Abstract
PURPOSE OF REVIEW:
The intent of this article is to review trends in multi-center neuroimaging trials and their value for research and implications for clinical treatment.
RECENT FINDINGS:
The rise in availability of magnetic resonance imaging for detecting disorders in the living brain has made it an attractive technology for assessing neural structure and function in a number of prominent diseases. Geographic factors underlying diseased populations coupled with complementary neuroimaging research programs have led to an increase in multi-center neuroimaging trials and consortia. Neuroimaging has become a major focus for multi-institutional research into a) progressive changes in brain architecture, b) proxy biomarkers of treatment response, and c) the effects of disease on patterns of cognitive activation and connectivity. Notable consortia and research trial studies have focused on Alzheimer's disease, pediatric brain cancer, and fetal alcohol syndrome, in addition to multi-institutional collaborative programs for mapping the normal brain. Such large-scale efforts necessitate close coordination of image data collection protocols, ontology development, computational requirements, concerted data archiving and sharing.
SUMMARY:
Multi-center neuroimaging trials, consortia, and collaboratives enable the acquisition of large-scale, purpose-driven datasets that can then be used by the broader community to model and predict clinical outcomes as well as guide clinicians in selecting treatment options for neurological disease.
Keywords: Neuroimaging, Multi-Center, Clinical Trials, Consortia, Collaboration, Data Sharing
Introduction
With the advent of modern neuroimaging technologies in the early 1990's, the proliferation of neuroimaging devices in neurological research centers around the world has made it possible for many centers to conduct leading edge brain research into many clinical syndromes [1,2,3]. However, it has also become recognized that investigators from across centers can pool efforts, resources, and data toward large-scale research collaborative programs aimed at refining understanding of frequently heterogeneous diseases of brain structure and function. Studies that could not be conducted in any one site, due to lack of subject numbers or given expertise, become possible in such collaborative efforts. Archives of raw and processed datasets may be examined jointly by investigators at federated centers or by unaffiliated researchers. With access to the large-scale research outcomes, clinicians can utilize this new information to direct patient treatment plans.
Multi-center collaborations, however, require careful coordination and division of labor, compliance with HIPAA regulations on data anonymization, etc. Additional considerations involve how differences in scanner make, model, manufacturer, field strength, field homogeneities, slew rates, and image reconstruction routines can influence the data and potentially lead to conflicting results. Computational resources are also important for how to maximize throughput toward results that can be put directly into clinical practice. In this article, we discuss several such multi-center efforts, note key clinical and research outcomes, and comment on the importance of careful coordination of such programs. Lastly, we discuss the potential that lay in large-scale archives of these data and how they may be potentially mined to identify unanticipated outcomes worthy of additional collaborative effort.
Rising Frequency of Multi-site Trials
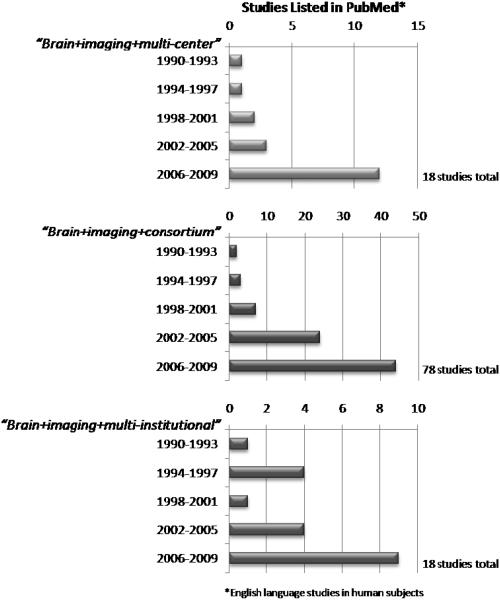
While an increasing number of individual research centers now enjoy the latest in neuroimaging technology in the form of Positron Emission Tomography (PET) and high-field magnetic resonance imaging (MRI), researchers have been quick to recognize the importance of pooling effort and resources toward clinically-relevant goals that could not be achieved at one center alone. The numbers of established multi-site clinical trials, collaborations, and research consortia has dramatically increased since the first studies using modern imaging methods first appeared (Figure 1). These efforts seek to leverage complementary approaches toward mapping brain function and structure and relating these to genomic influences, psychopharmacological interventions, and treatment outcomes.
Figure 1.
Multi-center clinical trials and research collaborations have resulted in increasing methods, primary research, and clinical outcome studies appearing the peer-reviewed literature. Exploration of PubMed citations, grouped into three year periods, reveals consistent increases in the numbers of publications from such efforts since the early 1990's. Examples of several leading multi-center programs are listed in Table 1.
Advantages of Multi-site Trials and Collaborations
Despite the fact that many neuroimaging centers may be well equipped to carry out focused studies individually, the greatest benefit in multi-center efforts is the ability to, in principle, obtain more neuroimaging data per unit time across a wider variety of the patient population in question. This provides a great number of clinical exemplars while also attempting to at once control issues of selectivity of patients while being able to more precisely examine patients having varying etiology. In the sense of greater data acquisition, an analogy exists with parallel computing where with multiple-CPUs it is possible to increase throughput manifold in the same amount of time. Other major advantages of multi-site neuroimaging trials include: 1) as a tool for cognitive neuroscience research into patterns of normal and altered brain function; 2) for prediction of which normal or slightly impaired individuals will develop alterations in brain anatomy and over what time period; 3) for early identification of neurological disease in at risk individuals (e.g. test sensitivity) and separation of certain forms of disease from others (e.g. disease specificity); 4) for monitoring the progression of disease across a geographically, culturally, and environmentally diverse population; and 5) for monitoring response to interventions and therapies. Functional imaging methods (fMRI, PET, and SPECT) have tended to be best suited toward tracking symptomatic therapy response, whereas anatomic imaging (structural MRI and diffusion tensor imaging) and amyloid PET [4] tend to be best suited toward disease modulation studies [5 **]. The additional potential for human neuroimaging with respect to pharmacological intervention or candidate gene studies is enormous [6 *]. By taking advantage of a wider variety of patient types, etiologies, and range of symptoms, multi-site studies can be better representative of the larger patient population with greater generality for population-level atlases of brain structure/function and, by their association with clinical variables, suggest treatment options with the widest possible efficacy. For the clinician, this translates into the greater ability to consider treatment options for patients against a number of demographic, case history, and etiological backgrounds.
Challenges in Multi-site Trial Coordination
While being geographically distributed is, from a patient demographics point of view, a principal strength of multi-center efforts, a number of challenges exist in coordinating such programs. These include issues concerning variation in scanner technologies across centers in which even the same make and model MRI system may demonstrate differing field inhomogeneity effects that can influence how data might be interpreted [7]. Even subtle distortions in scanner magnetic field homogeneity across sites can form an unwanted source of variance that must be controlled through the use of standardized phantom studies and strict quality assurance of imaging protocols [8 *]. Trial considerations also involve patient confidentiality and how investigators can have access to patient data and private health information stored at other institutes while also being compliant with Health Insurance Portability and Protection Act (HIPAA) privacy regulations [9]. At the same time, methods for ensuring seamless computational access for researchers to digital archives of neuroimaging and clinical data for use in comparative analysis and modeling necessitate careful examination. Factors include: 1) site-to-site networking requirements [10]; 2) user authentication protocols [11]; 3) means for rapid design and deployment of analysis workflows [12], data ontological definition, description, and data management [13], and large-scale data storage capabilities [14 *]. Regularly scheduled “all-hands” meetings supplemented with conference calls and web-meetings are necessary for measuring progress toward the stated aims of the trial. Multi-center trials, especially involving neuroimaging, are expensive, involving multiple investigators, imaging center hourly rates for scanning, travel, and other associated costs. Despite these challenges, the recent increase in the numbers of such multi-site trials and collaborations is indicative that policy makers and funding organizations recognize their benefits and understand them to be a worthy scientific, clinical, and financial investment.
Successful Examples in Multi-site Neuroimaging
Table 1 [15,16,17,18,19,20,21,22,23,24,25 **,26,27,28,29,30 *,31,32] describes a brief list of several ongoing or newly developed multi-site trials or collaborations working toward applying neuroimaging methods for mapping the normal brain and examining the brain in various neurological diseases. Here we summarize several major neuroimaging consortia:
Table 1.
Neuroimaging Trials or Collaborative Efforts Involving Multiple Research Sites
| Multi-Site Trial | Participating Institutions |
Subject Population |
Imaging Modalities |
URL | Citations |
|---|---|---|---|---|---|
|
Alzheimer's Disease Neuroimaging Initiative (ADNI) |
UCSF, NCIRE, VA – U. California, San Diego; Arizona State U.; Banner Good Samaritan Med Ctr; U. California, Davis; Mayo Clinic, Rochester, Minnesota; U. California, Berkeley |
Mild cognitive impairment; Alzheimer's disease; normal control subjects |
sMRI, PET |
www.adni-info.org www.loni.ucla.edu/ADNI/index.shtml |
[15,16,17,18,19,20,21,22,23] |
|
Collaborative Initiative on Fetal Alcohol Spectrum Disorder (CIFASD) |
San Diego State U.; Indiana U. Sch. Med.; U. California, San Diego; U. California, Los Angeles; U. New Mexico; Texas A&M; Indiana U.-Purdue U. Sch. Sci.; Ukrainian-American Birth Defects Program; U. North Carolina at Chapel Hill; Harvard; U of California, Davis; Folkhälsan Research Center- Helsinki; U. Cape Town, South Africa; Emory U.; Moscow Inst. Obs. and Gyn. |
Fetal alcohol syndrome children (with and without facial dysmorphia), normal control subjects |
sMRI, fMRI, DTI |
www.cifasd.org | [24] |
|
International Consortium for Brain Mapping (ICBM) |
U. California. Los Angeles; Montreal Neurological Inst.; U. Texas, San Antonio; Vogt Inst., Jülich, Germany |
Normal subjects; Methamphetamine patients; HIV; children; and Williams Syndrome |
sMRI; MRS; MRA; DTI/DWI; PET |
www.loni.ucla.edu/ICBM/ | [25 **,26,27] |
|
Functional Imaging Research in Schizophrenia Testbed (FIRSTBIRN) |
U. California, Irvine; U. California, San Diego; U. California, Los Angeles; Stanford U.; U. New Mexico; U. Minnesota; Massachusetts General Hosp.; Brigham and Women's Hosp.; Duke U.; U. North Carolina; U. Iowa |
Schizophrenia spectrum disorders, normal control subjects |
sMRI; fMRI; DTI |
www.nbirn.net/research/function | [28,29,30 *] |
|
Pediatric Brain Tumor Consortium (PBTC) |
Children's Memorial Hosp., Chicago; Duke U. Med. Ctr.; Texas Children's Cancer Ctr.; St. Jude Children's Research Hosp.; The Children's Hosp. of Philadelphia; Children's Hosp. of Pittsburgh; U. California, San Francisco U. California, San Francisco; Children's National Medical Ctr Children's National Medical Ctr.; NCI. |
Children with primary brain tumors |
sMRI; PET | www.pbtc.org | [31,32] |
Alzheimer's Disease Neuroimaging Initiative (ADNI)
The pathophysiologic process leading to neurodegeneration in Alzheimer's disease (AD) is thought to begin long before clinical symptoms develop. Existing therapeutics for AD improve symptoms, but increasing efforts are being directed toward the development of therapies to impede the pathologic progression of the disease. Although these medications must ultimately demonstrate efficacy in slowing clinical decline, there is an essential need for biomarker trials that will indicate whether a candidate disease-modifying therapeutic agent is actually altering the underlying degenerative process. A number of in vivo neuroimaging techniques, which can reliably and noninvasively assess aspects of neuroanatomy, chemistry, physiology, and pathology, hold promise as biomarkers. These neuroimaging measures appear to relate closely to neuropathological and clinical data, such as rate of cognitive decline and risk of future decline. As this work has matured, it has become clear that neuroimaging measures may serve a variety of potential roles in clinical trials of candidate neurotherapeutic agents for AD, depending in part on the question of interest and phase of drug development.
The highly successful NIA-funded Alzheimer's Disease Neuroimaging Initiative (ADNI) program was established to increase knowledge of the mechanisms of AD through the use of neuroimaging - thereby informing the development of treatment strategies aimed at slowing down or preventing neuronal death. ADNI has been instrumental in helping to identify clinical, neuroimaging, and biomarker outcome measures and longitudinal changes and the prediction of disease transitions. In particular, this has included neurodegeneration: the clinically and pathologically heterogeneous disease entity associated with slowly progressive neuronal loss in different anatomical and functional systems of the brain. Owing to increasing knowledge about the mechanisms leading to neurodegeneration as a result of ADNI, the development of treatments able to modify the neurodegenerative process will be easier. While more research is clearly needed to determine the continued value of newer neuroimaging modalities, i.e. diffusion, perfusion and functional MRI and MR spectroscopy, for clinical trials with neuroprotective drugs, the ADNI project can be considered to have been a highly successful first step in large-scale neuroimaging and the sharing of that information with a larger community studying the efficacy of leading-edge treatment.
Additionally, researchers at NIA-funded Alzheimer's Disease Research Centers (ADRC) have worked to translate research advances into improved diagnosis and care for Alzheimer's disease (AD) patients while, at the same time, focusing on the program's long-term goal--finding a way to cure and possibly prevent AD. Although each center has its own unique area of emphasis, a common goal of the ADCs is to enhance research on AD by providing a network for sharing new ideas as well as research results. Collaborative studies draw upon the expertise of scientists from many different disciplines. Some ADCs have satellite facilities, which offer diagnostic and treatment services and research opportunities in underserved, rural, and minority communities. The National Alzheimer's Coordinating Center at the University of Washington in Seattle, WA coordinates data collection and fosters collaborative research among the ADCs. Additionally, the National Cell Repository for Alzheimer's Disease maintains a database of family histories and medical records and provides genetic researchers with cell lines and/or DNA samples. Several of the ADRC's also conduct neuroimaging investigations but these data may not necessarily be widely available to researchers outside of each individual center.
Brain Function and Morphometry
The FIRST [29] and Morphometry [33] testbeds of the Biomedical Informatics Research Network (BIRN; www.nbirn.net) are multi-site projects funded by the NCRR and NIH to develop the methods for data collection, combination, and sharing from imaging protocols, for the following goals: 1) standardized calibration of equipment and imaging activation paradigms using geometric and human phantoms; 2) collection of MRI and/or fMRI data using a standardized protocol on clinical populations at different sites, while including the added value of each site's own methods; 3) combining MRI, fMRI, clinical and behavioral data into a federated database, leading to a deeper understanding of the functional neuroanatomy of the underlying disease than would be possible with any individual site's data. The eleven sites involved in the projects have people dedicated to the purposes of collecting calibration imaging data, developing analysis methods, determining experimental paradigms where needed, developing robust and expandable data storage and retrieval methods, populating a virtual data grid, and designing a searchable federated database of MRI and clinical data from multiple sites. Results from their examinations suggest that even previously obtained (legacy) structural data from across the multiple sites can be pooled to investigate questions of scientific interest [33]. In particular, statistical analyses suggested that a mixed-effects model employing site as a random effect best fits the data, accounting for site-specific effects while taking advantage of expected comparability of age-related effects. In combining samples from three research sites, significant age-related decline of hippocampal volume and right-dominant hippocampal asymmetry were detected in healthy elderly controls. These findings support the feasibility of combining legacy as well as new data from across multiple sites toward investigating novel scientific questions.
Fetal Alcohol Spectrum Disorder (FASD)
A recent neuroimaging thrust has been added to the Collaborative Initiative on Fetal Alcohol Spectrum Disorders (CIFASD). One of the driving goals of the entire CIFASD has been to determine if innovative techniques can be used to identify brain alterations, neurobehavioral deficits and facial characteristics and relationships between these variables to help define prenatal alcohol spectrum disorders (FASD). To help address this, brain mapping using high-resolution structural and functional MRI collected cross-sectionally and longitudinally from 80 FASD children evaluated across three distributed, multi-cultural data collection sites (San Diego, Los Angeles, and Cape Town). Their proposed longitudinal project seeks to highlight how an integrated approach relating neurobehavioral, functional and structural brain imaging data, and measures of facial (dis)morphology might yield important new insights on the complex nature of brain-behavior interactions and how they are altered by prenatal exposure to alcohol. Ultimately, this project, armed with neuroimaging outcomes, will enhance the capability for definitive FASD diagnoses that, in turn, will help clinicians manage and treat neurobehavioral deficits and associated secondary disabilities in FASD children.
Pediatric Brain Tumor
The Pediatric Brain Tumor Consortium (PBTC) was formed by the National Cancer Institute (NCI) in 1999 to improve the treatment of primary brain tumors in children. The PBTC's primary objective has been to rapidly conduct novel phase I and II clinical evaluations of new therapeutic drugs, new biological therapies, treatment delivery technologies and radiation treatment strategies in children from infancy to 21 years of age with primary central nervous system (CNS) tumors. They develop and coordinate innovative neuroimaging techniques and their use in early diagnosis of pediatric brain cancer [31], with resulting neuroimaging research being used to evaluate new treatment response criteria and the use of neuroimaging methods to understand regional brain effects [32,34].
While this is not an exhaustive list of ongoing multi-site efforts, it represents several examples of where neuroimaging has become a necessary and valuable part of collaborative research programs in the US. It might not be surprising that the successes of these and other programs have been replicated in countries around the world [35,36 *] with interest rising in international collaborative efforts toward mapping the brain and its disorders [37 *,38].
Future Needs
Throughout the coming decade of human brain imaging research, given the trends toward collaborative effort across multiple institutions, we can expect to see still more multi-site trials focused on particular neurological diseases and disorders. One such disease domain that is particularly poised for a multicenter collaboration is that of HIV and AIDS. In August 2008, the Center for Disease Control (CDC) published the first national HIV incidence of new HIV infection estimates using new technology and methodology that more directly measure the number of new HIV infections in the United States [39]. Their new analyses showed that in 2006, an estimated 56,300 new HIV infections occurred - a number that is considerably higher than the previous estimate of 40,000 annual new infections. However, though considerable progress has been made toward understanding the clinical response to various drugs designed to treat HIV/AIDS [40], the examination of the long term effects of the virus in the brain and its resistance to drugs via brain mapping technologies has only recently begun [41,42]. Whereas multi-center efforts have focused on magnetic resonance spectroscopic (MRS) neuroimaging in HIV/AIDS [43], a more broadly-based effort employing fMRI of cognitive functional changes, structural imaging of altered cortical thickness and other parameters, MRS, arterial spin labeling (ASL), as well as white matter imaging is sorely needed for assessing the effects of HIV on brain morphometry and function as well as the efficacy of extant and emerging HIV therapies.
While the potential for clinical and treatment understanding is enhanced in multi-site neuroimaging, considerable attention must be afforded to how such data might be efficiently organized, processed, and, firstly, made available to researchers at participating centers and then openly as a resource from which the entire neurological community might draw. The Laboratory of Neuro Imaging (LONI) has, for many years [44], been at the forefront of 1) providing the computational resources specifically tailored to support multi-center efforts [26,45], 2) providing a large-scale Image Data Archive (IDA) for use in the secure databasing of clinical research neuroimaging data [46], with 3) long term storage capabilities for petabyte levels of imaging study data. Still with advances in neuroimaging data acquisition methods that obtain more data per unit time, thereby making data computation considerations more serious, careful attention must be given to large-scale distributed Grid computing approaches [47] and their use in processing ever larger quantities of data. The importance of neuroimaging informatics cannot be overstated in the context of multi-center collaborations and will be an active area of work that will help to maximize the success of any neurological research domain using neuroimaging data.
Evaluation of Multi-Site Outcomes
For many clinicians, interpreting the outcomes from multi-site collaboratives can be challenging given the variety of neuroimaging terms being used, statistical methods being reported, and the sometimes lofty claims reported to justify the importance of the collaboration itself. Several factors are important to look for in evaluating the strengths of multi-site trials and the outcomes from research collaboratives: 1) Identifying whether the study has published or is using openly available human imaging protocols; 2) site-to-site quality assurance using standardized brain imaging phantoms; 3) whether the sites involved are using the same or differing scanner platforms; 4) performing a critical examination of whether the data were specifically obtained for the purposes of the study in question or if it is apparent that they were selected from a database for a secondary analysis; and 5) clear and understandable experimental goals and their relation to the multi-site effort should be articulated in the article. Each of these considerations will put the work into the overall context of the multi-site effort and guide the clinician in evaluating its quality, impact, and relevance to the patient-level (Table 2). Though multi-center efforts can frequently provide highly useful recommendations for the individual clinician, careful examinations of study quality and relevance remain important considerations before embarking on any new treatment programs based on their results.
Table 2.
Multi-Site Neuroimaging Evaluation Criteria
| Consideration | Factors | Rationale | Example |
|---|---|---|---|
| Study Description |
Has clear relationship to focus of multi-site effort. |
Some studies simply use the data but may only have peripheral importance to the main goals of the consortium effort. |
An a priori hypothesis involving a comparison between patients and controls during an fMRI task of complex cognition versus a methods paper describing a new analytic processing approach using existing data obtained during another study. |
| Technological Aspects |
Scanner make, model, field strength, and other factors that could vary across sites. |
Even the same make and model scanner can have considerable variability between related scans. This is compounded across scanner manufacturers. |
Data from one center in the study comes from a 1.5 Tesla scanner while data from another was obtained using a 3T Philips system. |
| Protocol Description |
Uses published or openly available image acquisition protocols. |
Imaging protocols are best when they have been subjected to peer-review by experts in pulse-sequence design and are openly available for use by the community. |
The authors report using a proprietary scan acquisition sequence, thus making it hard to validate results against the appropriateness of pulse- sequence parameters. |
| Imaging Quality Assurance |
Data quality across centers is maintained through the regular scanning of standard MR phantoms. |
Scanner distortions accumulate with time and the use of phantoms helps to ensure these are held to a minimum and differences in quality can be compared/equated across centers. |
One center reports using the BIRN agar-filled phantom to calibrate its data while another uses the ACR standard phantom. |
| Statistical Aspects |
Data are reported using well known and validated statistical packages/algorithms. |
While novel analytic approaches are worthwhile, statistical analyses using well understood and cited approaches will lend confidence to their interpretation and meaning. |
The authors report using Statistical Parametric Mapping v. 5 (SPM5) to analyze their fMRI data versus reporting results coming from software they wrote themselves. |
| Graphical Depiction of Results |
Figures from the studies show the results of neuroimaging comparisons clearly and concisely. |
Multi-site studies often include investigators expert in data visualization. However, many brain renderings are highly stylized with heavy doses of photo editing software used to embellish them. Care is required to keep “pretty pictures of the brain” in perspective. |
An overlay of significant BOLD activity on a T1 anatomical image versus interpolated false-color activation maps painted onto flattened surface representations of the cortex. Data from an individual patient may not appear the same. |
| Caveats and Alternatives |
The authors are forthcoming concerning limitations of their study or alternative explanations of results |
The experimental design may have confounds and the data may be open to multiple interpretations. This will have implications for extrapolation to the patient-level. |
The authors report anatomical changes in patients relative to controls but the selection of controls may have been biased across centers, thereby resulting in the reported effect. |
| Implications for the Patient |
In reviews of the multi-center effort, the PIs provide a perspective on what their results, findings, and outcomes mean for the treatment considerations of individual patients. |
Without eventually bring results into the context of the individual patient, multi-site efforts can fail to provide the information needed to guide treatment, suggest new clinical trials, or even justify the importance of the multi-center effort. |
The multi-center effort is, in and of itself, held up as a justification for the multi-center effort. |
Discussion
On a domestic or international scale, multi-site neuroimaging efforts offer the promise for gaining insights into major brain disease and the characterization of normal structure and function. Multi-site trials are an important element in the study of a disease or the process of evaluating an intervention. Linking together multiple sites facilitates the recruitment of large samples that yield high statistical power for both main analyses as well as secondary analyses of subgroups. Generalizability of results to the level of the population also is maximized. Because data come from multiple sites, investigators can explore how a treatment's effects vary across geographical diverse sites and how such variation relates to site characteristics, cultural and socio-economic factors. Such information can directly inform clinical decision making at the level of the patient and guide the selection of treatment options. Multi-site trials are costly, however, so the decisions that must be made, such as the population which to study and the expense of gathering neuroimaging data, have particular importance for policy makers and site PIs.
Many authors have noted how neuroimaging technologies has emerged over the past twenty years and changed forever how we view the brain in health and disease across temporal and spatial dimensions [48,49 *,50]. But, clearly, neuroimaging as a research site-specific tool cannot begin to fully probe the subtleties of diseases that may vary culturally and geographically, whose effects may vary by age or gender, or that may result from a highly variable exposure to environmental or pharmacological mechanisms. Leveraging expertise and technologies from across distributed institutions, with inherent variation in technological infrastructural, theoretical, and clinical emphasis is valuable toward building data resources of unprecedented size and scope. These may be mined and modeled to produce high-resolution maps of disease states in the brain and their responses to clinical intervention or treatment.
Coincident with developments in brain imaging technology and analysis approaches, the advantages for multi-site neuroimaging endeavors have resulted in a steadily increasing number of multi-site efforts in neurology and the brain sciences. Moving forward, we expect multi-site efforts to form a mainstay of major scientific exploration of the normal and diseased brain.
Conclusion
Multi-site efforts toward mapping the human brain in health and disease will continue to grow over the next decade. These important efforts will link the intellectual talent and resources of leading research centers toward better understanding of primary brain diseases, cancer, as well as the normal brain in development and aging. With more data being obtained with a view toward greater spatiotemporal precision, the informatics of neuroimaging will be an important consideration. Finally, these efforts are imperative for guiding treatment recommendations for neurological disorders domestically and internationally as well as at the level of the individual patient. Multi-center collaborations can be expected to strengthen understanding of brain diseases that affect all walks of life, all ages, and all cultures. This helps to translate neuroimaging trial outcomes directly into clinical applications.
Acknowledgements
The work was possible with support received from the National Center for Research Resources (NCRR; U54RR021813). The authors also wish to recognize the efforts of the members of the Laboratory of Neuro Imaging (LONI) and their collaborators.
References
Papers of particular interest have been highlighted as:
* of special interest
** of outstanding interest
- 1.Malhi GS, Lagopoulos J. Making sense of neuroimaging in psychiatry. Acta Psychiatr Scand. 2008;117:100–117. doi: 10.1111/j.1600-0447.2007.01111.x. [DOI] [PubMed] [Google Scholar]
- 2.Dickerson BC. Advances in functional magnetic resonance imaging: technology and clinical applications. Neurotherapeutics. 2007;4:360–370. doi: 10.1016/j.nurt.2007.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wishart HA, Saykin AJ, McAllister TW. Functional magnetic resonance imaging: emerging clinical applications. Curr Psychiatry Rep. 2002;4:338–345. doi: 10.1007/s11920-002-0081-y. [DOI] [PubMed] [Google Scholar]
- 4.Small GW, Kepe V, Ercoli LM, Siddarth P, Bookheimer SY, Miller KJ, Lavretsky H, Burggren AC, Cole GM, Vinters HV, et al. PET of brain amyloid and tau in mild cognitive impairment. N Engl J Med. 2006;355:2652–2663. doi: 10.1056/NEJMoa054625. [DOI] [PubMed] [Google Scholar]
- 5**.Chertkow H, Black S. Imaging biomarkers and their role in dementia clinical trials. Canadian Journal of Neurological Science. 2007;34:77–83. doi: 10.1017/s031716710000562x. An excellent paper describing the importance of biomarkers obtained from or measured using neuroimaging and their value in clinical trials. [DOI] [PubMed] [Google Scholar]
- 6*.Hariri AR, Weinberger DR. Imaging genomics. Br Med Bull. 2003;65:259–270. doi: 10.1093/bmb/65.1.259. An impotant review of fMRI studies that have established important physiological links between functional genetic polymorphisms and robust differences in information processing within distinct brain regions such as Alzheimer's disease, schizophrenia, and anxiety disorders. This article highlights the power of fMRI in exploring the functional impact of genetic variation in psychiatric illness. [DOI] [PubMed] [Google Scholar]
- 7.Friedman L, Glover GH, Krenz D, Magnotta V. Reducing inter-scanner variability of activation in a multicenter fMRI study: role of smoothness equalization. Neuroimage. 2006;32:1656–1668. doi: 10.1016/j.neuroimage.2006.03.062. [DOI] [PubMed] [Google Scholar]
- 8*.Friedman L, Glover GH. Report on a multicenter fMRI quality assurance protocol. J Magn Reson Imaging. 2006 doi: 10.1002/jmri.20583. Temporal stability during an fMRI acquisition is essential because the blood oxygen level-dependent (BOLD) effects of interest are only a few percent in magnitude. Likewise, certain distortions in T1 anatomical images could be misinterpreted as clinically relevant. Standardization of protocols across centers is an element of multi-site trials that cannot be over stated. [DOI] [PubMed] [Google Scholar]
- 9.Zhou Z, Liu BJ. HIPAA compliant auditing system for medical images. Comput Med Imaging Graph. 2005;29:235–241. doi: 10.1016/j.compmedimag.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 10.Keator DB, Grethe JS, Marcus D, Ozyurt B, Gadde S, Murphy S, Pieper S, Greve D, Notestine R, Bockholt HJ, et al. A national human neuroimaging collaboratory enabled by the Biomedical Informatics Research Network (BIRN) IEEE Trans Inf Technol Biomed. 2008;12:162–172. doi: 10.1109/TITB.2008.917893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smith K, Jajodia S, Swarup V, Hoyt J, Hamilton G, Faatz D, Cornett T. Enabling the sharing of neuroimaging data through well-defined intermediate levels of visibility. Neuroimage. 2004;22:1646–1656. doi: 10.1016/j.neuroimage.2004.03.048. [DOI] [PubMed] [Google Scholar]
- 12.Rex DE, Ma JQ, Toga AW. The LONI Pipeline Processing Environment. Neuroimage. 2003;19:1033–1048. doi: 10.1016/s1053-8119(03)00185-x. [DOI] [PubMed] [Google Scholar]
- 13.Temal L, Dojat M, Kassel G, Gibaud B. Towards an ontology for sharing medical images and regions of interest in neuroimaging. J Biomed Inform. 2008;41:766–778. doi: 10.1016/j.jbi.2008.03.002. [DOI] [PubMed] [Google Scholar]
- 14*.Wardlaw JM, Bath P, Sandercock P, Perry D, Palmer J, Watson G, Lloyd S, Geddes J, Farrall A. The NeuroGrid stroke exemplar clinical trial protocol. Int J Stroke. 2007;2:63–69. doi: 10.1111/j.1747-4949.2007.00092.x. NeuroGrid aims to develop and test grid technologies for collecting, analysing and interpreting, and secure archiving of neuroimaging data for large multicentre trials in common neurological disorders such as stroke. This paper details the goals of the project which are intended to appear as via online access as an integration of services, databases, and clinical meta-data, and the use of grid computing capapbility for processing and analysis. [DOI] [PubMed] [Google Scholar]
- 15.Boyes RG, Gunter JL, Frost C, Janke AL, Yeatman T, Hill DL, Bernstein MA, Thompson PM, Weiner MW, Schuff N, et al. Intensity non-uniformity correction using N3 on 3-T scanners with multichannel phased array coils. Neuroimage. 2008;39:1752–1762. doi: 10.1016/j.neuroimage.2007.10.026. ADNI Study. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jack CR, Jr., Bernstein MA, Fox NC, Thompson P, Alexander G, Harvey D, Borowski B, Britson PJ, J LW, Ward C, et al. The Alzheimer's Disease Neuroimaging Initiative (ADNI): MRI methods. J Magn Reson Imaging. 2008;27:685–691. doi: 10.1002/jmri.21049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yanovsky I, Thompson PM, Osher S, Hua X, Shattuck D, Toga AW, Leow A. Validating Unbiased Registration on Longitudinal MRI Scans from the Alzheimer's Disease Neuroimaging Initiative (ADNI). 5th IEEE International Symposium on Biomedical Imaging: From Nano to Macro; IEEE; Paris, France. May 14-17, 2008.2008. [Google Scholar]
- 18.Morra JH, Tu Z, Apostolova LG, Green AE, Avedissian C, Madsen SK, Parikshak N, Hua X, Toga AW, Jack CR, Jr., et al. Validation of a fully automated 3D hippocampal segmentation method using subjects with Alzheimer's disease mild cognitive impairment, and elderly controls. Neuroimage. 2008 doi: 10.1016/j.neuroimage.2008.07.003. The Alzheimer's Disease Neuroimaging Initiative. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hua X, Leow AD, Parikshak N, Lee S, Chiang MC, Toga AW, Jack CR, Jr., Weiner MW, Thompson PM. Tensor-based morphometry as a neuroimaging biomarker for Alzheimer's disease: An MRI study of 676 AD, MCI, and normal subjects. Neuroimage. 2008 doi: 10.1016/j.neuroimage.2008.07.013. The Alzheimer's Disease Neuroimaging Initiative. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jack C, Bernstein M, Fox N, Thompson P, Alexander G, Harvey D, Borowski B, Britson P, Whitwell J, Ward C, et al. The Alzheimer's disease neuroimaging initiative (ADNI): MRI methods. Journal of Magnetic Resonance Imaging. 2008;27:685–691. doi: 10.1002/jmri.21049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fletcher PT, Powell S, Foster NL, Joshi SC. Quantifying metabolic asymmetry modulo structure in Alzheimer's disease. Inf Process Med Imaging. 2007;20:446–457. doi: 10.1007/978-3-540-73273-0_37. [DOI] [PubMed] [Google Scholar]
- 22.Leow AD, Klunder AD, Jack CR, Jr., Toga AW, Dale AM, Bernstein MA, Britson PJ, Gunter JL, Ward CP, Whitwell JL, et al. Longitudinal stability of MRI for mapping brain change using tensor-based morphometry. Neuroimage. 2006;31:627–640. doi: 10.1016/j.neuroimage.2005.12.013. ADNI Preparatory Phase Study. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mueller SG, Weiner MW, Thal LJ, Petersen RC, Jack CR, Jagust W, Trojanowski JQ, Toga AW, Beckett L. Ways toward an early diagnosis in Alzheimer's disease: The Alzheimer's Disease Neuroimaging Initiative (ADNI) Alzheimers Dement. 2005;1:55–66. doi: 10.1016/j.jalz.2005.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moore ES, Ward RE, Wetherill LF, Rogers JL, Autti-Ramo I, Fagerlund A, Jacobson SW, Robinson LK, Hoyme HE, Mattson SN, et al. Unique facial features distinguish fetal alcohol syndrome patients and controls in diverse ethnic populations. Alcohol Clin Exp Res. 2007;31:1707–1713. doi: 10.1111/j.1530-0277.2007.00472.x. Cifasd. [DOI] [PubMed] [Google Scholar]
- 25**.Mazziotta JC, Woods R, Iacoboni M, Sicotte N, Yaden K, Tran M, Bean C, Kaplan J, Toga AW. The myth of the normal, average human brain--the ICBM experience: (1) subject screening and eligibility. Neuroimage. 2009;44:914–922. doi: 10.1016/j.neuroimage.2008.07.062. The International Consortium for Brain Mapping (ICBM) is among the oldest ongoing collaboratives for the mapping of the human brain using neuroimaging methods. This article describes the set of “normal” criteria for subject inclusion as well as associated exclusion criteria that they have developed over the course of this important multi-site effort. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mazziotta J, Toga AW, Evans A, Fox P, Lancaster J, Ziles K, Woods RP, Paus T, Simpson G, Pike B, et al. A probabilistic atlas and reference system for the human brain: International Consortium for Brain Mapping (ICBM) Philos Trans R Soc Lond B Biol Sci. 2001;356:1293–1322. doi: 10.1098/rstb.2001.0915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mazziotta JC, Toga AW, Evans A, Fox P, Lancaster J. A probabilistic atlas of the human brain: theory and rationale for its development. The International Consortium for Brain Mapping (ICBM) Neuroimage. 1995;2:89–101. doi: 10.1006/nimg.1995.1012. [DOI] [PubMed] [Google Scholar]
- 28.Kim DI, Mathalon DH, Ford JM, Mannell M, Turner JA, Brown GG, Belger A, Gollub R, Lauriello J, Wible C, et al. Auditory Oddball Deficits in Schizophrenia: An Independent Component Analysis of the fMRI Multisite Function BIRN Study. Schizophr Bull. 2009;35:67–81. doi: 10.1093/schbul/sbn133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Potkin SG, Ford JM. Widespread Cortical Dysfunction in Schizophrenia: The FBIRN Imaging Consortium. Schizophr Bull. 2009;35:15–18. doi: 10.1093/schbul/sbn159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30*.Friedman L, Stern H, Brown GG, Mathalon DH, Turner J, Glover GH, Gollub RL, Lauriello J, Lim KO, Cannon T, et al. Test-retest and between-site reliability in a multicenter fMRI study. Hum Brain Mapp. 2008;29:958–972. doi: 10.1002/hbm.20440. A study of the test-retest reliability between sites participating in a multi-center neuroimaging trial. Test-retest reliability was found to be high, but initially, between-site reliability was low, indicating a strong contribution from site and site-by-subject variance. Several factors were uncovered that can markedly improve between-site reliability, including increasing the size of the ROIs during processing, adjusting for smoothness differences, and inclusion of additional neuroimaging scanning runs. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mulkern RV, Forbes P, Dewey K, Osganian S, Clark M, Wong S, Ramamurthy U, Kun L, Poussaint TY. Establishment and results of a magnetic resonance quality assurance program for the pediatric brain tumor consortium. Acad Radiol. 2008;15:1099–1110. doi: 10.1016/j.acra.2008.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Poussaint TY, Phillips PC, Vajapeyam S, Fahey FH, Robertson RL, Osganian S, Ramamurthy U, Mulkern RV, Treves ST, Boyett JM, et al. The Neuroimaging Center of the Pediatric Brain Tumor Consortium-collaborative neuroimaging in pediatric brain tumor research: a work in progress. AJNR Am J Neuroradiol. 2007;28:603–607. [PMC free article] [PubMed] [Google Scholar]
- 33.Fennema-Notestine C, Gamst AC, Quinn BT, Pacheco J, Jernigan TL, Thal L, Buckner R, Killiany R, Blacker D, Dale AM, et al. Feasibility of multi-site clinical structural neuroimaging studies of aging using legacy data. Neuroinformatics. 2007;5:235–245. doi: 10.1007/s12021-007-9003-9. [DOI] [PubMed] [Google Scholar]
- 34.Gururangan S, Turner CD, Stewart CF, O'Shaughnessy M, Kocak M, Poussaint TY, Phillips PC, Goldman S, Packer R, Pollack IF, et al. Phase I trial of VNP40101M (Cloretazine) in children with recurrent brain tumors: a pediatric brain tumor consortium study. Clin Cancer Res. 2008;14:1124–1130. doi: 10.1158/1078-0432.CCR-07-4242. [DOI] [PubMed] [Google Scholar]
- 35.Hampel H, Burger K, Teipel SJ, Bokde AL, Zetterberg H, Blennow K. Core candidate neurochemical and imaging biomarkers of Alzheimer's disease. Alzheimers Dement. 2008;4:38–48. doi: 10.1016/j.jalz.2007.08.006. [DOI] [PubMed] [Google Scholar]
- 36*.Frisoni GB, Henneman WJ, Weiner MW, Scheltens P, Vellas B, Reynish E, Hudecova J, Hampel H, Burger K, Blennow K, et al. The pilot European Alzheimer's Disease Neuroimaging Initiative of the European Alzheimer's Disease Consortium. Alzheimers Dement. 2008;4:255–264. doi: 10.1016/j.jalz.2008.04.009. A detailed discussion of an early feasibility study of an Alzheimer's Disease Neuroimaging Initiative (ADNI) program for Europe. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37*.Van Horn J, Bandettini P, Cheng K, Egan G, Stenger V, Strother S, Toga A. New Horizons for the Next Era of Human Brain Imaging, Cognitive, and Behavioral Research: Pacific Rim Interactivity. Brain Imaging and Behavior. 2008;2:227–231. doi: 10.1007/s11682-008-9045-0. Since the late-1990's the Pacific Rim has quickly emerging as a community of brain imaging researchers with the potential for large-scale neuroimaging collaborations crossing national boundaries, cultures, and time zones. Such interactions may revolve around normal cognitive function, brain atlasing, or the examination of the brain in various neurological diseases that affect health worldwide (e.g. stroke, MS, Alzheimer's disease, etc) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grady C, Strother S. Pacific Rim Collaborations: A Canadian Perspective. Brain Imaging and Behavior. 2008;2:359–363. 10.1007/s11682-008-9035-2. [Google Scholar]
- 39.Hall HI, Song R, Rhodes P, Prejean J, An Q, Lee LM, Karon J, Brookmeyer R, Kaplan EH, McKenna MT, et al. Estimation of HIV incidence in the United States. Jama. 2008;300:520–529. doi: 10.1001/jama.300.5.520. HIV Incidence Surveillance Group. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hult B, Chana G, Masliah E, Everall I. Neurobiology of HIV. Int Rev Psychiatry. 2008;20:3–13. doi: 10.1080/09540260701862086. [DOI] [PubMed] [Google Scholar]
- 41.Chiang M-C, Dutton RA, Hayashi KM, Lopez OL, Aizenstein HJ, Toga AW, Becker JT, Thompson PM. 3D pattern of brain atrophy in HIV/AIDS visualized using tensor-based morphometry. Neuroimage. 2007;34:44–60. doi: 10.1016/j.neuroimage.2006.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thompson PM, Dutton RA, Hayashi KM, Lu A, Lee SE, Lee JY, Lopez OL, Aizenstein HJ, Toga AW, Becker JT. 3D mapping of ventricular and corpus callosum abnormalities in HIV/AIDS. Neuroimage. 2006;31:12–23. doi: 10.1016/j.neuroimage.2005.11.043. [DOI] [PubMed] [Google Scholar]
- 43.Lee PL, Yiannoutsos CT, Ernst T, Chang L, Marra CM, Jarvik JG, Richards TL, Kwok EW, Kolson DL, Simpson D, et al. A multi-center 1H MRS study of the AIDS dementia complex: validation and preliminary analysis. J Magn Reson Imaging. 2003;17:625–633. doi: 10.1002/jmri.10295. HIV MRS Consortium. [DOI] [PubMed] [Google Scholar]
- 44.Toga AW. The Laboratory of Neuro Imaging: what it is, why it is, and how it came to be. IEEE Trans Med Imaging. 2002;21:1333–1343. doi: 10.1109/TMI.2002.806432. [DOI] [PubMed] [Google Scholar]
- 45.MacKenzie-Graham A, Payan A, Dinov I, Van Horn JD, Toga AW. Neuroimaging Data Provenance Using the LONI Pipeline Workflow Environment. In: Moreau L, Foster I, editors. Provenance and Annotation of Data International Provenance and Annotation Workshop, IPAW; University of Utah; Salt Lake City, UT. 2008.2008. [Google Scholar]
- 46.Toga AW. Imaging databases and neuroscience. Neuroscientist. 2002;8:423–436. doi: 10.1177/107385802236971. [DOI] [PubMed] [Google Scholar]
- 47.Van Horn JD, Dobson J, Woodward J, Wilde M, Zhao Y, Voeckler J, Foster I. Grid-Based Computing and the Future of Neuroscience Computation. In: Senior C, Russell T, Gazzaniga MS, editors. Methods in Mind. MIT Press; 2006. pp. 141–170. [Google Scholar]
- 48.Rosen BR, Buckner RL, Dale A. Event-related functional MRI: Past, present, and future. Proceedings of the National Academy of Sciences USA. 1998;95:773–780. doi: 10.1073/pnas.95.3.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49*.Raichle ME. Functional Neuroimaging: A Historical and Physiological Perspective. In: Cabeza R, Kingstone A, editors. Handbook of Functional Neuroimaging of Cognition. MIT Press; 2001. pp. 3–26. A classic review of the history of functional neuroimaging. [Google Scholar]
- 50.Toga AW, Thompson PM. Maps of the brain. Anat Rec. 2001;265:37–53. doi: 10.1002/ar.1057. [DOI] [PubMed] [Google Scholar]