalizes centrosomal γ-tubulin and disorganizes microtubules (Fong et al., 2008). To investigate specifically the plus-end–associated function, we carried out rescue experiments of CDK5RAP2 knockdown by using the EB1-binding–deficient mutant CDK5RAP2(L938A/P939A). The construct was engineered to contain silent mutations within the siRNA-targeted sequence, making it unaffected by the siRNA used (Supplemental Figure 1A). The expression of CDK5RAP2(L938A/P939A) maintained the radial array of MTs in cells depleted of endogenous CDK5RAP2 (Supplemental Figure 1B). Although the mutant expression showed small rescuing effects on MT dynamics, MTs in the cells still displayed a significantly longer time in pause and a shorter time in growth than those in the control cells (Table 1). Therefore, EB1 binding and thus plus-end tracking are required for CDK5RAP2 to regulate MT dynamics.
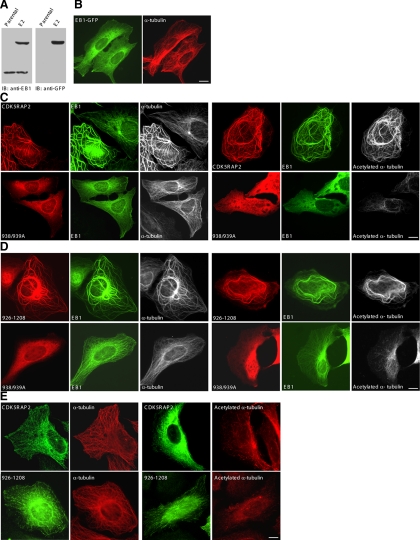
Next, we generated U2OS sublines stably expressing EB1-GFP at low levels; one of the selected clones, E2, expressed the protein at ∼1.5-fold of endogenous EB1 (Figure 6A). At this level, EB1-GFP decorated the entire network of MTs with enrichment at the plus ends (Figure 6B), which is in agreement with the previous reports (Mimori-Kiyosue et al., 2000a; Bu and Su, 2001; Ligon et al., 2003). Remarkably, transfection of CDK5RAP2 into E2 caused the formation of long MT bundles emanating from the centrosomes; both CDK5RAP2 and EB1 were enriched on the bundles (Figure 6C). In addition, the CDK5RAP2 expression gave rise to intense acetylation of MTs (Figure 6C), pointing to MT stabilization. In contrast, expression of the L938A/P939A mutant did not show such effects (Figure 6C). Transfection of the EB1-binding fragment 926-1208 and its L938A/P939A mutant yielded the same results as the respective full-length proteins (Figure 6D). Such effects of CDK5RAP2 expression also were observed using other selected EB1-GFP clones. In a control assay, expression of CDK5RAP2 or 926-1208 in parental U2OS did not induce MT bundling and acetylation (Figure 6E). Therefore, the interaction between CDK5RAP2 and EB1 generates notable impacts on MT structures in the transfected cells.
Figure 6.
The CDK5RAP2–EB1 complex induces MT bundling and acetylation. (A) The stable line of EB1-GFP (clone E2) and parental U2OS cells (Parental) were analyzed on immunoblots (IBs). (B) The stable line E2 was imaged for EB1-GFP and MTs (anti-α-tubulin). (C and D) The E2 line was transiently transfected with the wild type or the L938A/P939A mutant (938/939A) of FLAG-CDK5RAP2. The constructs transfected were the full-length protein (C) or the fragment 926-1208 (D). The cells were double stained for CDK5RAP2 (anti-FLAG) and MTs (anti-α-tubulin) or acetylated MTs (anti-acetylated α-tubulin). (E) CDK5RAP2 and 926-1208 were transiently expressed in parental U2OS. The cells were stained as above. Bars, 10 μm.
To eliminate the possibility that the formation of MT bundles in EB1-GFP stable lines transfected with CDK5RAP2 might be caused by cell apoptosis, we carried out the following assays. As a control, apoptosis of EB1-GFP stable cells was induced with staurosporine. First, we examined the morphology of nuclear DNA and did not find chromatin condensation and fragmentation in the stable cells transfected with CDK5RAP2 (Supplemental Figure 2, A and C). Second, we analyzed the subcellular localization of pericentrin, as it disperses from centrosomes in apoptotic cells (Moss et al., 2006; Sanchez-Alcazar et al., 2007). In CDK5RAP2-transfected stable cells, pericentrin was readily observed at the centrosomes (Supplemental Figure 2A). In contrast, pericentrin did not exhibit centrosomal localization in EB1-GFP cells undergoing apoptosis induced with staurosporine (Supplemental Figure 2B). Third, we analyzed caspase activation by using an antibody specifically recognizing active caspase-3. Active caspase-3 was undetectable in CDK5RAP2-transfected stable cells, whereas it was readily detected in apoptotic EB1-GFP cells (Supplemental Figure 2, C and D). Fourth, EB1-GFP stable cells transfected with CDK5RAP2 displayed MT patterns different from those of apoptotic cells. MT bundles were found throughout the cytoplasm in the coexpressing cells (Supplemental Figure 2, A and C), whereas MTs occurred only along the plasma membranes in apoptotic cells (Moss et al., 2006; Sanchez-Alcazar et al., 2007). We conclude that the coexpression of CDK5RAP2 and EB1 did not induce apoptosis.
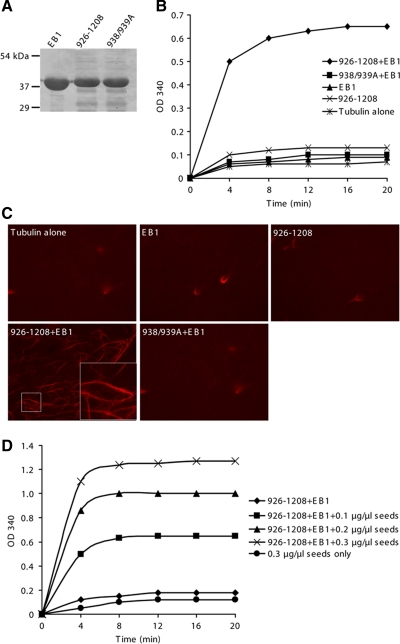
To gain insight into the mechanism of CDK5RAP2 action, we conducted MT assembly assays in vitro by using EB1, the EB1-binding fragment 926-1208, and its L938A/P939A mutant. These proteins were prepared by bacterial expression and purified (Figure 7A). The assembly assays were first conducted in the presence of MT seeds to test MT assembly by elongation from the seeds. The addition of either EB1 or 926-1208 into the assays did not promote MT polymerization; but the addition of both proteins dramatically enhanced the extent of polymerization (Figure 7B). When the L938A/P939A mutant was used instead of the wild type, the combination with EB1 failed to increase light scattering (Figure 7B). Clearly, the association of CDK5RAP2 with EB1 promotes MT polymerization. For morphological examination by microscopy, MTs were polymerized with a mixture of rhodamine-labeled and unlabeled tubulin. Long MT filaments and bundles were extensively formed in the sample containing both 926-1208 and EB1, but not in the others (Figure 7C). These results revealed that the CDK5RAP2–EB1 complex not only promotes polymerization but also bundles individual MTs.
Figure 7.
The CDK5RAP2–EB1 complex promotes MT polymerization. (A) The purified recombinant proteins were examined on a SDS-PAGE gel stained with Coomassie Blue. 938/939A, the L938A/P939A mutant of CDK5RAP2(926-1208). (B) MT assembly was performed in the presence of 0.1 μg/μl MT seeds. Recombinant proteins used included 926-1208 or 938/939A, 1.5 μM; and His6-EB1, 0.5 μM. (C) MTs were polymerized as specified in B from a mixture of rhodamine-labeled and unlabeled tubulin for examination by fluorescence microscopy. (D) MT assembly assays contained various amounts of MT seeds. CDK5RAP2(926-1208), 1.5 μM; His6-EB1, 0.5 μM.
We then performed MT assembly assays in the absence of MT seeds to explore MT-nucleating activity. However, the addition of both 926-1208 and EB1 had no significant effect on MT assembly (Figure 7D). Moreover, the combination of 926-1208 and EB1 displayed polymerizing activities dependent on the doses of MT seeds (Figure 7D). Under the assay conditions, the interaction between EB1 and the CDK5RAP2 fragment promoted MT elongation but did not induce nucleation.
DISCUSSION
In the present study, we have described the association of CDK5RAP2 with growing MT tips and the regulatory role of CDK5RAP2 in MT dynamics. At growing plus ends, +TIPs form various protein complexes with EB1, which acts as a mediator of the dynamic interaction network. EB1 is present at all growing MT ends and is required for a number of +TIPs to associate with the plus ends (Vaughan, 2005; Lansbergen and Akhmanova, 2006). Here, our data have demonstrated that CDK5RAP2 requires EB1 interaction for plus-end association. Therefore, CDK5RAP2 uses a hitchhiking mechanism, which is similar to several of the other +TIPs.
CDK5RAP2 belongs to a class of +TIPs that contain the basic and Ser-rich motif responsible for interaction with EB1 (Galjart, 2005; Akhmanova and Steinmetz, 2008). We have shown that CDK5RAP2 binds to the EB1 tail, a region that contains a protein-binding domain. Several structural studies have revealed that the carboxy-terminal region of EB1 homodimerizes through a coiled-coil domain to form a four-helix bundle (Hayashi et al., 2005; Honnappa et al., 2005; Slep et al., 2005). At the junction of the coiled-coil and four-helix bundle, a hydrophobic pocket comprising a cluster of conserved residues provides the protein-binding site. In the EB1 interaction with APC, the dipeptide Ile2805-Pro2806 within the basic and Ser-rich sequence of APC plays a critical role in EB1 association by interacting perhaps with the conserved residues of the hydrophobic pocket (Honnappa et al., 2005; Slep et al., 2005). We have found that Ala substitution for Leu938-Pro939, an equivalent to Ile2805-Pro2806 of APC, ablates the EB1-binding activity of CDK5RAP2, confirming that CDK5RAP2 binds to EB1 in a similar mode.
CDK5RAP2 is related to several proteins in lower organisms, including Drosophila centrosomin (Cnn) and Schizosaccharomyces pombe Mto1p and Pcp1p, in terms of γ-tubulin complex binding (Sawin et al., 2004; Fong et al., 2008). Cnn and Mto1p have been found to exist in satellite particles that move back-and-forth in MT-dependent manners (Megraw et al., 2002; Sawin et al., 2004). The back-and-forth moving behaviors of Cnn and Mto1p are different from CDK5RAP2 tracking of MT tips, which is a unidirectional movement. In addition, we did not find the basic and Ser-rich motif in the sequences of Cnn, Mto1p, and Pcp1p, implying that the tip-tracking function is not conserved in these proteins.
The EB1-binding motif found in human CDK5RAP2 is well conserved in the homologues from chimpanzee, bovine, and dog but not in those from rat and mouse. As a result, we have found that mouse CDK5RAP2 does not have EB1-binding and plus-end tracking properties. Therefore, the MT tip-associated function of CDK5RAP2 is a gain-of-function during the evolution of mammals. In line with this idea, CDK5RAP2 is a rapidly evolved protein with the molecular evolutionary rate significantly higher in primates than in rodents (Evans et al., 2006). CDK5RAP2 has been implicated to play a role in brain development, as its mutations cause primary microcephaly (Bond et al., 2005). Rodent brains have substantial differences in size and structure from the brains of primates, bovine, and dog. For example, the cerebral cortexes of primates, bovine, and dog have many grooves (i.e., sulci and fissures) to significantly enlarge the surface area compared with the smooth brains of rodents. It is plausible that CDK5RAP2 acts in brain development in part by associating with MT distal tips. Similarly, APC2 and EB1, +TIPs at the adherens junctions of Drosophila epithelial cells and male germline stem cells, regulate the orientation of mitotic spindles during the asymmetric division (Lu et al., 2001; Yamashita et al., 2003).
+TIPs are well positioned to regulate MT dynamics. The RNAi experiments from the current study revealed that CDK5RAP2 is a factor promoting MT growth and preventing MT pause, which is reminiscent of the EB1 effects on MTs (Rogers et al., 2002; Tirnauer et al., 2002; Busch and Brunner, 2004; Kita et al., 2006). The CDK5RAP2 function associated with MT dynamics is exerted largely by binding with EB1 and plus-end tracking, because the EB1-binding–deficient mutant rescued the dynamics defects only to a small extent. Moreover, the CDK5RAP2–EB1 complex promotes MT growth and induces MT bundling in vitro, and it stabilizes MTs in transfected cells via the formation of MT bundles. These observations suggest that CDK5RAP2 exists in one of the various complexes formed by EB1 to promote MT growth and dynamics at the plus ends.
EB1 has an intrinsic activity of promoting MT assembly, which can be stimulated upon binding APC or p150glued to the EB1 tail (Nakamura et al., 2001; Ligon et al., 2003; Hayashi et al., 2005). Our MT assembly experiments have shown that the CDK5RAP2–EB1 complex but not EB1 alone or in combination with the EB1-binding–deficient mutant of CDK5RAP2 promotes the elongation of MTs. Because CDK5RAP2 binds to the EB1 tail, the interaction presumably relieves EB1 autoinhibition, which is similar to the binding effects of APC and p150glued. It has been reported that EB1 and its fission yeast homologue Mal3, when applied alone, promote the spontaneous assembly of MTs (des Georges et al., 2008; Vitre et al., 2008). In our assays, EB1 alone did not exhibit any effect. We noted that the concentrations of EB1 and tubulin in our assembly assays are different from those used in these reports. Our assays were performed with 18 μM tubulin, which is below the critical concentration of tubulin, and 0.5 μM EB1, which is close to the physiological concentration (∼0.27 μM) measured from Xenopus eggs (Tirnauer et al., 2002). In the above-mentioned reports, 0.9 μM EB1 and 15 μM tubulin were used or a minimum of 2 μM Mal3 was applied to 4 μM in heterodimer concentration of S. pombe tubulin to stimulate MT assembly (des Georges et al., 2008; Vitre et al., 2008). Therefore, the discrepancy of the observed EB1 effects is probably due to the lower concentration of EB1 and the lower ratio of EB1 to tubulin in our assays. Indeed, EB1 displays concentration-dependent effects on MTs both in vitro and in transfected cells (Mimori-Kiyosue et al., 2000a; Bu and Su, 2001; Ligon et al., 2003; Vitre et al., 2008). Seemingly, EB1 can be auto-stimulatory if its concentration is high enough.
As a highly dynamic structure, MT cytoskeleton is capable of rapid remodeling in response to changes in the cellular environment. CDK5RAP2 is enriched at growing MT tips in cell protrusions. This and other results presented in the current study raise the possibility that CDK5RAP2 may participate in MT reorganization through the selective stabilization of MTs, thus playing a role in MT-dependent processes such as cell polarization and directional movement. It is envisaged that the plus-end accumulation of CDK5RAP2 is under tight regulation spatially and temporally. Such regulation of +TIPs would afford the guidance of MT reorganization and the sensitivity of the MT network to environmental cues.
Supplementary Material
ACKNOWLEDGMENTS
We thank Drs. Yuko Mimori-Kiyosue, Roger Y. Tsien, Richard A. Kahn (Emory University School of Medicine, Atlanta, GA), Carsten Janke, and Donald C. Chang (The Hong Kong University of Science and Technology, Hong Kong, China) for plasmid constructs. We also are indebted to Dr. Weichuan Yu and Tianwei Jiang (The Hong Kong University of Science and Technology) for assistance on protein sequence alignment. This work was supported by the Research Grants Council (General Research Fund and Collaborative Research Fund) and the University Grants Committee (Area of Excellence Scheme) of Hong Kong.
Abbreviations used:
- MT
microtubule.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E09-01-0009) on June 24, 2009.
REFERENCES
- Akhmanova A., Steinmetz M. O. Tracking the ends: a dynamic protein network controls the fate of microtubule tips. Nat. Rev. Mol. Cell Biol. 2008;9:309–322. doi: 10.1038/nrm2369. [DOI] [PubMed] [Google Scholar]
- Andersen J. S., Wilkinson C. J., Mayor T., Mortensen P., Nigg E. A., Mann M. Proteomic characterization of the human centrosome by protein correlation profiling. Nature. 2003;426:570–574. doi: 10.1038/nature02166. [DOI] [PubMed] [Google Scholar]
- Askham J. M., Moncur P., Markham A. F., Morrison E. E. Regulation and function of the interaction between the APC tumour suppressor protein and EB1. Oncogene. 2000;19:1950–1958. doi: 10.1038/sj.onc.1203498. [DOI] [PubMed] [Google Scholar]
- Askham J. M., Vaughan K. T., Goodson H. V., Morrison E. E. Evidence that an interaction between EB1 and p150(Glued) is required for the formation and maintenance of a radial microtubule array anchored at the centrosome. Mol. Biol. Cell. 2002;13:3627–3645. doi: 10.1091/mbc.E02-01-0061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieling P., Kandels-Lewis S., Telley I. A., van D. J., Janke C., Surrey T. CLIP-170 tracks growing microtubule ends by dynamically recognizing composite EB1/tubulin-binding sites. J. Cell Biol. 2008;183:1223–1233. doi: 10.1083/jcb.200809190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieling P., Laan L., Schek H., Munteanu E. L., Sandblad L., Dogterom M., Brunner D., Surrey T. Reconstitution of a microtubule plus-end tracking system in vitro. Nature. 2007;450:1100–1105. doi: 10.1038/nature06386. [DOI] [PubMed] [Google Scholar]
- Bond J., et al. A centrosomal mechanism involving CDK5RAP2 and CENPJ controls brain size. Nat. Genet. 2005;37:353–355. doi: 10.1038/ng1539. [DOI] [PubMed] [Google Scholar]
- Bu W., Su L. K. Characterization of functional domains of human EB1 family proteins. J. Biol. Chem. 2003;278:49721–49731. doi: 10.1074/jbc.M306194200. [DOI] [PubMed] [Google Scholar]
- Bu W., Su L. K. Regulation of microtubule assembly by human EB1 family proteins. Oncogene. 2001;20:3185–3192. doi: 10.1038/sj.onc.1204429. [DOI] [PubMed] [Google Scholar]
- Busch K. E., Brunner D. The microtubule plus end-tracking proteins mal3p and tip1p cooperate for cell-end targeting of interphase microtubules. Curr. Biol. 2004;14:548–559. doi: 10.1016/j.cub.2004.03.029. [DOI] [PubMed] [Google Scholar]
- Carvalho P., Tirnauer J. S., Pellman D. Surfing on microtubule ends. Trends Cell Biol. 2003;13:229–237. doi: 10.1016/s0962-8924(03)00074-6. [DOI] [PubMed] [Google Scholar]
- Ching Y. P., Qi Z., Wang J. H. Cloning of three novel neuronal Cdk5 activator binding proteins. Gene. 2000;242:285–294. doi: 10.1016/s0378-1119(99)00499-0. [DOI] [PubMed] [Google Scholar]
- des Georges A., Katsuki M., Drummond D. R., Osei M., Cross R. A., Amos L. A. Mal3, the Schizosaccharomyces pombe homolog of EB1, changes the microtubule lattice. Nat. Struct. Mol. Biol. 2008;15:1102–1108. doi: 10.1038/nsmb.1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans P. D., Vallender E. J., Lahn B. T. Molecular evolution of the brain size regulator genes CDK5RAP2 and CENPJ. Gene. 2006;375:75–79. doi: 10.1016/j.gene.2006.02.019. [DOI] [PubMed] [Google Scholar]
- Fong K. W., Choi Y. K., Rattner J. B., Qi R. Z. CDK5RAP2 is a pericentriolar protein that functions in centrosomal attachment of the gamma-tubulin ring complex. Mol. Biol. Cell. 2008;19:115–125. doi: 10.1091/mbc.E07-04-0371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu X., Choi Y. K., Qu D., Yu Y., Cheung N. S., Qi R. Z. Identification of nuclear import mechanisms for the neuronal CDK5 activator. J. Biol. Chem. 2006;281:39014–39021. doi: 10.1074/jbc.M512663200. [DOI] [PubMed] [Google Scholar]
- Galjart N. CLIPs and CLASPs and cellular dynamics. Nat. Rev. Mol. Cell Biol. 2005;6:487–498. doi: 10.1038/nrm1664. [DOI] [PubMed] [Google Scholar]
- Graser S., Stierhof Y. D., Nigg E. A. Cep68 and Cep215 (Cdk5rap2) are required for centrosome cohesion. J. Cell Sci. 2007;120:4321–4331. doi: 10.1242/jcs.020248. [DOI] [PubMed] [Google Scholar]
- Hayashi I., Ikura M. Crystal structure of the amino-terminal microtubule-binding domain of end-binding protein 1 (EB1) J. Biol. Chem. 2003;278:36430–36434. doi: 10.1074/jbc.M305773200. [DOI] [PubMed] [Google Scholar]
- Hayashi I., Wilde A., Mal T. K., Ikura M. Structural basis for the activation of microtubule assembly by the EB1 and p150Glued complex. Mol. Cell. 2005;19:449–460. doi: 10.1016/j.molcel.2005.06.034. [DOI] [PubMed] [Google Scholar]
- He L., Hou Z., Qi R. Z. Calmodulin Binding and Cdk5 Phosphorylation of p35 Regulate Its Effect on Microtubules. J. Biol. Chem. 2008;283:13252–13260. doi: 10.1074/jbc.M706937200. [DOI] [PubMed] [Google Scholar]
- Honnappa S., John C. M., Kostrewa D., Winkler F. K., Steinmetz M. O. Structural insights into the EB1-APC interaction. EMBO J. 2005;24:261–269. doi: 10.1038/sj.emboj.7600529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Z., Li Q., He L., Lim H. Y., Fu X., Cheung N. S., Qi D. X., Qi R. Z. Microtubule association of the neuronal p35 activator of Cdk5. J. Biol. Chem. 2007;282:18666–18670. doi: 10.1074/jbc.C700052200. [DOI] [PubMed] [Google Scholar]
- Jimbo T., Kawasaki Y., Koyama R., Sato R., Takada S., Haraguchi K., Akiyama T. Identification of a link between the tumour suppressor APC and the kinesin superfamily. Nat. Cell Biol. 2002;4:323–327. doi: 10.1038/ncb779. [DOI] [PubMed] [Google Scholar]
- Kita K., Wittmann T., Nathke I. S., Waterman-Storer C. M. Adenomatous polyposis coli on microtubule plus ends in cell extensions can promote microtubule net growth with or without EB1. Mol. Biol. Cell. 2006;17:2331–2345. doi: 10.1091/mbc.E05-06-0498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lansbergen G., Akhmanova A. Microtubule plus end: a hub of cellular activities. Traffic. 2006;7:499–507. doi: 10.1111/j.1600-0854.2006.00400.x. [DOI] [PubMed] [Google Scholar]
- Ligon L. A., Shelly S. S., Tokito M., Holzbaur E. L. The microtubule plus-end proteins EB1 and dynactin have differential effects on microtubule polymerization. Mol. Biol. Cell. 2003;14:1405–1417. doi: 10.1091/mbc.E02-03-0155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim A. C., Tiu S. Y., Li Q., Qi R. Z. Direct regulation of microtubule dynamics by protein kinase CK2. J. Biol. Chem. 2004;279:4433–4439. doi: 10.1074/jbc.M310563200. [DOI] [PubMed] [Google Scholar]
- Louie R. K., Bahmanyar S., Siemers K. A., Votin V., Chang P., Stearns T., Nelson W. J., Barth A. I. Adenomatous polyposis coli and EB1 localize in close proximity of the mother centriole and EB1 is a functional component of centrosomes. J. Cell Sci. 2004;117:1117–1128. doi: 10.1242/jcs.00939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu B., Roegiers F., Jan L. Y., Jan Y. N. Adherens junctions inhibit asymmetric division in the Drosophila epithelium. Nature. 2001;409:522–525. doi: 10.1038/35054077. [DOI] [PubMed] [Google Scholar]
- Megraw T. L., Kilaru S., Turner F. R., Kaufman T. C. The centrosome is a dynamic structure that ejects PCM flares. J. Cell Sci. 2002;115:4707–4718. doi: 10.1242/jcs.00134. [DOI] [PubMed] [Google Scholar]
- Mimori-Kiyosue Y., et al. CLASP1 and CLASP2 bind to EB1 and regulate microtubule plus-end dynamics at the cell cortex. J. Cell Biol. 2005;168:141–153. doi: 10.1083/jcb.200405094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimori-Kiyosue Y., Shiina N., Tsukita S. Adenomatous polyposis coli (APC) protein moves along microtubules and concentrates at their growing ends in epithelial cells. J. Cell Biol. 2000b;148:505–518. doi: 10.1083/jcb.148.3.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimori-Kiyosue Y., Shiina N., Tsukita S. The dynamic behavior of the APC-binding protein EB1 on the distal ends of microtubules. Curr. Biol. 2000a;10:865–868. doi: 10.1016/s0960-9822(00)00600-x. [DOI] [PubMed] [Google Scholar]
- Moss D. K., Betin V. M., Malesinski S. D., Lane J. D. A novel role for microtubules in apoptotic chromatin dynamics and cellular fragmentation. J. Cell Sci. 2006;119:2362–2374. doi: 10.1242/jcs.02959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura M., Zhou X. Z., Lu K. P. Critical role for the EB1 and APC interaction in the regulation of microtubule polymerization. Curr. Biol. 2001;11:1062–1067. doi: 10.1016/s0960-9822(01)00297-4. [DOI] [PubMed] [Google Scholar]
- Rogers S. L., Rogers G. C., Sharp D. J., Vale R. D. Drosophila EB1 is important for proper assembly, dynamics, and positioning of the mitotic spindle. J. Cell Biol. 2002;158:873–884. doi: 10.1083/jcb.200202032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusan N. M., Fagerstrom C. J., Yvon A. M., Wadsworth P. Cell cycle-dependent changes in microtubule dynamics in living cells expressing green fluorescent protein-alpha tubulin. Mol. Biol. Cell. 2001;12:971–980. doi: 10.1091/mbc.12.4.971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Alcazar J. A., Rodriguez-Hernandez A., Cordero M. D., Fernandez-Ayala D. J., Brea-Calvo G., Garcia K., Navas P. The apoptotic microtubule network preserves plasma membrane integrity during the execution phase of apoptosis. Apoptosis. 2007;12:1195–1208. doi: 10.1007/s10495-006-0044-6. [DOI] [PubMed] [Google Scholar]
- Sawin K. E., Lourenco P. C., Snaith H. A. Microtubule nucleation at non-spindle pole body microtubule-organizing centers requires fission yeast centrosomin-related protein mod20p. Curr. Biol. 2004;14:763–775. doi: 10.1016/j.cub.2004.03.042. [DOI] [PubMed] [Google Scholar]
- Shaner N. C., Campbell R. E., Steinbach P. A., Giepmans B. N., Palmer A. E., Tsien R. Y. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat. Biotechnol. 2004;22:1567–1572. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- Slep K. C., Rogers S. L., Elliott S. L., Ohkura H., Kolodziej P. A., Vale R. D. Structural determinants for EB1-mediated recruitment of APC and spectraplakins to the microtubule plus end. J. Cell Biol. 2005;168:587–598. doi: 10.1083/jcb.200410114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirnauer J. S., Grego S., Salmon E. D., Mitchison T. J. EB1-microtubule interactions in Xenopus egg extracts: role of EB1 in microtubule stabilization and mechanisms of targeting to microtubules. Mol. Biol. Cell. 2002;13:3614–3626. doi: 10.1091/mbc.E02-04-0210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasquez R. J., Howell B., Yvon A. M., Wadsworth P., Cassimeris L. Nanomolar concentrations of nocodazole alter microtubule dynamic instability in vivo and in vitro. Mol. Biol. Cell. 1997;8:973–985. doi: 10.1091/mbc.8.6.973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan K. T. TIP maker and TIP marker; EB1 as a master controller of microtubule plus ends. J. Cell Biol. 2005;171:197–200. doi: 10.1083/jcb.200509150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitre B., Coquelle F. M., Heichette C., Garnier C., Chretien D., Arnal I. EB1 regulates microtubule dynamics and tubulin sheet closure in vitro. Nat. Cell Biol. 2008;10:415–421. doi: 10.1038/ncb1703. [DOI] [PubMed] [Google Scholar]
- Yamashita Y. M., Jones D. L., Fuller M. T. Orientation of asymmetric stem cell division by the APC tumor suppressor and centrosome. Science. 2003;301:1547–1550. doi: 10.1126/science.1087795. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.