Abstract
In a genome-wide association study to identify loci associated with colorectal cancer (CRC) risk, we genotyped 555,510 SNPs in 1,012 early-onset Scottish CRC cases and 1,012 controls (phase 1.) In phase 2, we genotyped the 15,008 highest-ranked SNPs in 2,057 Scottish cases and 2,111 controls. We then genotyped the five highest-ranked SNPs from the joint phase 1 and 2 analysis in 14,500 cases and 13,294 controls from seven populations, and identified a previously unreported association, rs3802842 on 11q23 (OR = 1.1; P = 5.8 × 10-10), showing population differences in risk. We also replicated and fine-mapped associations at 8q24 (rs7014346; OR = 1.19; P = 8.6 × 10-26) and 18q21 (rs4939827; OR = 1.2; P = 7.8 × 10-28). Risk was greater for rectal than for colon cancer for rs3802842 (P < 0.008) and rs4939827 (P < 0.009). Carrying all six possible risk alleles yielded OR = 2.6 (95% CI = 1.75-3.89) for CRC. These findings extend our understanding of the role of common genetic variation in CRC etiology.
Colorectal cancer (CRC) is the third most common cancer and fourth-leading cause of cancer death worldwide. Lifetime risk in Western European and North American populations is around 5%. Both genetic and environmental factors contribute to disease etiology, with about one-third of disease variance attributed to inherited genetic factors1. Until very recently, the defined genetic contribution to CRC comprised rare, high-penetrance variants in a few genes (DNA mismatch repair genes2, APC, SMAD4, BMPR1A and MUTYH3). However, recent association studies have shown that common genetic variation in the 8q24 (refs. 4-6) and 18q21 (SMAD7)7 regions also contribute to CRC risk. To explore the role of common genetic variation in CRC etiology, we undertook a comprehensive, phased-design genome-wide association scan (GWAS), capitalizing on Scottish population characteristics. We selected early-onset cases for the genome-wide scan on the premise that these may be enriched for genetic contribution and so would provide enhanced power to detect associations. Controls were matched for age, sex and area of residence in phases 1 and 2.
In phase 1, we genotyped 1,012 early-onset CRC cases, comprising the youngest 10th percentile of CRC age distribution in Scotland, and matched controls using Illumina HumanHap300 and HumanHap240S arrays. We analyzed genotype data using a likelihood ratio test (LRT)8 with 2 degrees of freedom (genotypic model) to account for additive and dominant effects. Empirical significance thresholds were obtained by permuting case-control status 10,000 times9. For each permutation, we retained the largest test statistic from each chromosome and used it across all chromosomes to obtain chromosome-wise (Supplementary Table 1 online) and genome-wide significance thresholds (Supplementary Fig. 1 online). Phase 1 test statistics with 5% empirical genome-wide significance thresholds are shown in Supplementary Figure 2 online; none of the SNPs reached genome-wide significance (nonminally P = 1.12 × 10-7). There was no overall inflation of the test statistic (λ = 1.003), providing reassurance that systematic confounding factors are unlikely (Supplementary Fig. 3 online). Other process quality control measures are described in the Supplementary Note online.
From analysis of phase 1 data, we ranked SNPs by test statistic and selected the top 15,008 SNPs (P < 0.0272) for further analysis in phase 2. We determined the number of SNPs empirically, taking into account practical and financial constraints. We genotyped these 15,008 SNPs in 2,057 cases and 2,111 controls using the Illumina iSelect platform. After accounting for quality control measures (Supplementary Note), we included 13,450 SNP genotypes from 2,024 cases and 2,092 controls in the analysis. Joint analysis of phase 1 and 2 data again showed that none of the SNPs reached the genome-wide significance threshold obtained by permutation in phase 1 (Supplementary Fig. 4 and Supplementary Table 2 online). We estimated the Q value10 of each test (proportion of false positives incurred when the test is called significant) using phase 2 P values, and estimated the false-discovery rate to be approximately 40% for the top 300 ranked SNPs (Supplementary Fig. 5 online).
We took the five top-ranked SNPs from joint analysis of phase 1 and 2 data, equivalent to an empiric threshold of P < 10-5, for further analysis. In rank order by P value, the top SNPs in the combined phase 1 and 2 data were rs7014346 (8q24), rs4939827 (18q21), rs6533603 (4q25), rs3802842 (11q23.1) and rs9951602 (18q23). Unadjusted OR estimates using binary logistic regression in an additive genetic model are presented in Supplementary Table 2. rs7014346 (LRT = 26.64) reached chromosome-wise significance (P < 0.05), further replicating and refining the previous findings4-6 on the risk locus at 8q24. rs4939827 (LRT = 25.61) is located in intron 3 of SMAD7, replicating a recently reported association between this locus and CRC7.
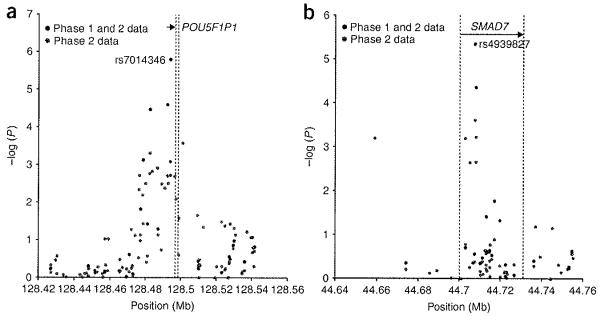
As the causative variants are unknown for rs7014346 (8q24) and rs4939827 (18q21), we undertook fine mapping by tagging all polymorphic HapMap CEU SNPs around these loci in phase 2 individuals (tagSNPs with r2 = 1 within the interval 50 kb on either side of the interval defined by rs7014346 and rs10505477 on 8q24, and of rs4939827 and rs12953717 on 18q21). Linkage disequilibrium (LD) plots for the 8q24, 18q23 and 11q23.1 regions are shown in Supplementary Figure 6 online. We used data for 94 SNPs successfully genotyped at 8q24 and 96 SNPs at 18q23 for fine mapping of the respective regions (Fig. 1). The association signal drops off sharply on either side of both rs7014346 and rs4939827. Next, we analyzed information from HapMap using IMPUTE11 for the 11q, 8q and 18q regions to estimate SNP genotypes that we did not type. SNPTEST was used to test for associations under a genotypic model. These analyses (Supplementary Fig. 7 online) show that rs7842552 is the top-ranking imputed SNP at the 8q24 locus (P = 3.84 × 10-7), rs4939827 remains the top-ranking SNP at 18q21 (P = 1.6 × 10-6) and rs3802842 indicates the peak of association at the 11q locus. Resequencing, tumor loss-of-heterozygosity (LOH) analysis and expression studies of genes within the regions delineated by fine mapping at 8q24 and 18q21 provided no additional insight into pathogenicity (Supplementary Note).
Figure 1.
Fine mapping of the 8q24 and 18q23 (SMAD7) loci. Graphs show -log10P against distance. Black dots correspond to the analysis of data generated from phase 1 and 2 individuals. Red dots are from the analysis of data from phase 2 individuals. rslDS are provided for the SNPs with peak evidence for association. These fine mapping data are further informed by results shown in Supplementary Figure 7 from IMPUTE and SNPTEST analysis of the loci on chromosomes 8q, 18q and 11q.
In phase 3, we genotyped eight additional independent case-control collections and tested for differences between populations. Genotyping was done using Taqman, Sequenom or Invader technology. Subjects were from Scotland, England (Cambridge), Canada (Ontario), Germany (Kiel and Heidelberg), Spain (Barcelona), Japan (Tokyo) and Israel (Haifa), comprising a total of 14,500 cases and 13,294 controls (Table 1). In a meta-analysis of all data to estimate pooled genetic effects (Table 2 and Fig. 2), we found that three of the five top-ranked associations replicated in phase 3 (rs7014346 on 8q24, rs4939827 on 18q21 and rs3802842 on 11q23), in agreement with our false-discovery rate estimate. Genotype counts and risk allele frequencies across populations for the five top-ranked SNPs are shown in Supplementary Table 3 online. We also tested for association at seven additional genotyped SNPs close to the replicated loci (Supplementary Tables 4 and 5 online).
Table 1. Smaples genotyped in phases 1, 2 and 3 from all populations.
| Population | Cases | Controls | Total | |
|---|---|---|---|---|
| Scotland GWAS | Scotland, phase 1 | 981 | 1,002 | 1,983 |
| Scotland, phase 2 | 2,023 | 2,092 | 4,115 | |
| Total GWAS | 3,004 | 3,094 | 6,098 | |
| Replication | Canada | 1,175 | 1,184 | 2,359 |
| DACHS | 1,373 | 1,480 | 2,853 | |
| England | 2,253 | 2,262 | 4,515 | |
| Israel | 1,789 | 1,771 | 3,560 | |
| Japan | 4,400 | 3,179 | 7,579 | |
| Kiel | 2,169 | 2,145 | 4,314 | |
| Scotland Replication | 937 | 941 | 1,878 | |
| Spain | 357 | 297 | 654 | |
| Total Replication | 14,453 | 13,259 | 27,712 | |
| Total samples | 17, 457 | 16,353 | 33,810 |
Table 2. SNPs genotyped in phase 3 and summary description from the meta-analysis of all available data.
| All samples |
Replication samples |
Replication samples of European ancestry |
||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP | Rank | Chr. | Position | Variant | Number of sets | Pheta | l2 (%) | Effects model | P | OR (95% CI) |
Number of sets | Pheta | l2 (%) | Effects model | P | OR (95% CI) |
Number of sets | Pheta | l2 (%) | Effects model | P | OR (95% CI) |
| rs6533603b | 3 | 4 | 113440251 | T | 9 | 0.001 | 68 | Random | 0.81 | 1.01 (0.95-1.07) |
8 | 0.36 | 9 | Fixed | 0.21 | 0.98 (0.94-1.01) |
7 | 0.44 | 0 | Fixed | 0.48 | 0.99 (0.95-1.03) |
| rs7014346b | 1 | 8 | 128493974 | A | 9 | 0.343 | 11 | Fixed | 8.60 × 10-26 | 1.19 (1.15-1.23) |
8 | 0.26 | 21 | Fixed | 1.85 × 10-20 | 1.19 (1.14-1.23) |
7 | 0.18 | 32 | Fixed | 4.44 × 10-16 | 1.19 (1.14-1.24) |
| rs7842552 | 8 | 128500876 | G | 8 | 0.026 | 56 | Random | 1.20 × 10-5 | 1.15 (1.08-1.23) |
7 | 0.02 | 59 | Random | 4.48 × 10-4 | 1.14 (1.06-1.23) |
6 | 0.01 | 65 | Random | 1.85 × 10-3 | 1.15 (1.05-1.25) |
|
| rs11213809 | 4 | 11 | 110640955 | A | 8 | 0.086 | 43 | Fixed | 7.93 × 10-8 | 1.11 (1.07-1.16) |
7 | 0.29 | 18 | Fixed | 6.79 × 10-4 | 1.08 (1.03-1.13) |
6 | 0.34 | 11 | Fixed | 2.98 × 10-4 | 1.09 (1.04-1.14) |
| rs3802842b | 11 | 110676919 | C | 9 | 0.050 | 48 | Fixed | 5.82 × 10-10 | 1.11 (1.08-1.15) |
8 | 0.18 | 32 | Fixed | 5.54 × 10-6 | 1.09 (1.05-1.13) |
7 | 0.43 | 0 | Fixed | 7.82 × 10-7 | 1.12 (1.07-1.17) |
|
| rs7947952 | 11 | 110686330 | T | 3 | 0.081 | 60 | Fixed | 0.04 | 1.06 (1.00-1.11) |
2 | 0.12 | 58 | Fixed | 0.81 | 1.01 (0.94-1.09) |
1 | - | - | - | 0.19 | 1.08 (0.96-1.21) |
|
| rs10749971 | 11 | 110694368 | G | 8 | 0.818 | 0 | Fixed | 6.71 × 10-6 | 1.09 (1.05-1.13) |
7 | 0.86 | 0 | Fixed | 9.46 × 10-4 | 1.08 (1.03-1.13) |
6 | 0.83 | 0 | Fixed | 7.67 × 10-4 | 1.08 (1.03-1.13) |
|
| rs4514461 | 11 | 110732153 | A | 8 | 0.072 | 46 | Fixed | 7.90 × 10-3 | 1.05 (1.01-1.09) |
7 | 0.20 | 30 | Fixed | 0.19 | 1.03 (0.99-1.07) |
6 | 0.21 | 30 | Fixed | 0.096 | 1.04 (0.99-1.09) |
|
| rs12799202 | 11 | 110749966 | G | 8 | 0.533 | 0 | Fixed | 4.98 × 10-4 | 1.07 (1.03-1.12) |
7 | 0.55 | 0 | Fixed | 0.01 | 1.06 (1.01-1.11) |
6 | 0.73 | 0 | Fixed | 0.003 | 1.08 (1.03-1.14) |
|
| rs4939827b | 2 | 18 | 44707461 | T | 9 | 0.336 | 12 | Fixed | 7.77 × 10-28 | 1.20 (1.16-1.24) |
8 | 0.25 | 22 | Fixed | 4.38 × 10-23 | 1.20 (1.16-1.24) |
7 | 0.17 | 33 | Fixed | 8.70 × 10-19 | 1.20 (1.15-1.25) |
| rs12953717 | 18 | 44707927 | T | 4 | 0.964 | 0 | Fixed | 3.10 × 10-11 | 1.18 (1.12-1.23) |
3 | 0.93 | 0 | Fixed | 2.07 × 10-7 | 1.18 (1.11-1.26) |
2 | 0.71 | 0 | Fixed | 0.002 | 1.18 (1.06-1.30) |
|
| rs9951602b | 5 | 18 | 74758767 | T | 9 | 0.059 | 47 | Fixed | 0.44 | 1.02 (0.97-1.07) |
8 | 0.04 | 53 | Random | 0.64 | 1.02 (0.94-1.10) |
7 | 0.03 | 56 | Random | 0.51 | 1.03 (0.94-1.13) |
The Mantel-Haenzsel method was used.
When there was no significant heterogeneity (Phet > 0.05), the Mantel-Haenszel method was used to estimate pooled allelic effects under a fixed effect model, A random effects model, the DerSimonian-Laird method, was used if there was significant heterogeneity.
SNPs identified through the GWAS.
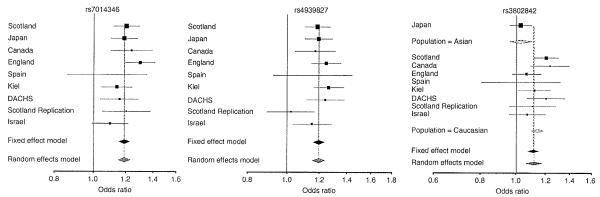
Figure 2.
Forest plot of effect size and direction for each of the three SNPs associated with CRC (rs7014346, rs4939827 and rs3802842). Symbol size indicates the weight of the study in the fixed effect model (the larger the symbol, the greater the weight), as in the output from the program R. rs3802842 was stratified by ethnic group, because most of the heterogeneity observed was due to differences between GWAS data from the Scottish and the Japanese populations.
rs7014346 is located on 8q24 and is in high linkage disequilibrium with SNPs that we previously reported (rs10505477 and rs6983267)4. However, rs7014346 gave the maximum association signal in the Scottish phase 1 and 2 data. rs7014346 is 3 kb upstream of POU5F1P1 and within intron 6 of the gene DQ515897. The association was independently replicated in all but the Spanish subjects (Supplementary Table 4) giving a combined P = 8.6 × 10-26. The lack of association in the Spanish cohort is most likely due to the small sample size, as there was no significant heterogeneity for rs7014346 across populations, and stratification tends to increase false positives rather than false negatives. Logistic regression analysis of the combined data showed that a genotypic model fit the data significantly better (P = 0.02) than an additive genetic model (Supplementary Table 6 online). Meta-analysis of the pooled data (Table 2 and Fig. 2) yielded ORs for populations of European ancestry of 1.25 (95% CI = 1.18-1.32) for AG and 1.38 (95% CI = 1.28-1.48) for AA genotypes. rs7014346 showed the peak association signal because rs7842552, identified by IMPUTE fine mapping, did not reach the same level of statistical support as rs7014346, and there was significant heterogeneity across study populations (P = 0.026).
rs4939827 is located within intron 3 of SMAD7 on chromosome 18q21. The combined P value for association with CRC was 7.77 × 10-28 (OR = 1.20). There was no heterogeneity among sample sets (P = 0.34; Table 2). The association replicated in all case-control collections individually, except the Spanish set again and the Scottish phase 3 samples (Fig. 2 and Supplementary Table 4). There was no evidence against an additive model for this SNP (Supplementary Table 6).
rs3802842 is located within a gene-rich region of chromosome 11q23, which adds complexity to attempts at identifying the causative variant. Within 100 kb of rs3802842, there are four ORFs (LOC120376, FLJ45803, C11orf53 and POU2AF1) and a sequence (rs12296076) identified as a polymorphic binding site target for miRNAs (see URLs section below) in high linkage disequilibrium. Of note, rs7014346 and rs3802842 were both close to genes encoding POU transcription factors. Hence, we genotyped five additional SNPs around rs3802842, notwithstanding that some SNPs showed only moderate statistical support (P < 0.03). However, after genotyping in multiple sample collections, we found that rs3802842 remained the best-supported SNP (Table 2). We observed substantial population-specific differences in risk at the 11q23 locus, with significantly different allelic effects between the Japanese and Scottish populations (P = 0.001) (Fig. 2). The difference in genetic effects at rs3802842 between Europeans and Japanese remained significant (P = 0.03), even when we excluded Scottish phase 1 data to avoid potential bias.
We did not find any evidence for gene-by-gene, sex, age, cancer stage, family history or cohort interactions (Supplementary Tables 6 and 7 online) with rs7014346, rs4939827 or rs3802842 in the populations of European ancestry. However, there were notable site-specific differences in risk associated with the 11q23 locus (rs3802842; P < 0.008) and the SMAD7 locus at 18q21 (rs4939827; P < 0.009) (Table 3 and Supplementary Fig. 8 online). The risk of rectal cancer was greater than for colonic cancer for both rs3802842 and rs4939827. It should also be noted that the differential effect on colon cancer risk and rectal cancer risk explains much of the population differences between Japanese and Caucasian populations for rs3802842, with colon cancer risk in particular driving the population difference.
Table 3. Meta-analysis of differences between colon and rectal cancer risk using data from all nine sample sets.
| Fixed effects model |
Random effects model |
Heterogeneity |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| SNP rsID | Reference allele | P | OR | 95% CI | P | OR | 95% CI | I2 | P | |
| Colon vs. Rectal | rs7014346 | A | 0.316 | 1.025 | 0.976-1.077 | 0.319 | 1.025 | 0.976-1.077 | 0.000 | 0.461 |
| rs4939827 | C | 0.009 | 1.068 | 1.017-1.123 | 0.009 | 1.068 | 1.017-1.123 | 0.000 | 0.566 | |
| rs3802842 | A | 0.008 | 1.070 | 1.018-1.126 | 0.008 | 1.070 | 1.018-1.126 | 0.000 | 0.822 | |
| Colon vs. Controls | rs7014346 | A | <10-15 | 1.200 | 1.156-1.247 | <10-15 | 1.201 | 1.156-1.247 | 0.000 | 0.742 |
| rs4939827 | T | 1.11 × 10-15 | 1.169 | 1.125-1.1214 | 1.11 × 10-15 | 1.169 | 1.125-1.214 | 0.000 | 0.583 | |
| rs3802842 | C | 10-15 | 1.088 | 1.046-1.132 | 3.90 × 10-3 | 1.097 | 1.030-1.169 | 55.87 | 0.020 | |
| Rectal vs. Controls | res7014346 | A | 2.53 × 10-2 | 1.175 | 1.123-1.229 | 2.83 × 10-6 | 1.166 | 1.094-1.244 | 45.67 | 0.065 |
| rs4939827 | T | <10-15 | 1.253 | 1.198-1.311 | <10-15 | 1.253 | 1.198-1.311 | 0.000 | 0.842 | |
| rs3802842 | C | 1.98 × 10-10 | 1.163 | 1.110-1.218 | 1.87 × 10-10 | 1.163 | 1.110-1.219 | 0.000 | 0.877 | |
Forest plots are shown in Supplementary Figure 9 online. For clarity of comparison, the reference alleles for colon versus rectal cancer risk are set to allow presentation of ORs in the same direction across all analyses.
Genome-wide association studies are beginning to unravel the genetic architecture underlying complex disease traits. In this study, we identify a previously unreported locus on 11q23, tagged by rs3802842, which is associated with CRC. Extending the previous observations made by ourselves4 and others5,6 at the chromosome 8q24 locus and at the SMAD7 locus7 on 18q21, we have fine mapped and further replicated these two associations, showing consistent effects across multi-ethnic populations. The variants are common in the general population, with risk allele frequencies in populations of European ancestry of 0.29, 0.37 and 0.52 for rs3802842, rs7014346 and rs4939827, respectively. The population attributable risks (PAR) in the Scottish population are estimated to be 6.5%, 9.6% and 3.3% for rs7014346, rs4939827 and rs3802842, respectively. In the Japanese population, the PAR was estimated to be 4.4% for rs 7014346 and 4% for rs4939827, primarily as a result of differences in allele frequency.
The observation that rs3802842 is associated with significantly different risk in Japanese compared to European samples is the first evidence for a population-specific CRC susceptibility allele. It is particularly noteworthy that the population difference is site-specific. The Japanese population does not show the increased risk of colonic cancer associated with rs3802842 that is observed in European populations, but it does show a similar risk of rectal cancer at that locus.
Although we urge caution in implementing models for predicting individual risk, such approaches incorporating multilocus genotypes could help identify high-risk subgroups within populations. Thus, for individuals who carry all six possible risk alleles at rs7014346, rs4939827 and rs3802842 (population frequency 0.005), the estimated OR is 2.6 (95% CI = 1.75-3.89). This underscores the potential for future risk profiling, even without identification of the causative variant12. However, large multinational cohort studies will be needed to validate such genetic risk predictive models.
In the context of genome-wide association studies, it is note-worthy that the associations that replicated across populations (rs7014346, rs4939827 and rs3802842) were ranked 449, 5,965 and 11,064 in our initial scan, respectively. Hence, follow-up of the 2.7% of putative associations from phase 1 seems appropriate. Some modeling suggests that a lower proportion taken forward to phase 2 is sufficient (<1%)13, but power to distinguish true from false positives using available tools (see URLs section below) may be overestimated. Study design for genome-wide scans is evolving and, as costs reduce, genotyping of large numbers of markers in large sample sets and avoiding phased designs altogether would be an ideal approach.
As well as providing risk estimates for population groups, identification of these loci provides new insights into disease causation. Despite extensive resequencing, we did not identify causative coding sequence variants in any of the genes at 8q24 (POU5F1P1, HsG57825 and DQ515897) or 18q21 (SMAD7). It seems likely that regulatory sequence variants or position effects underlie the associations detected here. Studies of the mechanisms by which these genetic associations impart CRC risk could inform the development of small molecule interventions for chemoprevention and chemotherapy.
METHODS
Study populations and genotyping
Phase 1 and 2 samples were collected in a prospective population-based study in Scotland (1999-2006). Cases were recruited soon after confirmed diagnosis of adenocarcinoma of large bowel (phase 1, aged ≤55 years; phase 2, aged <80 years), We genotyped individuals in phase 1 using Illumina HumanHap300 and HumanHap240S arrays on the Infinium platform, and we genotyped individuals in phase 2 using the Illumina iSelect custom panel. For individuals in phase 3 (described in Supplementary Methods online), we used Applied Biosystems (ABI) TaqMan assays exclusively to genotype subjects from the Scottish, English, German, Israeli and Spanish populations, TaqMan and Sequenom technologies for the Canadian samples and Invader assays for the Japanese samples. Call rates and departures from Hardy-Weinberg equilibrium (HWE) in control populations are shown in Supplementary Table 4, and quality control measures are described in the Supplementary Note.
Statistical methods
We analyzed phase 1 data using a likelihood ratio test (LRT) with 2 degrees of freedom (d.f.) to account for an additive and a dominant effect. Although we tested all SNPs under an allelic model, we did not use this for phase 2 SNP selection. Chromosome X SNPs were tested only with 2 d.f. in females and with 1 d.f. when combining male and female samples. We used only LRT statistics from females to select phase 2 SNPs. The mitochondrial and Y chromosome SNPs were tested with 1 d.f. and 2 d.f., respectively. Although only the first is strictly appropriate, only LRT with 2 d.f. was used as the selection criteria in phase 1. Thus, we applied more stringent selection criteria for selecting phase 2 SNPs on chromosome X, Y and mitochondrial DNA than for autosomal regions. Our approach to estimation of the false discovery rate and permutation testing to assess genome-wide and chromosome-wise empirical significant thresholds are both described in Supplementary Methods.
Testing differences in effects among populations
To test whether two different populations had different OR for a given SNP, we used a standar t-test on the natural log transformation of the ORs. Under asymptotic assumptions, the InOR is normally distributed with mean InOR and variance (). The statistic , where 1 and 2 indicate the two different populations, can be assumed to be normally distributed for large sample sizes.
Meta-analysis
We carried out the meta-analysis using the metabin option from the meta package of the R software. We used the Mantel-Haenszel method to estimate pooled allelic effects under a fixed effect model when there was not significant heterogeneity (PHet < 0.05). If there was significant heterogeneity, then we used a random effects model and the DerSimonian-Laird method. The OR was used as the summary measure. As we had used a genotypic model to select phase 3 SNPs, we also tested the same genetic model using logistic regression (fitting age, gender, sample set and genotype) on all but the Japanese case-control set, for which the raw phenotypic data was not available because of internal regulations in Japan. Nested models with and without the genotypes were compared using an analysis of deviance. Interaction terms were tested in the same fashion.
Fine mapping
LD plots for the 8q24, 18q21 and 11q23 regions are shown in Supplementary Figure 6. Detail on fine-mapping methodology is presented in Supplementary Methods. Briefly, we selected fine-mapping SNPs for the 8q24 and 18q21 regions using Phase 2 HapMap CEU data14. Haploview15 was used to select SNPs to tag (r2 = 1, MAF threshold = 0.00001) from all HapMap CEU SNPs between chromosome 8 positions 128426625 and 128543974 (50 kb centromeric of rs10505477 (ref. 4) and 50 kb telomeric of rs7014346). Including additional selected SNPs, there were 94 SNPs that successfully genotyped in 4,116 Scottish samples to tag 131 alleles at r2 ≥ 0.8 (mean maximum r2 = 0.999) using Haploview 4.0 tagger15. For 18q21, we selected SNPs from the interval between positions 44657461 and 44757927 (50 kb on either side of the interval between rs4939827 and rs12953717) and successfully genotyped 51 SNPs in 4,116 Scottish samples, tagging 64 alleles at r2 ≥ 0.8 (mean maximum r2 = 1) (Haploview 4.0 tagger). The 11q23 locus could not be formally fine mapped. However, as it encompasses a gene encoding a POU transcription gene family member (POU2AF1), we selected the six top-ranked SNPs < 100 kb from POU2AF1 and two SNPs located within the POU2AF1 gene itself. For further analysis, we used this genotyping data and HapMap data in the program IMPUTE11. We used genotyping data from phase 1 individuals along with HapMap data to estimate genotypes for SNPs that we did not genotype. SNPTEST was used to test for genotype-phenotype associations under a genotypic model. Details on IMPUTE and SNPTEST are presented in Supplementary Methods.
Resequencing, gene expression, tumor LOH and immunohistochemistry
Resequencing focused on regions at the 8q24 and 18q21 loci delineated by fine mapping and IMPUTE analysis. For 8q24 locus, there are three putative genes: POU5FIP1, HsG57825 and DQ515897. As POU5FIP1 is highly homologous to other POU genes, we designed chromosome 8-specific primers for all coding regions using a combination of primer design packages available through the University of California Santa Cruz and Primer3 (Supplementary Methods). Amplicons were sequenced and analyzed as described16 in 168 individuals (78 cases, 90 controls). Published expression data are available5, and we also found that POU5F1P1 is transcribed in blood leukocytes and colonic epithelium (data not shown). The associated SNPs at the chromosome 18q21 locus were both located within the genomic structure of SMAD7. Hence, we resequenced only exons and intron-exon boundaries of SMAD7 in 256 individuals (166 cases and 90 controls). Available annotation and expression data for 8q24 and 18q21 gene are described in Supplementary Methods.
We carried out LOH analysis in CRCs from up to 43 individuals heterozygous at rs7014346, rs4939827 and rs3802842, as well as the CA repeat marker, D18S58, at the 18q21 locus. LOH was assessed in relation to the risk allele. Tumor immunohistochemistry was done as described2 with minor modifications in 40 tumor and normal samples for which genotypes at rs7014346, rs4939827 and rs3802842 were previously defined.
ACKNOWLEDGMENTS
Edinburgh: We are grateful to all participants in these studies and to nursing and administrative staff on the COGS and SOCCS studies, We acknowledge the working arrangements with the Genotyping Core at the Wellcome Trust Clinical Research Facility, managed by L. Murphy, for sample preparations and genotyping (COGS, SOCCS, Scotland replication and LBC 1936 samples). We also thank departments in central Scottish NHS, including Cancer Registry, Scottish Cancer Intelligence Unit of ISD and the Family Practitioner Committee for population control recruitment. The work was funded by grants from Cancer Research UK (C348/A3758 and A8896, C48/A6361), Medical Research Council (G0000657-53203) and Scottish Executive Chief Scientist’s Office (K/OPR/2/2/D333, CZB/4/449), and a Centre Grant from CORE as part of the Digestive Cancer Campaign. J.P. was funded by an MRC PhD studentship. Research work at the Edinburgh Parallel Computing Centre was supported by the Scottish Funding Council through the ‘e-Science Data, Information and Knowledge Transformation 2′ (eDIKT2) project (SFC grant HR04019). The Lothian Birth Cohort 1936 phenotype and DNA collection was supported by Programme Grant number 251 and the Sidney De Haan Research Award from Research Into Ageing, and by the Disconnected Mind Award from Help the Aged. I.J.D. holds a Royal Society-Wolfson Research Merit Award. Sample collection, DNA extraction and phenotype data were collected at the Wellcome Trust Clinical Research Facility, Edinburgh.
Cambridge: We thank the SEARCH study team and all the participants in the study. P.D.P.P. is a Cancer Research UK Senior Clinical Research Fellow. T.K. is funded by the Foundation Dr Henri Dubois-Ferriere Dinu Lipatti.
Kiel: The study was supported by the German National Genome Research Network (NGFN) through the POPGEN biobank (BmBF 01GR0468) and the National Genotyping Platform. Further support was obtained through the MediGrid and Services@MediGrid projects (01Ak803G and 01IG07015B), SHIP is part of the Community Medicine Research net (CMR) of the University of Greifswald, Germany, which is funded by the Federal Ministry of Education and Research (grant no. ZZ9603), the Ministry of Cultural Affairs as well as the Social Ministry of the Federal State of Mecklenburg-West Pomerania.
Heidelberg: We wish to thank all participants and the staff of the participating clinics for their contribution to the data collection and B.Kaspereit, K. Smit and U. Eilber in the Division of Cancer Epidemiology, and U. Handte-Daub, S. Toth and B. Collins in the Division of Clinical Epidemiology and Aging Research, German Cancer Research Center for their excellent technical assistance. This study was supported by the German Research Council (Deutsche Forschungsgemeinschaft), grant numbers BR 1704/6-1, BR 1704/6-3 and CH 117/1-1, and by the German Federal Ministry for Education and Research, grant number 01 KH 0404.
Barcelona: The Bellvitge Colorectal Cancer Study has been funded by the Spanish Instituto de Salud Carlos III, FIS (grants 97/0787, 03/0114 and 05/1006), Ministry of Science and Education (SAF 06/06084) and Acción Transversal del Cáncer 2008.
London: We acknowledge Cancer Research UK Research funding and thank all those who participated in this study.
Michigan: Genotyping of Michigan samples was supported by NCI R01 CA81488, the Irving Weinstein Foundation and the University of Michigan Comprehensive Cancer Center Core Grant, P30 CA46592.
Tokyo: We thank members of the Rotary Club of Osaka Midosuji District (Japan) for collecting samples, and M. Kubo (RIKEN, Japan) for SNP genotyping. The study was supported by ‘Biobank Japan’, a project working toward personalized medicine.
Canada: We gratefully acknowledge the contribution of A. Belisle, V. Catudal and R. Fréchette. Cancer Care Ontario, as the host organization to the ARCTIC Genome Project, acknowledges that this project was funded by Genome Canada through the Ontario Genomics Institute, by Génome Québec, the Ministère du Dévelopement Économique et Régional et de la Recherche du Québec and the Ontario Institute for Cancer Research (B.W.Z., T.J.H., C.M.T.G. and S.G.).
Additional funding was provided by the National Cancer Institute of Canada (NCIC) through the Cancer Risk Assessment (CaRE) Program Project Grant. The work was supported through collaboration and cooperative agreements with the Colon Cancer Family Registry and PIs, supported by the National Cancer Institute, National Institutes of Health under RFA CA-95-011, including the Ontario Registry for Studies of Familial Colorectal Cancer (S.G.) U01 CA076783). The content of this manuscript does not necessarily reflect the views or policies of the National Cancer Institute or any of the collaborating institutions or investigators in the Colon CFR nor does mention of trade names, commercial products or organizations imply endorsement by the US Government or the Colon CFR.
Footnotes
URLs. miRNA-target interactions, http://www.patrocles.org; CaTS, http://www.sph.umich.edu/csg/abecasis/CaTS/.
Note: Supplementary information is available on the Nature Genetics website.
COMPETING INTERESTS STATEMENT
The authors declare competing financial interests: details accompany the full-text HTML version of the paper at http://www.nature.com/naturegenetics/
References
- 1.Lichtenstein P, et al. Enviornmental and heritable factors in the causation of cancer - Analyses of cohorts of twins from Sweden, Denmark, and Finland. N. Engl. J. Med. 2000;343:78–85. doi: 10.1056/NEJM200007133430201. [DOI] [PubMed] [Google Scholar]
- 2.Barnetson RA, et al. Identification and survival of carriers of multations in DNA mismatch-repair genes in colon cancer. N. Engl. J. Med. 2006;354:2751–2763. doi: 10.1056/NEJMoa053493. [DOI] [PubMed] [Google Scholar]
- 3.Tenesa A, et al. Association of MUTYH and colorectal cancer. Br. J. Cancer. 2006;95:239–242. doi: 10.1038/sj.bjc.6603239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zanke BW, et al. Genome-wide association scan identifies a colorectal cancer susceptibility locus on chromosome 8q24. Nat. Genet. 2007;39:989–994. doi: 10.1038/ng2089. [DOI] [PubMed] [Google Scholar]
- 5.Tomlinson I, et al. A genome-wide association scan of tag SNPs identifies a susceptibility variant for colorectal cancer at 8q24.21. Nat. Genet. 2007;39:984–988. doi: 10.1038/ng2085. [DOI] [PubMed] [Google Scholar]
- 6.Haiman CA, et al. A common genetic risk factor for colorectal and prostate cancer. Nat. Genet. 2007;39:954–956. doi: 10.1038/ng2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Broderick P, et al. A genome-wide association study shows that common alleles of SMAD7 influence colorectal cancer risk. Nat. Genet. 2007;39:1315–1317. doi: 10.1038/ng.2007.18. [DOI] [PubMed] [Google Scholar]
- 8.Sokal RR, Rohlf FJ. Biometry: the principles and Practice of Statistics in Biological Research. 3rd edn W.H. Freeman and Co.; New York: 1995. [Google Scholar]
- 9.Churchill GA, Doerge RW. Empirical threshold values for quantitative trait mapping. Genetics. 1994;138:963–971. doi: 10.1093/genetics/138.3.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. USA. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat. Genet. 2007;39:906–913. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- 12.Wray NR, Goddard ME, Visscher PM. Prediction of individual genetic risk to disease from genome-wide association studies. Genome Res. 2007;17:1520–1528. doi: 10.1101/gr.6665407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Skol AD, Scott LJ, Abecasis GR, Boehnke M. Joint analysis is more efficient than replication-based analysis for two-stage genome-wide association studies. Nat. Genet. 2006;38:209–213. doi: 10.1038/ng1706. [DOI] [PubMed] [Google Scholar]
- 14.Tenesa A, Dunlop MG. Validity of tagging SNPs across populations for association studies. Eur. J. Hum. Genet. 2006;14:357–363. doi: 10.1038/sj.ejhg.5201554. [DOI] [PubMed] [Google Scholar]
- 15.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 16.Farrington SM, et al. Germline susceptibility to colorectal cancer due to base-excision repair gene defects. Am. J. Hum. Genet. 2005;77:112–119. doi: 10.1086/431213. [DOI] [PMC free article] [PubMed] [Google Scholar]