Abstract
During metabolic evolution to improve succinate production in Escherichia coli strains, significant changes in cellular metabolism were acquired that increased energy efficiency in two respects. The energy-conserving phosphoenolpyruvate (PEP) carboxykinase (pck), which normally functions in the reverse direction (gluconeogenesis; glucose repressed) during the oxidative metabolism of organic acids, evolved to become the major carboxylation pathway for succinate production. Both PCK enzyme activity and gene expression levels increased significantly in two stages because of several mutations during the metabolic evolution process. High-level expression of this enzyme-dominated CO2 fixation and increased ATP yield (1 ATP per oxaloacetate). In addition, the native PEP-dependent phosphotransferase system for glucose uptake was inactivated by a mutation in ptsI. This glucose transport function was replaced by increased expression of the GalP permease (galP) and glucokinase (glk). Results of deleting individual transport genes confirmed that GalP served as the dominant glucose transporter in evolved strains. Using this alternative transport system would increase the pool of PEP available for redox balance. This change would also increase energy efficiency by eliminating the need to produce additional PEP from pyruvate, a reaction that requires two ATP equivalents. Together, these changes converted the wild-type E. coli fermentation pathway for succinate into a functional equivalent of the native pathway that nature evolved in succinate-producing rumen bacteria.
Keywords: biocatalyst, metabolic engineering, succinic acid
Succinate, a four-carbon dicarboxylic acid, is currently used as a specialty chemical in the food, agricultural, and pharmaceutical industries (1). Succinic acid can also serve as a starting point for the synthesis of many commodity chemicals used in plastics and solvents with a potential global market of $15 billion (2). Although succinate is primarily produced from petroleum, recent increases in costs have generated considerable interest in the fermentative production of succinate from sugars using Escherichia coli and other biocatalysts (2, 3).
The yield of succinate from glucose fermentation is primarily determined by carbon partitioning at the phosphoenolpyruvate (PEP) node (Fig. 1A). In rumen bacteria such as Anaerobiospirillum succiniciproducens (4), Actinobacillus succinogenes (5, 6) and Mannheimia succiniciproducens (7, 8), more than half of the phosphoenolpyruvate formed from glucose is carboxylated to oxaloacetate and converted to succinate, the primary fermentation product. However, requirements for complex nutrients by these bacteria increase both the cost and process complexity. Native strains of E. coli ferment glucose effectively in simple mineral salts medium but produce succinate only as a minor product (9). In E. coli, half of the PEP from glucose is metabolized directly to pyruvate by the PEP-dependent phosphotransferase system for glucose uptake. Most of the remaining PEP is used for ATP production by pyruvate kinases for ATP. To preserve redox balance, the resulting pyruvate is converted to formate, acetate, lactate, ethanol, and small amounts of succinate.
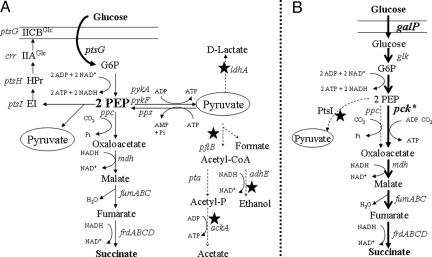
Fig. 1.
Comparison of mixed acid pathway for glucose fermentation in native E. coli and energy-conserving succinate production pathway in recombinant E. coli. (A) Mixed acid pathway: Phosphoenolpyruvate (PEP) is the primary source of energy for glucose uptake and phosphorylation. Succinate production is primarily determined by partitioning of carbon at the PEP node. In native E. coli, half of the pyruvate produced from PEP is used for this glucose uptake. The four solid stars indicate metabolic steps that have been blocked by constructed deletions. Dotted arrows indicate steps that are either nonfunctional or expected to have reduced carbon flow as a result of these deletions. Pyruvate (oval) is shown twice but represents a single metabolic pool. (B) Energy-conserving succinate production pathway: An acquired mutation in ptsI inactivating the PEP-dependent glucose uptake systems was functionally replaced by increased expression of the galP permease and glucokinase (glk). Additional mutations increased the expression of phosphoenolpyruvate carboxykinase (pck) sufficiently to make this enzyme the dominant route for carboxylation, increasing net production of ATP and increasing the pool of PEP available for succinate production. Bold solid arrows indicate the primary route for succinate production in KJ060 and related strains. Some intermediate steps in glycolysis have been omitted for clarity. G6P, glucose 6-phosphate; PEP, phosphoenolpyruvate.
Many investigators have described genetic engineering approaches to improve succinate production in E. coli by adding foreign genes (6, 10–13). The key to these improvements is increasing the carboxylation of PEP (and pyruvate) to a four-carbon dicarboxylic acid precursor of succinate. E. coli has four native carboxylation pathways that could potentially serve this function (Fig. 2). The carboxylation of PEP to oxaloacetate (OAA) by phosphoenolpyruvate carboxylase (ppc) is the primary fermentative route for succinate (Fig. 2A) (9, 14, 15). Energetically, the PPC-catalyzed reaction is strongly favored. Energy contained in PEP is lost in this reaction with the release of inorganic phosphate. Expression of genes encoding the other three carboxylation enzymes is repressed by glucose. These pathways are thought to function metabolically in the reverse direction (gluconeogenesis) during the oxidative metabolism of organic acids (4, 16–18).
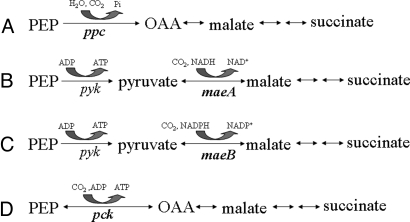
Fig. 2.
Carboxylation pathways potentially available for succinate production in E. coli. (A) PEP carboxylase (primary fermentative route). (B) NADH-linked malic enzyme (gluconeogenic). (C) NADPH-linked malic enzyme (gluconeogenic). (D) PEP carboxykinase (gluconeogenic). Genes encoding carboxylation activities are shown in bold. The energy in phosphoenolpyruvate is lost in the first pathway but conserved as ATP in the other three pathways. Pathway D is the dominant route for carboxylation in our succinate-producing strains of E. coli and in succinate-producing rumen bacteria. PEP, phosphoenolpyruvate; OAA, oxaloacetic acid.
Engineered strains of E. coli ATCC 8739 (KJ060 and KJ073) previously described by our laboratory (3, 19) produced high succinate yields without the use of foreign genes (700 mM succinate; 1.2 mol succinate per mol glucose). These yields are comparable to the best natural succinate-producing rumen bacteria such as Actinobacillus succinogenes (20). Strains KJ060 and KJ073 were developed by growth-based selection (metabolic evolution) after the deletion of the genes encoding alternative NADH-oxidizing pathways. During selection, improvements in growth depended on increased production of ATP for biosynthesis and succinate for redox balance.
In this communication, we describe the changes in metabolism that occurred during metabolic evolution of KJ060 and KJ073, which increased both ATP yield per glucose and succinate production. The gluconeogenic PEP carboxykinase (pck) was recruited (spontaneous mutation) to replace PEP carboxylase (ppc) as the primary carboxylation activity. The PEP-dependent phosphotransferase system was inactivated by a spontaneous point mutation and functionally replaced by the GalP permease (galP) and glucokinase (glk). With these changes, the net ATP yield during succinate production was doubled to 2.0 ATP per glucose. This resulting E. coli pathway is very similar to the energy-efficient pathway that has evolved in native succinate-producing rumen bacteria (5, 6).
Results
Recruitment of Gluconeogenic PEP Carboxykinase (pck) for Succinate Production.
The metabolic potential for E. coli includes four native carboxylation enzymes that could be used for succinate production (Fig. 2). The carboxylation of phosphoenolpyruvate (PEP) to oxaloacetate (OAA) by phosphoenolpyruvate carboxylase (ppc) is recognized as the primary pathway for the fermentative production of succinate in E. coli (Fig. 2A) (9, 15, 21). This reaction is essentially irreversible because of the energy loss associated with the release of inorganic phosphate. The three other carboxylation reactions are normally repressed by high concentration of glucose and are reported to function in the reverse direction for gluconeogenesis during oxidative metabolism of organic acids (4, 16–18). The second and third (Fig. 2 B and C) pathways use NADH-linked and NAPDH-linked malic enzymes (maeA and maeB, respectively) to catalyze the reversible carboxylation of pyruvate to malate. Both pathways allow energy to be conserved as ATP during the pyruvate formation from PEP. The fourth pathway uses PEP carboxykinase (pck) for the reversible carboxylation of PEP to OAA with the conservation of energy as ATP (Fig. 2D). Although PEP carboxykinase (Pck) typically functions only during gluconeogenesis in E. coli, an analogous PEP carboxykinase is present at very high levels in succinate-producing rumen bacteria, where it serves as the primary PEP carboxylating activity (5).
E. coli strain KJ073 was metabolically engineered by both targeted gene deletion and evolution for high-growth-rate and succinate production (3). Genes encoding each of the four carboxylating enzymes (ppc, maeA, maeB, and pck) were individually deleted in KJ073 to identify the primary pathway for succinate production (Table 1 and Table S1). Deletion of ppc, the gene encoding the primary carboxylating step in native E. coli, did not alter succinate production (strain XZ320). Deletion of the genes encoding malic enzymes (maeA and maeB) to produce strains XZ341 and XZ396, respectively, also had no effect on succinate production, consistent with their primary function in gluconeogenesis. Deletion of pck encoding PEP carboxykinase (strain XZ332), however, dramatically reduced cell growth, sugar metabolism, succinate production, and succinate yield. Complementation of this deletion mutant with the pck gene from strain KJ073 (plasmid pLOI4677) substantially improved succinate production to near that of KJ073 (Table 1).
Table 1.
Pck is critical for glucose fermentation to succinate by derivatives of KJ073
| Strain* | Genetic modification | Glucose consumed (mM) | Succinate (mM) | Yield† (mol/mol) | Acetate (mM) | Pyruvate (mM) |
|---|---|---|---|---|---|---|
| KJ073 | None | 301 ± 0 | 364 ± 25 | 1.21 ± 0.08 | 113 ± 8 | 3 ± 1 |
| XZ320 | KJ073, Δ ppc | 278 ± 0 | 348 ± 12 | 1.25 ± 0.04 | 100 ± 5 | — |
| XZ341 | KJ073, Δ maeA | 278 ± 0 | 339 ± 8 | 1.22 ± 0.03 | 106 ± 13 | — |
| XZ396 | KJ073, Δ maeB | 278 ± 0 | 337 ± 22 | 1.22 ± 0.08 | 116 ± 8 | — |
| XZ332 | KJ073, Δ pck | 92 ± 21 | 42 ± 11 | 0.46 ± 0.11 | 28 ± 5 | 32 ± 4 |
| XZ332 (pLOI4677) | Δ pck, pck overexpression | 273 ± 0 | 260 ± 36 | 0.95 ± 0.13 | 78 ± 4 | 32 ± 5 |
*All fermentations were performed in fleakers containing NBS medium with 5% glucose and 100 mM potassium bicarbonate.
†Yield was calculated as mol succinate produced per mol glucose metabolized.
Together, these results demonstrate that the gluconeogenic PEP carboxykinase was recruited during metabolic evolution to serve as the primary carboxylation reaction for fermentative succinate production in KJ073. Unlike the PEP carboxylase (Fig. 2A), PEP carboxykinase conserves energy providing an additional ATP per OAA (Fig. 2D). Because fermentative growth and ATP production are closely linked, the increase in ATP yield with PEP carboxykinase (Fig. 2D) could provide a competitive advantage during growth-based selection during the metabolic evolution steps leading to KJ073.
Comparison of Carboxylation Enzyme Activities in E. coli.
The activities of the four carboxylation enzymes were compared in wild-type strain ATCC 8739, KJ012 (deleted in primary fermentation pathways), and the evolved strains (Table 2). PEP carboxylase activities were similarly low in all, i.e., 0.02 to 0.03 U·mg protein−1. NADH-linked malic enzyme activity (MaeA; carboxylation direction) was also low, and NADPH-linked malic enzyme activity (MaeB; carboxylation direction) was undetectable. In contrast, PEP carboxykinase activity was 45-fold higher in KJ073 than in KJ012, an increase of more than 7 U·mg protein−1.
Table 2.
Comparison of carboxylation enzymes in engineered E. coli strains
| Strain | Cell yield (g/l) | Succinate (g/l·h) | Enzyme activity* [U·mg protein −1] |
pck mRNA | G to A change | |||
|---|---|---|---|---|---|---|---|---|
| Pck | Ppc | MaeA | MaeB | |||||
| E. coli ATCC | 2.0 ± 0.2 | 0.12 ± 0.01 | 0.30 ± 0.02 | 0.020 ± 0.002 | ND | <0.01 | 1.0 | No |
| KJ012‡ | 0.3 ± 0.1 | 0.04 ± 0.01 | 0.16 ± 0.01 | 0.025 ± 0.002 | 0.005 ± 0.001 | <0.01 | 0.2 | No |
| KJ017 | 1.7 ± 0.1 | 0.35 ± 0.03 | 0.70 ± 0.07 | 0.017 ± 0.001 | 0.012 ± 0.001 | <0.01 | 2.7 | No |
| KJ060 | 2.2 ± 0.1 | 0.90 ± 0.04 | 8.4 ± 0.7 | 0.030 ± 0.003 | 0.011 ± 0.002 | <0.01 | 7.2 | Yes |
| KJ071 | 1.5 ± 0.1 | 0.33 ± 0.04 | 0.68 ± 0.07 | 0.023 ± 0.002 | 0.010 ± 0.001 | <0.01 | 2.5 | No |
| KJ073 | 2.3 ± 0.1 | 0.82 ± 0.01 | 7.3 ± 0.5 | 0.027 ± 0.02 | 0.021 ± 0.001 | <0.01 | 8.4 | Yes |
| E. coli K12† | ND | ND | 0.14 | 0.14 | ND | ND | ND | ND |
| Actinobacillus succinogenes† | ND | ND | 4.7 | 0.01 | ND | ND | ND | ND |
*Pck, PEP carboxykinase; Ppc, PEP carboxylase; MaeA, NADH-linked malic enzyme; MaeB, NADPH-linked malic enzyme; ND, not determined. Crude extract was prepared from mid-log cells during fleaker fermentation.
†Data from VanderWerf et al. (5).
‡Poor growth.
PEP carboxykinase activities were also measured in the three intermediate strains isolated during the development of KJ073 (3). Large increases in PEP carboxykinase activity were correlated with improvements in cell yield and succinate production (Table 2). From KJ012 to KJ017 (metabolic evolution), PEP carboxykinase activity increased 4.3-fold, cell yield increased 5.7-fold, and succinate production increased 8.8-fold. From KJ017 to KJ060 (deletion of pflB followed by metabolic evolution), PEP carboxykinase activity increased 12-fold, cell yield increased 1.3-fold, and succinate production increased 2.6-fold. From KJ060 to KJ071 (deletion of mgsA followed by metabolic evolution), PEP carboxykinase activity decreased by 92%, cell yield decreased by 45%, and succinate production decreased by 63%. From KJ071 to KJ073 (deletion of poxB followed by metabolic evolution), PEP carboxykinase activity, cell yield, and succinate production were restored to levels equivalent to KJ060.
These results demonstrate a tight coupling between high PEP carboxykinase activity and fermentative growth. The PEP carboxykinase activities of the best succinate-producing E. coli strains were even higher than in the succinate-producing rumen bacterium, Actinobacillus succinogenes (Table 2).
Transcriptional Regulation of Carboxylating Genes.
The increase in PEP carboxykinase activity correlated with an increase in pck mRNA abundance (Table 2 and Fig. S1A).Transcript levels for ppc, maeA, and maeB were essentially unchanged, whereas pck transcript abundance increased in KJ073 as compared with the wild type, in agreement with the increase in PEP carboxykinase activity (Table 2).
Sequencing the pck gene revealed no difference in the coding or terminator regions of the pck gene from KJ073. However, a single point mutation (G to A transition at position −64 relative to the ATG start codon) was found in the upstream region (Table 2). All six strains were sequenced to identify the origin of this mutation during strain improvement. Although PEP carboxykinase activity was 4-fold higher in KJ017 than in KJ012, the G to A mutation in KJ073 was absent. The pck mutation (denoted pck*) was present only in KJ060 and KJ073 (high PEP carboxykinase activity) and absent in KJ071. Loss of this mutation in KJ071 was accompanied by a 10-fold decrease in PEP carboxykinase activity to a level equivalent to that of KJ017.
Introduction of the pck* point mutation (G to A) into KJ017, and KJ071 increased PEP carboxykinase activity by 9-fold (Table S2). Restoring the wild-type pck gene (A to G) in strains KJ060 and KJ073 decreased PEP carboxykinase activity by almost 8-fold. Together, these results demonstrate that the increase in PCK activity that occurred during the metabolic evolution of KJ017 to produce KJ060 is caused by a single nucleotide change in pck (G to A at −64 relative to the ATG start). Absence of this mutation in KJ017 suggests that the initial 4-fold increase in PEP carboxykinase activity observed in this strain is caused by another, yet-to-be-defined change or changes. Apparently, recruitment of the native gluconeogenic PCK for fermentative succinate production resulted from multiple mutations that affected transcription of the pck gene.
Changes in pck Regulation During Metabolic Evolution of KJ017 from KJ012.
The 4-fold increase in PEP carboxykinase activity that occurred during the development of KJ017 from KJ012 did not result from a mutation in pck and may involve a transcriptional regulator. The Cra protein has been shown to activate the expression of pck in E. coli (22). However, no mutation was found in the cra gene or upstream region. Deleton of cra in KJ017 and KJ060 did not affect PEP carboxykinase activity (Table S2).
The csrA system has also been reported to regulate the level of pck and other genes involved in glucose metabolism by altering mRNA stability (23–25). However, no mutation was found in the sequences (Table S1) of genes involved in this regulatory system (csrA, csrB, csrC, csrD, uvrY, or barA).
E. coli PEP carboxykinase activity is subject to glucose catabolite repression (26). Two Crp-binding sites have been identified in the promoter (27), quite distant from the point mutation in KJ060 and KJ073. The glucose phosphotransferase system (PTS) is involved in the regulation of catabolite repression, and inactivating PTS could reduce the repression by increased cAMP level. Genes associated with catabolite repression (cyaA, crp) and glucose uptake by the phosphotransferase system (ptsH, ptsI, crr, ptsG) were sequenced (upstream region through terminator). Only one mutation was found, a frame-shift mutation in the carboxy-terminal region of ptsI (single base deletion at position 1,673) in strains KJ060, KJ071, and KJ073. As this deletion was absent in KJ017, it cannot be responsible for the initial 4-fold increase in PEP carboxykinase activity in this strain (Table S2). Deletion of ptsI in KJ017 and in KJ060 did not affect PEP carboxykinase activity.
Potential changes in the catabolite repression of pck were investigated further by growing E. coli ATCC 8739, KJ012, KJ017, and KJ060 aerobically in LB medium with and without 5% glucose (Table 3). PEP carboxykinase activity has been reported to be maximal during late exponential phase of growth (26), and this was confirmed in our strains. In both ATCC 8739 and the starting strain for metabolic evolution (KJ012), PEP carboxykinase activities were strongly repressed (>70%) by the presence of glucose. Glucose-mediated repression was reversed by the addition of cyclic-AMP. In contrast, PEP carboxykinase activity was 4-fold higher in KJ017 than in KJ012 and was unaffected by the addition of glucose, indicating a loss of catabolite repression. PEP carboxykinase activity was even higher in KJ060 (5-fold that of KJ017) during growth in the absence of glucose, and increased further when glucose was included. In KJ060 and KJ073, glucose catabolite repression of pck has been replaced with glucose activation. Transcript abundance of cyaA, crp, ptsG, ptsI, and crr were compared among these strains (Fig. S1 B and C). Transcripts for cyaA, crp, and ptsG were generally higher in KJ017, KJ060, KJ071, and KJ073 than in KJ012 and ATCC 8739. Elevated levels of these proteins could mask catabolite repression by increasing the level of cyclic-AMP. This increase in cAMP level could be caused by inactivation of the native glucose PTS system.
Table 3.
Effect of glucose and cAMP on PEP carboxykinase (pck) and β-galactosidase (lacZ) activities*
| Strain | PEP carboxykinase activity† |
β-Galactosidase activity† |
|||||
|---|---|---|---|---|---|---|---|
| 5% Glu | No Glu | ratio‡ | Glu + cAMP | 5% Glu | No Glu | Ratio‡ | |
| 8739 | 0.14 ± 0.03 | 0.48 ± 0.06 | 3.4 | 0.55 ± 0.04 | 0.19 ± 0.01 | 0.56 ± 0.03 | 3.0 |
| KJ012 | 0.15 ± 0.02 | 0.46 ± 0.05 | 3.2 | 0.74 ± 0.06 | 0.29 ± 0.02 | 0.71 ± 0.05 | 2.4 |
| KJ017 | 0.59 ± 0.04 | 0.62 ± 0.04 | 1.1 | 1.0 ± 0.08 | 0.53 ± 0.03 | 0.71 ± 0.04 | 1.3 |
| KJ060 | 5.57 ± 0.43 | 2.78 ± 0.32 | 0.5 | 4.25 ± 0.48 | 0.69 ± 0.03 | 0.75 ± 0.05 | 1.1 |
| XZ642 | 0.12 ± 0.01 | 0.11 ± 0.02 | 1.0 | 0.11 ± 0.01 | — | — | — |
| XZ643 | 0.12 ± 0.01 | 0.12 ± 0.02 | 1.0 | 0.12 ± 0.01 | — | — | — |
*Cultures were grown aerobically in shaken flasks with Luria broth (37 °C, 200 rpm) and harvested at half maximal density (≈1 g dry wt/L). Activity for β-galactosidase was not measured in strains XZ642 and XZ643.
†Activity is expressed as U· mg protein −1.
‡Ratio is that of the activity without glucose to the activity with 5% glucose.
The loss of catabolite repression in these mutants was not limited to pck expression. β-Galactosidase activity was reduced by half in ATCC 8739 and KJ012 during growth with glucose (Table 3). This activity is normally repressed by glucose but was relatively unaffected in KJ017 and KJ060, consistent with general loss of Crp-mediated glucose regulation.
Deletion of the crp gene in KJ012 and KJ017 reduced the level of PEP carboxykinase activity in the resulting strains (XZ642 and XZ643, respectively) to that of the glucose-repressed parent, ATCC 8739 (Table 3). In these crp-deleted strains, activity was not affected by the addition of glucose or cAMP. Thus Crp can be regarded as an essential regulatory element for the 4-fold increase in pck expression observed in KJ017 and its derivatives. This Crp-mediated 4-fold increase in pck expression, together with the 10-fold increase in expression associated with the upsteam pck mutation, can account for the changes in the levels of PEP carboxykinase activity observed during the development of KJ060 and KJ073.
Other genes associated with cAMP-CRP regulation were also sequenced (Table S1), such as the predicted adenylate cyclase ygiF (14), a transcriptional co-activator for Crp sxy (14, 28), the cAMP phosphodiesterase cpdA (14, 29), and the global regulator of carbohydrate metabolism mlc (14, 30). However, none contained a mutation in the upstream, coding, or terminator regions.
Effects of the ptsI Mutation.
A frame-shift mutation in ptsI (single base pair deletion of position 1,673 bp) was found to have occurred during the metabolic evolution of KJ060 from KJ017. To investigate the significance of this mutation, the carboxy-terminal 175 bp of ptsI was deleted in KJ017 and KJ060 to produce XZ613 and XZ615, respectively (Table S1). This ptsI deletion had no effect on growth and PEP carboxykinase activities in KJ060 as expected. Deletion of the ptsI carboxy-terminus in KJ017 to produce XZ613 resulted in an inability to grow in New Brunswick Scientific (NBS) mineral salts containing glucose, consistent with loss of the primary uptake system for glucose (PEP-dependent phosphotransferase system). After several days of incubation, cultures of XZ613 began to grow and are presumed to be derivatives in which an alternative glucose transporter has been induced or activated (21).
The ptsI mutation found in KJ060, KJ071, and KJ073 would be expected to inactivate the PEP-dependent phosphotransferase system for glucose. Alternative glucose uptake systems such as GalP have been shown to restore glucose uptake in pts mutants (13, 31). Expression of galP was increased by 5-fold to 20-fold in these improved strains (Fig. S1B) as compared with the KJ012 and ATCC 8739, with a smaller increase in glucokinase (glk). Results from the deletion of individual transport genes confirmed that GalP serves as the dominant glucose transporter in evolved strains (Table S3).
The direct involvement of GalP in glucose metabolism was confirmed by deleting individual transport genes (galP, ptsG, manX) from KJ073. Deletion of ptsG and manX did not affect succinate production, whereas deletion of galP dramatically reduced both succinate production and growth. Replacing the mutant ptsI gene in KJ060 with a functional wild-type gene (XZ616) to restore PEP-dependent glucose transport was also detrimental, dramatically reducing growth in NBS-glucose medium. Restoring function to this transport system is presumed to compete with PEP carboxykinase, decreasing the PEP pool available for redox balance, ATP production, and succinate production. Thus, the loss of the PEP-dependent phosphotransferase system for glucose can be viewed as a beneficial event during strain development, enabled by the presence of an alternative transport systems such as GalP (and glucokinase).
Discussion
Carbon fluxes at the PEP node serve to limit the amount of succinate produced for redox balance during the anaerobic fermentation of glucose (Fig. 1A). In addition, these fluxes must provide sufficient energy (ATP) for growth, maintenance, and precursors for biosynthesis. Rumen bacteria that produce succinate as the dominant product use an energy-conserving PEP carboxykinase for OAA production (Fig. 2D) (5, 6). Native strains of E. coli produce succinate as a minor product and use PEP carboxylase (ppc) (Fig. 2A). Unlike PEP carboxykinase, PEP carboxylase is essentially irreversible because of the energy loss associated with release of inorganic phosphate. Previous studies have shown that overexpression of the native ppc gene in E. coli resulted in higher specific succinate production (32) and higher specific growth rate caused by increased carboxylation of PEP to oxaloacetate (33). However, because PEP is required for the native glucose transport system, overexpressing ppc also decreased the rate of glucose uptake without significantly increasing succinate yield (34, 35). This failure of the native PEP carboxylase to increase succinate yields diverted most research attention to a new metabolic design, overexpression of the pyruvate carboxylase from Lactobacillus lactis or Rhizobium etli as the carboxylating step (11, 35) and overexpression of PEP carboxykinase from Actinobacillus succinogenes (6) for the development of industrial biocatalysts, rather than pursuing further work with the native repertoire of E. coli genes.
Native E. coli strains have three alternative carboxylation pathways (Fig. 2) that typically function only in the reverse direction for gluconeogenesis (4, 16–18, 36). All three would allow the conservation of energy from PEP as ATP. With succinate as the sole route for NADH oxidation, growth-based selection (metabolic evolution) resulted in strains KJ017, KJ060, KJ071, and KJ073 with improvements in growth (cell yield) and succinate production. In these strains, the gluconeogenic PEP carboxykinase (pck) was recruited to serve as the primary carboxylation activity by transcriptional activation. The recruitment of PEP carboxykinase as the primary pathway for succinate production in E. coli was surprising. Previous studies have shown that increased expression of E. coli and A. succinogenes pck had no effect on succinate production (6, 32). A recent study demonstrated that increased expression of E. coli pck was detrimental for growth in minimal medium, decreasing the growth rate, the rate of glucose metabolism, and the yield of succinate (37).
Increased expression of pck in E. coli resulted in increased carbon flow into the four-carbon intermediate OAA for succinate production (redox balance), increased succinate production, and increased the net production of ATP for growth and maintenance. At least three mutations contributed to increased expression of pck: (i) loss of Crp-mediated glucose repression; (ii) gain of glucose activation; and (iii) a single base change in the upstream region of pck. Each of these events provided a basis for selection through metabolic evolution by increasing the level of PEP carboxykinase, increasing the flow of carbon into succinate, and increasing the conservation of metabolic energy as ATP. The combined action of these genetic events resulted in high levels of PEP carboxykinase (>7,000 U·mg protein−1 in KJ060 and KJ073), equivalent to rumen bacteria that have evolved to produce succinate as the primary fermentation product (5).
Additional energy-conserving changes were found in strains KJ060, KJ071, and KJ073, which may be essential for the recruitment of PEP carboxykinase as the primary route for fermentative succinate production. The PEP-dependent phosphotransferase system is the primary glucose uptake system in native strains of E. coli. During transport, half of the PEP produced from glucose is used for uptake and phosphorylation, limiting metabolic options for redox balance and ATP production. The improved strains contained a frame-shift mutation in ptsI that inactivated this uptake system. Expression of galP encoding a proton symport, which can transport glucose, was increased by up to 20-fold. Increased expression of GalP and the native glucokinase can functionally replace the glucose phosphotransferase system by using ATP rather than PEP for phosphorylation (13, 31, 38). This exchange provides an energy-efficient mechanism to increase the pool size of PEP available for carboxylation and to facilitate redox balance using succinate (Fig. 1B). All improved strains directed more than half of glucose carbon into four-carbon products (malate plus succinate) and required more than half of the PEP for redox balance. Pyruvate can be converted to PEP by ATP with the formation of PPi and AMP, but energy is wasted by this process. Thus, the ptsI mutation and expression of galP increased the energy efficiency of metabolizing glucose to succinate, providing a growth advantage during metabolic evolution.
Eliminating alternative routes for NADH oxidation other than the succinate together with metabolic evolution with selection for improvements in growth resulted in succinate-producing strains of E. coli (Fig. 1B) that are the functional equivalents of succinate-producing rumen bacteria such as Actinobacillus succinogenes and Mannheimia succiniciproducens (5, 39–41). OAA is produced using an energy-conserving PEP-carboxykinase. PEP is conserved to eliminate the need for energy-expensive regeneration (2-ATP equivalents) by using glucose permeases (and glucokinase) rather than the PEP-dependent phosphotransferase system for glucose uptake. Note that among rumen bacteria producing other fermentation products, the phosphotransferase system is widely used for glucose uptake (40). The most promising E. coli strains for succinate production, KJ060 and KJ073, produced ≈700 mM succinate with a yield of 1.2 mol succinate per mol glucose (3), comparable to the best natural succinate-producing rumen bacteria, Actinobacillus succinogenes (20).
Rational design of metabolic pathways based on current metabolic network models is a common method for engineering microorganisms for producing target compound with maximum yield. However, this method is not always the best strategy because of our limited understanding of the complicated metabolic network and dynamic kinetics of each reaction. This study of succinate production demonstrates that metabolic evolution can provide an excellent alternative method for strain improvement, through which currently unpredictable reactions can be selected to expand cellular metabolic capability.
Energy-conserving strategies that improved succinate production from glucose in E. coli could also be applied to other important problems in strain engineering. Glycerol is becoming an inexpensive feedstock because of the global increase in bio-diesel production. Being more reduced than glucose, each glycerol could be converted to succinate and could maintain redox balance. However, no net ATP would be produced during glycerol metabolism to succinate using the native energy-wasting carboxylation activity, PEP carboxylase. This problem should be solved by using the energy-conserving PEP carboxykinase and should allow the net formation of 1 ATP per succinate. In addition, the carboxylation product OAA is an important intermediate in cell metabolism that serves as a precursor for many other important fermentation products such as malic acid, fumaric acid, aspartic acid, lysine, threonine, and methionine, among others. The energy conservation strategies illustrated by this study could be used to develop and improve biocatalysts for the production of many industrially important chemicals.
Materials and Methods
Strains, Media and Growth Conditions.
Strains used in this study are listed in Table S1. Strains KJ012, KJ017, KJ060, KJ071, and KJ073 are derivatives of E. coli ATCC 8739, previously developed for succinate production (Table S1) (3). During strain construction, cultures were grown aerobically at 30, 37, or 39 °C in Luria broth (10 g·l−1 Difco tryptone, 5 g·l−1 Difco yeast extract, and 5 g·l−1 NaCl) containing 2% (wt/vol) glucose or 5% (wt/vol) arabinose. Ampicillin (50 mg·l−1), kanamycin (50 mg·l−1), or chloramphenicol (40 mg·l−1) were added as needed.
Genetic Methods.
Chromosomal genes were deleted seamlessly without leaving segments of foreign DNA, as described previously (19, 42). Red recombinase technology (Gene Bridges GmbH, Dresden, Germany) was used to facilitate chromosomal integration. Plasmids and primers used for gene deletions and cloning are listed in Table S1.
Fermentation.
For fermentative succinate production, strains were grown without antibiotics at 37 °C in NBS mineral salts medium (43) supplemented with 10% (wt/vol) glucose and 100 mM potassium bicarbonate unless stated otherwise. Preinocula for fermentation were grown by transferring fresh colonies into a 250-ml flask (100 ml NBS medium, 2% glucose, 120 rpm). After 16 h (37 °C, 120 rpm), this culture was diluted into a small fermentation vessel (fleaker, 500-ml volume) containing 300 ml NBS medium (10% glucose, 100 mM potassium bicarbonate; magnetic stirrer 200 rpm) to provide an inoculum of 0.033 g cell dry wt (CDW)·l−1. Fermentations were automatically maintained at pH 7.0 by adding base containing additional CO2 (2.4 M potassium carbonate in 1.2 M potassium hydroxide). Fermentation vessels were not gassed.
Effects of glucose and cAMP on PEP carbyoxykinase and β-galactosidase activities were measured in aerobically grown cultures (shaken flask, 250 ml volume) with 25 ml of Luria broth medium (37 °C, 250 rpm). Cells were grown in unsupplemented Luria broth and in Luria broth supplemented with either 5% glucose or 5 mM cAMP.
Enzyme Assay.
Cells were harvested by centrifugation (8,000 g for 5 min at 4 °C) during mid-log growth, washed with cold 100 mM Tris-HCl (pH 7.0) buffer, and resuspended in the same buffer (1 ml). After cellular disruption using a Fast Prep-24 (MP Biomedicals, Solon, OH), preparations were clarified by centrifugation (13,000 g for 15 min). Protein was measured by the BCA method (Pierce, Rockford, IL) using BSA as a standard
PEP carboxylase activity was measured as described by Cánovas and Kornberg (44). PEP carboxykinase activity was measured as described by Van der Werf et al. (5). Malic enzyme activity (carboxylation direction) was measured as described by Stols and Donnelly (18). The malic enzyme assay method was unsuitable for measurement of MaeA activity in wild-type E. coli because of the presence of lactate dehydrogenase. The activity of β-galactosidase was measured as described by Miller (45). In all assays, 1 unit of activity represents the amount of enzyme required to oxidize or reduce 1 μmol of substrate per min.
Real-Time Reverse Transcriptase—Polymerase Chain Reaction Analysis.
Cells were harvested during mid-log phase of growth by centrifugation after chilling in an ethanol bath containing dry ice and stored at −80 °C in RNALater (Qiagen). RNA was purified by using RNeasy Mini columns (Qiagen) and digestion with DNaseI (Invitrogen). cDNA was prepared with SuperScript II (Invitrogen) using 50 ng total RNA as template. Samples were analyzed using a Bio-Rad iCycler with SYBR Green RT-PCR (Bio-Rad). Absence of genomic contamination was confirmed by testing uncopied RNA. Transcript abundance was estimated using genomic DNA as a standard, and expression levels were normalized to that of the birA gene (46).
Analysis.
Cell dry weight was estimated by measuring the optical density at 550 nm (OD550) using a standard curve. Organic acids and glucose were measured by high-performance liquid chromatography (HPLC) (42).
Supplementary Material
Acknowledgments.
This research was supported by grants from the U.S. Department of Energy (FG02–96ER20222), the Florida High Technology Corridor Council, and Myriant Technologies, LLC.
Footnotes
Conflict of interest statement: L.O.I. is a consultant (Chief Science Officer) and minor shareholder in Myriant Technologies (formerly BioEnergy International), licensee of this technology from the University of Florida. Other authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0905396106/DCSupplemental.
References
- 1.Zeikus JG, Jain MK, Elankovan P. Biotechnology of succinic acid production and markets for derived industrial products. Appl Microbiol Biotechnol. 1999;51:545–552. [Google Scholar]
- 2.McKinlay JB, Vieille C, Zeikus JG. Prospects for a bio-based succinate industry. Appl Microbiol Biotechnol. 2007;76:727–740. doi: 10.1007/s00253-007-1057-y. [DOI] [PubMed] [Google Scholar]
- 3.Jantama K, et al. Combining metabolic engineering and metabolic evolution to develop nonrecombinant strains of Escherichia coli C that produce succinate and malate. Biotechnol Bioeng. 2008;99:1140–1153. doi: 10.1002/bit.21694. [DOI] [PubMed] [Google Scholar]
- 4.Samuelov NS, Lamed R, Lowe S, Zeikus JG. Influence of CO2-HCO3- Levels and pH on growth, succinate production, and enzyme-activities of Anaerobiospirillum Succiniciproducens. Appl Environ Microbiol. 1991;57:3013–3019. doi: 10.1128/aem.57.10.3013-3019.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.VanderWerf MJ, Guettler MV, Jain MK, Zeikus JG. Environmental and physiological factors affecting the succinate product ratio during carbohydrate fermentation by Actinobacillus sp. 130Z. Arch Microbiol. 1997;167:332–342. doi: 10.1007/s002030050452. [DOI] [PubMed] [Google Scholar]
- 6.Kim P, Laivenieks M, Vieille C, Zeikus JG. Effect of overexpression of Actinobacillus succinogenes phosphoenolpyruvate carboxykinase on succinate production in Escherichia coli. Appl Environ Microbiol. 2004;70:1238–1241. doi: 10.1128/AEM.70.2.1238-1241.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee PC, Lee SY, Hong SH, Chang HN. Isolation and characterization of a new succinic acid-producing bacterium, Mannheimia succiniciproducens MBEL55E, from bovine rumen. Appl Microbiol Biotechnol. 2002;58:663–668. doi: 10.1007/s00253-002-0935-6. [DOI] [PubMed] [Google Scholar]
- 8.Lee SJ, Song H, Lee SY. Genome-based metabolic engineering of Mannheimia succiniciproducens for succinic acid production. Appl Environ Microbiol. 2006;72:1939–1948. doi: 10.1128/AEM.72.3.1939-1948.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fraenkel DG. In: Escherichia coli and Salmonella: Cellular and Molecular Biology. Neidhardt FC, et al., editors. Washington, DC: ASM Press; 1996. pp. 189–198. [Google Scholar]
- 10.Gokarn RR, et al. The physiological effects and metabolic alterations caused by the expression of Rhizobium etli pyruvate carboxylase in Escherichia coli. Appl Microbiol Biotechnol. 2001;56:188–195. doi: 10.1007/s002530100661. [DOI] [PubMed] [Google Scholar]
- 11.Sanchez AM, Bennett GN, San KY. Efficient succinic acid production from glucose through overexpression of pyruvate carboxylase in an Escherichia coli alcohol dehydrogenase and lactate dehydrogenase mutant. Biotechnol Prog. 2005;21:358–365. doi: 10.1021/bp049676e. [DOI] [PubMed] [Google Scholar]
- 12.Vemuri GN, Eiteman MA, Altman E. Effects of growth mode and pyruvate carboxylase on succinic acid production by metabolically engineered strains of Escherichia coli. Appl Environ Microbiol. 2002;68:1715–1727. doi: 10.1128/AEM.68.4.1715-1727.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Q, Wu C, Chen T, Chen X, Zhao X. Expression of galactose permease and pyruvate carboxylase in Escherichia coli ptsG mutant increases the growth rate and succinate yield under anaerobic conditions. Biotechnol Lett. 2006;28:89–93. doi: 10.1007/s10529-005-4952-2. [DOI] [PubMed] [Google Scholar]
- 14.Keseler IM, et al. EcoCyc: A comprehensive database resource for Escherichia coli. Nucleic Acids Res. 2005;33:D334–D337. doi: 10.1093/nar/gki108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Unden G, Kleefeld A. In: Escherichia coli and Salmonella: Cellular and Molecular Biology. Bock A, et al., editors. Washington, DC: ASM Press; 2004. Chapter 3.4.5. [Google Scholar]
- 16.Kao KC, Tran LM, Liao JC. A global regulatory role of gluconeogenic genes in Escherichia coli revealed by transcriptome network analysis. J Biol Chem. 2005;280:36079–36087. doi: 10.1074/jbc.M508202200. [DOI] [PubMed] [Google Scholar]
- 17.Oh MK, Rohlin L, Kao KC, Liao JC. Global expression profiling of acetate-grown Escherichia coli. J Biol Chem. 2002;277:13175–13183. doi: 10.1074/jbc.M110809200. [DOI] [PubMed] [Google Scholar]
- 18.Stols L, Donnelly MI. Production of succinic acid through overexpression of NAD(+)-dependent malic enzyme in an Escherichia coli mutant. Appl Environ Microbiol. 1997;63:2695–2701. doi: 10.1128/aem.63.7.2695-2701.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jantama K, et al. Eliminating side products and increasing succinate yields in engineered strains of Escherichia coli C. Biotechnol Bioeng. 2008;101:881–893. doi: 10.1002/bit.22005. [DOI] [PubMed] [Google Scholar]
- 20.Guettler MV, Jain MK, Rumler D. Process for making succinic acid, microorganisms for use in the process and methods of obtaining the microorganisms. US patent 5. 1996;505 004. [Google Scholar]
- 21.Karp PD, et al. Multidimensional annotation of the Escherichia coli K-12 genome. Nucleic Acids Res. 2007;35:7577–7590. doi: 10.1093/nar/gkm740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saier MH, Jr., Ramseier TM. The catabolite repressor/activator (Cra) protein of enteric bacteria. J Bacteriol. 1996;178:3411–3417. doi: 10.1128/jb.178.12.3411-3417.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Babitzke P, Romeo T. CsrB sRNA family: Sequestration of RNA-binding regulatory proteins. Curr Opin Microbiol. 2007;10:156–163. doi: 10.1016/j.mib.2007.03.007. [DOI] [PubMed] [Google Scholar]
- 24.Pernestig AK, et al. The Escherichia coli BarA-UvrY two-component system is needed for efficient switching between glycolytic and gluconeogenic carbon sources. J Bacteriol. 2003;185:843–853. doi: 10.1128/JB.185.3.843-853.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Suzuki K, et al. Regulatory circuitry of the CsrA/CsrB and BarA/UvrY systems of Escherichia coli. J Bacteriol. 2002;184:5130–5140. doi: 10.1128/JB.184.18.5130-5140.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goldie H. Regulation of transcription of the Escherichia coli phosphoenolpyruvate carboxykinase locus: Studies with pck-lacZ operon fusions. J Bacteriol. 1984;159:832–836. doi: 10.1128/jb.159.3.832-836.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ramseier TM, et al. The global regulatory protein FruR modulates the direction of carbon flow in Escherichia coli. Mol Microbiol. 1995;16:1157–1169. doi: 10.1111/j.1365-2958.1995.tb02339.x. [DOI] [PubMed] [Google Scholar]
- 28.Cameron AD, Redfield RJ. Non-canonical CRP sites control competence regulons in Escherichia coli and many other gamma-proteobacteria. Nucleic Acids Res. 2006;34:6001–6014. doi: 10.1093/nar/gkl734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Imamura R, et al. Identification of the cpdA gene encoding cyclic 3′,5′-adenosine monophosphate phosphodiesterase in Escherichia coli. J Biol Chem. 1996;271:25423–25429. doi: 10.1074/jbc.271.41.25423. [DOI] [PubMed] [Google Scholar]
- 30.Plumbridge J. Regulation of gene expression in the PTS in Escherichia coli: The role and interactions of Mlc. Curr Opin Microbiol. 2002;5:187–193. doi: 10.1016/s1369-5274(02)00296-5. [DOI] [PubMed] [Google Scholar]
- 31.Yi J, Draths KM, Li K, Frost JW. Altered glucose transport and shikimate pathway product yields in E. coli. Biotechnol Prog. 2003;19:1450–1459. doi: 10.1021/bp0340584. [DOI] [PubMed] [Google Scholar]
- 32.Millard CS, Chao YP, Liao JC, Donnelly MI. Enhanced production of succinic acid by overexpression of phosphoenolpyruvate carboxylase in Escherichia coli. Appl Environ Microbiol. 1996;62:1808–1810. doi: 10.1128/aem.62.5.1808-1810.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Farmer WR, Liao JC. Reduction of aerobic acetate production by Escherichia coli. Appl Environ Microbiol. 1997;63:3205–3210. doi: 10.1128/aem.63.8.3205-3210.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chao YP, Liao JC. Alteration of growth yield by overexpression of phosphoenolpyruvate carboxylase and phosphoenolpyruvate carboxykinase in Escherichia coli. Appl Environ Microbiol. 1993;59:4261–4265. doi: 10.1128/aem.59.12.4261-4265.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gokarn RR, Eiteman MA, Altman E. Metabolic analysis of Escherichia coli in the presence and absence of the carboxylating enzymes phosphoenolpyruvate carboxylase and pyruvate carboxylase. Appl Environ Microbiol. 2000;66:1844–1850. doi: 10.1128/aem.66.5.1844-1850.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hong SH, Lee SY. Metabolic flux analysis for succinic acid production by recombinant Escherichia coli with amplified malic enzyme activity. Biotechnol Bioeng. 2001;74:89–95. doi: 10.1002/bit.1098. [DOI] [PubMed] [Google Scholar]
- 37.Kwon YD, Lee SY, Kim P. A physiology study of Escherichia coli overexpressing phosphoenolpyruvate carboxykinase. Biosci Biotechnol Biochem. 2008;72:1138–1141. doi: 10.1271/bbb.70831. [DOI] [PubMed] [Google Scholar]
- 38.Flores N, et al. Growth recovery on glucose under aerobic conditions of an Escherichia coli strain carrying a phosphoenolpyruvate: Carbohydrate phosphotransferase system deletion by inactivating arcA and overexpressing the genes coding for glucokinase and galactose permease. J Mol Microbiol Biotechnol. 2007;13:105–116. doi: 10.1159/000103602. [DOI] [PubMed] [Google Scholar]
- 39.Kim TY, et al. Genome-scale analysis of Mannheimia succiniciproducens metabolism. Biotechnol Bioeng. 2007;97:657–671. doi: 10.1002/bit.21433. [DOI] [PubMed] [Google Scholar]
- 40.Martin SA. Nutrient transport by ruminal bacteria: A review. J Anim Sci. 1994;72:3019–3031. doi: 10.2527/1994.72113019x. [DOI] [PubMed] [Google Scholar]
- 41.McKinlay JB, Shachar-Hill Y, Zeikus JG, Vieille C. Determining Actinobacillus succinogenes metabolic pathways and fluxes by NMR and GC-MS analyses of 13C-labeled metabolic product isotopomers. Metab Eng. 2007;9:177–192. doi: 10.1016/j.ymben.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 42.Zhang X, Jantama K, Moore JC, Shanmugam KT, Ingram LO. Production of L-alanine by metabolically engineered Escherichia coli. Appl Microbiol Biotechnol. 2007;77:355–366. doi: 10.1007/s00253-007-1170-y. [DOI] [PubMed] [Google Scholar]
- 43.Causey TB, Shanmugam KT, Yomano LP, Ingram LO. Engineering Escherichia coli for efficient conversion of glucose to pyruvate. Proc Natl Acad Sci USA. 2004;101:2235–2240. doi: 10.1073/pnas.0308171100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cánovas JL, Kornberg HL. Phosphoenolpyruvate carboxylase from Escherichia coli. Methods Enzymol. 1969;13:288–292. [Google Scholar]
- 45.Miller JH. A Short Course in bacterial genetics. NY: Cold Spring Harbor Laboratory Press; 1992. pp. 71–74. [Google Scholar]
- 46.Jarboe LR, Hyduke DR, Tran LM, Chou KJY, Liao JC. Determination of the Escherichia coli S-nitrosoglutathione response network using integrated biochemical and systems analysis. J Biol Chem. 2008;283:5148–5157. doi: 10.1074/jbc.M706018200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.