Abstract
Pichia pastoris (Pp) Pex8p, the only known intraperoxisomal peroxin at steady state, is targeted to peroxisomes by either the peroxisomal targeting signal (PTS) type 1 or PTS2 pathway. Until recently, all cargoes entering the peroxisome matrix were believed to require the docking and really interesting new gene (RING) subcomplexes, proteins that bridge these two subcomplexes and the PTS receptor-recycling machinery. However, we reported recently that the import of PpPex8p into peroxisomes via the PTS2 pathway is Pex14p dependent but independent of the RING subcomplex (Zhang et al., 2006). In further characterizing the peroxisome membrane-associated translocon, we show that two other components of the docking subcomplex, Pex13p and Pex17p, are dispensable for the import of Pex8p. Moreover, we demonstrate that the import of Pex8p via the PTS1 pathway also does not require the RING subcomplex or intraperoxisomal Pex8p. In receptor-recycling mutants (Δpex1, Δpex6, and Δpex4), Pex8p is largely cytosolic because Pex5p and Pex20p are unstable. However, upon overexpression of the degradation-resistant Pex20p mutant, hemagglutinin (HA)-Pex20p(K19R), in Δpex4 and Δpex6 cells, Pex8p enters peroxisome remnants. Our data support the idea that PpPex8p is a special cargo whose translocation into peroxisomes depends only on the PTS receptors and Pex14p and not on intraperoxisomal Pex8p, the RING subcomplex, or the receptor-recycling machinery.
INTRODUCTION
Peroxisomes are ubiquitous organelles of eukaryotic cells and function in diverse lipid metabolic pathways. Severe human peroxisomal biogenesis disorders are caused by defects in peroxisome biogenesis, making it imperative to understand how the biogenesis machinery functions (Steinberg et al., 2006). In addition, peroxisome biogenesis has several unique features that set it apart from the biogenesis of other subcellular organelles (Leon et al., 2006a). Unlike mitochondria and chloroplasts, peroxisomes do not contain their own DNA. Therefore, all peroxisomal matrix and membrane proteins are encoded by nuclear genes. They are synthesized on free ribosomes in the cytosol and many of them are posttranslationally targeted to peroxisomes (Subramani et al., 2000; Purdue and Lazarow, 2001).
Typically, the import of peroxisomal matrix proteins occurs via one of two pathways characterized by specific targeting signals (Subramani et al., 2000; Purdue and Lazarow, 2001). The majority of cargoes contain the peroxisomal targeting signal (PTS) type 1, which is a unique C-terminal tripeptide sequence (SKL or related variants) (Gould et al., 1989). Only a small number of cargoes carry the PTS2, which is a nonapeptide (typically RLX5HL and related variants) (Swinkels et al., 1991). So far, only two PTS2 cargoes, the β-oxidation enzyme β-ketoacyl CoA thiolase and Pex8p, have been found in yeast, in contrast to a relatively large number in plants (Reumann et al., 2004; Zhang et al., 2006). In addition, there might exist a third type of PTS, like those in alcohol oxidase in Pichia pastoris and acyl-CoA oxidase in Saccharomyces cerevisiae (Klein et al., 2002; Gunkel et al., 2004) whose import depends on the PTS1 receptor Pex5p, yet through completely distinct regions of interaction than those used by normal PTS1 cargoes.
The import of peroxisomal matrix proteins can be divided into five distinct steps: 1) receptor and cargo recognition in the cytosol, 2) docking of the receptor–cargo complex at the peroxisomal membrane, 3) translocation of the receptor-cargo complex across the peroxisomal membrane followed by cargo release, 4) export of the receptors from the peroxisome matrix to the membrane, and 5) recycling of receptors back to the cytosol for further rounds of import (Leon et al., 2006a; Platta and Erdmann, 2007). According to our current knowledge, the peroxisomal matrix protein import process requires the cooperation of >20 conserved peroxins, which are composed of PTS receptors (Pex5p, Pex7p, and coreceptors Pex18p/Pex20p/Pex21p), the docking subcomplex (Pex13p, Pex14p, and Pex17p), the really interesting new gene (RING) subcomplex (Pex2p, Pex10p, and Pex12p containing RING domains), proteins that bridge the docking and RING subcomplexes (Pex3p and Pex8p), as well as the receptor-recycling machinery (AAA ATPases, Pex1p and Pex6p, and the anchor proteins Pex15p/Pex26p for the latter, and the E2-like protein Pex4p, with its peroxisomal anchor protein Pex22p). Understanding exactly how these machineries work to orchestrate peroxisomal matrix protein import has been a preoccupation of the field.
In S. cerevisiae, the docking and RING subcomplexes assemble into the importomer by bridging via Pex8p, the only known predominantly intraperoxisomal peroxin at steady state, whereas in P. pastoris it is Pex3p that is proposed to bridge these two subcomplexes (Hazra et al., 2002; Agne et al., 2003; Rayapuram and Subramani, 2006). Pex8p is unusual in that it is not only a peroxin but also a matrix-localized cargo as well, containing both functional PTS1 and PTS2 sequences, which are used by two redundant import pathways, in Hansenula polymorpha and P. pastoris (Waterham et al., 1994; Zhang et al., 2006). In vitro experiments with H. polymorpha Pex8p indicate that this protein may be involved in PTS1 cargo release by inducing a conformational change of the receptor–cargo complex (Wang et al., 2003). However, it is uncertain whether Pex8p also plays a role in cargo release from the PTS receptors in vivo and whether the release of PTS2 cargoes from their receptor/s also uses Pex8p.
Deletion of any of the components of the peroxisomal import machinery (the importomer composed of the docking and RING subcomplexes, as well as the bridging proteins, and the distinct peroxisomal receptor-recycling machinery) eliminates the import of PTS1 as well as PTS2 cargoes, and the corresponding strains accumulate peroxisomal matrix proteins in the cytosol (Subramani et al., 2000). However, the import of Pex8p into peroxisomes does not follow all the rules for generic cargo (Zhang et al., 2006). The import of Pex8p into peroxisomes is indeed Pex14p dependent, but its translocation into the peroxisome matrix via the PTS2 pathway does not require the presence of the RING subcomplex or intraperoxisomal Pex8p. To date, the mechanism of protein translocation across the peroxisome membrane is the most elusive and complex aspect of peroxisome biogenesis because the minimal components of the translocation machinery have not yet been elucidated. Knowledge of the exact subunits and the composition of the translocon is necessary for a complete understanding of the structural and functional nature of this noncanonical translocon that transports folded and oligomeric proteins across the peroxisomal membrane (Leon et al., 2006a). To characterize whether the docking subcomplex itself might constitute the translocon instead of the entire importomer, and to define the minimum translocon, we analyzed the role of known peroxins in the import of Pex8p.
We show here that Pex13p and Pex17p are surprisingly not essential for the import of Pex8p into peroxisomes, although they do improve the efficiency of this process. Moreover, in accord with our former results in P. pastoris on the entry of Pex8p into the peroxisome matrix via the PTS2 pathway, we further confirmed that the entry of Pex8p by the PTS1 pathway does not require intraperoxisomal Pex8p or the RING subcomplex. In addition, as long as the PTS receptors/coreceptors are stable, Pex8p import into peroxisomes is also independent of the PTS receptor-recycling machinery. These results strongly indicate that, as a special cargo, the translocation of Pex8p across the peroxisomal membrane does not require the entire importomer; consequently, only one component of the docking subcomplex, Pex14p, in collaboration with the PTS receptors might constitute the minimum translocon for peroxisomal matrix protein import.
MATERIALS AND METHODS
Yeast Strains, Plasmids, and Culture Conditions
The P. pastoris strains and plasmids used are listed in Tables 1 and 2, respectively. Growth media components were as follow: rich medium YPD, 1% yeast extract, 2% peptone, 2% glucose; synthetic medium YNM, 0.67% yeast nitrogen base, 0.1% yeast extract, 0.5% (vol/vol) methanol; mineral oleate medium YNO, 0.67% yeast nitrogen base, 0.1% yeast extract, 0.2% (vol/vol) oleate, and 0.02% (vol/vol) Tween 40.
Table 1.
P. pastoris strains used in this study
| Name | Genotype | Reference |
|---|---|---|
| PPY301 | his4, arg4, Δpex1 (ARG4) | Heyman et al. (1994) |
| JC214 | his4, arg4, Δpex2 (ARG4) | Waterham et al. (1996) |
| STK108 | his4, arg4, Δpex4 (ARG4) | Koller et al. (1999) |
| PPY201 | his4, arg4, Δpex6 (ARG4) | Spong and Subramani (1993) |
| SSH6 | his4, pep4, prb1 Δpex13 (ZeoR) | Johnson et al. (2001) |
| SWS17D | his4, pep4, prb1 Δpex17 (KanR) | Synder et al. (1999) |
| SCM25 | his4, arg4, Δpex6 (ARG4) PPEX8-GFP-Pex8 (HIS4) | This study |
| SCM26 | his4, arg4, Δpex1 (ARG4) PPEX8-GFP-Pex8 (HIS4) | This study |
| SCM29 | his4, pep4, prb1 Δpex13 (ZeoR) PPEX8-GFP-PEX8 (HIS4) | This study |
| SCM30 | his4, pep4, prb1 Δpex17 (KanR) PPEX8-GFP-PEX8 (HIS4) | This study |
| SCM39 | his4, arg4, Δpex6 (ARG4) PPEX8-GFP-Pex8 (HIS4) PEX3-mRFP (ZeoR) | This study |
| SCM43 | his4, arg4, Δpex1 (ARG4) PPEX8-GFP-Pex8 (HIS4) PEX3-mRFP (ZeoR) | This study |
| SCM48 | his4, arg4, Δpex4 (ARG4) PPEX8-GFP-PEX8 (HIS4) | This study |
| SCM52 | his4, pep4, prb1 Δpex17 (KanR) PPEX8-GFP-PEX8 (HIS4) PEX3-mRFP ((ZeoR) | This study |
| SCM58 | his4, arg4, Δpex4 (ARG4) PPEX8-GFP-PEX8 (HIS4) PEX3-mRFP (ZeoR) | This study |
| SCM60 | his4, pep4, prb1 Δpex13 (ZeoR) PPEX8-GFP-PEX8 (HIS4) PEX3-mRFP (KanR) | This study |
| SCM81 | his4, arg4, Δpex2 (ARG4) Δpex20 (KanR) PPEX8-GFP-PEX8ΔAKL (HIS4) | This study |
| SCM85 | his4, arg4, Δpex2 (ARG4) Δpex20 (KanR) | This study |
| SCM91 | his4, arg4, Δpex2 (ARG4) Δpex20 (KanR) PPEX8-GFP-Pex8ΔAKL (HIS4) Pex3-mRFP (ZeoR) | This study |
| SCM93 | his4, arg4, Δpex2 (ARG4) Δpex20 (KanR) PPEX8-GFP-PEX8 (HIS4) | This study |
| SCM95 | his4, arg4, Δpex2 (ARG4) Δpex20 (KanR) PPEX8-GFP-Pex8 (HIS4) Pex3-mRFP (ZeoR) | This study |
| SCM98 | his4, arg4, Δpex4 (ARG4) PGAP-HA-Pex20(K19R) | This study |
| SCM100 | his4, arg4, Δpex6 (ARG4) PGAP-HA-Pex20(K19R) | This study |
| SNR2 | his4, arg4, Pex10-TAP (ZeoR) | This study |
| SNR12 | his4, arg4, Δpex8 (ARG4) Pex10-TAP (ZeoR) | This study |
| SSY5 | his4, arg4, Δpex20 (KanR) Δpex8 (ZeoR) | This study |
| SSY23 | his4, arg4, Δpex20 (KanR) Δpex8 (ZeoR) PPEX8-GFP-PEX8 (HIS4) | This study |
| SSY37 | his4, arg4, Δpex20 (KanR) Δpex8 (ZeoR) PPEX8-GFP-PEX8ΔAKL (HIS4) | This study |
| SSY43 | his4, arg4, Δpex20 (KanR) Δpex8 (ZeoR) PPEX8-GFP-PEX8 (HIS4) PEX3-mRFP (ARG4) | This study |
| SSY47 | his4, arg4, Δpex20 (KanR)Δpex8 (ZeoR) PPEX8-GFP-PEX8ΔAKL (HIS4) PEX3-mRFP (ARG4) | This study |
Table 2.
Plasmids used in this study
| Plasmid | Properties | Source |
|---|---|---|
| pCM121 | pIB1-based with HIS4 PPEX8-GFP-SKL | This study |
| pLZ119 | pIB1-based with HIS4 PPEX8-GFP-PEX8 | Zhang et al. (2006) |
| pLZ120 | pIB1-based with HIS4 PPEX8-GFP-PEX8ΔAKL | Zhang et al. (2006) |
| pLZ127 | pJC235 with ZeoR upstream of PPEX3-PEX3-mRFP | Zhang et al. (2006) |
| pJC235 | pIB1-derived with ARG4 PPEX3-PEX3-mRFP | Zhang et al. (2006) |
| pKSN215 | PPEX3-PEX3-mRFP with Kan Marker | Laboratory stock |
| pSEB47 | PEX20 knock out construct, KanR | Leon et al. (2006b) |
| pSY200 | PEX8 knock out construct, ZeoR | This study |
| pTW51 | pHIL-D2-based with HIS4 PAOX1-GFP-SKL | Wiemer et al. (1996) |
| pTW74 | pHIL-D2-based with HIS4 PGAPDH-GFP-SKL | Luers et al. (1998) |
Yeast cells were grown at 30°C in rich medium (YPD) to 1 OD 600/ml, washed with distilled H2O, and shifted either to synthetic methanol medium (YNM) for fluorescence microscopy, or to mineral oleate medium (YNO) for biochemical experiments.
Generation of the Δpex2 Δpex20 and Δpex8 Δpex20 Mutants
To generate the Δpex2 Δpex20 double deletion mutant (SCM85), a linear DNA fragment containing the Kanr gene flanked by the 5′ and 3′ region of the PEX20 gene was obtained from pSEB47 by digesting the plasmid with SalI and BglII (Leon et al., 2006b) and introduced into the Δpex2 strain by electroporation to replace the endogenous PEX20 gene. The double mutant strain was confirmed by polymerase chain reaction (PCR) analysis.
To disrupt PEX8 in Δpex20, 5′ and 3′ regions of the gene were amplified by PCR via OSY1/OSY2 (OSY1: CGTCTTGAAACGCTGGTATCCGTTTC, OSY2: CCAACTCG AGCATTAACAGGCACCTGAAGATAGGTA) and OSY3/OSY4 (OSY3: AAAGGAATTCG ATTTCTGTTGGATACATTGTGATTAGC, OSY4: TGCCGGATCCAGTGATGCTAGTTGT GGTTGATTATTG), respectively. The 5′ and 3′ fragments were transferred to pMYzeo (Yan et al., 2008) to generate pSY200. The pSY200 plasmid was linearized by digestion with ScaI and transformed into Δpex20 cells resulting in the Δpex8 Δpex20 strain (SSY5). The double mutant strain was confirmed by PCR analysis.
Subcellular Fractionation and Protease Protection
Subcellular fractionation from oleate-induced yeast cells was performed as described previously (Faber et al., 1998). Protease protection analysis was conducted with the P200 fraction isolated directly from the postnuclear supernatant (PNS) of oleate-grown cells. The pellet fraction was resuspended in ice-cold Dounce buffer [50 mM 2-(N-morpholino)ethanesulfonic acid, pH 6.0, 5 mM EDTA, 1% ethanol, and 1 M sorbitol] to a final concentration of 1 μg/μl. Freshly prepared proteinase K (80 μg) and trypsin (40 μg) was added to 200 μg of pellet fraction in the absence or presence of 0.5% Triton X-100, respectively. Aliquots were taken after incubation at room temperature for the indicated times. Trichloroacetic acid (final concentration, 12.5%) was added to terminate the reactions. Proteins were precipitated overnight on ice, washed three times with ice cold acetone, and resuspended in lysis buffer. Equal amounts of samples were subject to SDS-polyacrylamide gel electrophoresis (PAGE) and immunoblotting.
Flotation Gradients
The P200 fraction from a subcellular fractionation was resuspended in 0.5 ml of 65% (wt/wt) sucrose in Dounce buffer without sorbitol. In total, 2.25 ml of 50% (wt/wt) sucrose and 2.25 ml of 30% (wt/wt) sucrose (in Dounce buffer) were layered on top of the sample and spun for 18 h at 100,000 × g in an SW50.1 rotor (Beckman Coulter, Fullerton, CA). Ten 0.5-ml fractions were collected from the top, and equal volumes of fractions were analyzed by SDS-PAGE and Western blot analysis.
Fluorescence Microscopy
Cells were grown in YPD medium and switched to YNM during exponential phase. Images were captured on an Axioskop fluorescence microscope (AxioSkop 2 Plus, motorized; Carl Zeiss, Thornwood, NY) coupled to a cooled charge-coupled device monochrome camera (AxioCam MRM; Carl Zeiss) and analyzed using AxioVision 4 software.
Tandem Affinity Purification (TAP)-Tag Purification
Methanol grown cells were lysed in Dounce buffer containing protease inhibitor (protease inhibitor cocktail from Roche Applied Science, Indianapolis, IN; 5 μg/ml aprotinin, 5 μg/ml leupeptin). Cell debris and nuclei were eliminated by centrifugation at 4500 × g at 4°C for 20 min, and the supernatant was centrifuged to obtain the organelle fraction at 100,000 × g at 4°C for 30 min. The organelle enriched pellet fraction was resuspended in Dounce buffer containing 1% 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate (CHAPS) and incubated at 4°C for 1 h on a rotating machine for further solubilization. Unsolubilized material was removed by centrifugation at 100,000 × g for 30 min, and the solubilized protein was dialyzed overnight at 4°C against dialysis buffer (20 mM HEPES, pH 7.9, 50 mM KCl, 0.2 mM EDTA, 0.5 mM dithiothreitol [DTT], 25% glycerol, 5 μg/ml aprotinin, 5 μg/ml leupeptin, 1 mM phenylmethylsulfonyl fluoride, and protease inhibitor cocktail). The dialysate was mixed with immunoglobulin G (IgG) agarose beads in IPP150 buffer (10 mM Tris-Cl, pH 8.0, 150 mM NaCl, 0.5% CHAPS, 0.5 mM EDTA, and 1 mM DTT) for affinity chromatography. The first step purification was completed by removing the IgG-binding unit of protein A by using tobacco etch virus (TEV) enzyme (Promega, Madison, WI). The peroxisomal importomer was further purified using calmodulin beads and eluted in calmodulin elution buffer (10 mM Tris-Cl, pH 8.0, 150 mM NaCl, 0.5% CHAPS, 1 mM Mg-Acetate, 1 mM imidazole, and 5 mM EGTA).
RESULTS
Pex13p and Pex17p Are Not Essential for Targeting of Pex8p into the Peroxisome Matrix
In P. pastoris, the docking subcomplex consists of peroxins Pex13p, Pex14p, and Pex17p (Hazra et al., 2002; Agne et al., 2003). Pex14p is the initial binding site for the PTS receptors Pex5p and Pex7p, and our previous data showed that Pex14p is required for the import of Pex8p (Zhang et al., 2006), just as it is for all PTS1 and PTS2 cargoes. Consequently, we were interested in determining whether the other components of the docking subcomplex, Pex13p and Pex17p, are essential for the targeting of Pex8p to peroxisomes.
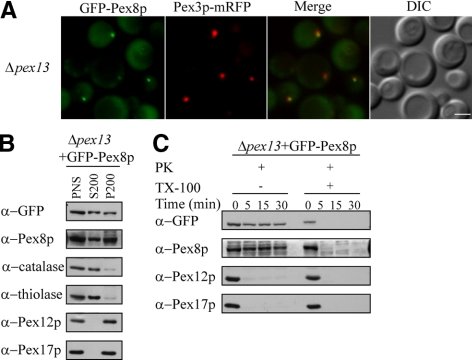
We introduced an amino-terminal green fluorescent protein (GFP)-tagged Pex8p, driven by its own promoter and known to complement Δpex8 cells, into Δpex13 cells and analyzed its targeting to peroxisomes by using fluorescence microscopy, subcellular fractionation, protease protection assays, and flotation gradients (Figure 1, A–C, and Supplemental Figure S2). Loss of PpPex13p eliminates the import of PTS1-, as well as PTS2-containing peroxisomal matrix proteins (Gould et al., 1996). However, in Δpex13 cells, GFP-Pex8p, which contains both PTS1 and PTS2, was localized partially to the cytosol and also to punctate structures that colocalized with a peroxisomal membrane marker Pex3p-mRFP (Figure 1A). In addition, differential centrifugation experiments confirmed that GFP-Pex8p was almost equally distributed in both the organelle pellet (P200) and cytosolic fractions (S200), whereas in wild-type cells GFP-Pex8p was almost exclusively in the peroxisomal fractions (Supplemental Figure S1A), strongly indicating that the import efficiency of Pex8p was affected but not abolished in Δpex13 cells. The behavior of the GFP-Pex8p mimicked that of endogenous Pex8p, which was also distributed between the cytosol and organelle pellet fractions (Figure 1B), demonstrating that the GFP-Pex8p reporter was not behaving aberrantly. In contrast, the targeting of catalase and thiolase, both markers for matrix protein import either via the PTS1 and PTS2 pathways, respectively, was almost abolished in the absence of Pex13p, but as expected, peroxisomal membrane markers Pex12p and Pex17p were sorted normally to the organelle pellet.
Figure 1.
GFP-Pex8p associates partially with peroxisomal remnants in Δpex13 cells. (A) Fluorescence microscopy analysis of methanol-grown Δpex13 cells coexpressing GFP-Pex8p and Pex3p-mRFP serving as a peroxisomal marker. DIC, differential interference contrast. (B) Equal proportions of 200,000 × g supernatant and pellet fractions from oleate-grown Δpex13 cells expressing GFP-Pex8p were separated by SDS-PAGE and immunoblotted with the indicated antibodies. (C) Protease protection assay of a P200 fraction isolated from oleate-grown Δpex13 cells expressing GFP-Pex8p. The organelle pellet (50 μg) was incubated at room temperature with 20 μg of proteinase K and 10 μg of trypsin for the indicated times, in the presence or absence of 0.5% Triton X-100. Equal proportions of each reaction were separated by SDS-PAGE followed by immunoblot analysis with the indicated antibodies. Bar, 2 μm.
To provide further evidence that GFP-Pex8p was targeted to the peroxisome matrix in Δpex13 cells, we performed protease protection experiments by using the P200 organelle pellet fractions of oleate-grown cells (Figure 1C). Similar to what was observed in the protease protection assay using wild-type cells (Supplemental Figure S1B), in the Δpex13 cells GFP-Pex8p, as well as endogenous Pex8p, were resistant to protease treatment in the absence of detergent and were degraded only after addition of Triton X-100. The peroxisomal membrane proteins Pex12p and Pex17p, serving as internal controls, were susceptible to proteases. In contrast, in the absence of Pex14p, both GFP-Pex8p and endogenous Pex8p were susceptible to protease treatment, consistent with our earlier report (Supplemental Figure S1D). Moreover, using flotation gradients, we showed that GFP-Pex8p and endogenous Pex8p were predominantly present in membranous remnants and not in protein aggregates in Δpex13 (Supplemental Figure S2).
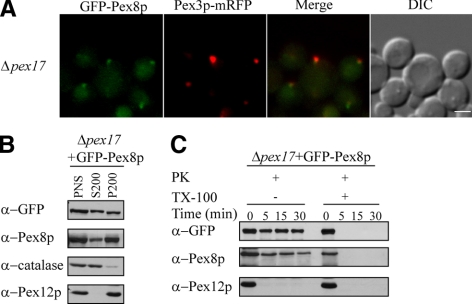
The Δpex17 strain is characterized by the cytosolic accumulation of peroxisomal matrix proteins by using PTS1 or PTS2 (Snyder et al., 1999). However, as shown by fluorescence microscopy and subcellular fractionation, the peroxisomal import of both GFP-Pex8p and endogenous Pex8p was partially impaired but not abolished (Figure 2, A and B). Under the same conditions, catalase import was impaired substantially but that of Pex12p was not, as expected (Figure 2B). In addition, protease protection assays showed that both GFP-Pex8p and endogenous Pex8p were able to translocate into the peroxisome matrix because they were resistant to protease treatment, whereas Pex12p was degraded under the same conditions (Figure 2C).
Figure 2.
GFP-Pex8p associates partially with peroxisomal remnants in Δpex17 cells. (A) Fluorescence microscopy analysis of methanol-grown Δpex17 cells coexpressing GFP-Pex8p and Pex3p-mRFP serving as a peroxisomal marker. (B) Differential centrifugation fractions of oleate-grown Δpex17 cells expressing GFP-Pex8p were immunoblotted with the indicated antibodies. (C) Protease protection assay of a P200 fraction isolated from oleate-grown Δpex17 cells expressing GFP-Pex8p followed by immunoblot analysis. Bar, 2 μm.
Import of Pex8p by the PTS1 Pathway Does Not Require the RING Subcomplex
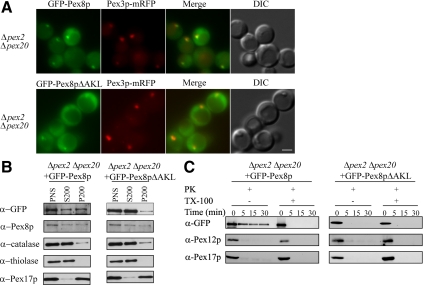
We showed previously, using Δpex2 cells, in which the other two components of the RING subcomplex, Pex10p and Pex12p, are either absent or down-regulated (Hazra et al., 2002), that the import of Pex8p by the PTS2 pathway does not require the RING subcomplex (Zhang et al., 2006). To characterize whether this is also true for the PTS1 pathway, we generated double-knockout mutant strains Δpex2 Δpex20 by deleting PEX20 in Δpex2 cells expressing either GFP-Pex8p or GFP-Pex8pΔAKL. Pex20p is a coreceptor and essential for the peroxisomal targeting of PTS2 proteins. GFP-Pex8p was targeted to the cytosol and also present partially in punctate peroxisomal remnants, which contained Pex3p-mRFP (Figure 3A, top). To our surprise, GFP-Pex8pΔAKL was largely cytosolic in Δpex2 Δpex20 cells, but in a small population of cells, it also associated with peroxisomal remnants (Figure 3A, bottom). In contrast, subcellular fractionation assays showed that GFP-Pex8p as well as endogenous Pex8p was almost evenly distributed in both the cytosol and organelle pellet fractions, whereas GFP-Pex8pΔAKL was predominantly in the cytosol fraction (Figure 3B). Other peroxisomal markers, catalase, thiolase, and Pex17p, behaved as expected (Figure 3B).
Figure 3.
Translocation of Pex8p into peroxisomes by the PTS1 pathway is Pex2p-independent. (A) Δpex2 Δpex20 strains expressing both functional GFP-Pex8p or GFP-Pex8pΔAKL and Pex3p-mRFP fusion proteins grown in synthetic medium (YNM) were visualized by fluorescence microscopy. (B) Differential centrifugation analysis of GFP-Pex8p and GFP-Pex8pΔAKL constructs expressed in oleate-grown Δpex2 Δpex20 cells using the indicated antibodies. (C) Protease protection assay of a P200 fraction isolated from oleate-grown Δpex2 Δpex20 cells expressing GFP-Pex8p or GFP-Pex8pΔAKL. The samples were analyzed by immunoblotting with the indicated antibodies. Bar, 2 μm.
To answer whether both GFP-Pex8p and GFP-Pex8pΔAKL are truly imported into the peroxisome matrix, we performed protease protection assays and found that GFP-Pex8p, but not GFP-Pex8pΔAKL, were protease protected under conditions where Pex12p and Pex17p were sensitive to protease (Figure 3C). The differential protease susceptibility of GFP-Pex8p, in contrast to that of GFP-Pex8pΔAKL also shows that the protease resistance of the former protein is not due to protein aggregation. These results indicate that some GFP-Pex8p is targeted into the peroxisome matrix in the Δpex2 Δpex20 cells, whereas GFP-Pex8pΔAKL was only associated with the cytosol-exposed surface of peroxisome remnants. Pex8pΔAKL is able to interact with full-length Pex5p as observed in yeast two-hybrid assays, through a domain in Pex5p distinct from the C-terminal tetratricopeptide repeat (TPR) domains (Zhang et al., 2006). This secondary binding site may explain why GFP-Pex8pΔAKL is able to associate with the surface of peroxisomes via Pex5p. The above-mentioned results demonstrate that the import of Pex8p into peroxisomes via the PTS1 pathway also does not depend on the RING subcomplex.
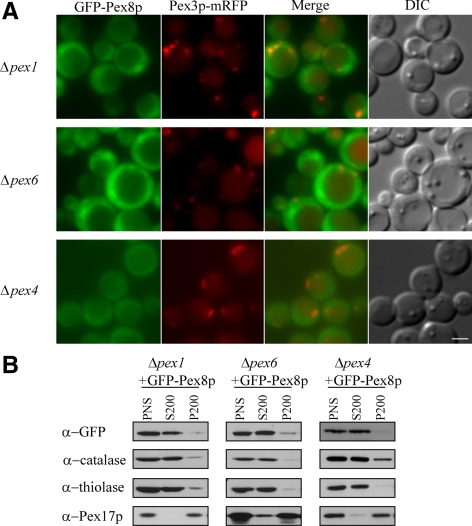
GFP-Pex8p Remains in the Cytosol in Δpex1, Δpex6, and Δpex4 Cells
To elucidate the complete cycle of receptor–cargo translocation followed by the return of the PTS receptors to the cytosol, we analyzed the role of the receptor-recycling machinery in the import of Pex8p. The importomer-associated receptor-recycling machinery, including the ubiquitin-conjugating enzyme, Pex4p, and its peroxisome-anchoring protein Pex22p, the AAA ATPase members Pex1p and Pex6p, and its peroxisome-anchoring protein, Pex15p (in yeast), are responsible for the recycling of the PTS1 receptor, Pex5p and the PTS2 coreceptor, Pex20p from the peroxisome surface back to the cytosol (Leon et al., 2006b; Platta and Erdmann, 2007). Pex5p and Pex20p are unstable and get degraded by the receptor accumulation and degradation in the absence of recycling (RADAR) pathway in the absence of any component of the receptor-recycling machinery (Leon et al., 2006b). In Δpex1, Δpex6 and Δpex4 cells, GFP-Pex8p accumulated in the cytosol (Figure 4A), which is expected because both Pex5p and Pex20p are rapidly degraded via the RADAR pathway. In these mutants, most of the catalase and thiolase were in the S200 fraction, but Pex17p was in the P200 fraction, as expected (Figure 4B).
Figure 4.
GFP-Pex8p remains in the cytosol in Δpex1, Δpex6, and Δpex4 cells. (A) Fluorescence microscopy analysis of methanol-grown cells (Δpex1, Δpex6, and Δpex4) coexpressing GFP-Pex8p and Pex3p-mRFP serving as a peroxisomal marker. (B) Differential centrifugation fractions of oleate-grown Δpex1, Δpex6, and Δpex4 cells expressing GFP-Pex8p were immunoblotted with the indicated antibodies. Bar, 2 μm.
Pex8p Targets to Peroxisome Remnants in Δpex4 and Δpex6 Cells If PTS Receptor Degradation via the RADAR Pathway Is Blocked
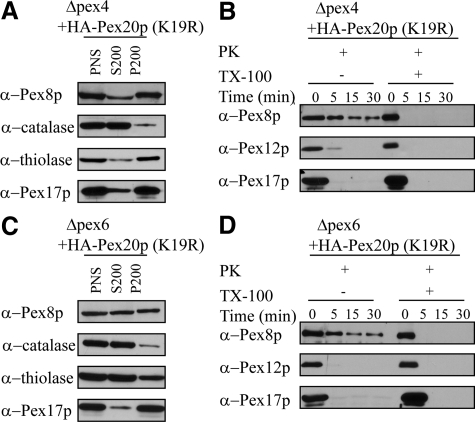
To determine whether the cytosolic mislocalization of GFP-Pex8p in the receptor recycling mutants was caused as a secondary effect of PTS-receptor instability, we overexpressed a receptor mutant, HA-Pex20p(K19R), under glyceraldehyde-3-phosphate dehydrogenase (GAPDH) promoter control. This mutant protein is stable and not down-regulated by the RADAR pathway in Δpex4 cells because the site of polyubiquitination (K19) on Pex20p is mutated (Leon et al., 2006b). As shown by subcellular fractionation assays, under such conditions the endogenous Pex8p (similar to PPY12 wild-type cells) was predominantly targeted to peroxisomes in Δpex4 cells through the PTS2 pathway because the degradation of Pex20p by the RADAR pathway was blocked (Figure 5A). It is noteworthy that thiolase, a PTS2 cargo, was also imported into peroxisomes because of the partially restored PTS2 pathway. In contrast, catalase, which depends on the PTS1 receptor Pex5p, still remained in the cytosol. Furthermore, protease protection assays showed that endogenous Pex8p was imported into the peroxisome matrix because it was resistant to protease treatment, whereas Pex12p and Pex17p were degraded under the same conditions (Figure 5B). It is interesting that similar overexpression of HA-Pex20p(K19R) in Δpex6 cells restored only partially Pex8p and thiolase import (Figure 5, C and D). Our results show that the import of Pex8p could be partially restored to some extent once the receptor instability by the RADAR pathway was blocked, proving that the entry of Pex8p into the peroxisomal matrix is not impaired as long as PTS receptors are stable and available for its targeting. Therefore, the receptor recycling machinery per se is not essential for peroxisomal matrix targeting of Pex8p.
Figure 5.
Pex8p is targeted to peroxisome remnants in Δpex4 and Δpex6 cells if the degradation of Pex20p by the RADAR pathway is blocked. (A and C) Differential centrifugation fractions of oleate-grown Δpex4 or Δpex6 cells expressing HA-Pex20p(K19R) were immunoblotted with the indicated antibodies. (B and D) Protease protection assay of a P200 fraction isolated from oleate-grown Δpex4 or Δpex6 cells expressing HA-Pex20p(K19R). The samples were analyzed by immunoblotting with the indicated antibodies.
The Import of Pex8p by the PTS1 Pathway Does Not Require Intraperoxisomal Pex8p
If Pex8p entry into the peroxisome matrix depends on the prior presence of intraperoxisomal Pex8p, then this raises the issue of how the first molecule of Pex8p entered peroxisomes during evolution. We hypothesized that the import of Pex8p by both PTS1 and PTS2 pathways might not depend on intraperoxisomal Pex8p. In a previous report, Zhang et al. (2006) showed that the import of Pex8p by the PTS1 pathway relied on pre-existing intraperoxisomal Pex8p by using the constructs GFP-Pex8pΔAKL and GFP-Pex8pPTS2m, bearing mutated PTS1 and PTS2 sequences, respectively. However, this conclusion is arguable for the GFP-Pex8pPTS2m construct, because disruption of the PTS2 signal resulted in a nonfunctional protein in vivo (data not shown). To investigate whether the import of Pex8p via the PTS1 pathway depends on intraperoxisomal Pex8p, we reinvestigated this issue by generating a double-knockout strain, Δpex8 Δpex20, expressing functional GFP-Pex8p. In these cells, Pex8p entry into peroxisomes must occur via the PTS1 pathway, because of the deletion of the PEX20 gene.
GFP-Pex8p was localized in punctate structures and colocalized with a peroxisomal membrane marker, Pex3p-mRFP (Figure 6A, top). Moreover, GFP-Pex8p could even complement the Δpex8 Δpex20 strain because large peroxisome clusters, instead of peroxisome remnants, were found. In contrast, upon deleting the C-terminal PTS1 tripeptide GFP-Pex8pΔAKL was largely cytosolic (Figure 6A, bottom). Only in a few cells, GFP-Pex8pΔAKL colocalized with tiny, Pex3p-mRFP–labeled peroxisome remnants. However, this fusion protein remains on the outer surface of peroxisomes (see below) because no peroxisome clusters could be observed anymore, and more importantly, such cells still had a growth defect in methanol medium (data not shown).
Figure 6.
Import of Pex8p by PTS1 pathways does not require intraperoxisomal Pex8p. (A) Fluorescence and DIC images of methanol-grown Δpex8 Δpex20 cells coexpressing GFP-Pex8p or GFP-Pex8pΔAKL and Pex3p-mRFP. (B) Differential centrifugation samples of oleate-grown Δpex8 Δpex20 cells expressing either GFP-Pex8p or GFP-Pex8pΔAKL were immunoblotted with the indicated antibodies. (C) Protease protection assay of a P200 fraction isolated from oleate-grown Δpex8 Δpex20 cells expressing GFP-Pex8p or GFP-Pex8pΔAKL. The samples were analyzed by immunoblotting with the indicated antibodies. Bar, 2 μm.
Subcellular fractionation assays showed that GFP-Pex8p was predominantly located in the organelle pellet fraction (P200) of Δpex8 Δpex20 cells, together with the peroxisomal markers catalase and Pex17p (Figure 6B). The restoration of catalase import into peroxisomes is yet another reflection that GFP-Pex8p can complement the PTS1-import deficient Δpex8 Δpex20 strain. In contrast, thiolase was mislocalized to the cytosol (S200 fraction) because the Δpex8 Δpex20 mutant strain is specifically defective in PTS2 import pathway (only after complementation with GFP-Pex8p). In contrast, GFP-Pex8pΔAKL, as well as catalase and thiolase, were predominantly localized in the cytosol in Δpex8 Δpex20 cells because both the PTS1 and PTS2 pathways were compromised. In addition, we performed protease protection experiments using the P200 organelle pellet fractions of oleate-grown cells (Figure 6C). In all cases, GFP-Pex8p was resistant to protease treatment in the absence of detergent, whereas GFP-Pex8pΔAKL, and the peroxisomal membrane proteins Pex12p and Pex17p were susceptible to proteases. In summary, these experiments demonstrate that the PTS1-dependent import of Pex8p into peroxisomes does not require intraperoxisomal Pex8p.
Pex8p Is Not Necessary for the Assembly of Importomer Subcomplexes
An interesting question regarding the import of Pex8p in the absence of pre-existing intraperoxisomal Pex8p is raised by the proposed function of Pex8p. If Pex8p is critical for the association of the docking and RING subcomplex into a larger import complex, i.e., importomer, as demonstrated in S. cerevisiae (Agne et al., 2003), how could Pex8p itself be imported in the absence of pre-existing intraperoxisomal Pex8p? The solution to this apparent paradox might lie in the fact that either the entire importomer, as defined by Agne et al. (2003), is not essential for Pex8p import, that Pex8p is not essential to hold the importomer subcomplexes together, or both. It is plausible, for example, that P. pastoris and S. cerevisiae may depend to varying extents on different proteins for the assembly of the importomer. Based on cross-linking and immunoprecipitation experiments in P. pastoris, Hazra et al. (2002) showed that Pex3p, instead of Pex8p, is required to link the docking and the RING subcomplexes, although Pex8p was found to be associated with the docking and RING subcomplexes in P. pastoris, too.
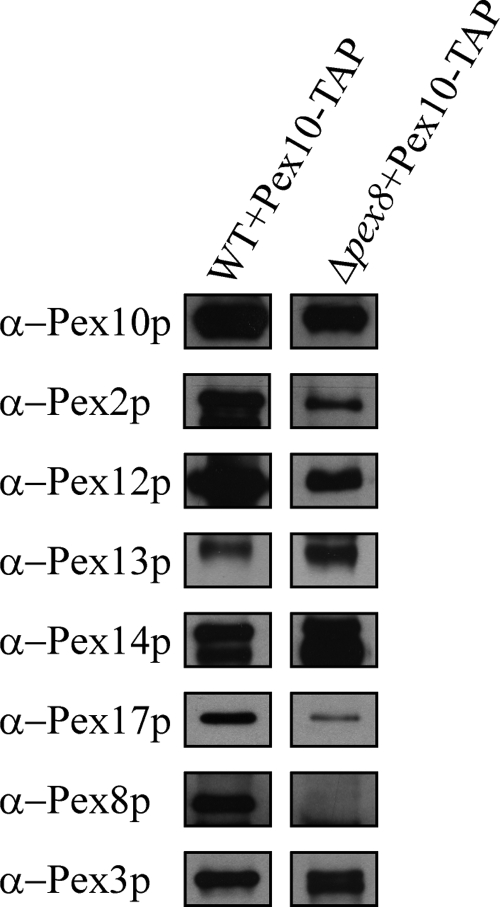
To provide an independent line of evidence that PpPex8p is not required for the interaction of the two subcomplexes, we isolated the peroxisome importomer by TAP-tag purification (Rigaut et al., 1999). We generated a construct containing Pex10p and a C-terminally fused IgG-binding unit of protein A, followed by a cleavage site for TEV protease and a calmodulin-binding protein (CBP) domain. This Pex10-TAP fusion protein was able to complement the Δpex10 strain (data not shown). To obtain pure and intact peroxisome importomers, we isolated crude peroxisome fractions and solubilized the membrane proteins with different detergents followed by IgG and calmodulin affinity chromatography. We found that the docking and RING subcomplexes together with Pex3p and Pex8p could be extracted when 1% CHAPS was added to the solubilization buffer, while using other detergents such as n-decyl-β-maltoside and n-dodecyl β-d-maltoside resulted in only partially solubilized importomer (data not shown). As shown in Figure 7, the RING finger proteins Pex2p, Pex10p, and Pex12p and the docking subcomplex components Pex13p, Pex14p, and Pex17p, as well as Pex8p and Pex3p were identified in Pex10-TAP-tag eluates of wild-type PPY12 cells. The presence of these Pex10-interacting proteins was also confirmed by parallel analyses using mass spectrometry (courtesy of Dr. John Yates, III, The Scripps Research Institute, La Jolla, CA), which also showed the specificity of the importomer purification because the integral membrane protein Pex22p was absent (data not shown). However, in the absence of Pex8p, all constituents of both the RING and docking subcomplexes, as well as Pex3p, still existed in Pex10-TAP-tag eluates. Therefore, these data suggest that Pex8p is not essential for bridging the docking and RING subcomplexes of the importomer in P. pastoris.
Figure 7.
Pex8p is not necessary for the assembling of importomer subcomplexes. Protein complexes were isolated from 1% CHAPS-solubilized membranes of wild-type cells and Δpex8 cells expressing Pex10p-TAP-tag via IgG-affinity chromatography and subsequent TEV protease cleavage followed by CBP-affinity chromatography. The TAP-tag eluates were analyzed by immunoblotting with the indicated antibodies.
DISCUSSION
The Import of Pex8p Requires Limited Peroxins Only
To the best of our knowledge, Pex8p is the only known peroxisomal matrix cargo that has an essential role in peroxisome biogenesis, which makes it an extraordinary peroxin (Waterham et al., 1994; Liu et al., 1995; Rehling et al., 2000; Smith and Rachubinski, 2001; Zhang et al., 2006). Unlike other cargoes that normally only have one PTS, PpPex8p has a PTS1 at the carboxy terminus and an unusual PTS2 in the middle of the protein. PpPex8p uses redundant pathways for its targeting to the peroxisomal matrix (Zhang et al., 2006). A conserved C-terminal PTS1 was found in the homologue of Pex8p in H. polymorpha and S. cerevisiae. Moreover, the N-terminal fragments of HpPex8p (1–16) and ScPex8p (1–112) alone are sufficient to direct a reporter protein into the peroxisomal matrix (Waterham et al., 1994; Rehling et al., 2000). These studies suggested the existence of a redundant pathway for peroxisomal import of Pex8p in lower eukaryotes.
With regard to the function of Pex8p in peroxisomal matrix protein import, several questions are raised: how does Pex8p itself get imported to the site where it performs its function? It is assumed that Pex8p has to be the first cargo that reaches peroxisome lumen because the lack of Pex8p results in mislocalization of peroxisomal matrix proteins to the cytosol (Waterham et al., 1994; Liu et al., 1995; Rehling et al., 2000; Smith and Rachubinski, 2001). This raises the possibility that the import of Pex8p is subject to special requirements of the import machinery and that these requirements may be less stringent in comparison with general cargoes.
Our previous studies showed that the translocation of Pex8p into peroxisomes is Pex14p dependent, but Pex2p and Pex8p independent via the PTS2 pathway, which is unusual for a cargo (Zhang et al., 2006). In exploring this in more detail, we found in this study that the two other components of the docking subcomplex, Pex13p and Pex17p, are not essential for the targeting of Pex8p into peroxisomes. We showed that Pex8p is delivered to the peroxisome matrix in Δpex13, as well as in Δpex17, cells albeit at a lower efficiency than in wild-type cells (Figures 1 and 2). These data suggest that both Pex13p and Pex17p are dispensable for docking and translocation of Pex8p into peroxisomes but that they may be necessary to enhance the efficiency of protein translocation into the peroxisome matrix.
Pex13p, like Pex14p, has been shown to interact at the surface of the peroxisome membrane with both the PTS1 and PTS2 receptors (Albertini et al., 1997; Stein et al., 2002). Therefore, Pex13p could also serve as a suitable candidate for a docking protein that interacts with the receptor–cargo complexes. However, studies from in vitro binding assays suggest that Pex14p is the initial docking factor that associates with the receptor–cargo complex. Otera et al. (2000) and Urquhart et al. (2000) found that in mammalian cells, cargo-bound Pex5p has a stronger affinity for Pex14p than for Pex13p, whereas cargo-free receptors interact more strongly with Pex13p, suggesting that Pex13p acts downstream of Pex14p.
Our result that the import of Pex8p into the peroxisomal matrix occurs in the absence of Pex13p, and the observation that Pex13p binds cargo-free receptors more strongly than cargo-loaded receptors, has added new evidence that within the import cascade, Pex13p plays a role downstream of Pex14p, after the translocation of receptor–cargo complexes into peroxisomes, and possibly even after cargo release.
Unlike Pex13p and Pex14p, which are found in all eukaryotic organisms, Pex17p only exists in lower eukaryotic cells (Kiel et al., 2006). Pex17p interacts with both receptors in a Pex14p-dependent manner (Huhse et al., 1998; Snyder et al., 1999). Therefore, Pex17p has been commonly accepted as one of the components of the docking subcomplex. However, its precise function in peroxisome biogenesis is still unknown. Regarding the import of Pex8p, Pex17p is not necessary for peroxisomal import of Pex8p, but it does play a role in the efficiency of Pex8p import.
In contrast to Pex8p, the import of catalase and thiolase was almost entirely blocked in Δpex13 and Δpex17 cells, indicating that Pex8p behaves differently from other PTS-containing cargoes. This conclusion was bolstered by our observation that in Δpex13 cells, when GFP-SKL was expressed at three different levels, from the GAPDH, AOX, or PEX8 promoters, it was mislocalized to the cytosol (Supplemental Figure S3). One possible explanation is that in these mutants, the lower efficiency in import of Pex8p, combined with the lack of another peroxin (e.g., Pex13p or Pex17p), leads to an accumulative import defect.
To extend our examination whether the import of Pex8p into peroxisome via the PTS1 pathway is also Pex2p-independent, we generated a Δpex2 Δpex20 strain, which has a compromised PTS2 import pathway (Figure 3). Although the import efficiency of GFP-Pex8p by the PTS1 pathway is decreased, it translocates into the peroxisome matrix because the fusion protein is protease protected as long as the peroxisome membranes remain intact. Hence, with respect to the import of Pex8p in the absence of the RING subcomplex, both the PTS1 and PTS2 pathways are redundant. Contributing factors to the inefficient import of GFP-Pex8p into peroxisomes of Δpex2 Δpex20 cells are the absence of the PTS2 pathway coupled with an inefficient PTS1 pathway caused by impaired Pex5p recycling from the peroxisomes to the cytosol in these cells (our unpublished observations).
Using fluorescence microscopy, we were surprised to find that GFP-Pex8pΔAKL was associated with peroxisomes given the fact that the entry of GFP-Pex8pΔAKL via both the PTS1 and PTS2 pathways was blocked. However, GFP-Pex8pΔAKL was not protease protected and therefore unable to translocate across the peroxisomal membrane (Figure 3C). According to the yeast two-hybrid data (Zhang et al., 2006), PpPex8pΔAKL interacts with the full-length Pex5p but not with its TPR domains, suggesting the N terminus of Pex5p has a second binding site for PpPex8p. Therefore, we assume that PpPex8pΔAKL was transported to the peroxisomal membrane in Δpex2 Δpex20 cells through its interaction with the N terminus of Pex5p, but this did not lead to translocation of Pex8pΔAKL into the peroxisome matrix. Consistent with this hypothesis, we found that GFP-Pex8pΔAKL was also associated with peroxisomes in some Δpex8 Δpex20 cells. However, as expected, GFP-Pex8pΔAKL just sits on the outside of the peroxisomal membrane because it cannot complement Δpex8 Δpex20 cells (Figure 6).
In the absence of components of the receptor recycling machinery, Pex5p and Pex20p are unstable and get degraded rapidly (Koller et al., 1999; Collins et al., 2000; Leon et al., 2006b). Here, we have shown that, as a consequence of the rapid degradation of recycling receptors, the import of Pex8p was almost eliminated because it relies on either the PTS1 or PTS2 pathway for targeting (Figure 4). However, the import of Pex8p could be almost fully restored upon the expression of HA-Pex20p(K19R) in Δpex4, which is recalcitrant to degradation by the RADAR pathway (Figure 5, A and B). This is an interesting result because stabilization of Pex20p after it has released the cargo, but has not yet been recycled from the peroxisome membrane to the cytosol, might not be expected to completely restore Pex8p and thiolase import. Indeed, this was the result we observed when HA-Pex20p(K19R) was overexpressed in another receptor recycling mutant, Δpex6 (Figure 5, C and D). Our interpretation of the disparity in the behaviors of the Δpex4 and Δpex6 cells with respect to the restoration of the Pex20p-dependent PTS2 import pathway is that some ubiquitin-conjugating enzyme other than Pex4p may be able to inefficiently allow some recycling of HA-Pex20p(K19R) when it is overexpressed and stabilized, whereas the loss of Pex6p cannot be compensated by another cellular component. As multifunctional proteins, Pex6p and Pex1p are involved not only in extraction of receptors from peroxisomal membrane for recycling but also play a role in the process of peroxisome maturation (Titorenko and Rachubinski, 2000). Regardless of the explanation, the suppression of the Pex8p import defect in Δpex4 and Δpex6 cells by overexpression of HA-Pex20p(K19R) suggests that the receptor recycling machinery is only indirectly involved in the import of Pex8p by maintaining the stability and recycling of receptors.
In summary, these studies also show that unlike most other PTS cargoes, Pex8p has redundant PTSs, enters peroxisomes via redundant pathways using a simpler basic machinery for its translocation, and finally does not require intraperoxisomal Pex8p (Figure 6). This makes it a special type of cargo that has evolved to become an important component for the peroxisomal import of other matrix cargoes. We do not exclude the possibility that trivial amounts of other PTS1 protein enter peroxisomes in the absence of Pex13p, Pex17p, and the RING complex.
Pex14p and the PTS Receptors Constitute the Minimal Matrix Translocation Machinery
Essentially, our data show that the import of Pex8p requires either PTS1 or PTS2 receptors, Pex14p and indirectly the peroxins that are responsible for the stability of the receptors. This gives reason to propose that receptor and Pex14p represent the minimal machinery for peroxisomal matrix protein import.
Three different models have been proposed to explain how cargo would be imported into the peroxisome matrix (Rayapuram and Subramani, 2006). In the first model, the docking as well as the RING subcomplexes cooperate together serving as the translocon. The receptor–cargo complex is translocated into the matrix after its simultaneous or sequential interaction with the docking and RING subcomplex. This first model is invalidated by the RING subcomplex- and Pex8p-independent peroxisomal entry of Pex8p, necessitating other models (Zhang et al., 2006). In the second model, the docking subcomplex itself represents the translocon, whereas Pex8p and the RING subcomplex are involved in posttranslocation events involving the PTS receptors. Recently, Erdmann and Schliebs (2005) proposed a third transient pore model, in which Pex5p is able to insert spontaneously and assembles into peroxisome membrane to build a translocon of variable size which accommodates different cargoes. In this model, the docking subcomplex might assist the receptor–cargo complex to insert into the peroxisome membrane. This reflects an assembly of the oligomeric receptor into a translocation pore, whereas the RING subcomplex is involved in the disassembly of the translocon and probably the activation of the receptor export pathway.
At present, we cannot be certain whether the minimal peroxisomal membrane translocon is comprised of Pex14p oligomers that allow cargo-bound PTS receptors across the membrane, or whether in view of the third model, PTS receptors along with Pex14p constitute the translocon. Pex14p has all the features required to form a transient pore or protein-conducting channel (Erdmann and Schliebs, 2005). First, it is an integral membrane protein (Komori et al., 1999; Hayashi et al., 2000; Jardim et al., 2000; Johnson et al., 2001) that is conserved across evolution from yeasts to humans. The N-terminal domain of Pex14p is strongly conserved and predicted to contain a transmembrane domain (TMD) (www.ch.embnet.org/software/TMPRED_form.html). Second, it associates with proteins destined to be translocated into the peroxisomal matrix (at least in an indirect manner through PTS receptors). In addition, Pex14p associates with the RING subcomplex via Pex3p or Pex8p (Johnson et al., 2001; Hazra et al., 2002; Agne et al., 2003). Last, but most importantly, Pex14p is able to transiently form homo-oligomers (Itoh and Fujiki, 2006; Cyr et al., 2008). In mammals, the GXXXG and AXXXA motifs in the TMD region of Pex14p are responsible for high molecular mass homo-oligomerization and might contribute to the formation of a hydrophilic channel. The association of cargo-loaded receptor with Pex14p could induce the oligomeric state of Pex14p in CHO cells or conformational changes in Leishmania donovani, which may indicate the formation of a transient pore or exposure of a pre-existing pore (Itoh and Fujiki, 2006; Cyr et al., 2008). Moreover, the size of the hydrophilic channel or translocon might be adjusted by the oligomerized state of Pex14p to import size-different cargoes.
To investigate whether the minimum translocon is composed of the import receptor, Pex14p, or both, these proteins need to be purified for incorporation into liposomes followed by direct demonstration of their channel-forming properties.
Supplementary Material
ACKNOWLEDGMENTS
We thank Shiyin Yao for constructing the Δpex8 Δpex20-related strains and for general technical help and Dr. John Yates, III for help with mass spectrometry of protein complexes. This work was supported by National Institutes of Health grant DK-41737 (to S. S.).
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E09-01-0037) on July 1, 2009.
REFERENCES
- Agne B., Meindl N. M., Niederhoff K., Einwachter H., Rehling P., Sickmann A., Meyer H. E., Girzalsky W., Kunau W. H. Pex8p: an intraperoxisomal organizer of the peroxisomal import machinery. Mol Cell. 2003;11:635–646. doi: 10.1016/s1097-2765(03)00062-5. [DOI] [PubMed] [Google Scholar]
- Albertini M., Rehling P., Erdmann R., Girzalsky W., Kiel J. A., Veenhuis M., Kunau W. H. Pex14p, a peroxisomal membrane protein binding both receptors of the two PTS-dependent import pathways. Cell. 1997;89:83–92. doi: 10.1016/s0092-8674(00)80185-3. [DOI] [PubMed] [Google Scholar]
- Collins C. S., Kalish J. E., Morrell J. C., McCaffery J. M., Gould S. J. The peroxisome biogenesis factors Pex4p, Pex22p, Pex1p, and Pex6p act in the terminal steps of peroxisomal matrix protein import. Mol. Cell. Biol. 2000;20:7516–7526. doi: 10.1128/mcb.20.20.7516-7526.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cyr N., Madrid K. P., Strasser R., Aurousseau M., Finn R., Ausio J., Jardim A. Leishmania donovani peroxin14 undergoes a marked conformational change following association with peroxin5. J. Biol. Chem. 2008;283:31488–31499. doi: 10.1074/jbc.M803529200. [DOI] [PubMed] [Google Scholar]
- Erdmann R., Schliebs W. Peroxisomal matrix protein import: the transient pore model. Nat. Rev. Mol. Cell Biol. 2005;6:738–742. doi: 10.1038/nrm1710. [DOI] [PubMed] [Google Scholar]
- Faber K. N., Heyman J. A., Subramani S. Two AAA family peroxins, PpPex1p and PpPex6p, interact with each other in an ATP-dependent manner and are associated with different subcellular membranous structures distinct from peroxisomes. Mol. Cell. Biol. 1998;18:936–943. doi: 10.1128/mcb.18.2.936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould S. J., Kalish J. E., Morrell J. C., Bjorkman J., Urquhart A. J., Crane D. I. Pex13p is an SH3 protein of the peroxisome membrane and a docking factor for the predominantly cytoplasmic PTS1 receptor. J. Cell Biol. 1996;135:85–95. doi: 10.1083/jcb.135.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould S. J., Keller G. A., Hosken N., Wilkinson J., Subramani S. A conserved tripeptide sorts proteins to peroxisomes. J. Cell Biol. 1989;108:1657–1664. doi: 10.1083/jcb.108.5.1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunkel K., van Dijk R., Veenhuis M., van der Klei I. J. Routing of Hansenula polymorpha alcohol oxidase: an alternative peroxisomal protein-sorting machinery. Mol. Biol. Cell. 2004;15:1347–1355. doi: 10.1091/mbc.E03-04-0258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi M., Nito K., Toriyama-Kato K., Kondo M., Yamaya T., Nishimura M. AtPex14p maintains peroxisomal functions by determining protein targeting to three kinds of plant peroxisomes. EMBO J. 2000;19:5701–5710. doi: 10.1093/emboj/19.21.5701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazra P. P., Suriapranata I., Snyder W. B., Subramani S. Peroxisome remnants in pex3Δ cells and the requirement of Pex3p for interactions between the peroxisomal docking and translocation subcomplexes. Traffic. 2002;3:560–574. doi: 10.1034/j.1600-0854.2002.30806.x. [DOI] [PubMed] [Google Scholar]
- Heyman J. A., Monosov E., Subramani S. Role of the PAS1 gene of Pichia pastoris in peroxisome biogenesis. J. Cell Biol. 1994;127:1259–1273. doi: 10.1083/jcb.127.5.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huhse B., Rehling P., Albertini M., Blank L., Meller K., Kunau W. H. Pex17p of Saccharomyces cerevisiae is a novel peroxin and component of the peroxisomal protein translocation machinery. J. Cell Biol. 1998;140:49–60. doi: 10.1083/jcb.140.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh R., Fujiki Y. Functional domains and dynamic assembly of the peroxin Pex14p, the entry site of matrix proteins. J. Biol. Chem. 2006;281:10196–10205. doi: 10.1074/jbc.M600158200. [DOI] [PubMed] [Google Scholar]
- Jardim A., Liu W., Zheleznova E., Ullman B. Peroxisomal targeting signal-1 receptor protein PEX5 from Leishmania donovani. Molecular, biochemical, and immunocytochemical characterization. J. Biol. Chem. 2000;275:13637–13644. doi: 10.1074/jbc.275.18.13637. [DOI] [PubMed] [Google Scholar]
- Johnson M. A., Snyder W. B., Cereghino J. L., Veenhuis M., Subramani S., Cregg J. M. Pichia pastoris Pex14p, a phosphorylated peroxisomal membrane protein, is part of a PTS-receptor docking complex and interacts with many peroxins. Yeast. 2001;18:621–641. doi: 10.1002/yea.711. [DOI] [PubMed] [Google Scholar]
- Kiel J. A., Veenhuis M., van der Klei I. J. PEX genes in fungal genomes: common, rare or redundant. Traffic. 2006;7:1291–1303. doi: 10.1111/j.1600-0854.2006.00479.x. [DOI] [PubMed] [Google Scholar]
- Klein A. T., van den Berg M., Bottger G., Tabak H. F., Distel B. Saccharomyces cerevisiae acyl-CoA oxidase follows a novel, non-PTS1, import pathway into peroxisomes that is dependent on Pex5p. J. Biol. Chem. 2002;277:25011–25019. doi: 10.1074/jbc.M203254200. [DOI] [PubMed] [Google Scholar]
- Koller A., Snyder W. B., Faber K. N., Wenzel T. J., Rangell L., Keller G. A., Subramani S. Pex22p of Pichia pastoris, essential for peroxisomal matrix protein import, anchors the ubiquitin-conjugating enzyme, Pex4p, on the peroxisomal membrane. J. Cell Biol. 1999;146:99–112. doi: 10.1083/jcb.146.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komori M., Kiel J. A., Veenhuis M. The peroxisomal membrane protein Pex14p of Hansenula polymorpha is phosphorylated in vivo. FEBS Lett. 1999;457:397–399. doi: 10.1016/s0014-5793(99)01087-x. [DOI] [PubMed] [Google Scholar]
- Leon S., Goodman J. M., Subramani S. Uniqueness of the mechanism of protein import into the peroxisome matrix: transport of folded, co-factor-bound and oligomeric proteins by shuttling receptors. Biochim. Biophys. Acta. 2006a;1763:1552–1564. doi: 10.1016/j.bbamcr.2006.08.037. [DOI] [PubMed] [Google Scholar]
- Leon S., Zhang L., McDonald W. H., Yates J., 3rd, Cregg J. M., Subramani S. Dynamics of the peroxisomal import cycle of PpPex20p: ubiquitin-dependent localization and regulation. J. Cell Biol. 2006b;172:67–78. doi: 10.1083/jcb.200508096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Tan X., Russell K. A., Veenhuis M., Cregg J. M. PER3, a gene required for peroxisome biogenesis in Pichia pastoris, encodes a peroxisomal membrane protein involved in protein import. J. Biol. Chem. 1995;270:10940–10951. doi: 10.1074/jbc.270.18.10940. [DOI] [PubMed] [Google Scholar]
- Luers G. H., Advani R., Wenzel T., Subramani S. The Pichia pastoris dihydroxyacetone kinase is a PTS1-containing, but cytosolic, protein that is essential for growth on methanol. Yeast. 1998;14:759–771. doi: 10.1002/(SICI)1097-0061(19980615)14:8<759::AID-YEA275>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- Otera H., Harano T., Honsho M., Ghaedi K., Mukai S., Tanaka A., Kawai A., Shimizu N., Fujiki Y. The mammalian peroxin Pex5pL, the longer isoform of the mobile peroxisome targeting signal (PTS) type 1 transporter, translocates the Pex7p.PTS2 protein complex into peroxisomes via its initial docking site, Pex14p. J. Biol. Chem. 2000;275:21703–21714. doi: 10.1074/jbc.M000720200. [DOI] [PubMed] [Google Scholar]
- Platta H. W., Erdmann R. Peroxisomal dynamics. Trends Cell Biol. 2007;17:474–484. doi: 10.1016/j.tcb.2007.06.009. [DOI] [PubMed] [Google Scholar]
- Purdue P. E., Lazarow P. B. Peroxisome biogenesis. Annu. Rev. Cell Dev. Biol. 2001;17:701–752. doi: 10.1146/annurev.cellbio.17.1.701. [DOI] [PubMed] [Google Scholar]
- Rayapuram N., Subramani S. The importomer—a peroxisomal membrane complex involved in protein translocation into the peroxisome matrix. Biochim. Biophys. Acta. 2006;1763:1613–1619. doi: 10.1016/j.bbamcr.2006.08.035. [DOI] [PubMed] [Google Scholar]
- Rehling P., Skaletz-Rorowski A., Girzalsky W., Voorn-Brouwer T., Franse M. M., Distel B., Veenhuis M., Kunau W. H., Erdmann R. Pex8p, an intraperoxisomal peroxin of Saccharomyces cerevisiae required for protein transport into peroxisomes binds the PTS1 receptor Pex5p. J. Biol. Chem. 2000;275:3593–3602. doi: 10.1074/jbc.275.5.3593. [DOI] [PubMed] [Google Scholar]
- Reumann S., Ma C., Lemke S., Babujee L. AraPerox. A database of putative Arabidopsis proteins from plant peroxisomes. Plant Physiol. 2004;136:2587–2608. doi: 10.1104/pp.104.043695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigaut G., Shevchenko A., Rutz B., Wilm M., Mann M., Seraphin B. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 1999;17:1030–1032. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]
- Smith J. J., Rachubinski R. A. A role for the peroxin Pex8p in Pex20p-dependent thiolase import into peroxisomes of the yeast Yarrowia lipolytica. J. Biol. Chem. 2001;276:1618–1625. doi: 10.1074/jbc.M005072200. [DOI] [PubMed] [Google Scholar]
- Snyder W. B., Koller A., Choy A. J., Johnson M. A., Cregg J. M., Rangell L., Keller G. A., Subramani S. Pex17p is required for import of both peroxisome membrane and lumenal proteins and interacts with Pex19p and the peroxisome targeting signal-receptor docking complex in Pichia pastoris. Mol. Biol. Cell. 1999;10:4005–4019. doi: 10.1091/mbc.10.12.4005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spong A. P., Subramani S. Cloning and characterization of PAS 5, a gene required for peroxisome biogenesis in the methylotrophic yeast Pichia pastoris. J. Cell Biol. 1993;123:535–548. doi: 10.1083/jcb.123.3.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein K., Schell-Steven A., Erdmann R., Rottensteiner H. Interactions of Pex7p and Pex18p/Pex21p with the peroxisomal docking machinery: implications for the first steps in PTS2 protein import. Mol. Cell. Biol. 2002;22:6056–6069. doi: 10.1128/MCB.22.17.6056-6069.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinberg S. J., Dodt G., Raymond G. V., Braverman N. E., Moser A. B., Moser H. W. Peroxisome biogenesis disorders. Biochim. Biophys. Acta. 2006;1763:1733–1748. doi: 10.1016/j.bbamcr.2006.09.010. [DOI] [PubMed] [Google Scholar]
- Subramani S., Koller A., Snyder W. B. Import of peroxisomal matrix and membrane proteins. Annu. Rev. Biochem. 2000;69:399–418. doi: 10.1146/annurev.biochem.69.1.399. [DOI] [PubMed] [Google Scholar]
- Swinkels B. W., Gould S. J., Bodnar A. G., Rachubinski R. A., Subramani S. A novel, cleavable peroxisomal targeting signal at the amino-terminus of the rat 3-ketoacyl-CoA thiolase. EMBO J. 1991;10:3255–3262. doi: 10.1002/j.1460-2075.1991.tb04889.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titorenko V. I., Rachubinski R. A. Peroxisomal membrane fusion requires two AAA family ATPases, Pex1p and Pex6p. J. Cell Biol. 2000;150:881–886. doi: 10.1083/jcb.150.4.881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urquhart A. J., Kennedy D., Gould S. J., Crane D. I. Interaction of Pex5p, the type 1 peroxisome targeting signal receptor, with the peroxisomal membrane proteins Pex14p and Pex13p. J. Biol. Chem. 2000;275:4127–4136. doi: 10.1074/jbc.275.6.4127. [DOI] [PubMed] [Google Scholar]
- Wang D., Visser N. V., Veenhuis M., van der Klei I. J. Physical interactions of the peroxisomal targeting signal 1 receptor pex5p, studied by fluorescence correlation spectroscopy. J. Biol. Chem. 2003;278:43340–43345. doi: 10.1074/jbc.M307789200. [DOI] [PubMed] [Google Scholar]
- Waterham H. R., Titorenko V. I., Haima P., Cregg J. M., Harder W., Veenhuis M. The Hansenula polymorpha PER1 gene is essential for peroxisome biogenesis and encodes a peroxisomal matrix protein with both carboxy- and amino-terminal targeting signals. J. Cell Biol. 1994;127:737–749. doi: 10.1083/jcb.127.3.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiemer E. A., Luers G. H., Faber K. N., Wenzel T., Veenhuis M., Subramani S. Isolation and characterization of Pas2p, a peroxisomal membrane protein essential for peroxisome biogenesis in the methylotrophic yeast Pichia pastoris. J. Biol. Chem. 1996;271:18973–18980. doi: 10.1074/jbc.271.31.18973. [DOI] [PubMed] [Google Scholar]
- Yan M., Rachubinski D. A., Joshi S., Rachubinski R. A., Subramani S. Dysferlin domain-containing proteins, Pex30p and Pex31p, localized to two compartments, control the number and size of oleate-induced peroxisomes in Pichia pastoris. Mol. Biol. Cell. 2008;19:885–898. doi: 10.1091/mbc.E07-10-1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Leon S., Subramani S. Two independent pathways traffic the intraperoxisomal peroxin PpPex8p into peroxisomes: mechanism and evolutionary implications. Mol. Biol. Cell. 2006;17:690–699. doi: 10.1091/mbc.E05-08-0758. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.