Abstract
For disease prognosis, the functional significance of the oncoviral integration locus in oncogenesis has remained enigmatic. The locus encodes several transcripts without protein products, but microRNAs (miRNAs) have recently been identified from a common oncoviral integration locus. miRNA is an endogenous, non-coding small RNA by which gene expression is suppressed. Although miRNA genes, such as let-7 in the nematode, have orthologs among animals, the relationship between miRNAs and tumorigenesis or tumor suppression has been mainly discovered in several human cancers. On the contrary, this review clearly demonstrates the potential for human tumorigenesis of both miRNA genes from oncoviral integration sites and other cellular onco-microRNA genes, and we conclude that alteration of the miRNA profile of cells can be defined as tumorigenic or tumor suppressive. Thus, we explain here that virally-pathogenic miRNAs could also be partly responsible for oncogenesis or oncogene suppression to confirm’ the RNA wave’, with the miRNAs hypothesized as a mobile and functional genetic element.
Keywords: Cancer, human genome, microRNA, non-coding RNA, oncovirus, tumor.
INTRODUCTION
It is well known that multiple copies of oncogenes are expressed in cancer cells. Chromosomal translocation is a common oncogene amplification mechanism in hematologic cancer. For example, the Philadelphia (Ph) chromosome is activated by chromosomal translocation in chronic myeloid leukemia, and the oncogene abl implicated in Abelson mouse leukemia is activated by the chromosomal translocation T(9;22) [1]. Burkitt’s lymphoma is caused by Epstein-Barr virus (EBV), and the avian myelocytomatosis (myc) oncogene is activated by chromosomal translocation T(8;14) (q24;q32) into the immunoglobulin gene locus, such as IGH at 14q32, IGK at 2p12, or IGL at 22q11, which was seen in 75-85% of cancer individuals [2, 3]. The oncogene chimera products may cause tumors; however, oncogenic effects could also mediated by the human MYC proto-oncogene [4, 5].
Further, about 16% of multiple myeloma cases exhibit plasmacytoma variant translocation gene (PVT1) region rearrangement independent of immunoglobulin loci. PVT1 and murine Pvt1 are common retroviral integration sites [2]. Murine leukemia virus (MLV) can integrate into the Pvt1 locus and induce T lymphomas in mouse and rat. The provirus acts on the integration site by adding tags of its own sequences. There are two types of oncoviral integration. The first type occurs when the provirus inserts near the proto-oncogene; the second type occurs when the provirus inserts into the oncogene itself.
The method of tagging a provirus without oncogenes has been used for identification of the cellular oncogene (c-onc) through its homology to the viral oncogene (v-onc). In this method, mice are infected with a retrovirus that excludes the oncogene. Genes neighboring the proviral integration site can be identified and classified as either proto-oncogenes or tumor suppressor genes. Using this method, both protein-coding genes and non-coding genes were discovered. Recently, microRNAs (miRNAs) from PVT1 have been identified as purported oncogenic miRNA (oncomirs) [6]. Further, using miRNA profiling of tumor cells and patient specimens, tumor suppressor miRNAs have also been identified. These discoveries regarding miRNA genes suggest that oncoviral integration should be reconsidered for tumorigenesis, and they highlight the importance of genome-wide research including miRNA gene location on different chromosomes. The concept of investigating miRNA genes more closely might be also extended to analysis of quantitative trait loci (QTL) [7, 8].
We recently introduced the RNA waves hypothesis (henceforth called RNA wave) that miRNA is a mobile and functional genetic element [9]. To confirm the resolution of RNA wave, this review first explains gene silencing pathway mediated by the miRNA gene, then uses oncogenic human herpesvirus 8 (HHV8) and retrotransposon I (including human immunodeficiency virus type 1 (HIV-1)) examples of the resident and genomic miRNA genes, respectively. Next, we focused on the relation between Knudson’s two-hit hypothesis on retinoblastoma (RB) as an early landmark [10] and RNA wave-based oncogenesis via oncoviruses. Finally, we discuss about the role of the miRNAs in tumorigenesis mechanisms and suggest the possibility of oncogenesis by viral miRNA inducing alteration of oncogenes or suppressor protein genes and oncogenic miRNA or suppressor miRNA gene expression.
BIOGENESIS AND FUNCTION OF THE CRUCIAL SMALL RNAs
In early 1990s, the phenomenon of RNA silencing was observed in an experiment involving pigmentation of the Petunia flower [11-13]. The exogenous gene transfer suppressed expression of the endogenous cognate gene, suggesting co-suppression of homologous genes in plants caused at least in part by chromosomal methylation. Napoli et al. [14] reported that the transgene induced small double stranded RNAs (dsRNA) and pigmentation was observed in the cells/organism by expression of dsRNA. This phenomenon is termed as RNA interference (RNAi).
In 1998, Mello and Fire reported that exogenous dsRNA induced green fluorescent protein (GFP) or β-galactosidase (β-gal) gene silencing in gfp or β-gal transgenic Caenorhabditis elegans [15]. When about 30 nucleotides (nts) of the complete paired dsRNA from gfp were transfected into nematode cells, the dsRNA bound Dicer-dependently to RNA-induced silencing complex (RISC). Further, the longer dsRNA was transfected and then the dsRNA was digested by RNase Dicer and incorporated into RISC (Fig. 1a).
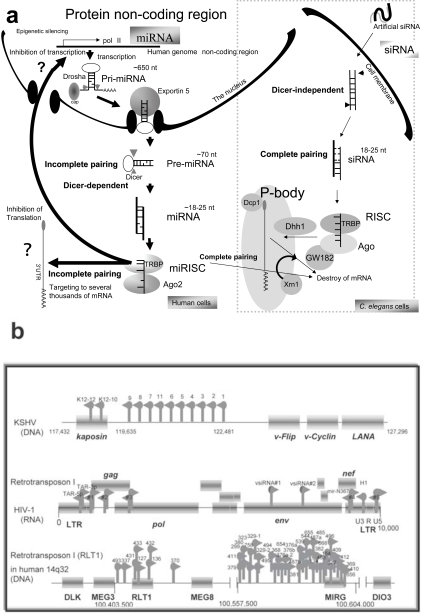
Fig. (1).
Human miRNA biogenesis and its sources. (a) Human miRNA genes are hidden in the protein non-coding regions and pri-miRNA involves from a long transcript precursor, which can be generated by Pol II or Pol III RNA promoters. The intronic miRNAs are transcribed by the Pol II promoters of the protein-coding genes. In the nucleus, the pri-miRNA is excised by Drosha RNase and the processed pre-miRNA is transported by Expotin-5 to the cytoplasm of the cell. The pre-miRNA is diced by Dicer RNase and then miRNAs are incorporated into a RNA-induced silencing complex. The matured miRNA suppresses translation and transcription by uncertain mechanisms. On the other hand, nematode siRNA can be transfected into the cells, where either strand of the siRNA can bind to the RISC independently of Dicer. In the case of complementarily-paired miRNA and siRNA-to-mRNA sequences, both small RNAs induce mRNA degradation in the P-body. Human Ago1 and Ago2 are localized in the P-body and associated with GW182. The N-terminal GW182 protein can interact with the PIWI domain of Ago1. Ago2 also localizes with the decapping enzyme Dcp1 for matured mRNA and the helicase Dhh1. mRNA degradation by siRNA can be performed by the exoribonuclease Xrn1. But usually miRNA is uncomplementarily paired to the 3’ UTR of target mRNA; therefore, miRNA is believed not to destroy mRNA. (b) A schematic representation of three miRNA sources in relation to the human genome. Twelve, 9 and 43 miRNAs in the KSHV (DNA virus), HIV-1 (RNA virus) genome, and human 14q32 chromosome, respectively, are represented by the gray arrows. Protein coding regions are represented as the gray bars. Ten miRNAs (1-9 and 11) of KSHV reside in the non-coding region and two (K12-10 and 12) are within the kaposin gene. Two miRNAs (MIRH1 and MIRN367) of HIV-1 were cloned and expression was detected by northern blotting. Two (TAR-3p and 5p) were observed with the artificial bio-assay and are involved in the secondary structured TAR region. Five (#1-#5) were predicted by the computing analysis. The sequences of miR-#4 is partially overlapped into those of miR-N367. Two viral siRNAs (vs iRNA#1 and #2) were detected by computation and northern blotting. The type I retrotransposon RTL1 is involved in the imprinted chromosome region. Five miRNAs (miR-127, miR-136, miR-431, -432, and -433) in the RTL1 gene were isolated. The antisense RTL1 transcript is maternally expressed, and it was initially reported in an ovine model as anti-PEG11. The maternal expression of the antisense transcript suggests to be controlled by these miRNAs.
Short interfering RNAs (siRNAs) and Argonute (Ago) protein target messenger RNA (mRNA) to processing bodies (P-bodies) in the cytoplasm. These cytoplasmic bodies are the sites for mRNA degradation [16]. Human Ago1 and Ago2 are co-localized with the P-body-associated phosphorylated autoimmune antigenic protein, GW182 [17]. The N-terminus of GW182 was found to interact with the PIWI domain of Ago1 [18]. In addition, human Ago2 co-localized with the decapping enzyme Dcp1a and Dcp2 for m7G-capped, matured mRNA and the helicase Dhh1 (Fig. 1a). Therefore, mRNA localization to P-bodies by siRNA is sufficient to cause translational repression with mRNA degradation by the exoribonuclease Xrn1 [16]. Later, endogenous siRNAs (19-30 nts) were cloned and it was shown in 2006 that, in the nematode, dominant function of endogenous RNA silencing is the transcriptional gene silencing (TGS) [19, 20]. Further, it has been reported that HIV-1 encodes viral siRNA precursor (Fig. 1b) [21].
Protein coding genes comprise only 1.5-2% of the whole genome. According to Watson and Crick’s theory, known as the Central Dogma, a protein coding gene is transcribed from genome DNA to mRNA, then the mRNA is translated to produce a polypeptide chain. The genetic information in DNA is copied and transmitted to daughter cells via DNA replication. Upon primary transcription, the mRNA is synthesized by RNA polymerase II (Pol II). The primary transcripts are extensively modified in the nucleus after transcription. The modifications of the mRNA usually consist of three events. The first is cap formation of 5’-terminus of the mRNA. The second is addition of a poly(A) tail at the 3’-terminus of the mRNA. The third is the removal of introns and the cap, and poly(A) tailing and the exons are produced during matured mRNA processing.
A secondary result of the Central Dogma theory was the concept that all necessary genetic information in contained within the protein-coding region. Therefore, genetic content in the non-coding region, except for introns and promoters, has been excluded from molecular biology studies. miRNAs can be generated from the non-coding region and can target to the 3’ untranslated region (UTR) of the mRNA. Further, miRNA is sometimes expressed from the intron. Therefore, splicing can be targeted by the intronic miRNA [22]. Thus, the Central Dogma is controlled by the miRNA genes because nearly 100% of coding transcripts are under the control of miRNAs and other short RNAs [23, 24]. While the host cell system following the Central Dogma is always used for viral protein synthesis, master regulator miRNA should control virus proliferation and pathogenesis [25, 26]. Although unknown protein coding genes as the open reading frames (orf) have only recently been established in computing analysis, it is now tenaciously understood that oncovirus has hidden miRNAs in their viral genome [27-32]. Computational analysis, northern blotting, and functional bio-analysis have showed the presence of HIV-1 miRNAs [33-35].
The miRNA genes are expressed as the master regulator of the Central Dogma in plants, worms, flies, mammals, and viruses, and they play an important role in developmental timing [36], morphologic changes [37], cell proliferation and death [38], hematopoiesis [39], nervous system control [40], pancreatic insulin secretion [41], adipogenesis [42], oncogenesis [43, 44] and viral disease [45-48]. Although small RNAs originate from many sources, including cellular genomes, viral replication intermediates, aberrant cellular RNAs, overexpressed transgenes, and transposons, more than 500 miRNA genes from human genome nearly have been listed up in miRBase. The miRNA genes are transcribed by Pol II as primary transcripts (pri-miRNA) from human as well as viral genomes [49] (see the left panel in Fig. 1a). Most miRNAs are likely to be transcribed by Pol II as the class II gene [50]. The primary transcripts of miRNA have experimentally been reported to contain a cap structure and poly (A) tail [51]. In the reverse genetic method, RNA polymerase III (Pol III)-expressing siRNA can induce cell toxicity, but Pol II-expressing miRNA cannot [52]. Therefore, the Pol II transcript from hairpin-structured pri-miRNA might be more relevant than Pol III for class III genes.
Although some pri-miRNA from intron (mirtron) does not contain a 5’-cap or a poly (A) tail [49], expression of miRNAs is commonly regulated by a promoter and an enhancer [53, 54]. In computational predictions with the CoVote method, most miRNAs have the same type of promoters as protein coding genes of C. elegans, H. sapiens, A thaliana and O. sativa [55]. Further, the K-Factor algorithm showed that the nucleotide composition of the miRNA upstream sequences is GC-rich (90%) [56]. However, the observation of expression of tens of thousands of non-coding RNAs (ncRNAs) suggests that the classical promoter-transcription-protein view may be incomplete because most of the ncRNAs identified have not been studied and the functions have not yet been investigated in genomic studies.
The pri-miRNAs are processed by RNase type III Drosha [57] and its cofactor DiGeorge syndrome critical region 8 (DGCR8) to pre-miRNA ~50-100 nts long [58]. The pre-miRNA is transported by exportin 5 (EXP5) from the nucleus to the cytoplasm [59, 60]. The exported pre-miRNAs are processed again by RNase type III Dicer and the trans-activator RNA-binding protein (TRBP) [61] and/or protein activator of protein kinase activated by dsRNA (PACT) [62]. The diced and matured miRNA duplex (~19-25 nts) is incorporated into the miRNA-induced silencing complex (miRISC) [63]. One strand of the duplex, miRNA*, and the other strand, including matured miRNA, are guided to the target mRNA [64]. In some cases, miRNA* is degraded during this step.
The miRNA generally have 5’-terminal monophosphates and a 2’, 3’ cis-diol at their 3’-terminus. Some 3’ modifications of miRNA have been reported [65, 66]. The two strands of miRNA from an incomplete paired dsRNA; therefore the numbers of miRNAs and their targets could be much greater than currently known.
The catalytic component of the RISC is the Ago protein, which has two domains of PIWI and the Piwi/Argonaute/Zwille (PAZ) for binding the guide strands [67, 68]. The PIWI and the 3’ UTR of the target mRNA were recognized by the bases 2-7 or 8 of the seed in the 5’ end of the miRNA as a guide. In the case of perfect complementary sequences, the target mRNA is sometimes degraded. The mechanisms are similar to those of exogenous siRNA (Fig. 1a). In the case of perfect sequence complementarity, translation of the target mRNA is blocked in the P body by the RNAi machinery [69].
Further, miRNA is sometimes associated with the RNA-induced transcriptional complex (RITS), which induces heterochromatin assembly and inhibits transcription of the target gene [70, 71]. It has been reported that miR-29b localizes in the nucleus and the hexanucleotide element directs the process of nuclear import [72]. This strongly suggests that miRNAs could play a role in the regulation of transcription. Although the exact mechanism of histone modification by small RNAs is unclear, mutations in Dicer, Ago and RNA-directed RNA polymerase (RdRP) result in defects of heterochromatin assembly in S. pombe [73]. RITS contains small RNA, a chromodomain protein (Chp), Ago and some unknown proteins. RITS also recruits the RdRP complex (RDRC) and Swi6 heterochromatin protein 1 (HP1)-family protein. Together with RTIS and RDRC, small RNAs facilitate heterochromatin modification targeting such as methylation of the lysine residue at position 9 of histone H3 (H3K9). Thus, miRNA targets not only the 3’ UTR, but also many other genes, and it is consequently speculated that miRNA could modulate almost all multi-gene expression [24]. In particular, miRNA could regulate several thousand genes, many more than the several hundred suggested in previous reports [74-76].
Although miRNA-mediated gene silencing mechanisms have been implicated in inhibition of translation, protein degradation, capping prevention, inhibition of ribosomal setup, inhibition of mRNA circularization and deadenylation plus decapping in post-initiation process [77, 78], as well as the above-mentioned mRNA processing by intronic miRNAs [79, 80], the effects of RITS interacting with miRNA also remain to be elucidated (Fig. 1a).
INVOLVEMENT OF VIRAL MIRNA GENES IN ONCOGENESIS
Kaposi’s sarcoma (KS) is a neoplasm with protean manifestations, so called opportunistic neoplasm as first described by Moritz Kaposi. This form of KS is designated as classic KS [81]. In early 1980s, KS was found to be one of the manifestations of human immunodeficiency virus type 1 (HIV-1), which causes human acquired immunodeficiency syndrome (AIDS), and this form is designated as epidemic KS [82]. Although HIV infection may be endemic in New York and California, where epidemic KS is also prevalent, epidemic KS is not caused by HIV-1. In 1994, the DNA sequence of a novel herpesvirus was identified in AIDS-associated KS [83]. The virus has been classified as a new human herpesvirus, HHV8 or Kaposi sarcoma-associated herpesvirus (KSHV).
Four KSHV proteins are produced in infected cells, and the proteins - kaposin, v-FLIP, v-Cyclin and the latency-associated nuclear antigen (LANA) - may be critical to KSHV pathogenesis [84]. Although the proteins’ functions have been investigated, the mechanism of oncogenesis is still not clear. miRNAs have also been identified within KSHV and a cluster of 12 miRNAs were found to be encoded in the KSHV latency-associated region (Fig. 1b) [85]. The KSHV miRNA cluster down-regulated SPP1, THBS1, S100A2, PRG1, ITM2A and RAB27A gene expression of HEK 293 cells [86]. S100A2 interacts with the p53 family proteins p67 and p73, and S100A2 can activate p53. Further, PRG1 is a granule proteoglycan secreted by hematopoietic cells, and it is involved in apoptosis. Samols et al. did not discuss the relationship between KS and S100A2 or PRG1, but their observations may be related with KS tumorigenesis.
According to computational and experimental analysis, HIV-1 encodes nine miRNAs (Fig. 1b) [34, 87-89]. HIV-1 infection is associated with many cancers, including the above-mentioned KS; primary central nervous system lymphoma (PCNSL); non-Hodgkin’s lymphoma (NHL); and carcinoma of the uterine cervix, oral, anus and testis. Although approximately 50% of PCNSL is related to EBV [90, 91], it is also known that HIV-1 directly stimulates B lymphocytes causing polyclonal hyperglobulinaemia in serum and follicular hyperplasia in lymphoid tissues. Overall 50% of outcomes remain poor understanding with PCNSL [92].
The HIV-1 miRNA gene (hiv1-miR-H1) is located in U3 region of the 3’-long terminal repeat (LTR) (Fig. 1b) and it down-regulates cellular apoptosis antagonizing transcription factor (AATF) gene expression. The AATF (or Che-1) interacts with Pol II and tumor suppressor retinoblastoma protein (RB) [93]. The AATF is also associated with an endogenous antagonist of prostate apoptosis-4 (Par-4). The Par-4 is reported to be involved in suppression of Bcl2 gene transcription [94]. Therefore, hiv1-miR-H1 (MIRH1) is likely to activate E2F activity and inhibit apoptosis. This may be a co-factor in the tumorigenesis of HIV-1 infection.
SOMATIC MUTATION OF THE RESIDENT miRNA
Human carcinogenesis is caused by the somatic mutations. These mutations may be induced by environmental agents, such as ionizing radiation (1.5-2.0 Gy) and/or occur spontaneously. Environmentally-induced mutation occurs as a break in single-stranded DNA, leading to damaged dsDNA, which plays a major role in carcinogenesis, according to the two-hit kinetics hypothesis (Fig. 2a) [95]. Expanding the hypothesis of two-hit kinetics to the level of chromosomal mutation in loss of a tumor suppressor gene, the first hit is heterozygotic mutation and the second hit is homozygotic one.
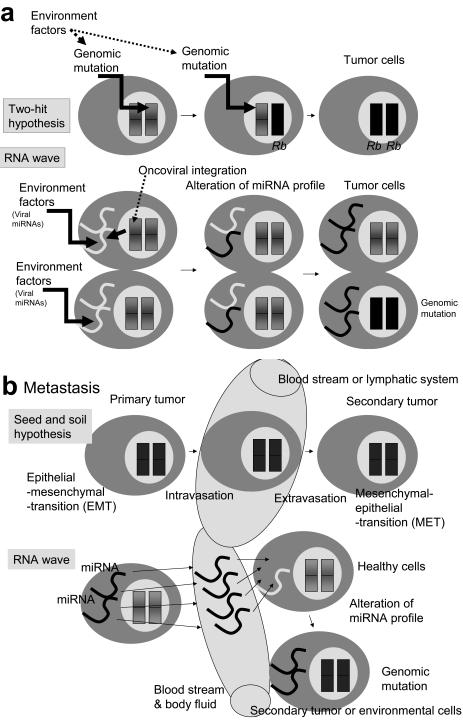
Fig. (2).
Incorporation of tumorigenesis and metastasis into the RNA wave. (a) Knudson’s two-hit hypothesis and the RNA wave. Sporadic retinoblastoma requires two hits of somatic mutation in homozygote gene by environmental factors [10]. The mutation of RNA is less stable than DNA and RNA has no repair enzymes. Environmental factors including viral miRNA or integrated proto-oncovirus miRNA can qualitatively and quantitatively change the profiles of resident miRNAs of cells. Deleted or ectopic miRNA alterations (called mutations) initiate tumorigenesis. In this case, the first hit to the resident miRNA genes, and the second hit to the genomic miRNA genes. A more sophisticated Knudson hypothesis may involve into more advanced RNA wave. (b) In the seed and soil hypothesis of metastasis, the primary tumor cells disseminate by intravasation to the blood or lymphatic system. This process requires an epithelial-mesenchymal transition (EMT) by the primary tumor cells, as well as primary tumors extravasation to distant tissues. There a mesenchymal-epithelial transition (MET) enables the primary tumors to change into the secondary tumors (metastasis). On the other hand, miRNAs such as miR-21 expressed in breast and ovarian cancer can easily be transported into the blood and possibly the lymphatic system. The cells in metastasis sites or their environment can alter profiles of miRNA expression by incorporation of the new miRNA into the cells. The change in the resident miRNAs may induce a change in the genomic miRNA and tumorigenesis via epigenetic chromatin modeling or chromosome mutation under the RNA wave. In this case, transfer of primary tumor cells with subsequent EMT and MET is not required at all.
The RB gene has been reported to be associated with chromosomal changes at 13q14 in homozygotic mutation [96]. Recently, in mouse models, ultraviolet B irradiation (50J) was reported to change the profile of miRNA in mouse NIH3T3 cells; miR-365 and oncogenic miR-21 were also upregulated [97]. Further, the radiation exposure resulted in significant alteration of miRNA expression in spleen and thymus tissues, and tumor suppressor miR-34a was increased, paralleled by a decrease in the expression of its target c-myc gene [98]. In human fibroblast cells, exposure of radiation (0.1-2.0 Gy) caused downregulation of miR-92b, a tumor suppressor (Table 1) [99]. These data suggest that miRNAs may act as specific regulators of cellular responses to environmental exposures, even if lower radiation doses than the level required to break of dsDNA, and miRNAs can contribute to cell tumorigenesis and/or repair (Fig. 2a).
Table 1.
Oncomir and Tumor Suppressor miRNA Profiles in Human Cancers
| Human | miRNA Involvement | |||
|---|---|---|---|---|
| Cancer | Oncomir | Suppressor miRNA | Oncogene Target | References |
| Oncoviral integration-induced | ||||
| Lymphomaa) | mir-1204 | [166] | ||
| (PVT-1) | ||||
| Erythroleukemiaa) | miR-17-92 | [165] | ||
| (Fli-3) | ||||
| T-cell leukemia | miR-106a | c-myc | [164] | |
| (Kis2) | ||||
| miR-19b-2 | ||||
| miR-92-2 | ||||
| miR-20b | ||||
| B-cell leukemia | miR-155 | miR-17-92 | [44] | |
| (BIC) | [142] | |||
| Non-oncoviral | ||||
| Brain cancer | miR-21 | miR-181 | [169] | |
| miR-221 | [135] | |||
| Breast cancer | miR-125b | [134] | ||
| miR-145 | ||||
| miR-21 | ||||
| miR-155 | ||||
| Chronic | miR-15 | BCL2 | [120, 170] | |
| lymphocytic | miR-16 | [119] | ||
| leukemia | ||||
| Gastric cancer | miR-34b | miR-128b | [171] | |
| miR-34c | miR-129 | |||
| miR-128c | miR-148 | |||
| miR-106b | [172] | |||
| miR-222 | ||||
| Colorectal | miR-143 | v-Ki-ras (KRAS) | [43] | |
| neoplasia | miR-145 | [173] | ||
| Pancreatic cancer | miR-21 | miR-375 | [174] | |
| miR-221 | ||||
| miR-222 | ||||
| miR-181a, b, d | ||||
| miR-155 | [145] | |||
| Hepatocellular | miR-18 | miR-199 | [175] | |
| carcinoma | miR-224 | miR-195 | ||
| miR-200 | ||||
| miR-125 | ||||
| miR-101 | v-fos | [176] | ||
| Lung cancer | miR-17-92 | let-7 | Ras | [136] |
| let-7a | [137] | |||
| [177] | ||||
| [138] | ||||
| Papillary | miR-222 | [130] | ||
| thyroid | miR-146 | [178] | ||
| carcinoma | miR-181 | |||
| Teasticular | miR-372 | [179] | ||
| germ cell | miR-373 | |||
| tumor | ||||
| Ovarian cancer | miR-21 | [116] | ||
| miR-141 | ||||
| miR-200a, b, c | ||||
| miR-203 | ||||
| miR-205 | ||||
| (In sera specimens) | ||||
Note:
mouse-model.
In addition, it is well known that RNA does not have repair mechanisms, therefore the resident miRNA is more susceptible to environmental stimulation than genomic DNA. The two-hit theory is involved in the RNA wave. Without dsDNA damage, the X-ray-susceptible resident miRNA genes that are somatically mutated and/or the quantitatively changed miRNA profiles (the first hit) can be integrated into the DNA genome via transposing RNA, like the exosomal miRNAs [100] and then the altered genomic miRNA gene expression (the second hit) affects the phenotypic alteration of cells. It has been shown that the miR-125b-1 gene is inserted into IGH allele in B-cell acute lymhpoblastic leukemia by unknown mechanisms [101]. This process, rather than integration itself, may epigenetically introduce to a defect into a chromosome [102].
Metastasis, the spread and growth of a primary tumour, is the most common cause of death for cancer patients. The primary tumor cells intravasate into the blood stream or lymphatic system and invade into new organ [103]. Then the secondary tumor (the seed) must be able to grow in the microenvirnment (the soil) of the secondary site. This is the seed and soil hypothesis. In this hypothesis, tumor cells switch their phenotype from epithelial-mesenchymal transition (EMT) to mesenchymal-epithelial transition (MET) (Fig. 2b) [104].
Matastasis-related miRNAs has been reported. For example, miR-21 oncogenic miRNA is upregulated in metastasis of hepatocellular carcinoma, B cell lymphoma, cholangiocyte, and glioblastoma, as well as breast, colorectal, gastric, lung, ovarian, uterine, cervical, prostate, oesophageal, and pancreatic cancers [105]. miR-21 suppresses PDCD4, MASPIN, PTEN, TPM1 and TIMP3 metastasis suppressors [106-110]. The importance of other miRNAs, including miR-10b (upregulated), miR-126 (downregulated) and miR-206 (downregulated) have also been demonstrated, and HOXD10 or SOX4 transcription factors are regulated by these miRNAs [111-115]. HOXD10 or SOX4 expression decreases cancer malignancy by inhibition of tumor cell migration and ECM remodeling. Recently, the metastatic and oncogenic miR-21 gene was observed in serum specimens of ovarian cancer [116].
In general, miRNA levels are similar among healthy individuals [117]. This finding supports the RNA wave that miRNA genes have mobile genetic element (MGE)-like mobility. Therefore, without transformation a primer tumor cell from EMT to MET, miRNA information can be sent to oncogenic-susceptible cells in the secondary site via the blood stream (Fig. 2b). Thus, new opportunities for metastasis may arise from a circulating wave of oncogenic miRNAs in body fluid.
ONCOGENIC AND TUMOR SUPPRESSOR miRNAs: CELL CYCLE ARREST INDUCED BY miRNAs
Proper regulation of the cell cycle is critical to regulation of cell growth and the prevention of cancer. Some miRNAs play an important role in initiation and progression of human cancer (Table 1). has-miR-15a and miR-16-1 are located at human chromosome 13q14, and deletion of this region causes B-cell chronic lymphocytic leukemia (CLL). An approximately 30 kb region of 13q14 contains both genes and approximate 68% CLL cases are the deleted mutation of this region [118].
Further, expression of miR-15a and miR-16-1 has been reported to be related to Bcl2 expression in CLL and to down-regulate bcl2 gene expression [119]. As a result of Bcl2 suppression, both genes induced apoptosis in tumor cells. Therefore, the miR-16 cluster inhibited the growth of CLL engrafts in nude mouse [120]. The miR-16 family negatively regulates cellular growth and the cell cycle to trigger G0/G1 accumulation via Cyclin D1 (CCND1) suppression [121, 122].
Retinoblastoma is one of genetic diseases, and approximately 40% of cases are familiar (MIM 180200) [10]. The two-hit hypothesis can be applied to RB. The RB gene as well as TP53 suppress tumor growth and is related to the cell cycle. RB is activated by dephosphorylation, and binds to cellular transcriptional factor E2F, and then cell cycle is stopped in G1/S check-point. CCND1 is cooperated to Cyclin kinase 4/6 (CDK4/6) complex and recruit RB [123-125]. While inactivation of RB is induced by phosphorylation of RB (Fig. 3a), deletion of miR-15 and miR-16 causes RB hyperphosphorylation and desregulation of cell growth. This suggests that miR-15 and miR-16 are regulators of the RB/E2F pathway. Therefore, miR-15a and miR-16-1 are tumor suppressors.
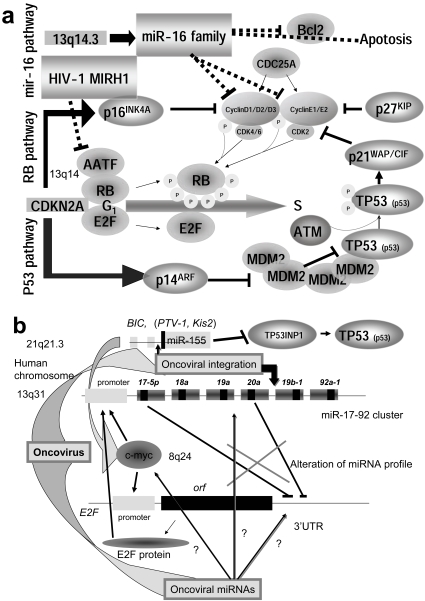
Fig. (3).
Master regulator miRNA of oncogenic and tumor suppressor. (a) The tumor suppressor has two pathways, RB and p53 pathway. The miR-16 family regulates the cell cycle G1/S by modulating CDK4/6-Cyclin D1/D2/D3 and CDK2-Cyclin E1/E2 complexes. Two complexes are related to RB phosphorylation, and the phosphorylated RB is released from E2F. Thus the activated E2F drives infinitive transcription from G1 into S. Another player in carcinogenesis is CDKN2A, which encodes p16INK4A and p14ARF. The former is transcribed from exons 1α, 2 and 3, and the latter is from exon 1β, 2 and 3 with different reading frames of the CDKN2A gene. p16 inhibits the CDK4/6-Cyclin D complex and p14 mediates G1 arrest by destabilizing the MDM2 protein. The MDM2 oncogene induces many sarcomas by binding to p53 (TP53). Therefore, p14 maintains the level of p53. p53 phosphorylated by ATM inhibits CDK2/Cyclin E complex via p21WAP/CIP activation with p27KIP. The main mechanism of tumor suppression is intervention by CDK4/6-Cyclin D and CDK2-Cyclin E. miR-16 can induce apoptosis by suppression of Bcl2. Thus, the miR-16 family plays an important role for tumor suppression. Although viral oncoproteins, such as adenovirus E1A, SV40 T antigen, as well as human papillomavirus E7, bind RB, the HIV-1 viral MIRH-1 was recently shown to suppress AATF which inhibits E2F activation by association with RB. Therefore, oncoviral MIRH1 may be tumorigenic. (b) Cell cycle transcription by E2F is also modulated by a c-myc-regulated miRNA cluster miR-17-92 [138]. In the upper part of the figure showing the miR-17-92 cluster, the large box represents the pre-miRNA and the small box shows the mature miRNA. MYC (c-myc) promotes the transcription of both miR-17-92 and E2F by binding at the cacgtg site on the miR-17-92 gene and the E2F promoter. miR-17-5p and miR-20a downregulate E2F gene expression. miR-17-92 results in the inhibition of apoptosis via the p14ARF pathway. miR-155 oncogenic miRNA in oncoviral integration site BIC activates c-myc and inhibits p53 by inactivation of TP53INP1. In some cases, oncoviral integration upregulates miR-155 expression, but in other cases, oncoviral integration inhibits miR-17-92 expression. Thus, oncoviral integration is tumorigenic. However, no relation between integration site and miRNA genes has been reported. The primary cause of altered miRNA expression in cancer cells is still unaccounted for. While miR-125b-1 insertion into IGH locus has been found in acute lymphoblastic leukemia, if the oncovirus takes the resident miRNA as the MGE, the mobile miRNA may be able to alter the profiles of miRNA in infected cells. Subsequently, hidden oncoviral miRNAs may have an important role for tumorigenesis in the cells.
E2F1, E2F2 and E2F3 are all activators of cell cycle progression [126-129]. During G1, none of E2F is associated with RB. Therefore, either inactivation of RB or E2F1 expression can promote apoptosis. The miR-17a and miR-20 genes inhibited the translation of the E2F1 mRNA. Further, the miR-17-92 cluster is regulated by MYC protein in human B-cell lymphoma [130]. These results suggest that miR-17a and miR-20 can act as tumor suppressive miRNA [131].
E2F1-3 expression was blocked by miR-20a via targeting of E2F1-3 mRNA at the 3’ UTR, and the promoter of the miR-17-92 cluster was responsive to the E2F1-3 transcriptional factors. This suggests that there is an auto-regulatory mechanism in which E2F1-3 gene expression is controlled by miR-20a, while the miRNA expression itself is modulated by E2F1-3 at the transcriptional level (Fig. 3b) [132]. The miR-17-92 cluster is located on human chromosome 13q31. It has been shown that this region is associated with several types of lymphoma and solid tumors [133]; however, individual miRNAs could not accelerate tumor formation. The miR-17-92 cluster from polycistron suppresses cell death and extensive apoptosis and promotes high proliferation of cells. These results suggest that oncogenicity requires a cooperative interaction among miRNAs in the cluster or a mutation in the MYC protein. The genomic miRNA genes could regulate cell cycle.
EPIGENETIC AND APOPTOSIS-RELATED miRNAs
The miR-21 gene was found to be upregulated in breast cnacer [135] and glioblastomas [135]. When expression of miR-21 was inhibited in glioblastomas, caspases were activated and an apoptotic pathway was enhanced [135]. Therefore, miR-21 may be an anti-apoptotic factor in human glioblastoma cells as well as a metastasis-related factor as mentioned above.
let-7 miRNA was reduced in lung cancer [136]. Although let-7 down-regulates RAS and MYC by targeting their mRNAs [137, 138], RAS and MYC over expression have been implicated in lung cancer. Therefore, it is suggested that regulation of let-7 levels may be a key to oncogenesis in human lung tissue. However, there is also evidence to the contrary. In some lung and colon cancers, let-7 was over expressed [139]. While some miRNA promoter regions contain putative p53 tumor suppressor (TP53) consensus sites, it is well known that the loss of p53 induces oncogenesis. The expression levels of let-7g, miR-181b and miR-200c were elevated in p53-deleted colon cancer [139]. The human let-7a-3 gene on chromosome 22q13.31 was associated with a CpG island, and elevated expression of let-7a-3 in a human lung cancer resulted in enhanced tumor phenotypes and oncogenic changes [140]. Although human cancer cells are partly characterized by chromosomal changes in genomic DNA methylation compared with nonmalignant cells, Saito et al. recently treated cancer cells with a DNA methyltransferase inhibitor, 5-aza-2’-deoxycytidine (DAC), and showed that miR-127 was affected by DNA hypermethylation [141]. These results suggest that miRNA may be partially responsible for epigenetic gene reactivation in tumor cells.
ONCOMIR IN OTHER CANCERS
Evaluation of breast cancer tissue using an miRNA microarray showed that miR-125b, miR-145, miR-21 and miR-155 were significantly suppressed in cancer tissues [134]. In the case of Burkitt’s lymphoma, the expression of miR-155 increases [142, 143]. BIC, a proto-oncogene, is known to be activated by retroviral provirus insertion in certain human B cell lymphomas as well as several other cancers; however, the molecular basis of BIC-related cancer is unknown. Recently it was reported that the miR-155 gene locates in the region of the BIC gene (See Fig. 3b) and miR-155 is thought to be responsible for the oncogenic activity of BIC via c-myc oncogene activation [142, 144]. Further, miR-155 suppressed TP53-induced nuclear protein 1 (TP53INP1) expression and indirectly inhibited TP53 activity in pancreatic tumor (Table 1) [145].
Although Salter et al. found that the miRNA profiles of normal and breast or lymphatic cancer tissues were significantly different, it has also been observed that miRNA expression pattern correlates with cancer stage, and the proliferation index is used for diagnosis as well as the prediction of chemotherapeutic response [146]. Further, miR-196a2 SNPs (rs11614913C/T) were related to survival of individuals with non-small cell lung cancer [147]. In addition, oncoviral integration stimulates miRNA expression at the integraiton site, and upregulation of oncogenic miRNA initiates tumorigenesis. Thus, it is suggested that combinational information of the resident miRNA genes, partially virally oncogenic miRNA genes, play an important role in tumorigenesis.
MECHANISMS OF TUMORIGENESIS BY VIRAL miRNAs
miRNA may also be the master regulator of tumorigenesis. In the case of the human genome, miRNAs are usually transcribed from the intergenic non-coding region, either from introns or, less frequently from exons of the protein coding region. The intergenic non-coding region is corresponding to approximate 98% of genomic information, including retrotransposable elements (TEs) such as long interspersed elements (LINEs), short interspersed elements (SINEs), Retrotransposons and transposons.
Transposons have been discovered by McClintock approximately 50 years ago [148], TEs have been found in 46.2% of the human genome, 34.2% of canine, 38.7% of murine, and 9.4% of avian genomes [149]. TEs are divided into class I and II. Class I elements are retroelements including retrotransposon, LINEs, and SINEs. The LTR retrotransposon is closely related to retroviruses, such as HIV-1, human RTL1/MART1, human endogenous retrovirus-K (HERV-K), Drosophila gypsy and Copia. Class II elements include DNA transposons. The Activator-Dissociation (Ac-Ds) family in maize, Tam in Antirrhinum, the P element in Drosophila, and the Tc1 element in C. elegans are well known. The recent discovered miniature inverted-repeat TE (MITE) has some properties of both class I and II elements. MITEs have been reported in plants, humans, Xenopus, the yellow fever mosquito and Aedes aegypti [150]. DNA hypomethylation is linked to activation of these TEs, L1 or Alu and tumor metastasis [151]. The activation of TE allows for them to be transcribed and translocated to other genomic alleles, and for them to induce genomic aberrance. There is a relation between hypomethylation of TEs in neuroendocrine tumor and lymph node metastasis [152].
Dobzhansky, who had a very special interest in gene mutation, famously said that nothing about TEs makes sense except in the light of evolution. But at the same time, he realized that a major piece of the mutation puzzle was still missing [153]. It is likely that the missing piece is miRNAs. Both type I and II TEs have hairpin structures, and the stem looping produces miRNAs. Further, genetic research on viroids has revealed that TEs are suppressed by RNA silencing [154]. The Tc1 can be suppressed in an RNAi-dependent manner, and retrotransposons of S. pombe have also been silenced via the RNAi pathway. The murine endogenous retrovirus-L (MuERV-L) and intracisternal A particle (IPA) retrotransposons have been silenced by Dicer expression [155]. From experiments on Dicer knockout (KO) human cells, L1 has been reported to be expressed [156].
Interestingly, miRNAs have been emerged from TEs. The retrotransposon-like gene 1 (RTL1/MART1) (approximate 1.3 kb, 5 exon and 4 intron) in the DLK1/MEG3-DIO3 imprinted domain (14q32) of the human genome includes at least five miRNAs (Fig. 1b) [157, 158]. In the DLK/MEG3 region, seven imprinted genes have been annotated; three genes, DLK1, RTL1 and DIO3, are paternal, and four genes, MEG3, anti-RTL1, MEG8 and MIRG are maternal. The paternally-expressed genes encode protein, but the maternally expressed genes represent non-coding RNA. Imprinting genes that implicate cell growth are often shown to aberrantly express in cancer [159, 160]. The miRNAs are transcribed only from the maternal chromosome in the mouse model, as are miR-127 and miR-136. RT-PCR showed that miR-136 is generated from a larger hot transcript in the anti-RTL gene, while miR-127 was only detected by northern blotting [161]. The generation of miR-136 was determined but miR-127 usually could not be detected.
In the embryo brain or in the placenta, imprinting miR-127 and RTL1 are co-expressed, suggesting that miR-127 can maternally regulate TE itself. Further, DIRS-1 and Skipper retrotransposons of the social amoeba encode miRNAs, suggesting that miRNAs in TE may generally regulate TE [162]. Genome-wide research for miRNAs in human cells failed to uncover miR-127 function for the brain and the placenta, therefore, the meaning of above results has not yet been understood at all. In 2005, Smalheiser and Torvik have examined with the RepeatMasker algorithm for prediction of pre-miRNA and showed that the miRNA precursors of several highly conserved miRNAs involved sequences of TEs [163]. Similar to miR-N367 (or other 6 miRNAs, TAR-3P, TAR-5P, #1, #4, #5, and H1) derived from HIV-1 3’-LTR (See Fig. 1b), LINE-2 miRNAs, miR-95 and miR-151 were mapped in the 3’ end of the LINE-2 sequence [163], and it is possible that both miRNAs target LINE-2-derived mRNA transcripts. Thus, we speculate that miRNAs might be hidden in TEs like oncoviruses in the chromosomal domain, and that the oncovirus-derived miRNAs might control the TEs themselves, while viral miRNAs might simultaneously have ability to regulate the expression of other cellular transcripts, such as those involved in tumorigenesis. This idea has involved into the RNA wave beyond the innate immune system.
As the proof of above speculation, murine radiation leukemia virus (RadLV) integration into the Kis2 locus gave rise to overexpression of ncRNA, which contains an miRNA cluster that includes miR-106a, -18b, -19b-2, -20b, -92-2 and -363. The overexpression of these miRNAs by oncoviral integration showed the oncogenic potential to murine NIH-3T3 cells [164]. The murine mir-106-363 cluster is homologous to the above mentioned miR-17-92 cluster in humans.
At the integration site of the Friend murine leukemia virus (F-MuLV), the Fli-3 locus encodes the miR-17-92 cluster. Induction of the miR-17-92 expression enhances erythroblast transformation by F-MuLV integration [165]. Further, the Pvt1 locus in murine encodes miR-1204, -1205, -1206, -1207-5p, -1207-3p and -1208. Except for miR-1206, all of these miRNAs have been expressed in T lymphocytes [166]. Although multiple myeloma cases without translocation of immunoglobulin gene have been observed, oncovirus integration into the Pvt1 locus has been shown to induce T lymphoma, and alteration of miRNA expression caused upregulation of 400kb downstream of the myc gene expression. However, there is no explanation for why integration of oncovirus without an oncogene induces alteration of miRNA expression. Further, no association has been detected between xenotropic murine leukemia virus-related virus (XMRV) interaction sites and proto-oncogene, tumor suppressor gene, and miRNA gene which have been implicated in human prostate cancers [167]. Since speculative viral miRNAs were documented in human T-cell leukemia (lymphotropic) virus-1 (HLTV-1) oncogenesis [168], these pathogenic RNAs may involve miRNAs. This can be predicted in silico. In fact, HIV-1 provirus is known to express MIRH1 at certain state of infection T lymphocyte as mentioned above. Including the viroid case, retrotransposons or endogenous retroviruses as proviruses would encode miRNAs as hidden miRNAs and the viral miRNA could target miRNA itself, the miRNA cluster of its integration site, the actual oncogenes, and/or the tumor suppressor genes.
CONCLUSIONS
The RNA wave consists of four concepts: 1) infection induces miRNA production in the virus and/or host; 2) the induced miRNAs have MGE-like mobility; 3) the mobile miRNAs can self-proliferate; and 4) cells contain both resident and genomic miRNAs. Based on those four concepts, it is possible that miRNA cannot only mediate RNA silencing to control expression MGEs, but also that miRNA may be necessary to incorporate MGEs into the genome for evolution. Therefore, the miRNA gene has both mobile and functional genetic elements. Although profiles of expression of oncomir and/or tumor suppressor miRNAs have shown that aberrant expression of the genomic miRNA results in tumorigenesis, which substances change miRNA expression and why have not been ascertained. Therefore, the primary cause of altered miRNA expression in cancer cells is still unaccounted for.
One of environmental factors based on the two-hit hypothesis could be oncovirus as the MGE inserting into the integration loci, which encodes the miRNA genes; as a result, the insertion would alter the expression profile of the miRNA genes. This process has been shown to induce tumorigenesis; however, if pathogenic resident miRNA were picked up into an oncovirus as a tag and then ectopically integrated into the chromosome, the integrated miRNA gene as would act like foreign DNA and could result in imposition of epigenetic regulation, like imprinting, that may occasionally induce tumorigenesis in host cells. Nevertheless, the pathogenic viral miRNA and activated endogenous retrotransposon miRNA should translocate and duplicate the miRNA genes within the host chromosome. Tumorigenesis may result from a “tsunami” of emerging oncogenic viral RNA codes in the oncoviral genome.
ACKNOWLEDGEMENTS
I thank Dr. C. Rowthon for his knowledgeable English editing.
ABBREVIATIONS
- AATF
= Apoptosis antagonizing transcription factor
- abl
= Abelson mouse leukemia
- Ac-Ds
= Activator-Dissociation
- Ago
= Argonaute
- AIDS
= Human acquired immunodeficiency syndrome
- β-gal
= β-galactosidase
- CDK4/6
= Cyclin kinase 4/6
- Chp
= Chromodomain protein
- CLL
= Chronic lymphocytic leukemia
- c-onc
= Cellular oncogene
- DGCR8
= DiGeorge syndrome critical region gene 8
- dsDNA
= Double-stranded DNA
- dsRNA
= Double-stranded RNA
- EBV
= Epstein-Barr virus
- EMT
= Epithelial-mesenchymal transition
- ES
= Embryonic stem
- EXP5
= Exportin 5
- F-MuLV
= Friend murine leukemia virus
- GFP
= Green fluorescent protein
- HERV-K
= Human endogenous retrovirus-K
- HIV-1
= Human immunodeficiency virus type 1
- H3K9
= Methylation of the lysine residue at position 9 of histone H3
- HLTV-1
= Human T-cell leukemia (lymphotropic) virus-1
- HP1
= Heterochromatin protein 1
- IG
= Immunoglobulin
- IPA
= Intracisternal A particle
- KO
= Knockout
- KS
= Kaposi’s sarcoma
- KSHV
= Kaposi sarcoma-associated herpesvirus
- LANA
= Latency-associated nuclear antigen
- LINE
= Long interspersed element
- LTR
= Long terminal repeat
- MET
= Mesenchymal-epithelial transition
- MGE
= Mobile genetic element
- miRISC
= miRNA-induced silencing complex
- miRNA
= microRNA
- MITE
= Miniature inverted-repeat TE
- MLV
= Murine leukemia virus
- mRNA
= Messenger RNA
- myc
= Avian myelocytomatosis
- NHL
= Non-Hodgkin’s lymphoma
- nts
= Nucleotides
- oncomir
= Oncogenic miRNA
- PACT
= Protein activator of protein kinase activated by dsRNA
- Par-4
= Prostate apoptosis-4
- PAZ
= Piwi/Argonaute/Zwille
- P-body
= Processing body
- PCNSL
= Primary central nervous system lymphoma
- pre-miRNA
= Precursor miRNA
- Ph
= Philadelphia
- pri-miRNA
= Primary miRNA
- Pol
= RNA polymerase
- PVT
= Plasmacytoma variant translocation
- QTL
= Quantitative trait loci
- RB
= Retinoblastoma (protein)
- RDRC
= RdRP complex
- RdRP
= RNA-directed RNA polymerase
- RISC
= RNA-induced silencing complex
- RITS
= RNA-induced transcriptional complex
- RNAi
= RNA interference
- SINE
= Short interspersed element
- siRNA
= Short interfering RNA
- TE
= Retrotransposable element
- TGS
= Transcriptional silencing
- TP53INP1
= Tumor protein 53-induced nuclear protein 1
- TRBP
= Trans-activator RNA (TAR)-binding protein
- UTR
= Untranslated region
- v-onc
= Viral oncogene
- XMRV
= Xenotropic murine leukemia virus-related virus
REFERENCES
- 1.Chissoe SL, Bodenteich A, Wang Y-F, et al. Sequence and analysis of the human ABL gene, the BCR gene, and regions involved in the Philadelphia chromosomal translocation. Genomics. 1995;27:67–82. doi: 10.1006/geno.1995.1008. [DOI] [PubMed] [Google Scholar]
- 2.Huppi K, Siwarski D. Chimeric transcripts with an open reading frame are generated as a result of translocation to the PVT-1 region in mouse B-cell tumors. Int J Cancer. 1994;59:848–51. doi: 10.1002/ijc.2910590623. [DOI] [PubMed] [Google Scholar]
- 3.Shtivelman E, Henglein B, Groitl P, Lipp M, Bishop JM. Identification of a human translocation unit affected by the variant chromosomal translocations 2;8 and 8;22 of Burkitt lymphoma. Proc Natl Acad Sci USA. 1989;86:3257–60. doi: 10.1073/pnas.86.9.3257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cory S, Graham M, Webb E, Corcoran L, Adams JM. Variant (6;15) translocation in murine plasmacytomas involve a chromosome 15 locus at least 72 kb from the c-myc oncogene. EMBO J. 1985;4:675–81. doi: 10.1002/j.1460-2075.1985.tb03682.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Palumbo AP, Boccadoro M, Battaglio S, et al. Human homologue of moloney leukemia virus integration-4 locus (MLVI-4), located 20 kilobases 3’ of the myc gene, is rearranged in multiple myelomas. Cancer Res. 1990;50:6478–82. [PubMed] [Google Scholar]
- 6.Beck-Engeser GB, Lum AM, Huppi K, et al. Pvt1-encoded microRNAs in oncogenesis. Retrovirology [serial on the Internet] 2008. Jan, [[cited 2008 January 14] 5:4 [about 14 screens]]. Available from: http://www.retrovirology.com/content/5/1/4 . [DOI] [PMC free article] [PubMed]
- 7.Bao L, Zhou M, Wu L, et al. PolymiRTS database: linking polymorphisms in microRNA target sites with complex traits. Nucleic Acids Res. 2007;36:D51–D54. doi: 10.1093/nar/gkl797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Street NR, Sjödin A, Bylesjö M, et al. A cross-species transcriptomics approach to identify genes involved in leaf development. [[cited 2008 December 5] 9: 589 [about 18 screens]];GMC Genom [serial on the Internet] 2008 Dec; doi: 10.1186/1471-2164-9-589. Available from: http://www.biomedcentral.com/1471-2164/9/589 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fujii YR. Formulation of new algorithmics for miRNAs. [[cited 2008 April 7] 2:37-43 [about 7 screens]];Open Virol J [serial on the Internet] 2008 Apr; doi: 10.2174/1874357900802010037. Available from: http://bentham.org/open/tovj/openaccess2.htm . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Knudson AG. Two genetic hits (more or less) to cancer. Nature Rev Cancer. 2001;1:157–62. doi: 10.1038/35101031. [DOI] [PubMed] [Google Scholar]
- 11.Jorgensen R. Altered gene expression in plants due to trans interactions between homologous genes. Trends Biotechnol. 1990;8:340–4. doi: 10.1016/0167-7799(90)90220-r. [DOI] [PubMed] [Google Scholar]
- 12.Van de Krol AR, Mur LA, Beld M, et al. Flavonoid genes in petunia: addition of a limited number of gene copies may lead to a suppression of gene expression. Plant Cell. 1990;2:291–9. doi: 10.1105/tpc.2.4.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baulcombe D. RNA silencing in plants. Nature. 2004;431:356–63. doi: 10.1038/nature02874. [DOI] [PubMed] [Google Scholar]
- 14.Napoli C, Lemieux C, Jorgensen R. Introduction of a chimeric chalcone synthase gene into petunia results in reversible co-suppression of homologous genes in trans. Plant Cell. 1990;2:279–89. doi: 10.1105/tpc.2.4.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fire A, Xu S, Montogomery M, et al. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–11. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 16.Fillman C, Lykke-Anderson J. RNA decapping inside and outside of processing bodies. Curr Opin Cell Biol. 2005;17:326–31. doi: 10.1016/j.ceb.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 17.Liu J, Rivas FV, Wohlschlegel J, et al. A role for the P-body component GW182 in microRNA function. Nat Cell Biol. 2005;7:1261–6. doi: 10.1038/ncb1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ding L, Spencer A, Morita K, Han M. The development timing regulator AIN-1 interacts with miRISCs and may target the Argonate protein ALG-1 to cytoplasmic P bodies in C. elegans. Mol Cell. 2005;19:437–47. doi: 10.1016/j.molcel.2005.07.013. [DOI] [PubMed] [Google Scholar]
- 19.Borsani O, Zhu J, Verslues PE, Sunka R, Zhu JK. Endogenous siRNAs derived from a pair of natural cis-antisense transcripts regulate salt tolerance in Arabidopsis. Cell. 2005;123:1279–91. doi: 10.1016/j.cell.2005.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee RC, Hammell CM, Ambros V. Interacting endogenous and exogenous RNAi pathways in Caenorhabditis elegans. RNA. 2006;12:589–97. doi: 10.1261/rna.2231506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bennasser Y, Le S-Y, Benkirane M, Jeang K-T. Evidence that HIV-1 encodes a siRNA and a suppressor of RNA silencing. Immunity. 2006;22:607–19. doi: 10.1016/j.immuni.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 22.Ying SY, Lin SL. Intronic microRNA (miRNA) Biochem Biophys Res Commun. 2005;326:515–20. doi: 10.1016/j.bbrc.2004.10.215. [DOI] [PubMed] [Google Scholar]
- 23.Miranda KC, Huynh T, Tay Y, et al. A pattern-based method for the identification of microRNA binding sites and their corresponding heteroduplexes. Cell. 2006;126:1203–17. doi: 10.1016/j.cell.2006.07.031. [DOI] [PubMed] [Google Scholar]
- 24.Rigoutsos I. Pyknons as putative novel and organism-specific regulatory motifs. In: Morris KV, editor. RNA and the regulation of gene expression. London: Horizon Scientific Press; 2008. pp. 149–63. [Google Scholar]
- 25.Cullen BR. Viruses and microRNAs. Nat Genet. 2006;38:S25–S30. doi: 10.1038/ng1793. [DOI] [PubMed] [Google Scholar]
- 26.Lin J, Cullen BR. Analysis of the interaction of primate retroviruses with the human RNA interference machinery. J Virol. 2007;81:12218–26. doi: 10.1128/JVI.01390-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yeung ML, Bennasser Y, Le SY, Jeang KT. siRNA, miRNA and HIV: promises and challenges. Cell Res. 2005;15:935–46. doi: 10.1038/sj.cr.7290371. [DOI] [PubMed] [Google Scholar]
- 28.Qi P, Han J, Lu Y, Wang C, Bu F. Virus-encoded microRNAs: future therapeutic targets? Cell Mol Immunol. 2006;3:411–9. [PubMed] [Google Scholar]
- 29.Scaria V, Hariharan M, Maiti S, et al. Host-virus interaction: a new role for microRNAs. [[cited 2006 October 11] 3: 68 [about 9 screens]];Retrovirology [serial on the Internet] 2006 Oct; doi: 10.1186/1742-4690-3-68. Available from: http://www.retrovirology.com/content/3/1/68 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Konstantinova P, Ter Brake O, Haasnoot J, De Haan P, Berkhout B. Trans-inhibition of HIV-1 by a long hairpin RNA expressed within the viral genome. [[cited 2007 March 1] 4: 15 [about 14 screens]];Retrovirology [serial on the Internet] 2007 Mar; doi: 10.1186/1742-4690-4-15. Available from: http://www.retrovirology.com/content/4/1/15 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ludwig LB. RNA silencing and HIV: hypothesis for the etiology of the severe combined immunodeficiency induced by the virus. [[cited 2008 September 11] 5: 79 [about 13 screens] ];Retrovirology [serial on the Internet] 2008 Sep; doi: 10.1186/1742-4690-5-79. Available from: http://www.retrovirology.com/content/5/1/79 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Corbeau P. Interfering RNA and HIV: reciprocal interference. [[cited 2008 September 26] 4: e1000162 [about 7 screens]];PLoS Pathog [serial on the Internet] 2008 Sep; doi: 10.1371/journal.ppat.1000162. Available from: http://www.plospathogens.org/article/info%3Adoi%2F10.1371%2Fjournal.ppat.1000162 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yeung ML, Bennasser Y, Jeang KT. miRNAs in the biology of cancers and viral infection. Curr Med Chem. 2007;14:191–7. doi: 10.2174/092986707779313417. [DOI] [PubMed] [Google Scholar]
- 34.Kaul D, Ahlawat A, Gupta SD. HIV-1 genome-encoded hiv1-mir-H1 impairs cellular responses of infection. Mol Cell Biochem. 2008;323:143–8. doi: 10.1007/s11010-008-9973-4. [DOI] [PubMed] [Google Scholar]
- 35.Fujii YR, Saksena NK. Viral infection-related microRNAs in viral and host genomic evolution. In: Morris KV, editor. RNA and the regulation of gene expression. London: Horizon Scientific Press; 2008. pp. 91–107. [Google Scholar]
- 36.Reinhart BJ, Slack FJ, Basson M, et al. The 21-Nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–6. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 37.Chen XM. A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science. 2004;303:2022–5. doi: 10.1126/science.1088060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brennecke J, Hipfner DR, Stark A, Russell RB, Cohen SM. Bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell. 2003;113:25–36. doi: 10.1016/s0092-8674(03)00231-9. [DOI] [PubMed] [Google Scholar]
- 39.Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303:83–6. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- 40.Krichevsky AM, King KS, Donahue CP, Khrapko K, Kosik KS. A microRNA array reveals extensive regulation of microRNAs during brain development. RNA. 2003;9:1274–81. doi: 10.1261/rna.5980303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Poy MN, Eliasson L, Krutzfeldt J, et al. A pancreatic islet-specific microRNA regulates insulin secretion. Nature. 2004;432:226–30. doi: 10.1038/nature03076. [DOI] [PubMed] [Google Scholar]
- 42.Xu PZ, Vernooy SY, Guo M, Hay BA, et al. The Drosophila microRNA mir-14 suppresses cell death and is required for normal fat metabolism. Curr Biol. 2003;13:790–5. doi: 10.1016/s0960-9822(03)00250-1. [DOI] [PubMed] [Google Scholar]
- 43.Michael MZ, O’Connor SM, Pellekaan NGV, et al. Reduced accumulation of specific microRNAs in colorectal neoplasia. Mol Cancer Res. 2003;1:882–91. [PubMed] [Google Scholar]
- 44.Metzler M, Wilda M, Busch K, Viehmann S, Borkhardt A. High expression of precursor microRNA-155/BIC RNA in children with Burkitt’s lymphoma. Genes Chromosomes Cancer. 2004;39:167–9. doi: 10.1002/gcc.10316. [DOI] [PubMed] [Google Scholar]
- 45.Couturier JP, Root-Bernstein RS. HIV may produce inhibitory microRNA (miRNAs) that block production of CD28, CD4 and some interleukins. J Theor Biol. 2005;235:168–84. doi: 10.1016/j.jtbi.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 46.Weinberg MS, Morris KV. Are viral-encoded microRNAs mediating latent HIV-1 infection? DNA Cell Biol. 2006;25:223–31. doi: 10.1089/dna.2006.25.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huang J, Wang F, Argyris E, et al. Cellular microRNAs contribute to HIV-1 latency in resting primary CD4+ T lymphocytes. Nat Med. 2007;13:1241–7. doi: 10.1038/nm1639. [DOI] [PubMed] [Google Scholar]
- 48.Triboulet R, Mari B, Lin Y-L, et al. Suppression of microRNA-silencing pathway by HIV-1 during virus replication. Science. 2007;315:1579–82. doi: 10.1126/science.1136319. [DOI] [PubMed] [Google Scholar]
- 49.Lee Y, Kim M, Han J, et al. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23:4051–60. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Aukerman M, Sakai H. Regulation of flowering time and floral organ identity by a microRNA and its APETALA2-like target genes. Plant Cell. 2003;15:2730–41. doi: 10.1105/tpc.016238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tam W. Identification and characterization of human BIC, a gene on chromosome 21 that encodes a noncoding RNA. Gene. 2001;274:157–67. doi: 10.1016/s0378-1119(01)00612-6. [DOI] [PubMed] [Google Scholar]
- 52.Lo HL, Chang T, Yam P, et al. Inhibition of HIV-1 replication with designed miRNAs expressed from RNA polymerase II promoters. Gene Ther. 2007;14:1503–12. doi: 10.1038/sj.gt.3303011. [DOI] [PubMed] [Google Scholar]
- 53.Houbaviy H, Dennis L, Jaenisch R, Sharp P. Characterization of a highly variable eutherian microRNA gene. RNA. 2005;11:1245–57. doi: 10.1261/rna.2890305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xie Z, Allen E, Fahlgren N, et al. Expression of Arabidopsis miRNA genes. Plant Physiol. 2005;138:2145–54. doi: 10.1104/pp.105.062943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhou X, Ruan J, Wang G, Zhang W. Characterization and identification of microRNA core promoters in four model species. PLoS Comput Biol. 2007;3:e37. doi: 10.1371/journal.pcbi.0030037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lee J, Li Z, Brower-Sinning R, John B. Regulatory circuit of human microRNA biogenesis. PLoS Comput Biol. 2007;3:e67. doi: 10.1371/journal.pcbi.0030067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lee Y, Ahn C, Han J, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–9. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 58.Shiohama S, Sasaki T, Noda S, Minoshima S, Shimizu N. Molecular cloning and expression analysis of a novel gene DGCR8 located in the DiGeorge syndrome chromosomal region. Biochem Biophys Res Commun. 2003;304:184–90. doi: 10.1016/s0006-291x(03)00554-0. [DOI] [PubMed] [Google Scholar]
- 59.Lund E, Guttinger S, Calado A, et al. Nuclear export of microRNA precursor. Science. 2003;303:95–8. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- 60.Yi R, Qin Y, Macara IG, Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003;17:3011–6. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bennasser Y, Jeang K-T. HIV-1 Tat interaction with Dicer: requirement for RNA. [[cited 2006 December 20] 3, 95 [about 6 screens]];Retrovirology [serial on the Internet] 2006 Dec; doi: 10.1186/1742-4690-3-95. Available from: http://www.retrovirology.com/content/3/1/95 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lee Y, Hur I, Park S-Y, et al. The role of PACT in the RNA silencing pathway. EMBO J. 2006;25:522–32. doi: 10.1038/sj.emboj.7600942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shwarz DS, Hutvanger G, Du T, Xu Z, Aronin N, Zamore PD. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- 64.Ro S, Park C, Young D, Snaders KM, Yan W. Tissue-dependent paired expression of miRNAs. Nucleic Acids Res. 2007;35:D1–D10. doi: 10.1093/nar/gkm641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yang Z, Ebright YW, Yu B, Chen X. HEN1 recognizes 21-24 nt small RNA duplexes and deposits methyl group onto the 2’ OH of the 3’ terminal nucleotide. Nucleic Acids Res. 2006;34:667–75. doi: 10.1093/nar/gkj474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Vagin VV, Sigova A, Li CJ, et al. A distinct small RNA pathway silences selfish genetic elements in the germline. Science. 2006;313:320–4. doi: 10.1126/science.1129333. [DOI] [PubMed] [Google Scholar]
- 67.Liu CG, Calin GA, Meloon B, et al. An oligonucleotide microchip for genome-wide microRNA profiling in human and mouse tissues. Proc Natl Acad Sci USA. 2004;101:9740–4. doi: 10.1073/pnas.0403293101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Meister G, Landthaler M, Patkaniowska A, et al. Human Argonaute 2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol Cell. 2004;15:185–97. doi: 10.1016/j.molcel.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 69.Liu J, Valencia-Sanchez MA, Hannon GJ, Parker R. MicroRNA-dependent localization of targeted mRNAs to mammalian P-bodies. Nat Cell Biol. 2005;7:719–23. doi: 10.1038/ncb1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schramke V, Allshire R. Hairpin RNAs and retrotransposon LTRs effect RNAi and chromatin-based gene silencing. Science. 2003;301:1069–74. doi: 10.1126/science.1086870. [DOI] [PubMed] [Google Scholar]
- 71.Klenov MS, Lavrov SA, Stolyarenko AD, et al. Repeat-associated siRNAs cause chromatin silencing of retrotransposons in the Drosophila melanogaster germline. Nucleic Acids Res. 2007;35:5430–8. doi: 10.1093/nar/gkm576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hwang HW, Wentzel EA, Mendell JT. A hexanucleotide element directs microRNA nuclear import. Science. 2007;315:97–100. doi: 10.1126/science.1136235. [DOI] [PubMed] [Google Scholar]
- 73.Volpe TA, Kidner C, Hall IM, et al. Regulation of heterochromatic silencing and histone H3 Lysine-9 methylation by RNAi. Science. 2002;297:1833–7. doi: 10.1126/science.1074973. [DOI] [PubMed] [Google Scholar]
- 74.Lim LP, Glasner ME, Yekta S, Burge CB, Bartel DP. Vertebrate microRNA genes. Science. 2003;29:1540. doi: 10.1126/science.1080372. [DOI] [PubMed] [Google Scholar]
- 75.Bentwich I, Avniel A, Karov Y, et al. Identification of hundreds of conserved and nonconserved human microRNAs. Nat Genet. 2005;37:766–70. doi: 10.1038/ng1590. [DOI] [PubMed] [Google Scholar]
- 76.Rajewsky N. microRNA target predictions in animals. Nat Genet. 2006;38(Suppl 38):S8–13. doi: 10.1038/ng1798. [DOI] [PubMed] [Google Scholar]
- 77.Eulalio A, Huntzinger E, Izaurralde E. Getting to the root of miRNA-mediated gene silencing. Cell. 2008;132:9–14. doi: 10.1016/j.cell.2007.12.024. [DOI] [PubMed] [Google Scholar]
- 78.Fujii YR. Symphony of AIDS: a microRNA-based therapy. In: Rossi JJ, Gaur RK, editors. Regulation of gene expression by small RNAs. Boca Raton: CRC Press; 2009. pp. 333–49. [Google Scholar]
- 79.Ronchetti D, Lionetti M, Mosca L, et al. An integrative genomic approach reveals coordinated expression of intronic miR-335, miR-342, and miR-561 with deregulated host genes in multiple myeloma. [[cited 2008 August 13] 1: 37 [about 9 screens]];BMC Med Genomics [serial on the Internet] 2008 Aug; doi: 10.1186/1755-8794-1-37. Available from: http://www.biomedcentral.com/1755-8794/1/37 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Barik S. An intronic microRNA silences genes that are functionally antagonistic to its host gene. Nucleic Acids Res. 2008;36:5232–42. doi: 10.1093/nar/gkn513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kaposi M. Idiopathiches multiples pigment sarcom der Haut. Arch Dermatol Syph. 1872;4:265–72. [Google Scholar]
- 82.Bayley AC, Downing RG, Cheing-song-Popov R, et al. HTLV-III serology distinguishes atypical and endemic Kaposi’s sarcoma in Africa. Lancet. 1985;1:359–61. doi: 10.1016/s0140-6736(85)91383-2. [DOI] [PubMed] [Google Scholar]
- 83.Chang Y, Cesarman F, Pessin MS, et al. Identification of new human herpes virus-like DNA sequences in AIDS-associated Kaposi’s sarcoma. Science. 1994;266:1865–9. doi: 10.1126/science.7997879. [DOI] [PubMed] [Google Scholar]
- 84.Dourmishev LA, Dourmishev AI, Palmeri D, Schwartz RA, Lukac DM. Molecular genetics of Kaposi’s sarcoma-associated herpes-virus (human Herpesvirus-8) epidemiology and pathogenesis. Microbiol Mol Biol Rev. 2003;67:175–212. doi: 10.1128/MMBR.67.2.175-212.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Pearce M, Matsumura S, Wilson AC. Transcripts encoding K12, v-FLIP, v-Cyclin, and the microRNA cluster of Kaposi’s sarcoma-associated herpesvirus originate from a common promoter. J Virol. 2005;79:14457–64. doi: 10.1128/JVI.79.22.14457-14464.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Samlos MA, Skalsky RL, Maldonado AM, et al. Identification of cellular genes targeted by KSHV-encoded microRNAs. PLoS Pathog. 2007;3:e65. doi: 10.1371/journal.ppat.0030065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yamamoto T, Omoto S, Mizuguchi M, et al. Double-stranded nef RNA interferes with human immunodeficiency virus type 1 replication. Microbiol Immunol. 2002;46:809–17. doi: 10.1111/j.1348-0421.2002.tb02768.x. [DOI] [PubMed] [Google Scholar]
- 88.Bennasser Y, Le S-Y, Yeung ML, Jeang K-T. HIV-1 encoded candidate micro-RNAs and their cellular targets. [[cited 2004 December 15] 1: 43[about 5 screens]];Retrovirology [serial on the Internet] 2004 Dec; doi: 10.1186/1742-4690-1-43. Available from: http://www.retrovirology.com/content/1/1/43 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Klase Z, Winograd R, Davis J, et al. HIV-1 TAR miRNA protects against apoptosis by altering cellular gene. [cited 2009 February 16];Retrovirology [serial on the Internet] 2009 Feb; doi: 10.1186/1742-4690-6-18. 6: 18 [about 17 screens]. Available from: http://www.retrovirology.com/content/ 6/1/18 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Doll D, List AF. Burkitt’s lymphoma in a homosexual. Lancet. 1982;1:1026–7. doi: 10.1016/s0140-6736(82)92031-1. [DOI] [PubMed] [Google Scholar]
- 91.Ziegler JL, Miner RC, Rosenbaum E, et al. Outbreak of Burkitt’s like-lymphoma in homosexual men. Lancet. 1982;2:631–3. doi: 10.1016/s0140-6736(82)92740-4. [DOI] [PubMed] [Google Scholar]
- 92.Levine A. Acquired immunodeficiency syndrome-related lymphoma. Blood. 1992;80:8–20. [PubMed] [Google Scholar]
- 93.Monaco L, Passananti C, Fanciulli M. Genomic structure and transcriptional regulation of Che-1 a novel partner of Rb. Gene. 2003;321:57–63. doi: 10.1016/s0378-1119(03)00834-5. [DOI] [PubMed] [Google Scholar]
- 94.Cheema SK, Mishra SK, Rangnekar VM, et al. Par-4 transcriptionally regulates Bcl-2 through a WT1-binding site on the bcl-2 promoter. J Biol Chem. 2003;278:19995–20005. doi: 10.1074/jbc.M205865200. [DOI] [PubMed] [Google Scholar]
- 95.Vilenchik MM, Knudson AG. Endogenous DNA double-strand breaks: production, fidelity of repair, and induction of cancer. Proc Natl Acad Sci USA. 2003;100:12871–6. doi: 10.1073/pnas.2135498100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Canenee WK, Dryia TP, Phillips RA, et al. Expression of recessive alleles by chromosomal mechanisms in retinoblastoma. Nature. 1983;305:779–84. doi: 10.1038/305779a0. [DOI] [PubMed] [Google Scholar]
- 97.Guo L, Huang ZX, Chen XW, et al. Differential expression profiles of microRNAs in NIH3T3 cells in response to UVB irradiation. Photochem Photobiol. 2008 doi: 10.1111/j.1751-1097.2008.00482.x. in press. [DOI] [PubMed] [Google Scholar]
- 98.Ilinytskyy Y, Zemp Fj, Koturbash I, Kovalchuk O. Altered microRNA expression patterns in irradiated hematopoietic tissues suggest a sex specific protective mechanism. Biochem Biophys Res Commun. 2008;377:41–5. doi: 10.1016/j.bbrc.2008.09.080. [DOI] [PubMed] [Google Scholar]
- 99.Maes OC, An J, Sarojini H, Wu H, Wang E. Changes in microRNA expression patterns in human fibroblasts after low-LET radiation. J Cell Biochem. 2008;105:824–34. doi: 10.1002/jcb.21878. [DOI] [PubMed] [Google Scholar]
- 100.Hunter MP, Ismail N, Zhang AI, et al. Detection of microRNA expression in human peripheral blood microvesicles. PLoS ONE. 2008;3:e3694. doi: 10.1371/journal.pone.0003694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sonoki T, Iwanaga E, Mitsuya H, Asou N. Insertion of microRNA-125b-1, a human homologue of lin-4, into a rearranged immunoglobulin heavy chain gene locus in a patient with precursor B-cell acute lymphoblastic leukemia. Leukemia. 2005;19:2009–10. doi: 10.1038/sj.leu.2403938. [DOI] [PubMed] [Google Scholar]
- 102.Lujambio A, Ropero S, Ballestar E, et al. Genetic unmasking of an epigenetically silenced microRNA in human cancer cells. Cancer Res. 2008;67:1424–9. doi: 10.1158/0008-5472.CAN-06-4218. [DOI] [PubMed] [Google Scholar]
- 103.Steeg PS. Tumor metastasis: mechanistic insights and clinical challenges. Nat Med. 2006;12:895–904. doi: 10.1038/nm1469. [DOI] [PubMed] [Google Scholar]
- 104.Fidler IJ. The pathogenesis of cancer metastasis: the ‘seed and soil’ hypothesis revivsited. Nat Rev Cancer. 2003;3:453–8. doi: 10.1038/nrc1098. [DOI] [PubMed] [Google Scholar]
- 105.Gregory PA, Bracken CP, Bert AG, Goodall GJ. MicroRNAs as regulators of epithelial-mesenchymal transition. Cell Cycle. 2008;7:1–6. doi: 10.4161/cc.7.20.6851. [DOI] [PubMed] [Google Scholar]
- 106.Zhu S, Wu H, Wu F, et al. MicroRNA-21 targets tumor suppressor genes invasion and metastasis. Cell Res. 2008;18:350–9. doi: 10.1038/cr.2008.24. [DOI] [PubMed] [Google Scholar]
- 107.Asangani IA, Rasheed SA, Nikolova JH, et al. MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene. 2008;27:2128–36. doi: 10.1038/sj.onc.1210856. [DOI] [PubMed] [Google Scholar]
- 108.Frankel LB, Christoffersen NR, Jacobsen A, et al. Programmed cell death 4 (PDCD4) is an important functional target of the microRNA miR-21 in breast cancer cells. J Biol Chem. 2008;283:1026–33. doi: 10.1074/jbc.M707224200. [DOI] [PubMed] [Google Scholar]
- 109.Lu Z, Lui M, Stribinskis V, et al. MicroRNA-21 promotes cell transformation by targeting the programmed cell death 4 gene. Oncogene. 2008;27:4373–9. doi: 10.1038/onc.2008.72. [DOI] [PubMed] [Google Scholar]
- 110.Meng FR, Henson R, Wehbe-Janek H, et al. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology. 2007;133:647–58. doi: 10.1053/j.gastro.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ma L, Teruya-Feldstein J, Weinberg RA. Tumor invasion and metastasis initiated by microRNA-10b in breast cancer. Nature. 2007;449:682–8. doi: 10.1038/nature06174. [DOI] [PubMed] [Google Scholar]
- 112.Tavazoie SF, Alarcon C, Oskarsson T, et al. Endogenous human microRNAs that suppress breast cancer metastasis. Nature. 2008;451:147–52. doi: 10.1038/nature06487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Dahiya N, Sherman-Baust CA, Wang TL, et al. MicroRNA expression and identification of putative miRNA targets in ovarian cancer. PLoS ONE. 2008;3:e2436. doi: 10.1371/journal.pone.0002436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Garzon R, Garofalo M, Martelli MP, et al. Distinctive microRNA sifnature of acute myeloid leukemia bearing cytoplasmic mutated nucleophosmin. Proc Natl Acad Sci USA. 2008;105:3945–50. doi: 10.1073/pnas.0800135105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Wang X, Tang S, Le SY, et al. Aberrant expression of oncogenic and tumor-suppressive microRNAs in cervical cancer is required for cancer cell growth. PLoS ONE. 2008;3:e2557. doi: 10.1371/journal.pone.0002557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Taylor DD, Gercel-Taylor C. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol Oncol. 2008;110:13–21. doi: 10.1016/j.ygyno.2008.04.033. [DOI] [PubMed] [Google Scholar]
- 117.Gilad S, Meiri E, Yogev Y, et al. Serum microRNAs are promising novel biomarkers. PLoS ONE. 2008;3:e3148. doi: 10.1371/journal.pone.0003148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Bottoni A, Piccin D, Tagliati F, et al. miR-15a and miR-16-1 down-regulation in pituitary adenomas. J Cell Physiol. 2005;204:280–5. doi: 10.1002/jcp.20282. [DOI] [PubMed] [Google Scholar]
- 119.Cimmino A, Calin GA, Fabbri M, et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci USA. 2005;102:13944–9. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Calin GA, Dumitru CD, Shimizu M, et al. Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc. Natl Acad Sci USA. 2002;99:15524–9. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Linsley PS, Schelter J, Burchard J, et al. Transcripts targeted by the microRNA-16 family cooperatively regulate cell cycle progression. Mol Cell Biol. 2007;27:2240–52. doi: 10.1128/MCB.02005-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Liu Q, Fu H, Sun F, et al. miR-16 family induces cell cycle arrest by regulating multiple cell cycle genes. Nucleic Acids Res. 2008;36:5391–404. doi: 10.1093/nar/gkn522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Marusyk A, DeGregori J. Building a better model of cancer. Cell Div. 2006;1:24. doi: 10.1186/1747-1028-1-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Moon N-S, Di Stefano L, Morris EJ, et al. E2F and p53 induce apoptosis independently during Drosophila development but intersect in the context of DNA damage. PLoS Genet. 2008;4:e1000153. doi: 10.1371/journal.pgen.1000153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Attwooll C, Denchi EL, Helin K. The E2F family: specific functions and overlapping interests. EMBO J. 2004;23:4709–4716. doi: 10.1038/sj.emboj.7600481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.DeGregori J. E2F and cell survival: context really is key. Dev Cell. 2006;9:442–4. doi: 10.1016/j.devcel.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 127.Adelman CA, Petrini JHJ, Attwooll CL. Modeling disease in the mouse: lessons from DNA damage response and cell cycle control genes. J Cell Biochem. 2006;97:459–73. doi: 10.1002/jcb.20701. [DOI] [PubMed] [Google Scholar]
- 128.Morris EJ, Ji J-Y, Yang F, et al. E2F1 represses β–catenin transcription and is antagonized by both pRB and CDK8. Nature. 2008;455:552–8. doi: 10.1038/nature07310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Paulson QX, Pusapati RV, Hong S, et al. Transgenic expression of E2F3a causes DNA damage leading to ATM-dependent apoptosis. Oncogene. 2008;27:4954–61. doi: 10.1038/onc.2008.138. [DOI] [PubMed] [Google Scholar]
- 130.He L, Thomson JM, Heman MT, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–33. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Woods K, Thomson JM, Hammond SM. Direct regulation of an oncogenic microRNA cluster by E2F transcription factors. J Biol Chem. 2007;282:2130–4. doi: 10.1074/jbc.C600252200. [DOI] [PubMed] [Google Scholar]
- 132.Sylveatre Y, De Guire V, Querido E, et al. An E2F/miR-20a auto-regulatory feed-back loop. J Biol Chem. 2006;282:2135–43. doi: 10.1074/jbc.M608939200. [DOI] [PubMed] [Google Scholar]
- 133.Hwang HW, Mendell JT. MicroRNAs in cell proliferation, cell death, and tumorigenesis. Br J Cancer. 2006;94:776–80. doi: 10.1038/sj.bjc.6603023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Iorio MV, Ferracin M, Liu CG, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065–70. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 135.Chan JA, Krichevsky AM, Kosik KS. MicroRNA-21 is an antiapoptoic factor in human glioblastoma cells. Cancer Res. 2005;65:6029–33. doi: 10.1158/0008-5472.CAN-05-0137. [DOI] [PubMed] [Google Scholar]
- 136.Takamizawa J, Konishi H, Yanagisawa K, et al. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 2004;64:3753–6. doi: 10.1158/0008-5472.CAN-04-0637. [DOI] [PubMed] [Google Scholar]
- 137.Johnson SM, Grosshans H, Shingara J, et al. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–47. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 138.O’Donnell KA, Wentzel EA, Zeller KI, et al. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–43. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- 139.Nakajima G, Hayashi K, Xi Y, et al. Non-coding microRNAs hsa-let-7g and hsa-miR-181b are associated with chemoresponse to S-1 in colon cancer. Cancer Genomics Proteomics. 2006;3:317–24. [PMC free article] [PubMed] [Google Scholar]
- 140.Brueckner B, Stresemann C, Kuner R, et al. The human let-7a-3 locus contains an epigenetically regulated microRNA gene with oncogenic function. Cancer Res. 2007;67:1419–23. doi: 10.1158/0008-5472.CAN-06-4074. [DOI] [PubMed] [Google Scholar]
- 141.Saito Y, Liang G, Egger G, et al. Specific activation of microRNA-127 with downregulation of the proto-oncogene BCL6 by chromatin-modifying drugs in human cancer cells. Cancer Cell. 2006;9:435–43. doi: 10.1016/j.ccr.2006.04.020. [DOI] [PubMed] [Google Scholar]
- 142.Eis PS, Tam W, Sun LP, et al. Accumulation of miR-155 and BIC RNA in human B cell lymphomas. Proc Natl Acad Sci USA. 2005;102:3627–32. doi: 10.1073/pnas.0500613102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Meltzer M, Wilda M, Busch K, Viehmann S, Borkhardt A. High expression of precursor microRNA-155/BIC RNA in children with Burkitt lymphoma. Genes Chromosomes Cancer. 2004;39:167–9. doi: 10.1002/gcc.10316. [DOI] [PubMed] [Google Scholar]
- 144.Tam W, Hughes SH, Hayward WS, Besmer P. Avian bic, a gene isolated from a common retroviral site in avian leukosis virus-induced lymphomas that encoding RNA, cooperates with c-myc in lymphomagenesis. J Virol. 2002;76:4275–86. doi: 10.1128/JVI.76.9.4275-4286.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Gironella M, Seux M, Xie M–J, et al. Tumor protein 53-induced nuclear protein 1 expression is repressed by miR-155, and its restoration inhibits pancreatic tumor development. Proc Natl Acad Sci USA. 2007;104:16170–5. doi: 10.1073/pnas.0703942104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Salter KH, Acharya CR, Walters KS, et al. An integrated approach to the prediction of chemotherapeutic response in patients with breast cancer. PLoS ONE. 2008;3:e1908. doi: 10.1371/journal.pone.0001908. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 147.Hu Z, Chen J, Tian T, et al. Genetic variants of miRNA sequences and non-small cell lung cancer survival. J Clin Invest. 2008;118:2600–8. doi: 10.1172/JCI34934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.McClintock B. Induction of instability at selected loci in maize. Genetics. 1953;38:579–99. doi: 10.1093/genetics/38.6.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Lindblad-Toh K, Wade CM, Mikkelsen TS, et al. Genome sequence, comparative analysis and haplotype structure of the domestic dog. Nature. 2005;438:803–19. doi: 10.1038/nature04338. [DOI] [PubMed] [Google Scholar]
- 150.Feshchotte C, Zhang X, Wesser SR. Miniature inverted-repeat transposable elements and their relationship to established DNA transposons. In: Craige NL, Craige R, Gellert M Lambowitz, editors. Mobile DNA II. Washington DC: ASM Press; 2002. pp. 997–1007. [Google Scholar]
- 151.Eden A, Gaudet F, Waghmare A, Jaenisch R. Chromosomal instability and tumors promoted by DNA hypomethylation. Science. 2003;300:489–92. doi: 10.1126/science.1083557. [DOI] [PubMed] [Google Scholar]
- 152.Choi IS, Estecio MR, Nagano Y, et al. Hypomethylation of LINE-1 and Alu in well-differential neuroendocrine tumors (pancreatic endocrine tumors and carcinoid tumors) Mod Pathol. 2007;20:802–10. doi: 10.1038/modpathol.3800825. [DOI] [PubMed] [Google Scholar]
- 153.Dobzhansky T. A homozygous translocation in Drosophila Melanogaster. Proc Natl Acad Sci USA. 1929;15:633–8. doi: 10.1073/pnas.15.8.633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Ding B, Itava A. Viroid: a useful model for studying the basic principles of intfeaction and RNA biology. Mol Plant Microbe Intract. 2007;20:7–20. doi: 10.1094/MPMI-20-0007. [DOI] [PubMed] [Google Scholar]
- 155.Soifer HS. Regulation of mammalian mobile DNA by RNA-based silencing pathway. In: Morris KV, editor. RNA and the regulation of gene expression. London: Horizon Scientific Press; 2008. pp. 110–31. [Google Scholar]
- 156.Yang N, Kazazian HH Jr. L1 retrotransposition is suppressed by endogenously encoded small interfering RNAs in human cultured cells. Nat Struct Mol Biol. 2006;13:763–71. doi: 10.1038/nsmb1141. [DOI] [PubMed] [Google Scholar]
- 157.Charlier C, Segers K, Wagenaar D, et al. Human-ovine comparative sequencing of a 250-kb imprinted domain encompassing the Callipyge (clpg) locus and identification of six imprinted transcripts: DLK1, DAT, GTL2, PEG11, antiPEG11, and MEG8. Genome Res. 2001;11:850–62. doi: 10.1101/gr.172701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Kircher M, Bock C, Paulsen M. Structural conservation vs functional divergence of maternally expressed microRNAs in the Dlk1/Gtl2 imprinting region. [Jul;[cited 2008 July 23] 9:346 [about 19 screens]];BMC Genomics [serial on the Internet] 2008 Jul; doi: 10.1186/1471-2164-9-346. Available from: http://www.biomedcentral.com/1471-2164/9/346 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Joyce JA, Schofield PN. Genomic imprinting and cancer. Mol Pathol. 1998;51:185–90. doi: 10.1136/mp.51.4.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Esteller M, Herman JG. Cancer as an epigenetic disease: DNA methylation and chromatin alterations in human tumors. J Pathol. 2002;196:1–7. doi: 10.1002/path.1024. [DOI] [PubMed] [Google Scholar]
- 161.Seitz H, Youngson N, Lin S-P, et al. Imprinted microRNA genes transcribed antisense to a reciprocally imprinted retrotransposon-like gene. Nat Genet. 2003;34:261–2. doi: 10.1038/ng1171. [DOI] [PubMed] [Google Scholar]
- 162.Hinas A, Reimegard J, Wanger EGH, Nellen W, Ambros VR, Soderbom F. The small RNA repertoire of Dictyostelium discoideum and its regulation by components of the RNAi pathway. Nucleic Acids Res. 2007;35:6714–26. doi: 10.1093/nar/gkm707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Smalheiser NR, Torvik VI. Mammalian microRNAs derived from genomic repeats. Trends Genet. 2005;21:322–6. doi: 10.1016/j.tig.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 164.Landais S, Landry S, Legault P, Rassart E. Oncogenic potential of the miR-106-363 cluster and tis implication in human T-call leukemia. Cancer Res. 2007;67:5699–707. doi: 10.1158/0008-5472.CAN-06-4478. [DOI] [PubMed] [Google Scholar]
- 165.Cui J-W, Li Y-J, Brown J, et al. Retroviral insertional activation of the Fli-3 locus in erythroleukemias encoding a cluster of microRNAs that convert Epo-induced differentiation to proliferation. Blood. 2007;110:2631–40. doi: 10.1182/blood-2006-10-053850. [DOI] [PubMed] [Google Scholar]
- 166.Beck-Engeser GB, Lum AM, Huppi K, et al. Pvt1-encoded microRNAs in oncogenesis. [[cited 2008 January 14] 5: 4 [about 14 screens]];Retrovirology [serial on the Internet] 2008 Jan; doi: 10.1186/1742-4690-5-4. Available from: http://www.retrovirology.com/content/5/1/4 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Kim S, Kim N, Dong B, et al. Integration site preference of Xenotropic murine leukemia virus-related virus, a new human retrovirus associated with prostate cnacer. J Virol. 2008;82:9964–77. doi: 10.1128/JVI.01299-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Bouzar AB, Willems LL. How HTLV-1 may subvert miRNAs for persistence and transformation. [[cited 2008 November 12] 5: 101[about 6 screens]];Retrovirology [serial on the Internet] 2008 Nov; doi: 10.1186/1742-4690-5-101. Available from: http://www.retrovirology.com/content/5/1/101 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Ciafre SA, Galardi S, Mangiola A, et al. Extensive modulation of a set of microRNAs in primary glioblastoma. Biochem Biophys Res Commun. 2005;334:1351–8. doi: 10.1016/j.bbrc.2005.07.030. [DOI] [PubMed] [Google Scholar]
- 170.Calin GA, Cimmino A, Fabbri M, et al. MiR-15a and miR-16-1 cluster functions in human leukemia. Proc Natl Acad Sci USA. 2008;105:5166–71. doi: 10.1073/pnas.0800121105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Katada T, Ishiguro H, Kuwabara Y, et al. microRNA expression profile in undifferentiated gastric cancer. Int J Oncol. 2009;34:537–42. [PubMed] [Google Scholar]
- 172.Kim Y–K, Yu J, Han TS, et al. Functional links between clustered microRNAs: suppression of cell-cycle inhibitors by microRNA clusters in gastric cancer. Nucleic Acids Res. 2009;37:1672–81. doi: 10.1093/nar/gkp002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Chen X, Gui X, Zhang H, et al. Role of miR-143 targeting KRAS in colorectal tumorigenesis. Oncogene. 2009;23:151–3. doi: 10.1038/onc.2008.474. [DOI] [PubMed] [Google Scholar]
- 174.Bloomston M, Frankel WL, Petrocca F, et al. MicroRNA expression patterns to differentiate pancreatic adenocarcinoma from normal pancreas and chronic pancreatitis. JAMA. 2007;297:1923–5. doi: 10.1001/jama.297.17.1901. [DOI] [PubMed] [Google Scholar]
- 175.Murakami Y, Yasuda T, Saigo K, et al. Comprehensive analysis of microRNA expression patterns in hematocellular carcinoma and non-tumorous tissues. Oncogene. 2006;25:2537–45. doi: 10.1038/sj.onc.1209283. [DOI] [PubMed] [Google Scholar]
- 176.Li S, Fu H, Wang Y, et al. MicroRNA-101 regulates expression of the v-fos FBJ murine osteosarcoma viral oncogene homolog (FOS) oncogene in human hepatocellular carcinoma. Hepatology. 2009;49:1194–202. doi: 10.1002/hep.22757. [DOI] [PubMed] [Google Scholar]
- 177.Hayashita Y, Osada H, Tatematsu Y, et al. A polycistronic microRNA cluster, miR-17-92, is overexpressed in human lung cancers and enhances cell proliferation. Cancer Res. 2005;65:9628–32. doi: 10.1158/0008-5472.CAN-05-2352. [DOI] [PubMed] [Google Scholar]
- 178.Pallante P, Visone R, Ferracin M, et al. MicroRNA deregulation in human thyroid papillary carcinomas. Endocr Relat Cancer. 2006;13:497–508. doi: 10.1677/erc.1.01209. [DOI] [PubMed] [Google Scholar]
- 179.Voorhoeve PM, le Sage C, Schrier M, et al. A genetic screen implicates miRNA-372 and miRNA-373 as oncogenes in testicular germ cell tumors. Cell. 2006;124:1169–81. doi: 10.1016/j.cell.2006.02.037. [DOI] [PubMed] [Google Scholar]