Abstract
MAL, a compact hydrophobic, four-transmembrane-domain apical protein that copurifies with detergent-resistant membranes is obligatory for the machinery that sorts glycophosphatidylinositol (GPI)-anchored proteins and others to the apical membrane in epithelia. The mechanism of MAL function in lipid-raft–mediated apical sorting is unknown. We report that MAL clusters formed by two independent procedures—spontaneous clustering of MAL tagged with the tandem dimer DiHcRED (DiHcRED-MAL) in the plasma membrane of COS7 cells and antibody-mediated cross-linking of FLAG-tagged MAL—laterally concentrate markers of sphingolipid rafts and exclude a fluorescent analogue of phosphatidylethanolamine. Site-directed mutagenesis and bimolecular fluorescence complementation analysis demonstrate that MAL forms oligomers via ϕxxϕ intramembrane protein–protein binding motifs. Furthermore, results from membrane modulation by using exogenously added cholesterol or ceramides support the hypothesis that MAL-mediated association with raft lipids is driven at least in part by positive hydrophobic mismatch between the lengths of the transmembrane helices of MAL and membrane lipids. These data place MAL as a key component in the organization of membrane domains that could potentially serve as membrane sorting platforms.
INTRODUCTION
The formation and maintenance of epithelial cell polarity relies on stringent regulation of intracellular transport and sorting processes. The apical plasma membrane (PM) domain is a robust yet sophisticated sphingolipid- and cholesterol-enriched protective barrier against harsh extracellular environments that maintains exchange and regulatory capacities (Schuck and Simons, 2004). Sphingolipid rafts have been postulated as lipid microdomains that serve as platforms for apical cargo sorting and targeting processes as well as transport-carrier formation (Simons and Ikonen, 1997). However, a major controversy surrounds the inconsistency between the observed nanoscale and short life span of lipid microdomains in biological membranes and their role in signaling or transport-platform formation (Munro, 2003; Sharma et al., 2004). Based on the copatching of cross-linked raft components and its dependence on cholesterol (Harder et al., 1998), it has been proposed that the extent of microdomain clustering may be modulated by protein oligomerization.
When considering the existence of larger and more stable membrane heterogeneities or domains, which consist of distinct lipids and proteins while excluding others, one has to identify how the system compensates for the loss in entropy (Garcia-Saez et al., 2007). The hypothesis that protein–protein and protein–lipid interactions facilitate both the up-scaling and temporal stability of such lipid domains could potentially put this debate to rest (Jensen and Mouritsen, 2004; Hancock, 2006). It is accepted that membrane lipids are more flexible in responding to applied external forces than embedded proteins (Engelman, 2005). Thus, it has been hypothesized that integral membrane proteins perturb the surrounding lipids to adjust the bilayer thickness to that of their transmembrane-spanning domains (Mitra et al., 2004; Kandasamy and Larson, 2006). This perturbation occurs mainly because exposure of hydrophobic moieties to the aqueous environment is energetically unfavorable. Hence, membrane lipids may yield by bending as well as by selective accumulation of lipids with matching tail lengths (Kuzmin et al., 2005). Alternatively, positively mismatched membrane proteins have been shown to respond to mismatch with the surrounding lipids by aggregating, to minimize the mismatched contact zones (Gil et al., 1997; Fernandes et al., 2003), although substantial aggregation requires additional binding inducement in the form of protein–protein interactions (Sperotto and Mouritsen, 1991). In general, liquid ordered domains are thicker than the surrounding liquid-disordered membrane. Moreover, long saturated fatty acyl tails have been shown to be a prerequisite for the association between sphingolipid rafts and glycophosphatidylinositol (GPI)-anchored proteins (Benting et al., 1999; Megha et al., 2007). A proposed offshoot of such protein–lipid mismatching interactions is that integral membrane proteins might be key determinants of membrane thickness. This has been demonstrated to be the case in organelles of the secretory pathway (Mitra et al., 2004). Membranes of secretory organelles increase in thickness in the anterograde direction. The apical epithelial PM is thickest, whereas the endoplasmic reticulum (ER) membrane is thinnest. Furthermore, membrane thickness is key in targeting secretory membrane proteins. For example, localization of Golgi glycosyltransferases is determined by the length of their membrane-crossing helices (Munro, 1998; Sprong et al., 2001).
MAL is a compact hydrophobic 17-kDa protein with four predicted transmembrane helices that localizes to the epithelium's apical membrane domain (Alonso and Weissman, 1987; Cheong et al., 1999; Puertollano and Alonso, 1999). In epithelia, it has been shown to be essential for the apical sorting of several proteins, including GPI-anchored proteins (Puertollano et al., 1999; Martin-Belmonte et al., 2000). MAL also copurifies with detergent-resistant membranes in cold Triton X-100 extraction. The predicted membrane-crossing helices of MAL constitute the MARVEL domain, which is found in ∼20 open reading frames in the human genome (Sanchez-Pulido et al., 2002). MAL2 and Bene are its closest homologues (de Marco et al., 2001; Marazuela et al., 2004a). The MARVEL domain is also found in the tight-junction-localized protein occludin and in the synaptic-membrane-localized synaptophysin and synaptogyrin. Thus, a common feature of proteins containing the MARVEL domain is localization to specialized domains within surface membranes.
In this study, we used spontaneously formed large clusters of MAL tagged with the tandem self-dimerizing red fluorescent protein HcRED. These synthetic ordered arrays of DiHcRED-MAL are formed in part via cross-linking of the upper and lower surface membranes, mediated by the HcRED moiety. We found apical lipid-raft markers to be enriched in the membrane within the clusters compared with the surrounding PM. A phospholipid fluorescent analogue was completely excluded from these structures. Site-directed mutagenesis of DiHcRED-MAL and bimolecular fluorescence complementation (BiFC) assay demonstrated an association between DiHcRED-MAL cluster formation and MAL's intrinsic propensity for self-oligomerization. Modulations of the PM by exogenously added cholesterol or long-chain ceramide were used to test the premise that MAL clustering, as well as its association with raft apical markers, are mediated, at least in part, by positive hydrophobic mismatching between the transmembrane helices of MAL and the hydrophobic tails of membrane lipids.
MATERIALS AND METHODS
Materials and Constructs
CTXB488 and C5-BODIPY-GM1 ganglioside were purchased from Invitrogen (Carlsbad, CA). N-Lignoceroyl-d-erythro-sphingosin and N-lauroyl-d-erythro-sphingosin were purchased from Avanti Polar Lipids (Alabaster, AL). Cholesterol, methyl-β-cyclodextrin (MβCD), filipin complex, and the protein synthesis inhibitor cycloheximide were purchased from Sigma-Aldrich (St. Louis, MO). Human MAL cDNA was subcloned into pECFP-C1 or pEYFP-C1 (Clontech, Palo Alto, CA) and into the tandem dimer far-red HcRED (DiHcRED) molecules in the DiHcRED-C1 vector kindly donated by Jan Ellenberg (European Molecular Biology Laboratory, Heidelberg, Germany), by using XhoI and SacII restriction sites. MAL-FLAG has been described previously (Puertollano and Alonso, 1999). GPI-mCytrin and GPI-mCFP were prepared as described previously (Glebov and Nichols, 2004). Dimyristoyl-farnesyl-green fluorescent protein (GFP) was a kind gift from Yoel Kloog (Tel-Aviv University, Tel-Aviv, Israel).
Cell Culture and Transfections
COS7 and Madin-Darby canine kidney (MDCK) cells were grown at 37°C in a 5% CO2-humidified atmosphere. Cell cultures were maintained in DMEM supplemented with 10% (vol/vol) fetal calf serum (FCS) and penicillin/streptomycin (Biological Industries, Bet-Haemek, Israel). FuGENE6 reagent (Roche Diagnostics, Mannheim, Germany) and Lipofectamine 2000 reagent (Invitrogen) were used for plasmid DNA transfections of subconfluent COS7 and MDCK cells, respectively. To produce stably expressing DiHcRED-MAL clones, transfected MDCK cells were grown in DMEM containing 500 μg/ml G418 (Sigma-Aldrich). Confocal laser-scanning microscopy (CLSM) experiments were carried out from 18 to 24 h after transfection.
Immunofluorescence Microscopy
MDCK cells stably expressing DiHcRED-MAL were grown on transwell filters (Corning, Corning, NY) for 3–4 d, fixed in a 2% (vol/vol) formaldehyde solution, and sequentially incubated for 1 h at 37°C with primary rat R40.76 anti-zona occludens (ZO) 1 monoclonal antibody (mAb) (kindly provided by Yoram Altschuler, The Hebrew University of Jerusalem, Jerusalem, Israel) and Cy3-modified secondary anti-rat antibodies diluted in phosphate-buffered saline (PBS) containing 5% FCS and 0.2% (vol/vol) saponin. Transwell filters were then cut and mounted for CLSM.
Quantification of DiHcRED-MAL Expression in COS7 Cells
An identical number of COS7 cells were plated at 60% confluence on a 35-mm glass-bottomed microwell dish (MatTek, Ashland, MA) and on a 35-mm tissue culture dish (Corning). Cells were transfected with DiHcRED-MAL by using FuGENE6. After 18 h, all cells in the microwell dish were labeled with 1 μg/ml Alexa-488–labeled cholera toxin B subunit (CTXB488) to estimate both cell number and transfection efficiency. Data were calculated by averaging 15 fields under 0.7× zoom by using a 25× numerical aperture (NA) 0.8 objective. The cells grown on the regular 35-mm dish were lysed using 1% (vol/vol) Triton X-100 and 60 mM octyl-glucoside. Western blot analysis was performed on the cell lysate and on a range of known concentrations of purified HcRed protein (Clontech). Membranes were probed with primary polyclonal anti-HcRed and secondary horseradish peroxidase-conjugated anti-rabbit antibodies and developed by enhanced chemiluminescence. Films were scanned and the bands were quantified using ImageJ software (National Institutes of Health, Bethesda, MD). The data were plotted and fitted to an exponential equation using KaleidaGraph software (Synergy Software, Essex-Junction, VT) (R2 = 0.998).
Immunofluorescence Cross-linking Experiments
COS7 cells were grown on glass coverslips and transfected with MAL-FLAG or cotransfected with MAL-FLAG and GPI-anchored mCytrin. After 24 h, cells were washed twice with ice-cold DMEM containing 50 mM HEPES, pH 7.2, and 1% FCS and incubated for 45 min at 4°C in the same buffer with the addition of 10 μg/ml anti-FLAG M2 mAb (Sigma-Aldrich). Secondary Cy3-tagged anti-mouse antibody (5 μg/ml) was added for 30 min at 4°C, washed, and the cells were fixed in 2% (vol/vol) formaldehyde either immediately or after a 10-min incubation at 4°C with 1 μg/ml CTXB488. Coverslips were then mounted for CLSM.
Membrane Labeling with CTXB488 or C5-BODIPY-GM1
COS7 cells at 60% confluence in Lab-Tek chambered glass coverslips (Nalge Nunc International, Naperville, IL) were transfected with DiHcRED-MAL by using FuGENE6 and after 18–24 h, labeled with 1 μg/ml CTXB488 or 5 nM C5-BODIPY-GM1 as a bovine serum albumin (BSA) complex, or with 0.1 μg/ml DiIOc18.
Cholesterol Loading and Filipin Staining
Addition of cholesterol (final concentration, 10 mM) to cells was carried out using cholesterol-saturated MβCD at an ∼6:1 M ratio (Christian et al., 1997) prepared in DMEM containing 10% FCS and 20 mM HEPES, pH 7.4. For cholesterol preloading, the cholesterol-saturated MβCD was added 4–5 h after transfection with DiHcRED-MAL. CLSM was performed 20 h after transfection using a 25 × 0.8 NA oil immersion objective. Filipin staining was performed as described previously (Watari et al., 1999) on fixed cells expressing DiHcRED-MAL by using 5 μg/ml filipin.
Addition of Ceramides and 1,2-Ditetradecanoyl-sn-glycero-3-phosphoethanolamine (Pacific Blue-DMPE)
N-Lignoceroyl-d-erythro-sphingosin or N-lauroyl-d-erythro-sphingosin (1 μmol) in chloroform:methanol (2:1, vol/vol) was dried in an Eppendorf tube. Ethanol (20 μl) was added, followed by sonication in a bath sonicator and addition of 500 μl of BSA at 37°C in PBS. The tubes were sonicated for 5–10 min at 37°C before addition of 200 nmol of dissolved lipid per surface area of 1.8 cm2 in a Lab-Tek chamber.
Live Cell Microscopy
Fluorescence images were obtained using a confocal microscope (LSM model PASCAL or 510 META with an Axiovert 200 microscope; Carl Zeiss MicroImaging, Jena, Germany). Fluorescence emissions resulting from 405-nm excitation for cyan fluorescent protein (CFP) or filipin, 488-nm excitation for GFP or Alexa-488, 488- or 514-nm excitation for yellow fluorescent protein (YFP), and 543-nm excitation for DiHcRED or Cy3 were detected using filter sets supplied by the manufacturer. Cells were kept at 37°C on the microscope stage by using an electronically controlled airstream incubator. Long time-lapse image sequences were captured using a 25× 0.8 NA Plan-Neofluar objective. In addition, the autofocusing function integrated into the “advanced time series” macro set (Carl Zeiss MicroImaging,) was used.
Laurdan Labeling and Image Analysis
Images were obtained with a microscope (DM IRE2; Leica, Wetzlar, Germany), equipped with photon-multiplier tubes and acquisition software (Leica). Laurdan fluorescence was excited at 800 nm with a multiphoton laser system (Verdi/Mira 900; Coherent, Auburn, CA). Laurdan-intensity images were recorded simultaneously, and emissions were in the range of 400–460 and 470–530 nm (Gaus et al., 2003). Microscopy calibrations were performed as described previously (Gaus et al., 2003). For confocal microscopy, a helium-neon laser was used to excite DiHcRED (excitation, 543 nm; emission, 550–620 nm). For fixed cells, a 100× oil objective, NA 1.4, was used; for live cells, a 63× water objective, NA 1.3, was used, and images were recorded at room temperature. The general polarization (GP) values, defined as I(400–460) − I(470–530)/I(400–460) + I(470–530), were calculated as described previously (Gaus et al., 2003). GP values range from −1 to +1, with ordered membrane domains typically being >0.3. GP images were pseudocolored (Photoshop; Adobe Systems, Mountain View, CA), and the mean GP value of the PM and DiHcRED-MAL clusters was determined for n individual images, as described previously (Gaus et al., 2006).
Site-directed Mutagenesis
Mutagenesis was carried out on the DiHcRED-MAL plasmid by using a QuickChange (Stratagene, La Jolla, CA) mutagenesis kit. The following forward primer sequences were used: 5′-CCTGGGTCACCTTGGGAGCAGCCTACCACTG-3′ (D88G), 5′-CTTCACCACCTTGCCCGGCTTGCTCTTCATCTTTG-3′ (D24G), 5′-CGTGTCTGTGTTCTCCTTCGTGGCCACC-3′ (C62S), 5′-CACCATCACGATGCAACCCGGCTTCACCTACAGGC-3′ (D115P), 5′-CTACAGGCACTACCATATCAACATTGCTGC-3′ (E124I), 5′-CTTCACCACCTTGCCCGACTTGTTCATCTTTGAGTTTATCTTCG-3′ (Δ26L), 5′-CCTTGATCATCCTGTACATTGGAGCCCACGGTGGAG-3′ (Δ74I), 5′-GCCTACCACTGCACCGCTCTCTTTTACCTCAGC-3′ (Δ96A), 5′-GAAAACATTGCTGCCGTGTTCTCCTACATAGCC-3′ (Δ129V); 5′-GCTTCTCGGTCGCTACCACCTTGCC-3′ (F19A); 5′-CTGCCCAGTGGCGCTTCGGTCGCTAC-3′ (F16A/F19A), 5′-GGTCCAGGGCGCTGTGATGTTCGTGTCTGTG-3′ (W54A), and 5′-CAGGGCGCTGTGATGGCTGTGTCTGTGTTCTGC-3′ (W54A/F57A).
RESULTS
In this study, we put forward the idea that MAL is a central polypeptide constituent in the formation and maintenance of the lipid-raft–based sorting platform (Hancock, 2006). This hypothesis foretells two fundamental properties for MAL: the first property is the capacity to oligomerize via protein–protein interactions to nucleate and form the putative supramolecular membrane scaffold, or assembly, of functional membrane domains; and the second property is the ability to interact with membrane lipids to facilitate their lateral segregation (Sperotto and Mouritsen, 1991).
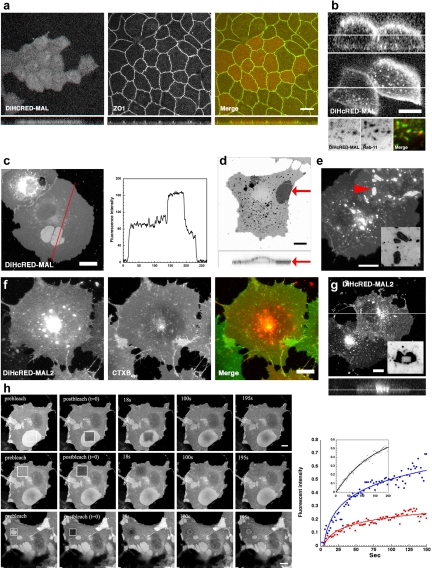
MAL was studied by following the expression of fluorescent protein (FP)-tagged chimeras in both polarized MDCK epithelial cells and nonpolarized COS7 cells. The latter cells do not express endogenous MAL. The tandem dimer of the far-red FP HcRED (DiHcRED) (Wilmann et al., 2005), mCFP, or mYFP was attached to the cytosolic N terminus of human MAL, its homologue MAL2 (de Marco et al., 2002), as well as the MARVEL domain containing protein occludin. In transwell filters, polarized MDCK cells stably expressed fluorescently tagged MAL (Figure 1, a and b) and MAL2 (data not shown) that efficiently arrived at the apical PM. In addition, three-dimensional analysis of polarized MDCK cells expressing DiHcRED-MAL demonstrated additional localization to intracellular organelles (Figure 1b). Colocalization with Rab11-YFP (Figure 1b, bottom) identified these membranes as an endocytic recycling compartment (Marazuela et al., 2004b).
Figure 1.
Expression, localization, and aggregate formation of DiHcRED-MAL and DiHcRED-MAL2 in COS7 cells. (a) Three-dimensional immunofluorescence reconstruction analysis of confocal z-sections of polarized MDCK cells. A stable clone expressing DiHcRED-MAL (left and red in colored image on the right) was fixed and labeled as described in Materials and Methods, 3 d after plating on transwell filters. Cells were labeled with primary anti-ZO1 rat mAb and secondary Cy3-labeled anti-rat antibody (center and green in colored image on the right); bar, 10 μm. (b) Fluorescence x-z (top) and x-y (bottom) sections of polarized MDCK cells. Images of a stable clone expressing DiHcRED-MAL were imaged as described in Materials and Methods, 3 d after plating on a transwell filter; bar, 10 μm. Bottom panel are images of a living cell coexpressing DiHcRED-MAL (red) and Rab11-YFP (green) (c) Transient coexpression of DiHcRED-MAL in COS7 cells results in the formation of large clusters in the PM. Red line marks the line plot of fluorescence intensity on the right-hand side; bar, 10 μm. (d) Inverted three-dimensional reconstruction of a COS 7 cell expressing DiHcRED-MAL (xy top and xz bottom). Top red arrow indicates the horizontal position of the 90° side view at the bottom; bar, 10 μm. (e) Transient coexpression of DiHcRED-occludin in COS7 cells results in the formation of clusters in the PM; bar, 10 μm. Inset is an inverted enlarged image of the area marked by red arrowhead. (f) Overexpression of DiHcRED-MAL2 (red) in surface-labeled COS7 cells. Cells expressing DiHcRED-MAL2 were labeled with 1 μg/ml Alexa488-modified cholera toxin B subunit (CTXB488, green) as described in Materials and Methods; bar, 10 μm. (g) An xy (top) and xz (bottom) view of three-dimensional reconstruction analysis of confocal z-sections of COS7 cells expressing DiHcRED-MAL2; bar, 10 μm. Inset is an inverted image of a cell expressing DiHcRED-MAL2, 2 min after a rectangle was photobleached in the midst of an intracellular aggregate. (h) Fluorescence recovery after photobleaching analysis of OC dynamics. A square was photobleached within an OC (top), over the PM (middle) or over an entire OC (bottom) in COS7 cells expressing DiHcRED-MAL; bars, 10 μm (also see Supplemental Movie 1). Graph on the right shows analysis of fluorescence intensities within the photobleached regions of interest over the PM (blue), within an OC (red) or over an entire OC (inset).
The steady-state distribution of DiHcRED-MAL in COS7 cells included two distinct populations within the PM (Figure 1c). A significant fraction of the PM-localized molecules were observed in distinct flat, round-edged, and bright homogeneous fluorescent clusters. These seemingly aggregate-like structures of DiHcRED-MAL localized to and were embedded within the PM. The fluorescence intensity of the clustered DiHcRED-MAL was approximately twice that of the surrounding PM. These data were also supported by a three-dimensional reconstruction of confocal z-sections of a DiHcRED-MAL-expressing cell (Figure 1d). Such PM clusters were also observed with DiHcRED-occludin (Figure 1e), although they were smaller, less abundant, and localized to areas of PM overlap between cells. PM localization was also observed in the transient expression of fluorescently tagged DiHcRED-MAL2 in COS7 cells 18–20 h after transfection (Figure 1f). However, in this case, the overexpressed protein exhibited a strong propensity to aggregate intracellularly. Three-dimensional reconstructed digital images of DiHcRED-MAL2–expressing cells demonstrated that the large fluorescent perinuclear aggregates were indeed intracellular (Figure 1g). A photobleached rectangle in the middle of one of these aggregates did not recover after 2 min (Figure 1g, inset).
Photobleaching inside a DiHcRED-MAL cluster (Figure 1h) or of an entire cluster resulted in slow but nevertheless significant recovery compared with that of the PM, indicating that rather than aggregates, these integral membrane protein structures are dynamic ordered assemblies that actively exchange with the rest of the surrounding membrane (also see Supplemental Movie 1). Thus, based on these observations, we named these PM-localized intramembrane protein assemblies ordered clusters (OCs), to distinguish them from regular aggregates. As demonstrated by the plot in Figure 1h, recovery of DiHcRED-MAL within the OCs was much slower than that in the surrounding PM. In addition, DiHcRED-MAL within the OCs had a twofold larger immobile fraction. The dynamic nature of the OCs was demonstrated by time-lapse analysis of COS7 cells incubated in the presence of the protein-synthesis inhibitor cycloheximide (Supplemental Movie 2). This experiment showed the time-dependent changes in OC shape and size up to complete dissociation resulting from net turnover in the presence of the protein synthesis inhibitor. Although protein–aggregate formation is frequently a direct result of overexpressing a foreign protein, we focused on the DiHcRED-MAL OCs based on the assumption that their formation is associated with, and therefore might shed light on, the mechanism of MAL-induced membrane domains. We estimated the average number of DiHcRED-MAL molecules per cell to be 1.1·× 108. This number was obtained by quantifying a standard curve of different amounts of purified DiHcRED and DiHcRED-MAL from cell lysate with a calculated number of cells after Western analysis (data not shown). Quantitative analysis of the total DiHcRED-MAL fluorescence partitioning between PM and OCs, measured using digital images of 10 cells expressing DiHcRED-MAL, demonstrated that the OCs contain on average 14.5 ± 7.6% of the total DiHcRED-MAL.
Characterization of DiHcRED-MAL Clusters
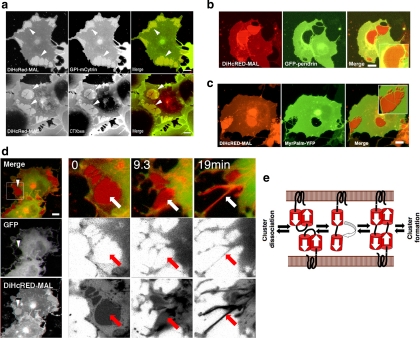
To elucidate the membrane composition within the DiHcRED-MAL clusters, we analyzed the relative distribution of two bona fide lipid-raft and apical markers within and outside the OCs. COS7 cells were cotransfected with DiHcRED-MAL and GPI-mCytrin (Glebov and Nichols, 2004). Figure 2a (top) shows that GPI-mCytrin is enriched within the OCs relative to the surrounding PM. Considering that the net residual lipid surface area within the OC is to a great extent smaller than the surrounding membrane, GPI-mCytrin concentration is much higher than depicted by the increase in fluorescence intensity. Similarly, DiHcRED-MAL OCs were enriched more than threefold with the raft marker GM1 ganglioside, based on binding of CTXB488 (Nichols, 2003) (Figure 2a, bottom).
Figure 2.
Characterization of DiHcRED-MAL OCs. (a) Confocal images of living COS7 cells transiently cotransfected with DiHcRED-MAL (left-hand images and red) and GPI-mCytrin (center image and green, top), or transfected with DiHcRED-MAL and labeled with 1 μg/ml CTXB488 (center image and green, bottom) added 10 min before confocal image acquisition as described in Materials and Methods. Arrowheads indicate OCs; bars, 10 μm. (b) OCs are inaccessible to GFP-tagged integral membrane proteins. Confocal images of COS7 cells coexpressing DiHcRED-MAL (red) and GFP-Pendrin (green) (also see Supplemental Figure 1). Inset is enlarged area containing an OC; bars, 10 μm. (c) OCs are inaccessible to membrane-associated cytosolic proteins. Confocal images of COS7 cells coexpressing DiHcRED-MAL (red) and myristoyl-palmitoylated anchored YFP (YFP-MyrPalm, green). Inset is enlarged area containing an OC; bar, 10 μm. (d) OCs are made up of cross-linked upper and lower PM. Time-lapse images of living COS7 cells coexpressing DiHcRED-MAL (red) and a cytosolic GFP (green) were captured using a confocal microscope after addition of 40 nM latrunculin B. White arrowheads and white and red arrows indicate clusters that dissociated during the experiment (also see Supplemental Move 3); bar, 10 μm. (e) Scheme of possible events leading to upper and lower PM cross-linking and OC formation.
We then coexpressed DiHcRED-MAL in COS7 cells with various fluorescently tagged integral or membrane-associated proteins. Figure 2, b and c (also see Supplemental Figure S1) demonstrates that the OCs are tight protein assemblies, because the membrane within the OCs seems to be inaccessible to most transmembrane- or cytosolic-membrane-associated proteins. These data can be interpreted as an indication of high DiHcRED-MAL density in the OCs. The lack of any detectable cytosolic membrane-associated markers beneath the OCs is demonstrated in Figure 2d (left, white arrowhead) for cells coexpressing DiHcRED-MAL and a cytosolic soluble GFP. These data led us to assume that these clusters consist of cross-linked DiHcRED-MAL molecules from opposing basal and upper surface membranes (Figure 1d). To test this hypothesis, we subjected the cells coexpressing DiHcRED-MAL and cytosolic soluble GFP to actin depolymerization by latrunculin B. The rationale was that if the OCs are made up of adjacent cross-linked surface membranes, then actin depolymerization-induced cell widening will cause their dissociation. Latrunculin B did induce dissociation of the OCs and their transformation into tubular membranes (Figure 2d; also see Supplemental Movie 3). These data support the premise that the DiHcRED-MAL OCs are formed by cross-linking of the upper and lower surface membranes by the tandem HcRED dimers, as reported for the interaction of ER-retained anti-parallel GFP-tagged proteins (Snapp et al., 2003). Cross-linking of the upper and lower surface membranes may potentially be initiated by the formation of intermolecular dimers made up of two closed or open conformation tandem dimers from opposing surface membranes, as demonstrated in the scheme in Figure 2e. Such a structure would prevent access to a fluid phase cytosolic marker. That OCs are seen only in flat cells in which the upper and lower surface membranes are in proximity, as in COS7 or U2OS (data not shown), is consistent with the data in Figure 2d.
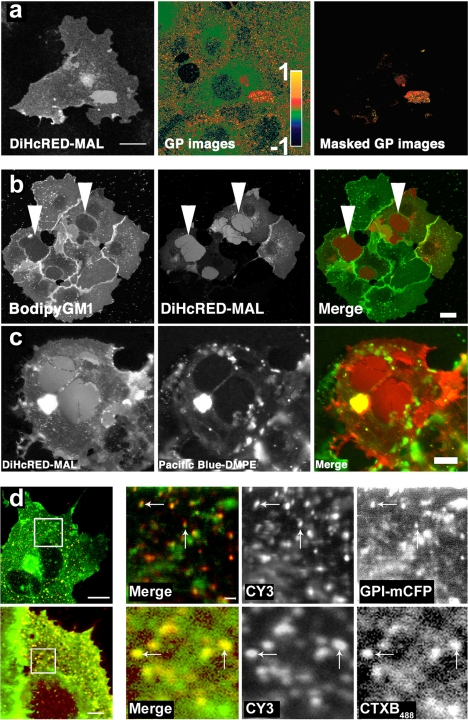
To further study the properties of the membrane within the OCs, we carried out multiphoton analysis of cells expressing DiHcRED-MAL colabeled with the fluorescent probe laurdan. The laurdan fluorescent lipid reports on the degree of membrane ordering via changes in its emission spectra (Gaus et al., 2006). We expected an accumulation of raft markers to increase the degree of membrane ordering within the OCs. Figure 3a demonstrates that the membrane within the OCs is in a more highly ordered state than the surrounding PM (GP values for clusters: 0.413 ± 0.033; GP at PM: 0.316 ± 0.059; n = 27; p < 0.001). These data demonstrate that the membrane within the cluster is both thicker and more ordered relative to the average value of the surrounding membrane. It also lends support to the premise that the intramembrane MAL moiety is involved in OC formation. The concentration of raft lipids and the increased laurdan GP values both indicate that the membrane within the OC is thicker than the surrounding PM. To confirm this, we analyzed the distribution of two exogenously added fluorescent lipid analogues in cells expressing DiHcRED-MAL. Figure 3b shows that the fluorescent short-chain analogue BODIPY-GM1 (Pagano et al., 2000) is not concentrated in the OCs, as were the endogenous GM1 sphingolipids. We propose that the short and bulky C5-BODIPY fatty acyl moiety is unable to interact with MAL like the long saturated fatty acyl tails of endogenous glycosphingolipids do. This observation identified the intramembrane fatty acyl moiety of GM1, rather than its carbohydrate head group, as a key determinant of its MAL-mediated segregation. The fluorescent phospholipid analogue Pacific Blue-DMPE was also excluded from DiHcRED-MAL OCs (Figure 3c). The fluorophore in this phospholipid analogue is attached to the phosphoethanolamine head group of the DMPE phospholipids. This finding suggests that the OCs select for lipids with fatty acyl chains longer than 14 carbons.
Figure 3.
Characterization of the lipid membrane within OCs and the concentration of GM1 and GPI-FP by antibody cross-linked MAL. (a) The membrane within the OCs is more ordered (thicker) than the surrounding membrane. Laurdan-labeled DiHcRED-MAL–expressing COS7 cells were imaged, and intensity values were converted to GP values presented as pseudocolors with ordered (1) to fluid (−1) domains ranging from red to green, respectively, as described in Materials and Methods. The masked GP images show the GP values of the highly ordered DiHcRED-MAL clusters; bar, 20 μm. (b) OCs exclude short-chain C5-BODIPY-GM1. Confocal images of living COS7 cells transiently transfected with DiHcRED-MAL (center image and red) and labeled with C5-BODIPY-GM1 complexed to BSA (left image and green) as described in Materials and Methods; bar, 10 μm. (c) OCs exclude Pacific Blue-DMPE. Confocal images of living COS7 cells transiently transfected with DiHcRED-MAL (left image and red) and labeled with Pacific Blue-modified DMPE complexed to BSA as described in Materials and Methods (center image and green); bar, 10 μm. (d) Localization of GPI-mCFP and CTXB488 to antibody-mediated cross-linked FLAG-tagged MAL (MAL-FLAG). COS7 cells plated on glass coverslips were transiently cotransfected with MAL-FLAG (CY3 and red in merged image) and GPI-mCFP (green in merged image) or transfected with MAL-FLAG incubated with primary anti-FLAG and secondary Cy3-tagged anti-mouse antibodies, fixed with 2% formaldehyde, treated with antibodies as described in Materials and Methods, and further labeled with 1 μg/ml CTXB488 after fixation. White boxes on the left-hand side were scanned with a 16× zoom to visualize cross-linked MAL-FLAG with the highest possible resolution. Arrows indicate MAL-FLAG clusters and cross-linked MAL-FLAG; bars, 10 μm.
Next, we sought to establish that the MAL moiety is exclusively responsible and sufficient for mediation of the lateral concentration of GPI-FP and GM1 in the absence of DiHcRED tag. To this end, we used antibody cross-linking of extracellularly FLAG-tagged MAL (Puertollano and Alonso, 1999) (MAL-FLAG) with primary anti-FLAG and secondary Cy3-modified anti-mouse antibodies (Figure 3d) (Lachmanovich et al., 2003). We found that similar to DiHcRED-MAL OCs, cross-linking of MAL-FLAG mediated by divalent antibodies results in a local increase in the concentrations of GPI-FP and GM1.
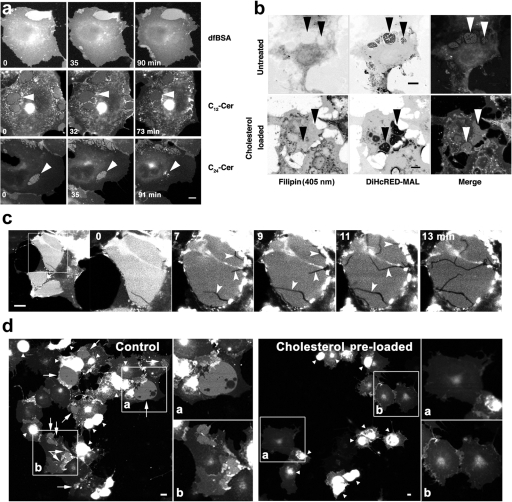
To expand upon our finding that interaction of the MAL transmembrane domains (TMDs) with the tails of membrane lipids results in the concentration of apical lipids, we examined whether changing the membrane composition might alter the dynamic steady state of the OCs. We added exogenous cholesterol or ceramides to modify the membrane's composition. It is important to note that, unlike a well-defined liposome-based experimental system, the use of a biological membrane in an intact cell limits our ability to provide an unequivocal description of how the externally added lipid exerts its effect. However, it is reasonable to assume that one of the effects of increasing the level of cholesterol, as well as of long-chain ceramides, will be a shift toward a liquid-ordered state and an increase in average membrane thickness. The effects of exogenously added ceramides are summarized in Figure 4a and in Supplemental Movie 4. COS7 cells expressing DiHcRED-MAL were imaged after the addition of a solution of concentrated defatted BSA alone (Figure 4a) or as a complex with the short- or long-chain ceramides N-lauroyl-d-erythro-sphingosin (C12-Cer) or N-lignoceroyl-d-erythro-sphingosin (C24-Cer), respectively, to determine the effects on OCs. C24-Cer clearly shifted the steady state of DiHcRED-MAL by inducing the dissociation of OCs after ∼90 min. The extended time required for C24-Cer to exert its effect was presumably a result of its slow and inefficient exchange between the BSA carrier protein and the COS7 membrane. These data support the hypothesis that OC formation, as well as lateral concentration of apical lipids, are at least partially mediated by positive hydrophobic mismatching between the relatively long membrane-crossing helices of MAL and the lipids of the COS7 cell membrane.
Figure 4.
Effect of exogenously added ceramides or cholesterol on OCs. (a) Addition of ceramide-BSA complexes to living COS7 cells expressing DiHcRED-MAL. Confocal images of living COS7 cells incubated for 1–2 h in the presence of defatted (df) BSA (top), C12-Cer-BSA complex (middle), or C24-Cer (bottom). Arrowheads point to OCs (also see Supplemental Movie 6); bar, 10 μm. (b) Cholesterol levels before (top) and after (bottom) cholesterol addition. COS7 cells expressing DiHcRED-MAL (center inverted images and red) and untreated (top) or incubated for 1 h in the presence of 10 mM cholesterol-saturated MβCD (bottom) were fixed and labeled with filipin (left inverted images and green) as described in Materials and Methods. The filipin channels for top and bottom panels were scanned with 26 and 5% of 405-nm laser power, respectively. Arrowheads point to OCs; bars, 10 μm. (c) Cholesterol-induced change in intensity (from 0 to 13 min) and fractures indicated by arrowheads in DiHcRED-MAL clusters. Living cells were imaged as described in Materials and Methods 20 h after transfection with DiHcRED-MAL. Cholesterol-saturated MβCD was added (final concentration, 10 mM), and images were captured at 30-s intervals for 90 min (see also Movie 5 in supplementary material). (d) Cholesterol preloading blocks DiHcRED-MAL cluster formation. Images of living cells 20 h after addition of cholesterol-saturated MβCD (final concentration, 10 mM) and transfection with DiHcRED-MAL. White arrows and arrowheads point to clusters and other aggregates, respectively. Insets (a and b) are magnified twofold on the right-hand side of each image; bars, 10 μm.
Based on the premise that addition of cholesterol shifts the membrane toward a more ordered state, we analyzed the effect of exogenously added cholesterol on DiHcRED-MAL OC formation as well as on pre-existing OCs. Using the cholesterol-sequestering fluorescent agent filipin to label COS7 cells expressing DiHcRED-MAL, the OCs seemed to be deficient in cholesterol compared with the surrounding PM (Figure 4b, bottom). Incubation with cholesterol-saturated MβCD increased total PM levels of cholesterol, as found in the OCs as well. Addition of cholesterol to pre-existing OCs of living cells resulted in the formation of rapidly growing and branching fractures within the clusters, a sudden decrease in their fluorescence intensity, and changes in cluster shape that tended toward reduced surface area and occasionally resulted in complete dissociation (Figure 4c and Supplemental Movie 5). Furthermore, preloading COS7 cells with cholesterol before transfection with DiHcRED-MAL expression vector completely prevented OC formation (Figure 4d). The addition of cholesterol to an intact biological membrane has multiple and complex effects that may depend, for example, on the overall quantity added. Nonetheless, our results with cholesterol are consistent with the prediction that increased levels of cholesterol would reduce the positive hydrophobic mismatching interactions between MAL TMDs and the PM lipids, presumably by increasing the membrane's overall thickness via a shift toward the liquid-ordered phase. Complementary to the effect of changing membrane lipid composition on OCs, we proceeded to perturbation by mutagenesis of the MAL moiety.
Mutagenesis of DiHcRED-MAL and BiFC Analysis
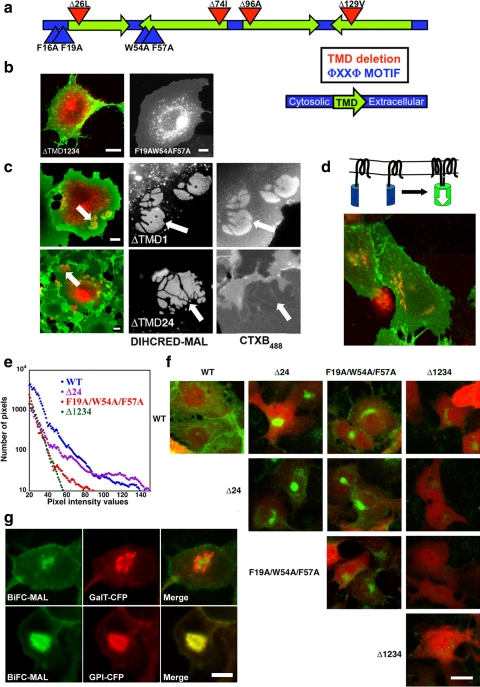
A possible explanation for the function of MAL is that both lateral segregation of lipids and protein aggregation (OC formation) are facilitated at least in part by positive hydrophobic mismatching between the length of the MAL's TMDs and the average width of the hydrophobic fatty acyl core of the membrane (Fernandes et al., 2003; Jensen and Mouritsen, 2004; Engelman, 2005; Hancock, 2006). To further pursue this hypothesis, we applied site-directed mutagenesis to the DiHcRED-MAL chimera. Mutagenesis and deletions were carried out according to the following strategies: 1) deletion of a single amino acid in each TMD or combinations of single amino acid deletions in two, three, or all four TMDs; and 2) mutagenesis of bulky amino acids within the ϕXXϕ intramembrane protein–protein interaction motif (Sal-Man et al., 2007). The locations of the key mutations and deletions are shown in the scheme in Figure 5a. All of the mutations were expressed in COS7 cells and were tested for arrival at the PM, OC formation, and concentration of CTXB488 within the OCs. Arrival of a mutant protein at the PM signified proper folding followed by proper secretory trafficking and targeting. Effects on OC formation and concentration of CTXB488 were interpreted as perturbations of either protein–protein or protein–lipid interactions. Most of the TMD-deletion mutants exhibited aberrant folding and trafficking and thus did not efficiently arrive at the PM, as exemplified in Figure 5b (left) by the intracellular localization of ΔTMD1234 (surface-labeled with CTXB488). Nevertheless, two mutations did produce interesting phenotypes: two single amino acid deletions within the second and fourth TMDs (ΔTMD24) and the ϕXXϕ to AXXA mutation. Complete elimination of both ϕXXϕ motifs at positions 16–19 and 54–57 (F16A, F19A, W54A, F57A, Figure 5a) in DiHcRED-MAL caused severe folding problems based on its cytosolic localization that indicates failure to be cotranslationally inserted into the ER membrane (data not shown). Elimination of a single ϕXXϕ motif at positions 16–19 or 54–57 had no apparent effect on arrival at the PM or OC formation. The mutant F19A/W54A/F57A efficiently arrived at the PM, but OC formation was abolished (Figure 5b, right). Figure 5c demonstrates the phenotype of ΔTMD24. This mutant efficiently arrived at the PM and formed OCs, as did ΔTMD1 (Figure 5c, top). However, ΔTMD24 essentially excluded GM1 from the OCs, in contrast to other OC-forming mutants of DiHcRED-MAL or the wild type. Thus, in the ΔTMD24 mutant, cluster formation was uncoupled from the lateral concentration of lipids (Figure 5c, bottom).
Figure 5.
Analysis of MAL self-oligomerization by using mutagenesis and BiFC analysis. (a) Scheme showing the relative locations of the various mutations and deletions in MAL. (b) Expression of DiHcRED-MAL mutations. Confocal images of living COS7 cells transiently transfected with DiHcRED-MALΔTMD1234 (left, red) and labeled with 1 μg/ml CTXB488 (left, green) or DiHcRED-MALF19A/W54A/F57A (right); bars, 10 μm. (c) OCs of DiHcRED-MALΔTMD24 exclude rather than concentrate GM1. Confocal images of living COS7 cells transiently transfected with DiHcRED-MALΔTMD1 (top, red and center image) or with DiHcRED-MALΔTMD24 (bottom, red and center image) labeled with 1 μg/ml CTXB488 (green and right images). Arrows point to CTXB488 segregating or excluding OCs; bars, 10 μm. (d) Scheme demonstrating the principle of BiFC for YN- and YC-tagged MAL. Image under the scheme: BiFC analysis of self-oligomerization of MAL. MDCK cells were transfected with the YN- and YC-tagged wild-type MAL and GalT-CFP (red). (e) Quantitative BiFC analysis of self-oligomerization of MAL. A histogram of the distribution of pixel intensity in MDCK cells cotransfected with cytosolic cerulean as an expression-level indicator and either the YN- and YC-tagged MAL wild type (blue; average CFP fluorescence, 27 ± 17), ΔTMD24 (violet; average CFP fluorescence, 21 ± 17), F19A/W54A/F57A (red; average CFP fluorescence, 21 ± 16), or ΔTMD1234 (green; average CFP fluorescence, 40 ± 31). All images in the panel were captured with identical parameters of zoom, laser power, laser attenuation, offset, and photomultiplier gain. (f) BiFC analysis of self-oligomerization of MAL. MDCK cells were transfected with combinations of the YN- and YC-tagged MAL wild type (WT), ΔTMD24, F19A/W54A/F57A or ΔTMD1234 and cytosolic cerulean (red) as an expression-level indicator. All images in the panel were captured with identical parameters of zoom, laser power, laser attenuation, offset and photomultiplier gain; bar, 10 μm. (g) Colocalization of YN/YC-MAL dimers (green) with GalT-CFP (red, top) or GPI-CFP (red, bottom) in MDCK cells; bar, 10 μm.
Overall, these data demonstrate that mutations in the ϕXXϕ motifs as well as shortening of the TMDs of the DiHcRED-MAL chimera perturb OC formation and protein–lipid interactions, respectively. This lends support to the premise that OC formation, as well as lateral concentration of lipids, are mediated by interactions within the hydrophobic core of the membrane between MAL TMDs and the lipid fatty acyl tails. Moreover, the exclusive effect of shortening two TMDs on lipid concentration but not on OC formation suggests that OC formation is at least in part associated with MAL's intrinsic propensity for self-oligomerization, mediated by the recently characterized ϕXXϕ motif (Sal-Man et al., 2007). Consequently, we sought to substantiate MAL's propensity for self-oligomerization in epithelial cell membranes as well as independently of the DiHcRED moiety-mediated OCs. We therefore used BiFC analysis (Kerppola, 2006), a method aimed at identifying interacting proteins by their ability to generate a functional fluorescent protein from two complementary halves tagged to each one (Figure 5d, scheme). This analysis is carried out in intact cells and therefore, apart from demonstrating the interaction between two proteins, it provides information on the interacting proteins' intracellular localization. Figure 5d demonstrates BiFC fluorescence at the PM of MAL tagged with the C (YC) or N (YN) terminus halves of enhanced (E)YFP coexpressed with GalT-CFP in MDCK cells.
Wild-type MAL and three other mutant MAL proteins were tagged with the C (YC) or N (YN) terminus halves of EYFP. The three mutations YC/YN-MALF19A/W54A/F57A, YC/YN-MALΔTMD24, and YC/YN-MALΔTMD1234 served to separately estimate the contributions of MAL self-oligomerization, protein-lipid interactions and the irreversible spontaneous association of the YC and YN moieties, respectively. All combinations of YN- and YC-tagged pairs were coexpressed in MDCK cells together with soluble CFP marker as a reference for expression level. Figure 5e compares the histograms of the BiFC fluorescence from pairs of the wild-type and the three aforementioned mutations. The fluorescence intensity of the wild-type MAL YC/YN pair is the highest, whereas that of the YC/YN-MALΔTMD1234 pair is the weakest. As shown in Figure 5f, significantly bright YFP fluorescence at the PM resulting from fluorescence complementation was detected in MDCK cells expressing the YC- and YN-tagged wild-type MAL. A reduced amount of fluorescence was associated with all of the combinations that included YN/YC-MALΔTMD24 or YN/YC-MALF19A/W54A/F57A as one member of the pair. For these combinations, the yellow fluorescence localized to intracellular membranes rather than to the PM (Figure 5f). Higher levels of expression of the wild-type BiFC pair resulted in intense PM and Golgi labeling. Moreover, YC/YN-MAL dimers frequently aggregated within the Golgi apparatus, where the BiFC pairs colocalized with GPI-FP and were segregated from the Golgi marker GalT-CFP (Figure 5g). These data strongly support earlier findings that MAL can self-oligomerize and that its oligomers attract GPI-anchored proteins. Moreover, the oligomerization occurs while MAL is traversing the secretory pathway.
The Intracellular Distribution of DiHcRED-MAL in MDCK Cells
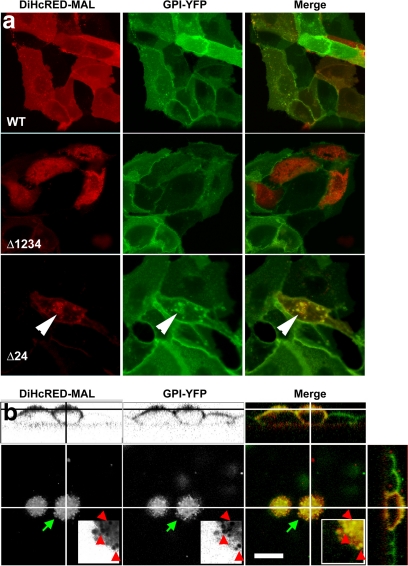
The work presented in this study was carried out with MAL tagged with several types of fluorescent proteins, mainly DiHcRED-MAL. Although we were not able to directly assay its function, overexpression of the DiHcRED-MALΔTMD24 (Figure 6a, bottom) or DiHcRED-MALF19A/W54A/F57A (data not shown) in stable GPI-YFP–expressing MDCK cells resulted in partial retention of GPI-YFP in intracellular compartments. The ER-retained DiHcRED-MALΔTMD1234 had no effect on GPI-YFP distribution, probably because it did not interfere with the endogenous MAL. Wild-type DiHcRED-MAL colocalized with the surface-expressed GPI-YFP (Figure 6a, top) or with the apically targeted GPI-YFP in filter-grown polarized MDCK cells (Figure 6b). Together, these data indicate that even in a background of endogenous MAL, GPI-YFP trafficking is affected by the transiently overexpressed DiHcRED-tagged mutants.
Figure 6.
Intracellular localization of GPI-YFP in MDCK cells coexpressing wild-type or mutant DiHcRED-MAL. (a) Colocalization of WT (top), ΔTMD1234 (middle), or ΔTMD24 (bottom) DiHcRED-MAL and stably expressed GPI-YFP in MDCK cells. Arrowheads point to intracellular pools of GPI-YFP colocalized with ΔTMD24. (b) Colocalization of GPI-YFP and DiHcRED-MAL in polarized MDCK cells. Three-dimensional reconstruction of a confocal z-section of polarized MDCK cells stably expressing GPI-YFP and DiHcRED-MAL grown on transwell filters. Lines crossing images show positions of 180° and 90° sections. Green arrows show area enlarged threefold in the inset. Red arrowheads show coincident concentrations of DiHcRED-MAL and GPI-YFP; bar, 10 μm.
DISCUSSION
In this study, we analyzed clusters of DiHcRED-MAL formed by its heterologous overexpression in flat cells. These highly organized protein assemblies were generated by cross-linking of the upper and lower surface membranes, mediated by the tandem red fluorescent DiHcRED moiety. The details of DiHcRED-MAL OC formation are not completely understood. However, it is reminiscent of the previously reported organized smooth ER, known to consist of cubic-phase membranes generated by weakly dimerizing antiparallel FP-tagged ER proteins (Snapp et al., 2003; Almsherqi et al., 2006). The MAL tetra-spanning moiety is important in OC formation. The DiHcRED-tagged MAL homologue—MAL2—chimera with shorter predicted TMDs did not generate OCs. In contrast, OCs were observed for the MARVEL-domain–containing, DiHcRED-tagged occludin, which contains 23–25 amino acids in each predicted TMD (data not shown). In this study, DiHcRED-MAL OCs were used for two purposes. The first was based on OC consisting of a highly dense intramembrane protein structure and thus the residual lipids within it represent MAL's proximal PM lipid environment. The second was based on OCs being part of a dynamic steady state of DiHcRED-MAL in the PM of COS7 cells. Perturbations in this steady state were generated by both mutagenesis of DiHcRED-MAL and modulation of the membranes by using exogenously added lipids.
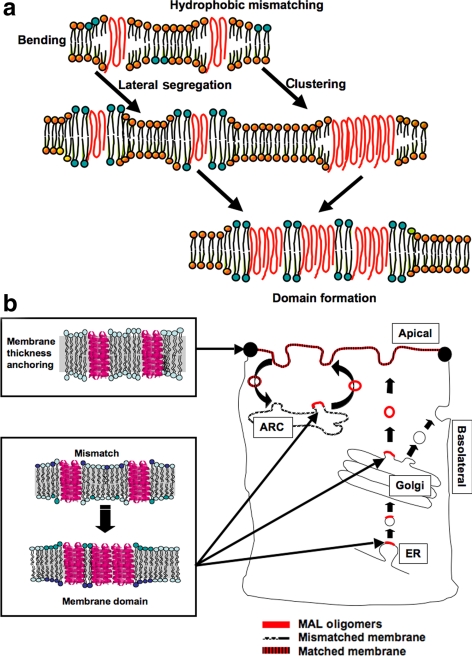
The current amended version of the Singer–Nicolson model attributes a major role to proteins in effecting membrane organization (Engelman, 2005). Proteins are involved in fusion, fission, and curvature induction, and in the current view, they are also involved in maintaining or using the nonhomogeneous nature of biological membranes. Membranes are thus composed of dynamic segregated lipid domains that vary in thickness and composition and have distinct biological functions. The hypothesis that protein–lipid interactions are involved in the up-scaling and temporal stabilization of the highly dynamic heterogeneities known as sphingolipid rafts originated from the copatching of raft components (Harder and Simons, 1997) is now largely accepted (Hancock, 2006; Lingwood et al., 2008) (Figure 7a). However, examples of actual candidate interacting proteins have not yet been suggested. The data in this study provide evidence of a potential role for MAL in the formation and stabilization of membrane domains that may potentially serve as apical sorting platforms, and as a possible membrane-thickness–stabilizing entity (Figure 7b).
Figure 7.
Facilitation of protein clustering and domain formation by positive hydrophobic mismatch between MAL and membrane lipids. (a) Scheme demonstrating the possible consequence of positive hydrophobic mismatch between MAL TMDs and the width of the hydrophobic core of the membrane bilayer. (b) Spontaneous MAL oligomerization (red bars) is facilitated during transport between thin-membrane organelles. In the apical membrane, where alleviated mismatch promotes monomer formation, MAL participates in determining membrane thickness. ARC, apical recycling compartment.
Evidence of MAL Self-Oligomerization
There are several lines of evidence supporting MAL's propensity for homo-oligomerization. The first is the actual formation of dynamic OCs by overexpression of the DiHcRED-MAL chimera in flat (COS7 or U2OS) cells. Site-directed mutagenesis revealed that aromatic amino acids within the two ϕXXϕ motifs are essential for OC formation. Moreover, this same mutant exhibited impaired dimerization in BiFC analysis compared with the wild type. No effect on OC formation was observed for the ΔTMD24 mutant with two shortened TMDs. However, the ΔTMD24 OCs did not attract GM1, indicating that OC formation can be uncoupled from the ability to laterally concentrate lipids. Interpretation of the BiFC analysis is limited by the stability of the product dimer, which does not represent the probable dynamic nature of MAL oligomers. In intracellular membranes, YN-MAL and YC-MAL dimers frequently formed aggregates, which were usually associated with higher expression levels. Altogether, the BiFC data confirm that MAL forms oligomers and that their formation occurs during secretory transport. The degree of MAL oligomerization depends on numerous factors that affect local membrane concentration, such as transport rate and expression level, as well as membrane content and average thickness, which establish the degree of hydrophobic mismatch.
Lateral Segregation of Lipids
MAL is mostly embedded in the PM and according to most of the TMD-predicting algorithms, its TMDs are relatively long (22-23 amino acids). Lipid inclusion in rafts is strongly associated with, and potentially dependent on the length of the fatty acyl tails. The possible role of positive hydrophobic mismatching in the MAL-lipid interaction is supported first by the exclusion of short-chain BODIPY-GM1 and Pacific Blue-DMPE from the OCs; and second, in addition to the laurdan spectral analysis, by the inability of ΔTMD24 mutant OCs to concentrate GM1. An alternative explanation for the latter results might be that each of the two deletions induced a 100° turn of two of the membrane-crossing helices, which could potentially change the protein–lipid interphase that favors lipids with long saturated fatty acyl tails. MAL's ability to attract GPI-FPs was demonstrated here in various systems that were independent of the OCs. At a high level of expression in these cells, BiFC dimers had already formed in the Golgi apparatus and frequently aggregated there as well.
Mechanism of MAL and the MARVEL Domain
Approximated calculations based on previously published values for line-tension energies of exposed hydrophobic amino acids of TMDs (Kuzmin et al., 2005) indicate that alleviation of positive mismatch (between MAL TMDs and lipid tails) by cluster formation easily compensates for the loss of entropy (Kozlov, personal communication). Thus, we propose that our findings on MAL illustrate a potential mechanism by which sphingolipid rafts may form a functional protein and lipid sorting domain. Positive hydrophobic mismatching between MAL TMDs and membranes of the appropriate thickness could be one of the forces promoting propagation and stabilization of the otherwise dynamic and nanoscale raft domains. This process coincides with the self-oligomerization of MAL. Our hypothesis also predicts that the sorting function of MAL will be specific to membranes of smaller average thickness, as well as to membranes comprising a lipid mixture with highly variable hydrophobic-core width. Because the ER, the Golgi, and the basolateral membranes are thinner, on average, than the apical membrane, it is reasonable to assume that the sorting function of MAL is executed early on in its secretory transport (Keller et al., 2001). Little data exist on the composition of endocytic organelle membranes. However, one might assume that the MAL-positive apical recycling compartment contains lipids with highly variable fatty acyl lengths as it communicates with many intersecting intracellular pathways (Ang et al., 2004). MAL-mediated domain formation probably constitutes only the initial step in the multistage maturation process of apical-sorting domains. Along the way, formation of the apically targeted carrier undoubtedly involves diverse types of interactions mediated by a sundry of transport machinery proteins. The novelty of the data presented here does not lie in the introduction of a novel mechanism or concept to explain how membrane domains are formed or stabilized but rather in the identification and characterization of a candidate protein with a distinct task in this cellular process. We show that MAL fulfills all of the requirements for mediating the formation and maintenance of the apical-sorting-membrane domain. Similarly, MAL meets the requirements to operate as a protein determining apical membrane thickness.
Finally, MAL TMDs constitute the MARVEL domain, which is associated with mostly unknown proteins but also with the better known occludin, synaptophysin, and synaptogyrin. Future study should focus on how the MARVEL domain–lipid interaction is used for diverse cellular tasks (Anton et al., 2008; Zech et al., 2009).
Supplementary Material
ACKNOWLEDGMENTS
Many thanks to M. M. Kozlov, D. Andelman (Tel-Aviv University), and K. Simons (CBG MPI, Dresden, Germany) for critical discussions; and to Yoel Kloog, B. Nichols (Medical Research Council-Laboratory of Molecular Biology, Cambridge, United Kingdom), Y. Altschuler, and J. Ellenberg for reagents. This work is supported by the Israel Science Foundation grant 679/05 and by the United States-Israel Binational Science foundation grant 281/05 (to K. H.). This work was supported in part by grants from the Ministerio de Educación y Ciencia (BMC2003-03297; BFU2006-01925; GEN2003-20662-C07-02) and the Comunidad de Madrid (GR/SAL/0096/2004) (to M.A.A.).
Abbreviations used:
- ER
endoplasmic reticulum
- FP
fluorescent protein
- GP
general polarization
- GPI
glycophosphatidylinositol
- OC
ordered cluster
- PM
plasma membrane
- TMD
transmembrane domain.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E09-02-0142) on June 24, 2009.
REFERENCES
- Almsherqi Z. A., Kohlwein S. D., Deng Y. Cubic membranes: a legend beyond the Flatland* of cell membrane organization. J. Cell Biol. 2006;173:839–844. doi: 10.1083/jcb.200603055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso M. A., Weissman S. M. cDNA cloning and sequence of MAL, a hydrophobic protein associated with human T-cell differentiation. Proc. Natl. Acad. Sci. USA. 1987;84:1997–2001. doi: 10.1073/pnas.84.7.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ang A. L., Taguchi T., Francis S., Folsch H., Murrells L. J., Pypaert M., Warren G., Mellman I. Recycling endosomes can serve as intermediates during transport from the Golgi to the plasma membrane of MDCK cells. J. Cell Biol. 2004;167:531–543. doi: 10.1083/jcb.200408165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anton O., Batista A., Millan J., Andres-Delgado L., Puertollano R., Correas I., Alonso M. A. An essential role for the MAL protein in targeting Lck to the plasma membrane of human T lymphocytes. J. Exp. Med. 2008;205:3201–3213. doi: 10.1084/jem.20080552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benting J., Rietveld A., Ansorge I., Simons K. Acyl and alkyl chain length of GPI-anchors is critical for raft association in vitro. FEBS Lett. 1999;462:47–50. doi: 10.1016/s0014-5793(99)01501-x. [DOI] [PubMed] [Google Scholar]
- Cheong K. H., Zacchetti D., Schneeberger E. E., Simons K. VIP17/MAL, a lipid raft-associated protein, is involved in apical transport in MDCK cells. Proc. Natl. Acad. Sci. USA. 1999;96:6241–6248. doi: 10.1073/pnas.96.11.6241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christian A. E., Haynes M. P., Phillips M. C., Rothblat G. H. Use of cyclodextrins for manipulating cellular cholesterol content. J. Lipid. Res. 1997;38:2264–2272. [PubMed] [Google Scholar]
- de Marco M. C., Kremer L., Albar J. P., Martinez-Menarguez J. A., Ballesta J., Garcia-Lopez M. A., Marazuela M., Puertollano R., Alonso M. A. BENE, a novel raft-associated protein of the MAL proteolipid family, interacts with caveolin-1 in human endothelial-like ECV304 cells. J. Biol. Chem. 2001;276:23009–23017. doi: 10.1074/jbc.M009739200. [DOI] [PubMed] [Google Scholar]
- de Marco M. C., Martin-Belmonte F., Kremer L., Albar J. P., Correas I., Vaerman J. P., Marazuela M., Byrne J. A., Alonso M. A. MAL2, a novel raft protein of the MAL family, is an essential component of the machinery for transcytosis in hepatoma HepG2 cells. J. Cell Biol. 2002;159:37–44. doi: 10.1083/jcb.200206033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelman D. M. Membranes are more mosaic than fluid. Nature. 2005;438:578–580. doi: 10.1038/nature04394. [DOI] [PubMed] [Google Scholar]
- Fernandes F., Loura L. M., Prieto M., Koehorst R., Spruijt R. B., Hemminga M. A. Dependence of M13 major coat protein oligomerization and lateral segregation on bilayer composition. Biophys. J. 2003;85:2430–2441. doi: 10.1016/s0006-3495(03)74666-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Saez A. J., Chiantia S., Schwille P. Effect of line tension on the lateral organization of lipid membranes. J. Biol. Chem. 2007;282:33537–33544. doi: 10.1074/jbc.M706162200. [DOI] [PubMed] [Google Scholar]
- Gaus K., Gratton E., Kable E. P., Jones A. S., Gelissen I., Kritharides L., Jessup W. Visualizing lipid structure and raft domains in living cells with two-photon microscopy. Proc. Natl. Acad. Sci. USA. 2003;100:15554–15559. doi: 10.1073/pnas.2534386100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaus K., Zech T., Harder T. Visualizing membrane microdomains by Laurdan 2-photon microscopy. Mol. Membr. Biol. 2006;23:41–48. doi: 10.1080/09687860500466857. [DOI] [PubMed] [Google Scholar]
- Gil T., Sabra M. C., Ipsen J. H., Mouritsen O. G. Wetting and capillary condensation as means of protein organization in membranes. Biophys. J. 1997;73:1728–1741. doi: 10.1016/S0006-3495(97)78204-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glebov O. O., Nichols B. J. Lipid raft proteins have a random distribution during localized activation of the T-cell receptor. Nat. Cell Biol. 2004;6:238–243. doi: 10.1038/ncb1103. [DOI] [PubMed] [Google Scholar]
- Hancock J. F. Lipid rafts: contentious only from simplistic standpoints. Nat. Rev. Mol. Cell Biol. 2006;7:456–462. doi: 10.1038/nrm1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harder T., Scheiffele P., Verkade P., Simons K. Lipid domain structure of the plasma membrane revealed by patching of membrane components. J. Cell Biol. 1998;141:929–942. doi: 10.1083/jcb.141.4.929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harder T., Simons K. Caveolae, DIGs, and the dynamics of sphingolipid-cholesterol microdomains. Curr. Opin. Cell Biol. 1997;9:534–542. doi: 10.1016/s0955-0674(97)80030-0. [DOI] [PubMed] [Google Scholar]
- Jensen M. O., Mouritsen O. G. Lipids do influence protein function—the hydrophobic matching hypothesis revisited. Biochim. Biophys. Acta. 2004;1666:205–226. doi: 10.1016/j.bbamem.2004.06.009. [DOI] [PubMed] [Google Scholar]
- Kandasamy S. K., Larson R. G. Molecular dynamics simulations of model trans-membrane peptides in lipid bilayers: a systematic investigation of hydrophobic mismatch. Biophys. J. 2006;90:2326–2343. doi: 10.1529/biophysj.105.073395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller P., Toomre D., Diaz E., White J., Simons K. Multicolour imaging of post-Golgi sorting and trafficking in live cells. Nat. Cell Biol. 2001;3:140–149. doi: 10.1038/35055042. [DOI] [PubMed] [Google Scholar]
- Kerppola T. K. Design and implementation of bimolecular fluorescence complementation (BiFC) assays for the visualization of protein interactions in living cells. Nat. Protoc. 2006;1:1278–1286. doi: 10.1038/nprot.2006.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzmin P. I., Akimov S. A., Chizmadzhev Y. A., Zimmerberg J., Cohen F. S. Line tension and interaction energies of membrane rafts calculated from lipid splay and tilt. Biophys. J. 2005;88:1120–1133. doi: 10.1529/biophysj.104.048223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachmanovich E., Shvartsman D. E., Malka Y., Botvin C., Henis Y. I., Weiss A. M. Co-localization analysis of complex formation among membrane proteins by computerized fluorescence microscopy: application to immunofluorescence co-patching studies. J. Microsc. 2003;212:122–131. doi: 10.1046/j.1365-2818.2003.01239.x. [DOI] [PubMed] [Google Scholar]
- Lingwood D., Ries J., Schwille P., Simons K. Plasma membranes are poised for activation of raft phase coalescence at physiological temperature. Proc. Natl. Acad. Sci. USA. 2008;105:10005–10010. doi: 10.1073/pnas.0804374105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marazuela M., Acevedo A., Garcia-Lopez M. A., Adrados M., de Marco M. C., Alonso M. A. Expression of MAL2, an integral protein component of the machinery for basolateral-to-apical transcytosis, in human epithelia. J. Histochem. Cytochem. 2004a;52:243–252. doi: 10.1177/002215540405200212. [DOI] [PubMed] [Google Scholar]
- Marazuela M., Martin-Belmonte F., Garcia-Lopez M. A., Aranda J. F., de Marco M. C., Alonso M. A. Expression and distribution of MAL2, an essential element of the machinery for basolateral-to-apical transcytosis, in human thyroid epithelial cells. Endocrinology. 2004b;145:1011–1016. doi: 10.1210/en.2003-0652. [DOI] [PubMed] [Google Scholar]
- Martin-Belmonte F., Puertollano R., Millan J., Alonso M. A. The MAL proteolipid is necessary for the overall apical delivery of membrane proteins in the polarized epithelial Madin-Darby canine kidney and Fischer rat thyroid cell lines. Mol. Biol. Cell. 2000;11:2033–2045. doi: 10.1091/mbc.11.6.2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megha, Sawatzki P., Kolter T., Bittman R., London E. Effect of ceramide N-acyl chain and polar headgroup structure on the properties of ordered lipid domains (lipid rafts) Biochim. Biophys. Acta. 2007;1768:2205–2212. doi: 10.1016/j.bbamem.2007.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra K., Ubarretxena-Belandia I., Taguchi T., Warren G., Engelman D. M. Modulation of the bilayer thickness of exocytic pathway membranes by membrane proteins rather than cholesterol. Proc. Natl. Acad Sci. USA. 2004;101:4083–4088. doi: 10.1073/pnas.0307332101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro S. Localization of proteins to the Golgi apparatus. Trends Cell Biol. 1998;8:11–15. doi: 10.1016/S0962-8924(97)01197-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro S. Lipid rafts: elusive or illusive? Cell. 2003;115:377–388. doi: 10.1016/s0092-8674(03)00882-1. [DOI] [PubMed] [Google Scholar]
- Nichols B. J. GM1-containing lipid rafts are depleted within clathrin-coated pits. Curr. Biol. 2003;13:686–690. doi: 10.1016/s0960-9822(03)00209-4. [DOI] [PubMed] [Google Scholar]
- Pagano R. E., Watanabe R., Wheatley C., Dominguez M. Applications of BODIPY-sphingolipid analogs to study lipid traffic and metabolism in cells. Methods Enzymol. 2000;312:523–534. doi: 10.1016/s0076-6879(00)12937-4. [DOI] [PubMed] [Google Scholar]
- Puertollano R., Alonso M. A. MAL, an integral element of the apical sorting machinery, is an itinerant protein that cycles between the trans-Golgi network and the plasma membrane. Mol. Biol. Cell. 1999;10:3435–3447. doi: 10.1091/mbc.10.10.3435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puertollano R., Martin-Belmonte F., Millan J., de Marco M. C., Albar J. P., Kremer L., Alonso M. A. The MAL proteolipid is necessary for normal apical transport and accurate sorting of the influenza virus hemagglutinin in Madin-Darby canine kidney cells. J. Cell Biol. 1999;145:141–151. doi: 10.1083/jcb.145.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sal-Man N., Gerber D., Bloch I., Shai Y. Specificity in transmembrane helix-helix interactions mediated by aromatic residues. J. Biol. Chem. 2007;282:19753–19761. doi: 10.1074/jbc.M610368200. [DOI] [PubMed] [Google Scholar]
- Sanchez-Pulido L., Martin-Belmonte F., Valencia A., Alonso M. A. MARVEL: a conserved domain involved in membrane apposition events. Trends Biochem. Sci. 2002;27:599–601. doi: 10.1016/s0968-0004(02)02229-6. [DOI] [PubMed] [Google Scholar]
- Schuck S., Simons K. Polarized sorting in epithelial cells: raft clustering and the biogenesis of the apical membrane. J. Cell Sci. 2004;117:5955–5964. doi: 10.1242/jcs.01596. [DOI] [PubMed] [Google Scholar]
- Sharma P., Varma R., Sarasij R. C., Ira, Gousset K., Krishnamoorthy G., Rao M., Mayor S. Nanoscale organization of multiple GPI-anchored proteins in living cell membranes. Cell. 2004;116:577–589. doi: 10.1016/s0092-8674(04)00167-9. [DOI] [PubMed] [Google Scholar]
- Simons K., Ikonen E. Functional rafts in cell membranes. Nature. 1997;387:569–572. doi: 10.1038/42408. [DOI] [PubMed] [Google Scholar]
- Snapp E. L., Hegde R. S., Francolini M., Lombardo F., Colombo S., Pedrazzini E., Borgese N., Lippincott-Schwartz J. Formation of stacked ER cisternae by low affinity protein interactions. J. Cell Biol. 2003;163:257–269. doi: 10.1083/jcb.200306020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperotto M. M., Mouritsen O. G. Mean-field and Monte Carlo simulation studies of the lateral distribution of proteins in membranes. Eur. Biophys. J. 1991;19:157–168. doi: 10.1007/BF00196342. [DOI] [PubMed] [Google Scholar]
- Sprong H., van der Sluijs P., van Meer G. How proteins move lipids and lipids move proteins. Nat. Rev. Mol. Cell Biol. 2001;2:504–513. doi: 10.1038/35080071. [DOI] [PubMed] [Google Scholar]
- Watari H., Blanchette-Mackie E. J., Dwyer N. K., Glick J. M., Patel S., Neufeld E. B., Brady R. O., Pentchev P. G., Strauss J. F., 3rd Niemann-Pick C1 protein: obligatory roles for N-terminal domains and lysosomal targeting in cholesterol mobilization. Proc. Natl. Acad. Sci. USA. 1999;96:805–810. doi: 10.1073/pnas.96.3.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilmann P. G., Petersen J., Pettikiriarachchi A., Buckle A. M., Smith S. C., Olsen S., Perugini M. A., Devenish R. J., Prescott M., Rossjohn J. The 2.1A crystal structure of the far-red fluorescent protein HcRed: inherent conformational flexibility of the chromophore. J. Mol. Biol. 2005;349:223–237. doi: 10.1016/j.jmb.2005.03.020. [DOI] [PubMed] [Google Scholar]
- Zech T., Ejsing C. S., Gaus K., de Wet B., Shevchenko A., Simons K., Harder T. Accumulation of raft lipids in T-cell plasma membrane domains engaged in TCR signalling. EMBO J. 2009;28:466–476. doi: 10.1038/emboj.2009.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.