Abstract
The protein kinase haspin/Gsg2 plays an important role in mitosis, where it specifically phosphorylates Thr-3 in histone H3 (H3T3). Its protein sequence is only weakly homologous to other protein kinases and lacks the highly conserved motifs normally required for kinase activity. Here we report structures of human haspin in complex with ATP and the inhibitor iodotubercidin. These structures reveal a constitutively active kinase conformation, stabilized by haspin-specific inserts. Haspin also has a highly atypical activation segment well adapted for specific recognition of the basic histone tail. Despite the lack of a DFG motif, ATP binding to haspin is similar to that in classical kinases; however, the ATP γ-phosphate forms hydrogen bonds with the conserved catalytic loop residues Asp-649 and His-651, and a His651Ala haspin mutant is inactive, suggesting a direct role for the catalytic loop in ATP recognition. Enzyme kinetic data show that haspin phosphorylates substrate peptides through a rapid equilibrium random mechanism. A detailed analysis of histone modifications in the neighborhood of H3T3 reveals that increasing methylation at Lys-4 (H3K4) strongly decreases substrate recognition, suggesting a key role of H3K4 methylation in the regulation of haspin activity.
Keywords: ATP binding, histone modification, phosphorylation mechanism, germ cell-specific gene 2 (Gsg2), mitosis
Eukaryotic protein kinases (ePKs) constitute a large group of enzymes that regulate a vast diversity of cellular processes. Kinase activity depends on a number of highly conserved sequence motifs required for ATP/Mg2+ binding and catalysis. About 10% of human ePKs lack one or more essential catalytic motifs and have been initially classified as inactive pseudokinases (1). However, a number of such proteins are active kinases, including haspin [haploid germ cell–specific nuclear protein kinase, encoded by Germ cell–specific gene 2 (Gsg2)]. Haspin lacks both the conserved ATP/Mg2+ binding motif Asp-Phe-Gly (DFG), which is replaced by Asp-Tyr-Thr (DYT), and the Ala-Pro-Glu (APE) motif usually found at the C terminus of the activation segment (2). In addition, haspin shares only weak sequence homology with ePKs and contains a highly divergent kinase domain with several unique inserts (2, 3). Haspin mRNA is abundant in testis and also present in somatic cells (4, 5), and orthologues are found throughout eukaryotic phyla (2). Ectopically expressed haspin localizes to the nucleus in interphase and to the chromosomes in mitosis (4, 6), while depletion of haspin leads to premature chromatid separation, failure of chromosome alignment, and a block in mitosis in a prometaphase-like state (6, 7).
Haspin autophosphorylates in vitro (4, 6, 8) and phosphorylates its only currently known substrate, histone H3, at threonine-3 (H3T3ph) both in vitro and in cells (6). H3T3 phosphorylation occurs specifically during mitosis and is particularly prominent at inner centromere regions (6, 7, 9). Recently, H3T3ph has been suggested to relieve an inhibitory effect of histone H3 on Aurora B, leading to activation of this key mitotic kinase (10).
To understand the phosphorylation mechanism of haspin despite the absence of key catalytic motifs, we determined high-resolution crystal structures of human haspin in complex with ATP and the ATP mimetic inhibitor 5-iodotubercidin, a potent inhibitor of haspin. We further defined the influence of the histone H3 N terminus on haspin substrate recognition.
Results
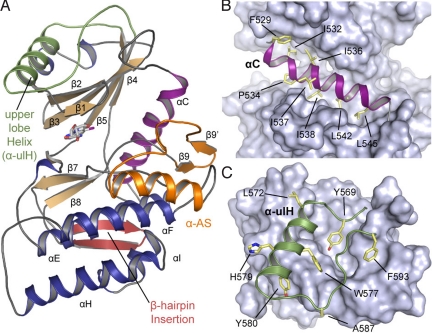
The structure of the haspin kinase domain (from Lys-470 to the C-terminal end, Lys-798) was initially solved by single-wavelength anomalous dispersion (SAD), and subsequent structures were determined by molecular replacement. All structures were refined to low R/Rfree values and showed acceptable deviations from standard bond length and geometry [supporting information (SI) Table S1]. All structures were well defined and revealed the typical bilobed kinase domain architecture, although lacking the ePK helix αG and containing a number of previously unseen structural features (Fig. 1) also highlighted in an alignment with cdk2 (Fig. S1).
Fig. 1.
Structural features of haspin. (A) Overall structure of haspin showing the location of haspin-specific inserts, as well as the main secondary structure elements. The ulH helix is highlighted in green, the activation segment is in yellow, and the hairpin insert is in brown. (B) Detailed view of helix αC (magenta) embedded by hydrophobic residues shown in stick representation. (C) The ulH insert. Conserved aromatic residues that interact with the upper lobe are shown and labeled.
Stable Positioning of Helix αC.
Helix αC is typically a highly mobile structural element in kinases that is moved into the correct position for catalysis by various regulatory mechanisms (11). Intriguingly, in haspin, αC is immobilized by a large number of hydrophobic contacts that tightly embed this helix (Fig. 1B). These hydrophobic interactions are largely conserved in haspin orthologues, suggesting that this property is conserved in haspins (Fig. S2). A salt bridge between the αC glutamate (Glu-535) and the active site lysine residue (Lys-511) found in all haspin structures is a hallmark of active kinases and indicates the correct positioning of αC for catalysis. αC also is tightly linked to the activation segment by hydrophobic contacts provided by a unique β sheet in the activation loop. Low overall temperature factors in αC and the embedded nature of this helix suggest that the movement and plasticity of αC helix in haspin are restricted.
A Helical Insert in the Upper Lobe Stabilizes the P-Loop.
The most remarkable feature of the upper lobe architecture is an insert that forms an additional helix [called the upper lobe helix (ulH)] connected to β5 by a large loop (Tyr-569 to Phe-593). This insert is conserved in all haspin orthologues (except the divergent Saccharomyces cerevisiae homologues Alk1 and Alk2) and binds to a deep cradle formed by the 5β-strand network typically found in protein kinases. The conformation of this insert is stabilized by a core of highly conserved hydrophobic and aromatic residues (Fig. 1C). Interestingly, the loop connecting the ulH helix to β5 interacts with the entire length of the phosphate-binding “P-loop,” forming a network of polar interactions involving main chain contacts and Lys-489, Glu-492, Glu-497, and Asn-588 (Fig. S3A). The P-loop is typically a very mobile, glycine-rich region that is often disordered in crystal structures even when the cofactor or an inhibitor is bound. In haspin, the P-loop is well defined and is stabilized by the ulH insert in a conformation suitable for ATP binding. It is noteworthy that part of the PKA P-loop (i.e., Lys-47, Thr-48 of β1, and Arg-56 of β2) interacts with a C-terminal regulatory tail and thus functions as a “gate” for ATP binding (12). Moreover, in AGC kinase family proteins such as Aurora A and Aurora B, the N-lobe β-barrel surface interacts with the regulatory proteins TPX2 (13) and the IN-box segment of the inner centromere protein INCENP (14), increasing the kinase activity. Similarly, the haspin ulH insert, which lies in the cradle formed by β strands, might potentially bind to the N-terminal domain or other proteins, perhaps positively or negatively regulating its activity. In addition, when all 3 haspin structures are superimposed, the rigid nature of the P-loop becomes evident and reveals small differences in upper lobe orientation showing that the P-loop and the ulH insert move together as a rigid body (Fig. S3A).
A β Hairpin Anchors the αC C-Terminal Loop.
A haspin-specific β-hairpin insert (Lys-663–Cys-679) located between the β7 and β8 strands packs against the lower lobe and makes numerous main chain contacts with a loop region C-terminal to αC, as well as a hydrogen bond between the partially conserved residues Asn-670 and Asn-551 (Fig. S3B). A number of hydrophobic anchor points present in the β hairpin are conserved as well, but because most interactions are mediated by main chain contacts, there is little sequence conservation in this region. Interestingly, a β-hairpin insert located between strands β7 and β8 also is present in Cdc2-like kinases (CLKs) (15), but there it is oriented differently and packs against the opposite side of the lower kinase lobe (Fig. S4).
Haspin Has an Atypical Activation Segment Architecture.
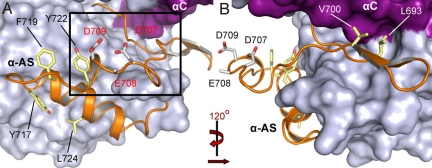
In most kinases, the activation segment is a regulatory element of ≈35 residues located between the 2 conserved motifs, DFG and APE (11). Typically this element consists of an ATP/Mg2+-binding site (DFG motif), a short β strand (β9), the activation loop, and the P + 1 loop. The activation segment shows considerable conformational diversity, but structural comparisons reveal that 2 invariable anchor points formed by the DFG motif and the central portion of the P + 1 loop and helix αEF lock the termini of this regulatory element (11). For a large number of protein kinases, the unphosphorylated activation segment is disordered or locked into an inactive conformation, while phosphorylation of one or more residues in the segment stabilizes this region in an active conformation suitable for the binding of substrates. The haspin activation segment differs in several ways from those of other protein kinases (Fig. 2). First, the DFG motif is changed to DYT. Second, there is an additional β strand (β9′) that forms an antiparallel sheet with β9. This leads to shortening of the activation loop and reorientation of the segment structure. The additional β sheet links the activation segment with αC by largely hydrophobic contacts. Third, the β6 strand typically found in ePKs is missing. This strand usually forms an antiparallel β sheet with β9, a function that has been assumed in haspin by β9′. Fourth, a large, highly conserved aromatic core stabilizes the conformation of the entire segment. Fifth, the catalytic loop HRD motif arginine (Arg-648), which typically interacts with phosphate moieties in serine/threonine kinases that require activation segment phosphorylation for activity, forms a hydrogen bond with the backbone of the activation loop tip, linking the segment to the catalytic loop. Finally, the missing APE motif and P + 1 loop are replaced by a large helical insert (αAS; Gln-718–Lys-727) (Fig. 2). The entire activation segment is well structured, suggesting that it is in a constitutively active conformation suitable for substrate binding. The only possible phosphorylation site in the activation segment, the partially conserved Ser-705, is oriented toward the solvent, but no autophosphorylation of the recombinant protein is observed at this position. The unusual activation segment architecture, including a cluster of acidic residues (Asp-707, Glu-708, and Asp-709) and the missing P + 1 loop and APE motif, results in a significantly altered substrate binding site. This view is reinforced by the absence of the αG helix that forms part of the substrate-binding site in other kinases. Electrostatic potentials projected onto the surface of haspin reveals a highly negatively charged channel between the 2 kinase lobes that most likely serves as a binding groove for the basic histone H3 tail (Fig. S1B).
Fig. 2.
The haspin activation segment. (A) The kinase is shown in a surface representation, with the activation segment shown as a ribbon diagram. Conserved aromatic residues that form a core and stabilize the conformation of the segment are shown in a stick representation. (B) Segment rotated by 120 degrees to provide a detailed view.
ATP Binding in the Absence of a Classical DFG Motif.
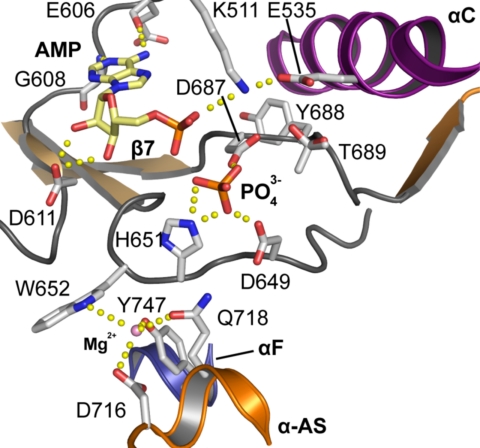
To address functional consequences of the altered DFG motif, we solved the structure of haspin in complex with ATP/Mg2+ (Fig. 3). The structure revealed that, similar to typical ePKs (16, 17), the adenosine moiety interacts with haspin by forming hydrogen bonds between the N1 and N6 amino groups and the backbone of the kinase hinge residues Glu-606 and Gly-608. The sugar moiety also retains a C2′-endo conformation. In ePKs, the α-phosphate is coordinated by the conserved active site lysine residue in the β3 strand. Indeed, the α-phosphate forms a polar contact with Lys-511 of haspin. Although the distance from Lys-511 to the closest oxygen atom of the ATP phosphate (3.5 Å) is too great for efficient hydrogen bonding, the low activity of a Lys511Ala mutant confirms the importance of this residue in haspin (Fig. 4A). Unfortunately, the β- and γ-phosphate moieties were not observed in the electron density maps and were assumed to be either disordered or hydrolyzed, and no Mg2+ ions were detected in the vicinity of the DYT motif. However, in a co-crystal structure with the nucleoside-based inhibitor iodotubercidin, a phosphate ion present in the crystallization solution was bound at a position expected to harbor the ATP γ-phosphate. The isolated phosphate group is directly coordinated by Asp-687 of the DYT, His-651 in the catalytic loop, and, unusually, the HRD motif Asp-649, suggesting that in haspins, the catalytic loop plays an important role in cofactor binding. The binding modes of ADP/PO43− between PKA and haspin are compared in Fig. S5. His-651 is highly conserved within the haspin family and lies in a position occupied by lysine in most serine/threonine kinases (2). The mutation His651Ala renders haspin largely inactive, confirming its importance for catalytic activity (Fig. 4A). Interestingly, we observed a metal-binding site that coordinates a Mg2+ ion in the interface of the catalytic loop, helix αF, and the activation segment (Asp-716, Gln-718, Trp-652, and Tyr-747) (Fig. 3).
Fig. 3.
Interaction of ATP with the haspin active site. Detailed view of interactions formed by the ATP phosphate moieties as well as the Mg2+-binding site. The figure represents an overlay of the iodotubercidin phosphate complex and the AMP complex to show all relevant observed interactions of the cofactor with haspin.
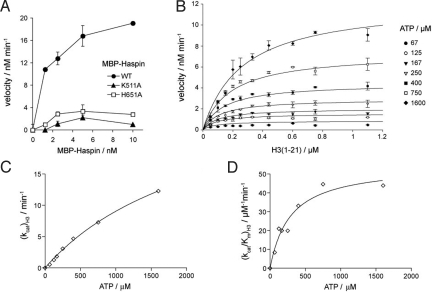
Fig. 4.
Enzyme activity data. (A) Phosphorylation of 1 μM histone H3 (1–21) peptide as a function of the concentration of WT and K511A and H651A mutants of MBP-haspin. Reactions were conducted with 200 μM ATP for 7 min. The mean and range of duplicate determinations are shown. (B) Initial reaction velocities as a function of histone H3 (1–21) concentration, at various fixed ATP concentrations. Reactions were carried out for 5 min with 1 nM MBP-haspin. (C and D) The data in (B) were further analyzed by the method of replots (see text). Hyperbolic curves indicate a random sequential mechanism (19). Also see Fig. S7.
We used differential scanning fluorimetry to determine the relative strength of cofactor binding to haspin. In this assay, binding affinities are measured indirectly as a function of melting temperature increase (ΔTm). Several studies have shown that ΔTm correlates linearly with binding constants of ligands (18). Nucleotides demonstrated no interaction with haspin in the absence of divalent ions (Fig. S5A), but significant ΔTm was observed in the presence of Mg2+, Mn2+, or Ca2+ ions, with the highest ΔTm for ATP. Ions alone also stabilized haspin, most likely due to the metal-binding site detected in the interface of the catalytic loop, helix αF, and the activation segment, suggesting a role of divalent ions in stabilizing the haspin active state.
Enzymatic Properties of Haspin.
The structure of the haspin kinase domain revealed features suggesting an active conformation. Consistent with this, both the His6-tagged protein kinase domain of haspin and an MBP fusion protein of full-length haspin expressed in E. coli were active kinases in vitro. The presence of the N-domain altered the kinetics of H3 peptide phosphorylation, increasing the KATP ≈5-fold, lowering the KH3 6-fold, and increasing kcat ≈15-fold (Table S2). Thus, the N-domain has the potential to modulate haspin activity in cells by influencing the conformation of the kinase domain and perhaps making direct interactions with the H3 substrate.
The unusual features of the haspin structure led us to conduct a more detailed kinetic analysis to gain insight into its reaction mechanism. We measured initial velocities of phosphorylation by full-length haspin and the kinase domain alone as a function of histone H3 (1–21) peptide concentration, at several fixed concentrations of ATP (Fig. 4B). The datasets were subject to global nonlinear least squares fitting to equations for 3 standard kinetic mechanisms, as described in ref. 19. All statistical measures modestly favored a random mechanism (Fig. S7A). To more fully distinguish between mechanisms, we used the method of replots (19). Both kcat and kcat/Km for each substrate were hyperbolically dependent on the concentration of the other substrate (Figs. 4 C and D and S7B), inconsistent with ping-pong and rapid equilibrium ordered mechanisms but supporting a rapid equilibrium random or steady-state ordered mechanism. Finally, product inhibition studies of MBP-haspin using ADP and H3 (1–21) T3ph peptides suggested a rapid equilibrium random mechanism (Fig. S7 C and D). Thus, haspin follows a random sequential mechanism in which ATP or H3 peptide can bind first, and binding of the first substrate decreases affinity for the second substrate.
Substrate Specificity of Haspin.
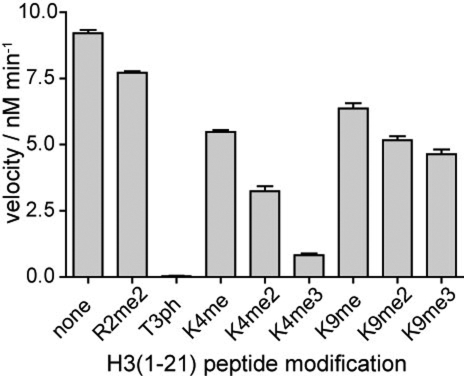
The surface electrostatics and the shape of haspin indicate the possibility of basic substrates like histone tails binding to a narrow, highly negatively charged channel located between the 2 kinase lobes. To examine the effect of posttranslational modifications of the histone tail, we determined the efficiency with which haspin catalyzed phosphorylation of H3 tail peptides with various preexisting modifications. While Arg-2 dimethylation and Lys-9 monomethylation, dimethylation, or trimethylation had little effect on phosphorylation, increasing methylation of the side chain of Lys-4 resulted in progressively reduced substrate recognition. The trimethylated substrate (K4me3) was poorly recognized as suitable haspin substrate (Figs. 5 and S8).
Fig. 5.
Effect of histone H3 modifications surrounding Thr-3 on haspin activity. Kinase reactions were carried out with 6 nM MBP-haspin, 1 μM H3 (1–21) peptide, and 120 μM ATP for 4 min. Similar results were obtained with the kinase domain alone (see Fig. S8).
Discussion
The structure of haspin reveals several structural features that are highly conserved in haspin-like kinases but not previously seen in other kinases. The main haspin inserts (helix ulH, the additional activation segment β9′ strand, and αAS helix, as well as the β-hairpin insert) appear to secure usually mobile structural elements, including the αC helix and P-loop, providing a novel means of stabilizing an active kinase conformation. This finding, coupled with the kinetic analysis of haspin activity in vitro, indicates constitutive kinase activity and is consistent with the absence of identifiable activation loop phosphorylation and the previous observation that both hypophosphorylated haspin immunoprecipitated from interphase cells and hyperphosphorylated haspin from mitotic cells have kinase activity (6). The question of how the activity of haspin is prevented from phosphorylating H3 in interphase cells is an interesting one. The N-domain of haspin is extensively phosphorylated during mitosis (6), and it is possible that in cells, this modulates access of haspin to its targets or the interaction of haspin with putative inhibitory proteins that regulate kinase activity, perhaps through binding to elements in the upper lobe, such as the uIH helix. In vitro assays reveal that the N-domain of haspin influences the kinetics of histone peptide phosphorylation; thus, another possibility is that regulatory proteins induce conformational changes in the N-domain to control the activity of the kinase domain.
The cofactor ATP binds to the haspin active site, forming typical interactions with the kinase hinge region. In one structure, an isolated phosphate moiety presumably mimicking the γ-phosphate interacts directly with the DYT motif aspartate, not via Mg2+ ions as expected. However, in vitro experiments showed that haspin requires Mg2+ for activity. Given the stabilizing effect of divalent metals on ATP binding, it is likely that Mg2+ normally bridges the β- and γ-phosphates with the DYT motif, and the isolated phosphate moiety seen in our structure may represent a hydrolyzed phosphate ion before its transfer to substrate. Other interesting features of the ATP–phosphate interactions are the direct hydrogen bonds formed to Asp-649 and His-651. His-651 is highly conserved in, and characteristic of, haspins (2), while the equivalent residue in most serine/threonine ePKs is a lysine (HRDxK). In PKA, this residue, Lys-168, forms interactions with the γ-phosphate of ATP, as well as with the HRD aspartate, the activation loop, and the substrate peptide. This residue is critical for phosphoryl transfer, and its mutation in PKA abolishes activity (20, 21). In haspin, His-651 is likely to directly coordinates the γ-phosphate of ATP, and the mutation His651Ala severely curtails activity, suggesting that His-651 fulfills a role similar to Lys-168 in PKA. In contrast, protein kinases of the divergent RIO/piD261/Bud32 superfamily typically contain serine or threonine at this position. The mutation Thr163Ala does not substantially alter the activity of piD261 (22), and Ser-220 in Rio2 does not directly contact ATP (23). Despite haspin's unique structural properties, kinetic analysis has shown that it utilizes a sequential reaction mechanism like those of ePKs that have been analyzed (24).
The most striking structural differences were observed in the haspin activation segment. The additional β9′ strand links the activation segment tightly to αC by numerous conserved hydrophobic contacts. In addition, the absence of αG helix and the substitution of the P + 1 loop and the helix αEF by a large helical insert results in a significantly changed substrate-binding surface. It is noteworthy that the atypical Rio kinases from Archaeoglobus fulgidus, which lack classical substrate-binding sites and the highly conserved catalytic motifs, also contain a highly electronegatively charged cleft between the 2 lobes possibly involved in the recognition of basic substrates (25). Similarly, the surface electrostatics of haspin suggests that the basic histone tail inserts into a narrow groove formed between the upper lobe and the activation segment. Haspin substrates identified in future are likely to feature phosphorylation sites imbedded in protruding basic regions that can enter this narrow binding site.
Interestingly, we found that H3T3 phosphorylation is modulated by methylation levels of the neighboring lysine residue (H3K4), with decreasing enzymatic activity on peptides with increasing methylation of H3K4. The bulkiness of K4 methylation likely compromises H3 tail interactions with the narrow substrate-binding groove of haspin. This finding is consistent with the observation that H3T3ph was detected adjacent to H3K4me, but not to H3K4me2 or H3K4me3, among histone peptides from mitotic cells (26). In mitosis, H3K4me2, H3K4me3, and H3T3ph have distinct localizations at centromeres (7, 27). It is possible that the influence of H3K4 methylation on haspin activity contributes to the creation of distinct zones of chromatin modification that have different functional attributes. In vertebrates, H3K4 dimethylation and trimethylation also occur at transcriptionally active genes (28). Cross-talk between histone modifications is only partially understood, but the strong negative influence, particularly of H3K4me3, on haspin catalytic activity suggests that the H3K4 methylation mark is a significant regulator of haspin activity.
Haspin plays an important role during mitosis. It appears to be vital for maintaining chromosome cohesion, because depletion of haspin leads to a loss of cohesin association, premature chromatid separation, and the failure of normal chromosome segregation (6, 7). Mitotic kinases have been recognized as appealing targets for the development of antimitotic drugs for cancer therapy. Although no direct link between haspin and cancer has been established, H3T3ph is reported to play an important role in the activation of the oncology target Aurora B (10). Because humans, and most other organisms, have a single haspin gene with low sequence similarity to ePKs, haspin is a particularly interesting target for the development of specific inhibitors. Indeed, haspin-selective inhibitors have been identified by high-throughput screening of chemical libraries (29), and screening using 88 Ser/Thr kinases showed that the inhibitor iodotubercidin bound to only one other kinase with low nM affinity (30). Iodotubercidin's high potency, cell permeability, and selectivity for haspin make this compound an interesting potential tool for studying haspin's function and a good starting point for future inhibitor development.
Materials and Methods
Purification of Haspin.
The kinase domain (Lys-470–Lys-798) of human haspin was amplified from a synthetic codon-optimized gene and cloned into the T7 expression vector pNIC28-Bsa4 containing a TEV-cleavable N-terminal His6 tag. Vector-transformed Escherichia coli (BL21 Rosetta) was grown and purified as described in ref. 31, but with the addition of 750 mM NaCl before concentration of haspin by ultrafiltration. For selenomethionine labeling, the cells were grown in MD medium with 40 mg/L of selenomethionine for 12 h at 18 °C, and the protein was purified as the native protein.
Crystallization.
Crystallization was carried out at 4 °C using matrix-seeded sitting drops and initial crystals of low quality. The 5-iodotubercidin complex (I) was grown using equal volumes of protein (10 mg/mL) with 1 mM of the ligand and reservoir solution [0.1 M SPG (pH 7.0) and 60% MPD]. SeMet crystals were grown using 0.15 M NH4citrate (pH 5), 25% PEG3350, and 0.1 M Mes (pH 6.0). Crystals of the AMP complex (II) were grown using 0.2 M Li2SO4, 0.1 M Hepes (pH 7.5), and 25% PEG 3350. Crystals with 5-iodotubercidin and PO43− (III) were grown using a 2:1 ratio of protein and 0.4 M (NH4)H2PO4.
Data Collection and Structure Solution.
Crystals were cryoprotected using well solution supplemented with MPD (I) or ethylene glycol (II and III) and flash-frozen in liquid nitrogen. Data for I were collected at the Swiss Light Source (X10SA). Initial phases were determined using SAD. Selenium sites were located with SHELXD (32), and phases were calculated with autoSHARP (33). The model was built using BUCCANEER (34) and was manually completed in COOT (35). Data for II and III were collected with a Rigaku FR-E generator. Both structures were solved by molecular replacement using the model of I in PHASER (36) and refined using REFMAC5 (37). The models and structure factors have been deposited with PDB [accession codes 2VUW (I), 3DLZ (II), and 3IQ7 (III)].
Enzymology.
His6-tagged haspin kinase domain (Lys-470–Lys-798) and full-length MBP-haspin were produced as described (6, 29). Point mutations were introduced using QuikChange II XL (Stratagene). H3 (1–21)-GGK-biotin peptides were obtained from Millipore and Abgent, and were quantified by HABA/avidin assay (Sigma). Radiometric P81 filter binding kinase assays with γ-[33P]-ATP (PerkinElmer) were conducted as described in ref. 29. Haspin phosphorylated only histone H3 (1–21) peptides that were not prephosphorylated at Thr-3, indicating a single phosphorylation site. Endpoint assays were conducted within the linear initial velocity phase determined from reaction progress curves (Fig. S6). Data were analyzed by nonlinear least squares fitting using Graphpad Prism and SigmaPlot 10 and by the method of replots, as described in ref. 19. A thermal stability assay using differential scanning fluorimetry for nucleotide and cation binding also was performed, as described in ref. 31.
Supplementary Material
Acknowledgments.
The Structural Genomics Consortium is a registered charity (number 1097737) that receives funds from the Canadian Institutes for Health Research, the Canadian Foundation for Innovation, Genome Canada through the Ontario Genomics Institute, GlaxoSmithKline, Karolinska Institutet, the Knut and Alice Wallenberg Foundation, the Ontario Innovation Trust, the Ontario Ministry for Research and Innovation, Merck & Co., Inc., the Novartis Research Foundation, the Swedish Agency for Innovation Systems, the Swedish Foundation for Strategic Research, and the Wellcome Trust. This work was also supported by grants from the National Institutes of Health (R01 CA122608) and the American Cancer Society (RSG-05–134-01-GMC) (to J.M.G.H).
Footnotes
The authors declare no conflicts of interest.
This article is a PNAS Direct Submission.
Data deposition: Structural coordinates of haspin have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 2VUW, 3DLZ, and 3IQ7).
This article contains supporting information online at www.pnas.org/cgi/content/full/0901989106/DCSupplemental.
References
- 1.Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 2.Higgins JM. Structure, function and evolution of haspin and haspin-related proteins, a distinctive group of eukaryotic protein kinases. Cell Mol Life Sci. 2003;60:446–462. doi: 10.1007/s000180300038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Higgins JM. Haspin-like proteins: A new family of evolutionarily conserved putative eukaryotic protein kinases. Protein Sci. 2001;10:1677–1684. doi: 10.1110/ps.49901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tanaka H, et al. Identification and characterization of a haploid germ cell–specific nuclear protein kinase (Haspin) in spermatid nuclei and its effects on somatic cells. J Biol Chem. 1999;274:17049–17057. doi: 10.1074/jbc.274.24.17049. [DOI] [PubMed] [Google Scholar]
- 5.Higgins JM. The Haspin gene: Location in an intron of the integrin alphaE gene, associated transcription of an integrin alphaE-derived RNA and expression in diploid as well as haploid cells. Gene. 2001;267:55–69. doi: 10.1016/s0378-1119(01)00387-0. [DOI] [PubMed] [Google Scholar]
- 6.Dai J, Sultan S, Taylor SS, Higgins JM. The kinase haspin is required for mitotic histone H3 Thr 3 phosphorylation and normal metaphase chromosome alignment. Genes Dev. 2005;19:472–488. doi: 10.1101/gad.1267105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dai J, Sullivan BA, Higgins JM. Regulation of mitotic chromosome cohesion by Haspin and Aurora B. Dev Cell. 2006;11:741–750. doi: 10.1016/j.devcel.2006.09.018. [DOI] [PubMed] [Google Scholar]
- 8.Nespoli A, et al. Alk1 and Alk2 are two new cell cycle–regulated haspin-like proteins in budding yeast. Cell Cycle. 2006;5:1464–1471. doi: 10.4161/cc.5.13.2914. [DOI] [PubMed] [Google Scholar]
- 9.Polioudaki H, et al. Mitotic phosphorylation of histone H3 at threonine 3. FEBS Lett. 2004;560:39–44. doi: 10.1016/S0014-5793(04)00060-2. [DOI] [PubMed] [Google Scholar]
- 10.Rosasco-Nitcher SE, Lan W, Khorasanizadeh S, Stukenberg PT. Centromeric Aurora-B activation requires TD-60, microtubules, and substrate priming phosphorylation. Science. 2008;319:469–472. doi: 10.1126/science.1148980. [DOI] [PubMed] [Google Scholar]
- 11.Huse M, Kuriyan J. The conformational plasticity of protein kinases. Cell. 2002;109:275–282. doi: 10.1016/s0092-8674(02)00741-9. [DOI] [PubMed] [Google Scholar]
- 12.Johnson DA, Akamine P, Radzio-Andzelm E, Madhusudan M, Taylor SS. Dynamics of cAMP-dependent protein kinase. Chem Rev. 2001;101:2243–2270. doi: 10.1021/cr000226k. [DOI] [PubMed] [Google Scholar]
- 13.Bayliss R, Sardon T, Vernos I, Conti E. Structural basis of Aurora-A activation by TPX2 at the mitotic spindle. Mol Cell. 2003;12:851–862. doi: 10.1016/s1097-2765(03)00392-7. [DOI] [PubMed] [Google Scholar]
- 14.Sessa F, et al. Mechanism of Aurora B activation by INCENP and inhibition by hesperadin. Mol Cell. 2005;18:379–391. doi: 10.1016/j.molcel.2005.03.031. [DOI] [PubMed] [Google Scholar]
- 15.Bullock AN, et al. Kinase domain insertions define distinct roles of CLK kinases in SR protein phosphorylation. Structure. 2009;17(3):352–362. doi: 10.1016/j.str.2008.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gassel M, et al. Mutants of protein kinase A that mimic the ATP-binding site of protein kinase B (AKT) J Mol Biol. 2003;329:1021–1034. doi: 10.1016/s0022-2836(03)00518-7. [DOI] [PubMed] [Google Scholar]
- 17.Zhou T, et al. Crystal structure of the TAO2 kinase domain: Activation and specificity of a Ste20p MAP3K. Structure. 2004;12:1891–1900. doi: 10.1016/j.str.2004.07.021. [DOI] [PubMed] [Google Scholar]
- 18.Bullock AN, et al. Structural basis of inhibitor specificity of the human protooncogene proviral insertion site in moloney murine leukemia virus (PIM-1) kinase. J Med Chem. 2005;48:7604–7614. doi: 10.1021/jm0504858. [DOI] [PubMed] [Google Scholar]
- 19.Liu M, et al. Kinetic studies of Cdk5/p25 kinase: Phosphorylation of tau and complex inhibition by two prototype inhibitors. Biochemistry. 2008;47:8367–8377. doi: 10.1021/bi800732v. [DOI] [PubMed] [Google Scholar]
- 20.Gibbs CS, Zoller MJ. Rational scanning mutagenesis of a protein kinase identifies functional regions involved in catalysis and substrate interactions. J Biol Chem. 1991;266:8923–8931. [PubMed] [Google Scholar]
- 21.Madhusudan, Akamine P, Xuong NH, Taylor SS. Crystal structure of a transition state mimic of the catalytic subunit of cAMP-dependent protein kinase. Nat Struct Biol. 2002;9:273–277. doi: 10.1038/nsb780. [DOI] [PubMed] [Google Scholar]
- 22.Facchin S, et al. Structure-function analysis of yeast piD261/Bud32, an atypical protein kinase essential for normal cell life. Biochem J. 2002;364(Pt 2):457–463. doi: 10.1042/BJ20011376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.LaRonde-LeBlanc N, Guszczynski T, Copeland T, Wlodawer A. Autophosphorylation of Archaeoglobus fulgidus Rio2 and crystal structures of its nucleotide–metal ion complexes. FEBS J. 2005;272:2800–2810. doi: 10.1111/j.1742-4658.2005.04702.x. [DOI] [PubMed] [Google Scholar]
- 24.Adams JA. Kinetic and catalytic mechanisms of protein kinases. Chem Rev. 2001;101:2271–2290. doi: 10.1021/cr000230w. [DOI] [PubMed] [Google Scholar]
- 25.LaRonde-LeBlanc N, Wlodawer A. Crystal structure of A. fulgidus Rio2 defines a new family of serine protein kinases. Structure. 2004;12:1585–1594. doi: 10.1016/j.str.2004.06.016. [DOI] [PubMed] [Google Scholar]
- 26.Garcia BA, et al. Modifications of human histone H3 variants during mitosis. Biochemistry. 2005;44:13202–13213. doi: 10.1021/bi050906n. [DOI] [PubMed] [Google Scholar]
- 27.Sullivan BA, Karpen GH. Centromeric chromatin exhibits a histone modification pattern that is distinct from both euchromatin and heterochromatin. Nat Struct Mol Biol. 2004;11:1076–1083. doi: 10.1038/nsmb845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barski A, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 29.Patnaik D, et al. Identification of small molecule inhibitors of the mitotic kinase haspin by high-throughput screening using a homogeneous time-resolved fluorescence resonance energy transfer assay. J Biomol Screen. 2008;13:1025–1034. doi: 10.1177/1087057108326081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fedorov O, et al. A systematic interaction map of validated kinase inhibitors with Ser/Thr kinases. Proc Natl Acad Sci USA. 2007;104:20523–20528. doi: 10.1073/pnas.0708800104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Scheeff ED, Eswaran J, Bunkoczi G, Knapp S, Manning G. Structure of the pseudokinase VRK3 reveals a degraded catalytic site, a highly conserved kinase fold, and a putative regulatory binding site. Structure. 2009;17:128–138. doi: 10.1016/j.str.2008.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sheldrick GM. A short history of SHELX. Acta Crystallogr A. 2008;64:112–122. doi: 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- 33.Vonrhein C, Blanc E, Roversi P, Bricogne G. Automated structure solution with autoSHARP. Methods Mol Biol. 2007;364:215–230. doi: 10.1385/1-59745-266-1:215. [DOI] [PubMed] [Google Scholar]
- 34.Cowtan K. The Buccaneer software for automated model building, 1: Tracing protein chains. Acta Crystallogr D. 2006;62:1002–1011. doi: 10.1107/S0907444906022116. [DOI] [PubMed] [Google Scholar]
- 35.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 36.McCoy AJ, Grosse-Kunstleve RW, Storoni LC, Read RJ. Likelihood-enhanced fast translation functions. Acta Crystallogr D. 2005;61:458–464. doi: 10.1107/S0907444905001617. [DOI] [PubMed] [Google Scholar]
- 37.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum likelihood method. Acta Crystallogr D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.