Abstract
Transcriptional regulation is a fundamental process for regulating the flux of all metabolic pathways. For the last several decades, the lac operon has served as a valuable model for studying transcription. More recently, the switch that controls the operon has also been successfully adapted to function in mammalian cells. Here we describe how, using directed evolution, we have created a novel switch that recognizes an asymmetric operator sequence. The new switch has a repressor with altered headpiece domains for operator recognition, and a redesigned dimer interface to create a heterodimeric repressor. Quite unexpectedly, the heterodimeric switch functions better than the natural system. It can repress more tightly than the naturally occurring switch of the lac operon; it is less leaky and can be induced more efficiently. Ultimately these novel repressors could be evolved to recognize eukaryotic promoters and used to regulate gene expression in mammalian systems.
Keywords: Directed Evolution, Repression, Heterodimer, Chimeric Operator
INTRODUCTION
The viability of a cell often depends upon its ability to successfully regulate a variety of cellular processes. In virtually all organisms critical cellular events are controlled by modulating gene expression. Jacob and Monod were the first to suggest that a molecular switch could enhance or repress gene expression and as a consequence regulate cellular processes1. To understand how molecular switches function, two classical systems have been extensively analyzed; the epigenetic switch from the bacteriophage lambda, which controls phage life cycle and development, and the switch that regulates lactose metabolism in E.coli. In both instances, the switch is a two component system that is composed of a repressor protein and an operator DNA site. When the repressor binds to the operator a complex is established that physically blocks transcription by an RNA polymerase. The switch from bacteriophage lambda functions as a two state device; transcription is either on or off. The molecular switch that regulates the lac operon is slightly more complex since it behaves more like a potentiometer or dimmer switch. Inducer molecules potentiate the stability of the repressor operator complex and as a consequence the level of transcription is modulated.
The switch that regulates the lac operon has been well characterize (for a recent review see Wilson et al) 2. The repressor is a 360 amino acid protein that has a modular structure. It contains an NH2-terminal or “headpiece” domain (~60 residues) that binds specifically to operator DNA and a COOH-terminal “core” domain that binds inducers. The monomeric repressor self associates into a dimer of dimers, where a dimeric repressor molecule recognizes the operator using the classical helix-turn-helix (HTH) motif. The HTH provides the scaffolding for specific side chains that recognize the bases in the major groove of the operator. The operator of the lac operon, a short stretch of DNA (~17 base pairs), is composed of two nearly identical half sites that is located between the end of the lacI gene and the beginning of the lacZ gene 3. The COOH-terminal “core” domain consists of two sub-domains, an N-terminal domain (NTD) and a C-terminal domain (CTD). The inducer molecule binds to a pocket located at the interface of these two domains. When bound to the repressor, the NTDs adopt a conformation that is different from the conformation when bound to DNA. In contrast, the CTDs are structurally invariant and are responsible to stabilizing the dimeric structure of the repressor 4.
Although regulatory mechanisms are somewhat more complex in eukaryotes than in prokaryotes, the molecular switch of the lac operon has been adapted to control gene expression in mammalian systems. Hu and Davidson were among the first to exploit the switch of the lac operon to control reporter gene expression reversibly in mammalian cells 5. Building upon these results, Figge et al. demonstrated that when the lac repressor was inserted into a mammalian cell it was able to gain access to the mammalian chromosome and regulate a stably integrated reporter gene 6. In addition, Itzhaki et al. incorporated the lac operator into a mammalian promoter by homologous recombination and demonstrated that this bacterial switch could regulate the expression of endogenous loci 7. Even more dramatically, Scrable’s laboratory adapted the lac regulatory system to control gene expression in the mouse; demonstrating this bacterial switch can be used to regulate complex events in higher organisms8.
Inspired by the observation that the switch of the lac operon can be used to regulate gene expression in a variety of systems, we embarked on a project to create a novel repressor that could recognize altered operator sequences that were distinct from and not recognized by the wild type repressor. Although proteins containing zinc finger domains have been previously generated to recognize pre-selected target sequences9; 10, the lac repressor is equipped with the ability to repress as well as induce transcription repeatedly based on environmental conditions. Therefore, we hypothesized that the bacterial switch that regulates the lac operon could be genetically altered to regulate endogenous gene expression in mammalian systems by directly recognizing and repressing a mammalian promoter. Unlike classical operator sites, these target sequences are not symmetric making repression by a homodimeric lac repressor impossible. To circumvent this obstacle, we created a heterodimeric repressor where each monomer recognizes a distinct operator half site. The unique heterodimeric repressors have two different DNA binding domains as well as a dimer interface that cannot homodimerize. Interestingly, these novel repressors function better than the wild type repressor; they bind more tightly to the chimeric operator than the wild type repressor binds to the natural operator thereby reducing the basal level of transcription, and have an increased dynamic range such that the difference between the on and off (induced and repressed) states is more pronounced.
RESULTS
CREATING A SYMMETRIC SWITCH WITH ALTERED SPECIFICITY
In bacteria, many repressors homologous to the lactose repressor regulate transcription of inducible genes 11. These proteins, referred to as the LacI/GalR family, have similar structures and regulate transcription in an analogous fashion. All of the LacI/GalR family members have a headpiece domain that contains a helix -turn-helix (HTH) motif, which recognizes its operator site. The sequence conservation in the HTH region suggests that these proteins bind to similar operators. Discrimination between the different operators is most likely due to a few non-conserved residues on the helix-turn-helix motif. In addition to the HTH, all members of this family use a hinge helix for binding to the operator. Both the purine and lac repressors recognize the central portion of the operator by placing a pair of helices in the minor grove of the operator 12; 13. Members of the LacI/GalR family also have a C-terminal domain that is responsible for both effector binding and oligomerization. These domains have significant sequence homology, suggesting that that the architecture of the C-terminal domain is also well conserved even though members of the LacI/GalR superfamily respond to a variety of effector molecules. Transcription regulated by these repressors is modulated by allosteric effectors, including galactose, fructose, maltose, ribulose, hypoxanthine, xanthine, and spectrum of β-galactosides. These molecules bind to the core of the repressors and either act as inducers or as co-repressors. Each metabolite functions either by disrupting or stabilizing the repressor-operator complex. Even though the effector binding sites have similar architecture, the scaffolding is draped with different amino acid side chains to create the unique specificity. Not only are the repressor molecules in the Lac/GalR superfamily conserved but the operator sequences also possess noticeable homology. In fact all of the operator sequences that are recognized by this superfamily are pseudo palindromic. The primary differences in the operators are localized to the peripheral regions while the central base pairs are highly conserved. For example, the operators of the lac and gal operons differ only at positions two and four in the operator sequence yet these differences are sufficient for specificity 14.
Intrigued by the high level of conservation and the subtle differences that lead to distinct DNA binding patterns, Muller- Hill’s laboratory attempted to dissect the ‘code’ defining protein-DNA recognition within the family of proteins that contain the helix-turn-helix domain 14; 15; 16. By systematically altering each base in the operator and each residue in the recognition helix, they demonstrated that the first two residues on the recognition helix specifically recognize base pairs four and five of the lac operator, and that residue six recognizes the sixth base of the operator. Structural studies have confirmed that the most critical residues for specific operator recognition are Y17, Q18, and R22. 13; 17. These three amino acids protrude from the recognition helix of the repressor and form specific interactions with the bases in the major groove by contacting bases 4, 5, and 6 of the operator. A closer look at the recognition helix of LacI/GalR family members suggests that the recognition helix of the gal repressor differs from lac repressor at two of the three positions, valine replaces tyrosine and an alanine replaces glutamine. It was hypothesized and shown that a mutant lac repressor containing two amino acid substitutions, Y17V, Q18A, binds and represses a ‘gal-like’ hybrid operator containing a G:C to A:T change at position 4 in each half site of the symmetric lac operator. 14 In addition, this ‘gal-like’ operator(412) could discriminate wild type repressor binding.
There are many mutants of the lac repressor that bind to the “gal-like” operator. A library of ~203 or 8000 mutant repressors was created by introducing all twenty amino acids into the repressor at the three critical positions. Functional mutants were identified by selecting molecules that block transcription of a reporter gene, GFPmut3.1, whose expression is regulated by a “gal-like” operator embedded in the lac promoter (Figure 1). Using Fluorescence-Activated Cell Sorting (FACS), several altered repressors were identified and their relative ability to repress transcription was measured in vivo. Although the mutant Y17V,Q18A binds to the “gal-like” operator more tightly than the wild type repressor, mutant repressors with sequences,Y17T,Q18A and Y17I,Q18A, bind to this operator nearly 3 times more tightly than the originally identified mutant. A ‘gal-like’ operator (412), which results for a single change can be recognized by mutant repressors with amino acid changes at the first two positions on the recognition helix. Next we explored if there were mutant repressors in the library that could recognize a two base substitution.
Figure 1. Operator sequences used in reporter plasmids.
The operator sequences are aligned and numbered with respect to the central base pair. The top sequence is the natural (0R1) operator of the lac operon. The second sequences represents the ideal symmetric lac operator (212) followed by the third operator which corresponds to the gal-like operator (412). The 411 operator diverges the gal-like operator further at position 6. The final two operators are chimeric operators composed of the 212 and 411 half sites. The numerical identifiers associated with each operator correspond to the identity of the bases found in the right half site of each fully symmetric operator (1/2/3/4 = A/C/G/T respectively)
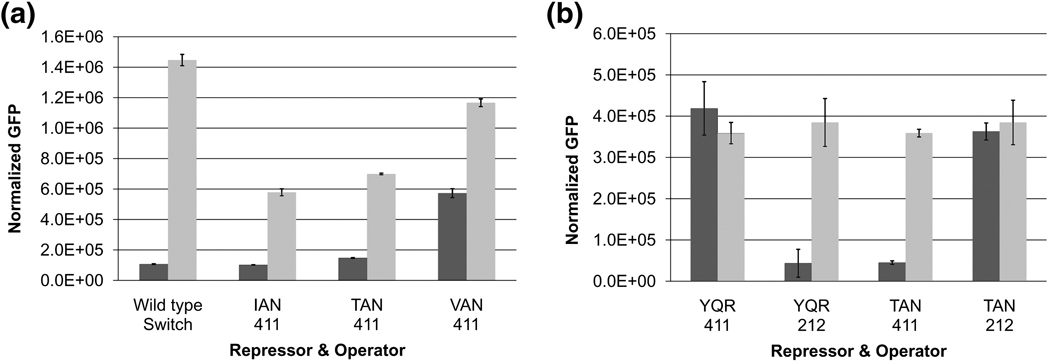
Both the lac and gal repressors have an arginine at position 22. The lac repressor uses this amino acid to recognize position 6 within the operator. This interaction is critical for operator recognition and changing the arginine to any other amino acid effectively destroys operator binding 18.We introduced an additional G:C to T:A substitution at position 6 of each half site, creating an even more divergent operator and verified that this operator (411) cannot be recognized by either the wild type repressor or the previously indentified mutants that bind to the ‘gal-like’ operator. Again we sorted our library of altered repressors and identified mutants that bound to this new operator and repressed transcription of GFP. Repression of several dozen mutants was measured in vivo and repressors with a sequence Y17T, Q18A, R22N (TAN) and Y17I, Q18A, R22N (IAN) bind to this altered operator with the greatest affinity and specificity while maintaining full inducibility. These mutants bind to this artificially created operator (411) and repress as well or better than the naturally occurring switch of the lac operon. Furthermore, neither mutant is able to repress the symmetric lac operator (212) or the ‘gal-like’ operator (412) (Figure 2a & b). By introducing two changes in the operator half sites, we have identified two novel repressors with altered specificity that retain the functional efficiency equivalent to the natural switch.
Figure 2. In vivo characterization of various combinations of repressors and operators.
Dark shading = no IPTG, Light shading = 2.5 mM IPTG (a) Both the IAN and TAN mutants are capable of repressing the 411 operator with approximately the same affinity as the wild type switch. (b) The two changes in the operator sequences, 212 and 411, allow for significant divergence of the half sites such that the wild type (YQR) and TAN mutant repressors specifically repress their respective operators.
CONSTRUCTING AN ASYMMETRIC SWITCH
Virtually all proteins that regulate transcription recognize operator sequences that are pseudo symmetric. The Lac/Gal family members recognize these palindromic operators by forming homo-dimers that bring together two identical DNA binding domains. Although each monomer contains a DNA binding domain as well as an effector binding site, a monomer can only recognize a half site of 6–8 base pairs, which greatly limits specificity and binding affinity. Therefore, dimerization allows the repressor to bind a larger stretch of DNA, increasing both the specificity and affinity of the interaction. In addition, specificity of the repressor operator interaction is further increased by introducing a small degree of asymmetry in the operator 18. While asymmetry would be advantageous for designing a novel switch, it comes at the cost of reduced binding affinity 19. To overcome this limitation we hypothesized that a chimeric operator containing two unique half sites could be recognized by a heterodimeric repressor containing two different DNA binding domains.
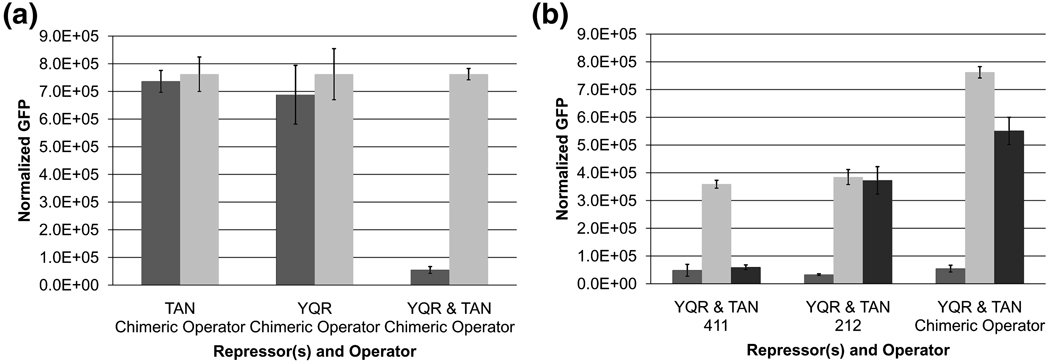
To create a chimeric operator the left half site of the 212 operator was combined with the right half site of the 411 operator and this operator was placed in the reporter system to control expression of GFP (Figure 1). Neither the homodimeric TAN mutant nor the wild type dimeric repressor is capable of repressing GFP when the reporter plasmid contains the chimeric operator. However, when the two repressors were co expressed we observe repression from the chimeric operator (Figure 3a). Since the individual homodimers cannot repress transcription from the chimeric operator, we suspect that when both repressors are co-expressed a hetero-dimer forms that can block expression of the reporter gene. The heterodimer can also be readily induced with the addition of IPTG. The chimeric switch not only shows tight repression but also full induction, comparable to the natural switch. However, these repressors have identical dimerization interfaces such that co-expression results in a mixed population of homo and heterodimeric repressors (Figure 3b). Naturally, the only way to make a unique heterodimer is by altering the dimer interface.
Figure 3. In vivo repression and induction analysis.
Dark shading = no IPTG, Light shading = 2.5 mM IPTG (a) Neither the wild type (YQR) nor the TAN mutant repressors are capable of repressing the chimeric operator in their homodimeric state. Only when the two mutants are co-expressed can repression be achieved. (b) When both repressors contain the wild type dimer interface however, a mixed population of dimeric repressors exists, as evidenced by the repression of each of the three operators analyzed. When the Y282A point mutation is introduced into the wild type repressor gene (Black bars), homodimerization (repression of the 212 operator) and heterodimerization (repression of the Chimeric operator) is lost.
CREATING AN INTERGENIC INTERACTION SUPPRESSOR
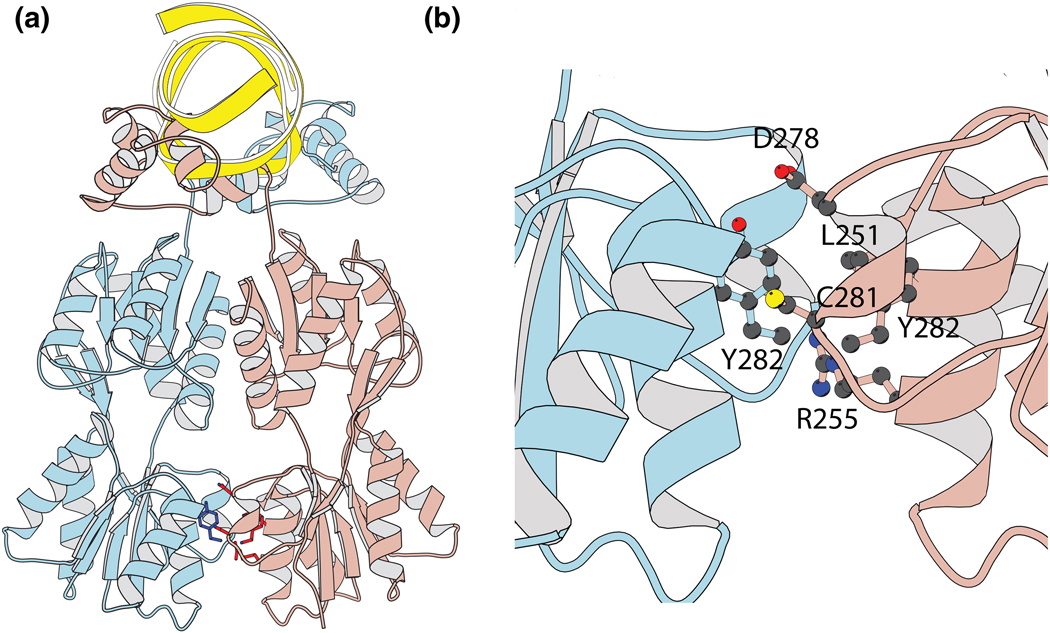
The intact repressor monomer folds into four discrete functional units; an NH2-terminal domain, a hinge region, a sugar binding or core domain, and a COOH-terminal helix (Figure 4). The core of the repressor is composed of two sub-domains that are topologically similar but it is the CTD that is responsible for dimerization. Several amino acid substitutions on the surface of the CTD effect the ability of the repressor to bind DNA presumably by altering the monomer-dimer equilibrium. The amino acid substitution at position 282 has been well characterized and Y282 A/D/S shift the repressor equilibrium to the monomeric state 20; 21. Each of these mutants fold properly and has the ability to bind to inducer ligand but have lost their ability to interact with the operator DNA. Random mutagenesis and in vivo selection experiments identified 22 intragenic suppressor revertants, several of which were located in the proximity of Y282 while others appear spread along the dimer interface and at the interface between the N and C-terminal sub domains21. Based upon this plasticity at the interface we hypothesized that a unique heterodimeric repressor could be identified by creating intergenic interaction suppressors to the Y282A/D or S point mutations.
Figure 4. Location of residues mutated in redesign of the dimer interface.
(a) Structure of the repressor containing 4 domains (b) Residues L251, R255, D278, C281 and Y282 on the repressor make intergenic interactions with residue Y282 on the dimer interface.
The Y282A/S/D mutations were introduced into the wild type repressor and as anticipated these mutants no longer bind and repress transcription of the reporter gene. Similarly when the TAN mutant is co-expressed with each monomeric YQR mutant, the heterodimer cannot form and there is no repression from the chimeric operator. Not only do the mutants prevent homo dimers from forming but the point mutations also interfere with heterodimerization (Figure 3b). At the dimer interface of the wild type repressor there are five amino acids from the second monomer that are in close proximity of Y282. Residues 251, 255, 278, 281, and 282 of one monomer associate with tyrosine 282 on the first monomer (Figure 4). To find intergenic interaction suppressors, we created a library where all twenty amino acids were introduced at these five positions in the second monomer. This library was then introduced into cells that contained one of the monomeric repressor mutants and was sorted using FACS to identify intergenic interaction suppressors capable of restoring the repressed phenotype. From these selection experiments, we identified several dozen mutant repressors that could suppress the deleterious effect of the Y282 point mutations by restoring repression of the chimeric operator.
There were many suppressor mutants that had the ability to restore a functional phenotype by complementing changes at position 282 (Table 1). Although many revertants were found that could complement Y282S and Y282A, complementation of the Y282D proved more difficult. Overall the plasticity at each of the 5 mutated sites was quite remarkable (Figure S2). Of the 5 sites, positions 251 and 255 displayed a high level of conservation with more than half of the mutants maintaining the wild type amino acids of leucine and arginine. In a number of the mutants the chemical nature at these two positions were swapped; position 251 becoming basic and position 255 hydrophilic. All but one mutant contained some combination of a basic and apolar residue at these two positions suggesting that the two residues may interact synergistically. Surprisingly, position 278 (D) displayed the least conservation. In fact of the 5 sites randomized, an acidic residue was never identified. Similar to the relationship between residues 251 and 255, positions 278, 281 and 282 consistently contained some combination of polar, hydrophilic and aromatic amino acids, suggesting that they may also function synergistically. However, we could not observe a general pattern to understand how the intergenic revertant mutants compliment the corresponding Y282A/S/D mutant repressors.
Table 1. Sequences and Repression Ratios of intergenic interaction repressors.
The table contains the sequence of each of the mutants identified in the selection experiments. They are organized into groups depending upon which monomer mutation they are capable of suppressing. Repression data is also presented for each mutant. The repression ratio is defined as the ratio of the fluorescent signal when the repressor is absent divided by the signal when it is present for a given reporter construct. Each ratio is further normalized to the repression ratio of the wild type switch.
| Repressor | Suppresses | 251 | 255 | 278 | 281 | 282 | Heterodimeric Repression |
Homodimeric Repression |
|---|---|---|---|---|---|---|---|---|
| Wild Type | L | R | D | C | Y | - | 100.0 | |
| 4a1 | Y282 A | K | L | G | M | C | 3.5 | 3.7 |
| 4a2 | Y282 A | L | R | C | C | Y | 23.9 | 20.8 |
| 4a3 | Y282 A | L | R | W | C | C | 40.8 | 161.5 |
| 4a4 | Y282 A | L | R | F | Y | Y | 35.4 | 316.8 |
| 4a6 | Y282 A | L | R | C | Y | F | 38.4 | 55.0 |
| 4a7 | Y282 A | L | R | V | C | Y | 29.7 | 73.7 |
| 4a8 | Y282 A | L | R | I | Y | F | 112.2 | 164.2 |
| 4a9 | Y282 A | L | R | S | C | Y | 78.0 | 225.8 |
| 4a10 | Y282 A | L | R | W | V | T | 175.1 | 30.3 |
| 4a11 | Y282 A | K | R | L | F | S | 99.2 | 271.7 |
| 4a12 | Y282 A | L | R | I | V | F | 52.8 | 35.6 |
| 4a13 | Y282 A | L | R | L | M | T | 90.7 | 39.5 |
| 4a14 | Y282 A | L | R | L | V | M | 59.7 | 30.9 |
| 4a15 | Y282 A | L | R | I | F | Y | 53.4 | 9.2 |
| 4a17 | Y282 A | F | R | L | V | T | 72.2 | 72.2 |
| 4a18 | Y282 A | K | L | S | Y | T | 36.8 | 36.8 |
| 4a19 | Y282 A | K | L | N | H | W | 249.1 | 70.8 |
| 4a20 | Y282 A | M | R | N | C | F | 154.3 | 4.1 |
| 4d2 | Y282 D | L | L | N | Y | F | 13.2 | 169.8 |
| 4d3 | Y282 D | L | R | F | C | T | 17.1 | 324.6 |
| 4d5 | Y282 D | L | R | S | C | Y | 11.3 | 96.2 |
| 4d7 | Y282 D | L | R | F | C | T | 20.7 | 345.0 |
| 4d8 | Y282 D | L | R | N | Y | F | 21.6 | 247.2 |
| 4d9 | Y282 D | L | R | F | C | T | 18.4 | 300.7 |
| 4d11 | Y282 D | H | G | W | K | W | 9.5 | 83.1 |
| 4s17 | Y282 S | L | R | L | C | M | 117.2 | 98.5 |
| 4s19 | Y282 S | M | R | S | C | L | 23.6 | 42.6 |
| 4s20 | Y282 S | L | R | I | C | F | 103.6 | 327.1 |
| 4S21 | Y282 S | K | F | V | Y | W | 108.6 | 4.0 |
| 4s22 | Y282 S | L | R | N | M | F | 98.7 | 92.1 |
| 4S23 | Y282 S | L | R | V | V | Y | 44.7 | 6.0 |
| 4s24 | Y282 S | L | R | W | Y | S | 88.4 | 288.0 |
| 4S25 | Y282 S | K | M | T | H | W | 188.8 | 4.0 |
| 4S26 | Y282 S | M | R | C | V | L | 35.0 | 7.3 |
| 4s27 | Y282 S | L | R | I | V | S | 76.7 | 164.2 |
| 4s28 | Y282 S | L | R | N | F | F | 172.7 | 212.8 |
| 4S29 | Y282 S | K | L | T | W | T | 263.8 | 11.4 |
| 4S30 | Y282 S | K | L | H | M | W | 96.7 | 4.1 |
| 4S31 | Y282 S | K | W | T | Q | W | 253.5 | 4.2 |
| 4s33 | Y282 S | M | R | I | F | F | 79.3 | 45.5 |
| 4s34 | Y282 S | L | R | V | C | Y | 218.4 | 142.5 |
| 4S35 | Y282 S | K | C | V | Y | T | 119.3 | 8.5 |
| 4S36 | Y282 S | K | K | S | Y | W | 169.5 | 5.1 |
| 4s37 | Y282 S | W | R | I | V | A | 208.6 | 162.8 |
| 4S38 | Y282 S | K | L | T | C | M | 135.5 | 4.2 |
| 4s39 | Y282 S | L | R | L | V | T | 182.6 | 34.4 |
| 4s40 | Y282 S | K | L | S | W | T | 326.8 | 42.1 |
| 4y1 | C281S Y282L | L | R | N | N | Y | 106.2 | 219.6 |
| 4y2 | C281S Y282L | L | R | N | N | F | 54.7 | 223.3 |
| 4y3 | C281S Y282L | L | R | N | M | L | 41.1 | 209.6 |
| 4y4 | C281S Y282L | L | R | N | W | Y | 100.2 | 207.4 |
| 4y5 | C281S Y282L | L | R | N | C | F | 94.8 | 266.1 |
| 4y6 | C281S Y282L | L | R | N | F | F | 78.5 | 234.3 |
| 4y7 | C281S Y282L | L | R | S | C | F | 43.2 | 232.1 |
| 4y8 | C281S Y282L | L | R | V | V | F | 56.4 | 130.8 |
| 4y9 | C281S Y282L | L | L | N | Y | Y | 45.7 | 114.5 |
| 4y10 | C281S Y282L | L | R | F | Y | F | 29.7 | 259.1 |
ANALYSIS OF INTERGENIC INTERACTION SUPPRESSOR MUTANTS
In theory the best heterodimeric repressors would recognize the chimeric operator with high affinity and have a protein-protein interface that is specific and cannot homodimerize. Since repression involves linked equilibria, in vivo repression assays provide an excellent means to examine the strength and specificity of the repressor interfaces. To determine the relative association strength between each intergenic interaction suppressor pair, the levels of GFP transcription were monitored in the presence and absence of IPTG. In addition, the ability of each suppressor mutant to self associate was determined by measuring repression of the symmetric 411 operator. Repression was measured for all of the potential heterodimers and a two dimensional plot (Figure 5) illustrates that some of the mutants identified can self associate as well as compliment the Y282A/S/D monomer. In fact, only a small fraction of intergenic suppressor mutants (9) are truly monomeric, functioning only in the presence of the Y282A/S monomer.
Figure 5. In vivo characterization of identified heterodimers.
Repression analysis represents the relative ability of each repressor to associate as either a homo-dimer or hetero-dimer depending on the operator used in analysis. The ability of each mutant to heterodimerize with its complimentary monomer (◊ Y282A, - Y282D, ▲ Y282S, ■ C281S/Y282L) is shown on the X axis. Each mutant was assayed in the presence of its complimentary monomer and the chimeric operator. The repression ratio is defined by the ratio between the fluorescent signal when the repressor is absent divided by the signal when the repressor is present. For each sample, the measured repression ratio was normalized to the repression ratio for the wild type switch (●). In an independent assay, the isolated intergenic suppressor mutants were analyzed for their ability to homodimerize and thus repress transcription from the 411 operator. The relative repression ratios compared to the wild type switch are plotted on the Y-axis.
To characterize the regulatory efficiency of the unique heterodimeric repressors identified, the four repressors displaying the highest propensity to heterodimerize were further analyzed in vivo. As a first step, we analyzed the specificity of operator recognition by introducing each of the plasmids containing the heterodimeric repressors into cells containing GFP reporters with either the chimeric operator (411:212) or the symmetric operator counterparts (212 or 411). GFP fluorescence was measured under repressing conditions and none of the heterodimers were able to significantly repress the symmetric operators (Figure 6a). Operator recognition was limited to the chimeric operator demonstrating that heterodimeric repressors containing the two distinct DNA binding domains possess specificity in operator recognition. In addition to specificity, each of the heterodimeric switches (repressor and chimeric operator) were found to function better than the natural switch: they repress tighter and are induced more efficiently (Figure 6b & c). Quite surprisingly, two of the heterodimeric repressors associate with the chimeric operator more tightly than the wild type repressor associates with the ‘ideal’ lac operator (212) (Figure 6b). Previously we have observed that tighter repression can be achieved by either altering the sequence of the repressor or the operator; however, it usually comes at the expense of poor inducibility 18. Here, by altering both the DNA binding domains and the dimer interface, we have been able to increase repression yet maintain inducibility.
Figure 6. Characterization of the top functioning heterodimeric repressors.
(a) Specificity of operator recognition was determined by measuring the repression ratio (GFP signal in absence of repressor/GFP signal with repressor). The heterodimeric repressors cannot repress either the 212 or 411 operators. (b) Fluorescent signals were measured for the top functioning heterodimeric repressors when GFP was controlled by the chimeric operator. All of the hits repress transcription better than the wild type switch. Two of the hits function better than the wild type switch using the ‘ideal’ operator. (c) Dark shading = no IPTG, Light shading = 2.5 mM IPTG. Unlike the wild type switch using the ‘ideal’ operator, each of the heterodimeric switches are capable of full induction. They therefore represent true ‘ideal’ switches since the increase in repression does not limit inducibility. (d) The increased repression seen for the heterodimeric repressor with the chimeric operator (black bar) is also seen when the heterodimers are altered to contain two wild type DNA binding domains (grey bars) and incubated with the natural OR1 operator. Repression for this construct is increased further when the ‘ideal’ symmetric (212) operator is used. Abbreviations- Het1: Y282A & 4A20, Het 2: Y282S & 4S29, Het 3: Y282S & 4S31, Het4: Y282S & 4S36
The tighter repression of the heterodimeric repressor can result from either increasing the binding affinity between the repressor head pieces and the chimeric operator or by strengthening the monomer-monomer interface since repression is a function of the linked equilibria. The contribution that the two equilibria make to the increased repression was determined by constructing a heterodimeric repressor with two identical DNA binding domains containing a sequence that corresponds to the wild-type repressor. This construct was introduced into cells containing reporters with the natural OR1 and ‘ideal’ symmetric (212) operators and the repression of GFP fluorescence was measured (Figure 6d). The heterodimeric mutants with identical DNA binding domains can repress transcription of the natural OR1operator as effectively of the heterodimeric mutants repress transcription of the promoter containing a chimeric operator(411:212). This would suggest that the increased repression strength is due to alterations at the dimer interface since the interface mutations represent the only differences between the wild type repressor and the heterodimeric constructs. In addition, when these heterodimers containing two wild type DNA binding domains were analyzed in the presence of the symmetric (212) operator, repression was increased even further. Similar to previous studies however, the increased repression for this operator came once again at the expense of inducibility 18.
CONCLUSION
Our ultimate goal remains to create a repressor that can bind directly to a sequence within a mammalian promoter and block transcription of an endogenous gene. The ability of model development and disease has to a large extent relied on traditional knockout strategies. This technique has been used successfully to identify the role of specific genes in a variety of organisms. However, this technology is limited and cannot be used to study genes that are essential due to problems of embryonic lethality. Although the Cre/ loxP recombination system 22, can circumvent some of these problems, recombination cannot be reversed and the problem remains that the deletion of essential genes is by definition lethal. The ability to modulate gene expression with a reversible switch could circumvent these problems. Itzhaki et al. demonstrated that by incorporating the lac operator into a mammalian promoter by homologous recombination it is possible to regulate the expression of endogenous loci7. Using a heterodimeric repressor it should be possible to create a reversible switch that can regulate the expression of endogenous genes by recognizing the endogenous promoter itself. Here we have taken the first step and demonstrated that by altering the amino acids that recognize the operator and the amino acids that are responsible for forming a dimeric repressor, we could create unique heterodimeric repressors that can recognize a DNA sequence that is void of symmetry and do not resemble any classical operator sequence. With the heterodimeric constructs at our disposal we can now mutate each of the DNA binding domains further to find combinations of mutations which are capable of repressing pre-selected target sequences. In addition, we serendipitously found that altering the protein-protein interface improves upon the repression of this switch, which might be a consequence of altering the monomer- dimer equilibrium. While creating a heterodimeric repressor is only the first step towards our goal of targeting mammalian promoters directly, the creation of these switches also afford us the opportunity to analyze the mechanistic details of the switch with asymmetric changes to the repressor. In addition to altering the residues involved in DNA recognition on each repressor monomer, directed mutagenesis can also be used on residues lining the ligand pocket to help further elucidate the pathways involved in ligand recognition and the allosteric response of induction.
EXPERIMENTAL PROCEDURES
Reporter Construction
The reporter plasmids used in these experiments were derived from the pBD1200 plasmid described previously (Daber 2009). To introduce each of the operators into the pBD1200 vector, full circle PCR was used (list of primers in supplement). After sequence validation the reporter plasmids were transformed into DH5alpha E.coli for selection experiments and mutant characterization.
Recognition Helix Library Construction
To identify mutant repressors which recognize the altered operators, the mutant repressor library with residues 17, 18 and 22 randomized was utilized. A complete description of the construction of the library can be found elsewhere (Daber 2009).
Repressor Co-expression Plasmid Construction
The first step in creating coexpression plasmids was the removal of the C-terminal tetramerization domain from each of the individual repressors (Repressor A: pBD21002, Repressor B: pBD21114 – primer sequences available upon request). The individual plasmids containing each gene were subjected to inverse PCR mutagenesis using primers BD.Lib.011 and BD.Lib.012 which engineered a stop codon after residue 332. Afterwards, a combination of restriction sites surrounding the core domain of the Repressor B gene was created to allow for the interface library to be inserted. A naturally occurring KasI site was located after the core region, so for the repressor A gene (pBD21010), this site was silently removed using inverse PCR mutagenesis (BD.Lib.013, BD.Lib.014). Subsequently, a HindIII site was introduced into the repressor B gene (pBD21005) upstream of the core domain using the same technique (BD.Lib.015, BD.Lib.016). Since the interface library was to be screened against several point mutations at residue 282 of repressor A, the pBD21004 plasmid was further altered to produce four different monomeric repressor A plasmids (pBD21006, pBD21007, pBD21008). Once all of these preparatory steps were completed the 3 modified plasmids bearing each of the of the repressor A mutants as well as the repressor A plasmid containing the dimeric wild type repressor were digested with DrdI. There is only 1 DrdI site in the non essential region of these vectors, but the asymmetric nature of the DrdI restriction site allows for directional cloning of a given insert. The modified Repressor B gene bearing the wt core domain surrounded by the HindIII and KasI restriction sites (pBD21005) was amplified (BD.lib.017, BD.lib.018) to introduce the directional DrdI sites before the promoter region and after the repressor stop codon. After digestion and gel purification the insert and target vectors were ligated and transformed into chemically competent DH5alpha cells. Upon sequence validations, each of the 4 plasmids (pBD22000, pBD22001, pBD22002, pBD22003) was ready for control analysis and insertion of the interface library cassette.
Intergenic Suppressor Repressor Library Construction
To search for mutations complementing the interface of various monomeric lactose repressor mutants, a site directed approach was used to create the repressor interface library. To generate the library of mutants at positions L251, R255, D278, C281 and Y282, an overlap extension PCR strategy was developed (2). Overall 4 fragments were generated which included the coding region for the entire lactose repressor. The first fragment did not introduce any mutations and included the bases encoding residues 1 to 250 (BD.Lib.003, BD.Lib.004). The second fragment involved a PCR reaction using 2 primers (BD.Lib.005, BD.Lib.006) resulting in the randomization of residues 251 and 255. This fragment contained a 20 base pair overlap with the last 20 bases of fragment 1. Fragment 3 was generated like the cassette generated previously (Daber 2009). Codons encoding residues 278, 281 and 282 were randomized as 5’ NNK using a Klenow fill in reaction with the template oligonucleotide (BD.Lib.007) and a complimentary oligo (BD.Lib.008). Similar to Fragment 2, Fragment 3 contained a 20 base pair overlap with the final bases of the second fragment. The fourth fragment was generated via conventional PCR with the primers BD.Lib.009 and BD.Lib.010. After each amplification reaction the products were gel purified and cleaned (Qiagen gel Extraction kit). To assemble the full length gene from the substituent fragments, equimolar quantities of each fragment were pooled in the presence of primers flanking both ends of the full length gene (BD.Lib.003, BD.Lib.010). Upon amplification the full length gene was gel purified and extracted (Qiagen Gel extraction Kit).
Once the randomized cassette library was generated for the second monomer, it was introduced into each of the dual repressor plasmids (pBD22000, pBD22001, pBD22002, pBD22003). Each of the target plasmids along with the library were digested with HindIII and KasI, gel purified then ligated with a ratio of vector to insert of 1:3. The ligation products were then desalted (Qiagen PCR Purification kit) and electroporated into competent DH5alpha cells. A small fraction (5%) of cells was plated to determine the size of each library while the remaining transformants were used to inoculate 25 mL overnight cultures. The plasmid libraries were isolated via the plasmid preparation procedure after the cultures had reached saturation.
Fluorescent Activated Cell Sorting
Similar to previous selection experiments (Daber 2009) Fluorescent Activated Cell Sorting was used to separate bacterial cells into various populations based upon the intensity of their fluorescent phenotype. In short, the first step involved transforming desired combinations of reporter and repressor libraries into cell lines containing the opposite plasmid. A small aliquot of these cells were plated to determine the number of total transformants while the remaining cells were used to inoculate a 10 mL overnight culture. The following morning a 25 mL culture was inoculated 1/100 with the saturated overnight culture. Once these cultures reached an optical density (A600) of 0.6, the samples were transferred to a 4°shaker for 30 minutes. After stalling cell growth, 1 OD of cells was pelleted at 3,000 rpm for 5 minutes at 4C. The cell pellet was then resuspended into sterile 5.0 mL of PBS, transferred into a sterile falcon tube and stored on ice. Glycerol slabs were prepared with 500 uL of the remaining culture for future studies.
Sorting was conducted at the University of Pennsylvania Flow Cytometry and Cell Sorting Facility. To establish proper population gates, positive (unrepressed) and negative (repressed) control samples were analyzed. The library samples were then sorted into each of these populations and the cells were collected in 5 mL falcon tubes containing 1.0 mL of 50% PBS and 50 % LB. Cells displaying an intermediate phenotype were also collected. Samples were then returned to storage on ice until appropriate volumes were plated on LB plates (150 × 20mm). Two rounds of sorting were conducted for the intergenic suppressor selection experiments. The population of cells showing the repressed phenotype were collected, grown overnight in LB to saturation, then sorted again before plating any cells on agar plates. After colonies became visible on plates, they were chosen to inoculate overnight cultures for plasmid preparation and sequencing.
Isolating the Monomeric Repressor Genes
Once heterodimeric repressors were selected for, it was necessary to isolate the second repressor gene of the dual repressor plasmid in order to characterize its ability to form homodimers. To remove the first repressor gene, inverse PCR mutagenesis was conducted with primers flanking this monomer resulting in a deletion mutation (Forward Primer: BD004 , Reverse Primer: BD005). The recovered plasmid maintained the origin of replication, the Choramphenicol resistance marker as well as the coding sequence for the second repressor.
In vivo Repression and Induction Assay
To analyze the phenotypes of various repressor – operator combinations an in vivo fluorescent assay was used. To quantitate the level of fluorescence and therefore indirectly measure the degree of transcription, cells were grown and analyzed in a Perkin Elmer Victor3 plate reader. In short combinations of repressors and operators were transformed and colonies were selected for overnight culture growth. Depending upon the goal of the assay, the combination of samples as well as the time point during growth at which the samples were analyzed varied. Since repression is defined as the ratio of the reporter signal in the absence of repressor divided by the signal when repressor is present, a repression assay required both of the aforementioned samples be grown. For an induction assay, a no repressor sample was not required. A sample containing the repressor and reporter were grown in the presence and absence of IPTG and the signals were compared. Fluorescence was monitored for cultures at all points during their growth curves and the largest dynamic range between the repressed and induced samples occurred prior to reaching stationary phase. In general, all assays were conducted by growing each triplicate samples in either 500 uL or 1 mL of LB in a 96 well plate, aliquoting 200 uL samples at the selected time point then measuring fluorescence (495nm excitation wavelength, 510nm emission wavelength) and optical density (A590) on a Perkin Elmer Victor3 Plate reader. To normalize the signal for each sample, the signal from the blank sample was subtracted and the resulting fluorescent signals were normalized to cell OD. The normalized signals from each of the replicates were averaged and the standard deviations were calculated (shown by error bars on plots).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Jacob F, Monod J. Genetic Regulatory Mechanisms in the Synthesis of Proteins. J. Mol. Biol. 1961;3:318–356. doi: 10.1016/s0022-2836(61)80072-7. [DOI] [PubMed] [Google Scholar]
- 2.Wilson CJ, Zhan H, Swint-Kruse L, Matthews KS. The lactose repressor system: paradigms for regulation, allosteric behavior and protein folding. Cellular & Molecular Life Sciences. 2007;64:3–16. doi: 10.1007/s00018-006-6296-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gilbert W, Muller-Hill B. Isolation of the Lac Repressor. Proc. Natl. Acad. Sci. USA. 1966;56:1891–1898. doi: 10.1073/pnas.56.6.1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bell CE, Lewis M. The Lac repressor: a second generation of structural and functional studies. Current Opinion in Structural Biology. 2001;11:19–25. doi: 10.1016/s0959-440x(00)00180-9. [DOI] [PubMed] [Google Scholar]
- 5.Hu MC, Davidson N. The inducible lac operator-repressor system is functional in mammalian cells. Cell. 1987;48:555–566. doi: 10.1016/0092-8674(87)90234-0. [DOI] [PubMed] [Google Scholar]
- 6.Figge J, Wright C, Collins CJ, Roberts TM, Livingston DM. Stringent regulation of stably integrated chloramphenicol acetyl transferase genes by E. coli lac repressor in monkey cells. Cell. 1988;52:713–722. doi: 10.1016/0092-8674(88)90409-6. [DOI] [PubMed] [Google Scholar]
- 7.Itzhaki JE, Gilbert CS, Porter AC. Construction by gene targeting in human cells of a "conditional' CDC2 mutant that rereplicates its DNA. Nature Genetics. 1997;15:258–265. doi: 10.1038/ng0397-258. [DOI] [PubMed] [Google Scholar]
- 8.Cronin CA, Gluba W, Scrable H. The lac operator-repressor system is functional in the mouse.[see comment] Genes & Development. 2001;15:1506–1517. doi: 10.1101/gad.892001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Segal DJ, Dreier B, Beerli RR, Barbas CF., 3rd Toward controlling gene expression at will: selection and design of zinc finger domains recognizing each of the 5'-GNN-3' DNA target sequences. Proc Natl Acad Sci U S A. 1999;96:2758–2763. doi: 10.1073/pnas.96.6.2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beerli RR, Segal DJ, Dreier B, Barbas CF., 3rd Toward controlling gene expression at will: specific regulation of the erbB-2/HER-2 promoter by using polydactyl zinc finger proteins constructed from modular building blocks. Proc Natl Acad Sci U S A. 1998;95:14628–14633. doi: 10.1073/pnas.95.25.14628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weickert MJ, Adhya S. A family of bacterial regulators homologous to Gal and Lac repressors. Journal of Biological Chemistry. 1992;267:15869–15874. [PubMed] [Google Scholar]
- 12.Schumacher MA, Choi KY, Zalkin H, Brennan RG. Crystal Structure of LacI Member, PurR, Bound to DNA : Minor Groove Binding by Alpha Helices. Science. 1994;266:763–772. doi: 10.1126/science.7973627. [DOI] [PubMed] [Google Scholar]
- 13.Lewis M, Chang G, Horton NC, Kercher MA, Pace HC, Schumacher MA, Brennan RG, Lu P. Crystal structure of the lactose operon repressor and its complexes with DNA and inducer.[see comment] Science. 1996;271:1247–1254. doi: 10.1126/science.271.5253.1247. [DOI] [PubMed] [Google Scholar]
- 14.Lehming N, Sartorius J, Niemoller M, Genenger G, v Wilcken-Bergmann B, Muller-Hill B. The interaction of the recognition helix of lac repressor with lac operator. EMBO Journal. 1987;6:3145–3153. doi: 10.1002/j.1460-2075.1987.tb02625.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lehming N, Sartorius J, Oehler S, von Wilcken-Bergmann B, Muller-Hill B. Recognition helices of lac and lambda repressor are oriented in opposite directions and recognize similar DNA sequences. Proceedings of the National Academy of Sciences of the United States of America. 1988;85:7947–7951. doi: 10.1073/pnas.85.21.7947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lehming N, Sartorius J, Kisters-Woike B, von Wilcken-Bergmann B, Muller-Hill B. Mutant lac repressors with new specificities hint at rules for protein--DNA recognition. [erratum appears in EMBO J 1990 May;9(5):1674] EMBO Journal. 1990;9:615–621. doi: 10.1002/j.1460-2075.1990.tb08153.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaptein R, Boelens R, Chuprina VP, Rullmann JA, Slijper M. NMR and nucleic acid-protein interactions: the lac repressor-operator system. Methods in Enzymology. 1995;261:513–524. doi: 10.1016/s0076-6879(95)61022-7. [DOI] [PubMed] [Google Scholar]
- 18.Daber R, Lewis M. 2009 unpublished. [Google Scholar]
- 19.Sadler JR, Sasmor H, Betz JL. A perfectly symmetric lac operator binds the lac repressor very tightly. Proceedings of the National Academy of Sciences of the United States of America. 1983;80:6785–6789. doi: 10.1073/pnas.80.22.6785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chakerian AE, Matthews KS. Characterization of Mutations in the Oligomerization Domain of Lac Repressor Protein. The Journal of Biological Chemistry. 1991;266:22206–22214. [PubMed] [Google Scholar]
- 21.Swint-Kruse L, Elam CR, Lin JW, Wycuff DR, Shive Matthews K. Plasticity of quaternary structure: twenty-two ways to form a LacI dimer. Protein Science. 2001;10:262–276. doi: 10.1110/ps.35801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gu H, Zou YR, Rajewsky K. Independent control of immunoglobulin switch recombination at individual switch regions evidenced through Cre-loxP-mediated gene targeting. Cell. 1993;73:1155–1164. doi: 10.1016/0092-8674(93)90644-6. [DOI] [PubMed] [Google Scholar]