Summary
Using expression cloning, we have identified an H2-M3-restricted epitope of the intracellular bacterial pathogen Listeria monocytogenes. Picomolar concentrations of an amino-terminal N-formylated hexapeptide, fMIGWII, targeted cells for lysis by CD8+ cytotoxic T cells, while the nonformylated peptide was approximately 100-fold less active. The sequence of the 185 aa protein source of this epitope predicts a transmembrane protein that retains its N terminus and assumes an Nout–Cin topology. This membrane orientation offers an explanation for the protection of the epitope from deformylases present in the bacterial cell and suggests an explanation for the ability of phagocytes to present H2-M3-restricted bacterial epitopes via a vacuolar TAP-independent mechanism.
Introduction
Genes within the murine major histocompatibility complex (MHC) encode an assortment of antigen-presenting molecules. The conventional and highly polymorphic MHC class II and MHC class Ia alleles encode membrane glycoproteins that bind antigenic peptide epitopes and present these epitopes to CD4+ or CD8+ T lymphocytes, respectively. The peptide epitopes presented to CD4+ T cells are largely processed from exogenous antigens and load MHC class II molecules within acidified vacuolar compartments of antigen-presenting cells (APCs). Conversely, the peptide epitopes presented to CD8+ cytotoxic T lymphocytes (CTLs) are generally processed from endogenous proteins by cytosolic or nuclear proteasomes. These cytosolic peptides are transported via the heterodimeric transporter associated with antigen processing (TAP) into the endoplasmic reticulum (ER) lumen, where they associate with the peptide-binding groove of nascent MHC class Ia molecules. These antigen presentation pathways thus limit the presentation of most antigens derived from extracellular and vacuolar pathogens to CD4+ T cells, whereas antigenic peptides from viruses and cytosolic intracellular bacteria are presented to CD8+ CTL (Germain and Margulies, 1993; Wolf and Ploegh, 1995).
The gram-positive bacterium Listeria monocytogenes is a facultative intracellular pathogen. Virulent Listeria strains express listeriolysin O (LLO), a hemolysin that facilitates escape of phagocytosed bacteria into the host cell cytosol (Tilney and Portnoy, 1989). Once in the cytosol, expression of other Listeria virulence factors, such as the ActA gene product and a phospholipase C, enable Listeria to spread from cell to cell without returning to the extracellular milieu (Kocks et al., 1992; Vazquez et al., 1992). These and other bacterial gene products that access the host cytosol thus enter the conventional MHC class Ia pathway of antigen processing and presentation. Two secreted Listeria proteins have previously been shown to provide peptide epitopes for MHC class Ia–restricted CD8+ CTL: LLO (residues 91–99) and p60 (residues 217–225 and 449–457) (Pamer et al., 1991; Pamer, 1994; Sijts et al., 1996). CTLs specific for the LLO and p60 epitopes lyse infected syngeneic target cells in vitro (Pamer et al., 1991; Sijts et al., 1996), and adoptive transfer of CTLs specific for LLO(91–99) and p60(217–225) protects syngeneic mice against subsequent Listeria challenge in vivo (Harty and Bevan, 1992; Harty and Pamer, 1995).
Mice infected with Listeria have long been known to develop CD8+ CTL responses that are not restricted by conventional polymorphic MHC class Ia alleles (De Libero and Kaufmann, 1986). Such MHC-unrestricted CTLs adoptively transfer protection to allogeneic hosts (Kaufmann et al., 1988; Lukacs and Kurlander, 1989) and demonstrate antigen-specific lysis of both syngeneic and allogeneic macrophage targets that are Listeria infected or pulsed with heat-killed Listeria monocytogenes (HKLm) (De Libero and Kaufmann, 1986; Kaufmann et al., 1988; Lukacs and Kurlander, 1989; Brown et al., 1992). This MHC-unrestricted pattern of CTL recognition was explained by the finding that an MHC class Ib gene with limited polymorphism, H2-M3, restricts the presentation of an unidentified Listeria-derived peptide to CTL effectors (Pamer et al., 1992). Similarly, the M3 molecule (encoded by H2-M3) was implicated in presentation of a detergent-solubilized Listeria membrane antigen to MHC-unrestricted CTLs (Kurlander et al., 1992). Presentation both of this solubilized antigen and of unfractionated HKLm to CTLs had previously been shown to be inhibited by Brefeldin A, chloroquine, ammonium chloride, monensin, and pepstatin A (Brown et al., 1992). Thus, the pathway used for processing and presentation of these complex Listeria antigens to H2-M3-restricted CTLs may incorporate features of the MHC class II pathway of antigen processing and presentation.
H2-M3wt is the allele of H2-M3 present in most common inbred mouse strains and is expressed in a variety of tissues (Smith et al., 1994; Fischer Lindahl et al., 1991; Wang et al., 1991). H2-M3 was originally identified due to its role in CTL responses to a maternally transmitted minor histocompatibility antigen (Mta) (Fischer Lindahl et al., 1991). Studies with Mta-specific CTLs revealed that the M3 molecule presents a hydrophobic peptide derived from the N terminus of a mitochondrial protein: the ND1 subunit of NADH dehydrogenase (Loveland et al., 1990; Wang et al., 1991). The N-terminal formylmethionine residue is essential for efficient presentation of the ND1 peptide to Mta-specific CTLs (Shawar et al., 1990; Smith et al., 1994), and peptides that initiate with N-formylmethionine efficiently block target cell lysis by these CTLs (Shawar et al., 1990, 1991; Vyas et al., 1992). Since only eubacteria, including mitochondria and chloroplasts, initiate protein synthesis with N-formylmethionine, these findings suggest that M3 may have evolved to play a specialized role in host defense against bacterial pathogens (Shawar et al., 1990, 1994; Pamer et al., 1993).
A molecule specialized for presentation of antigens from bacterial pathogens might well have evolved to load antigens from the intracellular compartments where bacteria reside. Consistent with loading of M3 in a vacuolar compartment, we provide evidence that phagocytes present heat-killed bacteria to H2-M3-restricted CTLs independent of an intact TAP transporter. We also describe the use of an expression cloning strategy to identify the first pathogen-derived epitope presented by M3. This epitope is derived from the hydrophobic N terminus of a novel Listeria membrane protein which is predicted to assume an Nout–Cin topology. We discuss the significance of this membrane topology in terms of the selection of bacterial epitopes for presentation by M3, and the possible mechanisms for loading of the M3 molecule in a vacuolar compartment.
Results
Previous reports of H2-M3-restricted presentation of Listeria antigen to T cell receptor (TCR)αβ CD8+ T cells have noted that targeting activity can be detected in the supernatant of Listeria cultures (Pamer et al., 1992), or that heat-killed organisms can provide antigen to macrophages (Kurlander et al., 1992). This is unlike the situation with conventional MHC class Ia–restricted presentation of LLO and p60 epitopes, which requires infection of cells with viable LLO-producing bacteria (Brunt et al., 1990; Pamer et al., 1991). To generate new CTL lines that could recognize exogenous bacterial antigen, we used HKLm to stimulate splenocytes from a C57BL/6 mouse previously primed by sublethal infection with viable virulent Listeria. CD8+ cytolytic clones were isolated by limiting dilution and the Cb10 clone (which expresses TCRαβ) was used for further study. The Cb10 CTL clone recognizes a hydrophobic epitope in natural peptide extracts of Listeria-infected macrophages, which elutes at fraction 40 (Fr40) on RP–HPLC. In addition, low molecular mass material in Listeria culture supernatants (LmSN) also contains targeting activity for the Cb10 CTL clone (see below). Supernatants of Escherichia coli, Salmonella typhimurium, or Bacillus subtilis, or sonicates of these cultures, did not contain targeting activity.
H2-M3 Restricts Cb10 CTL Responses
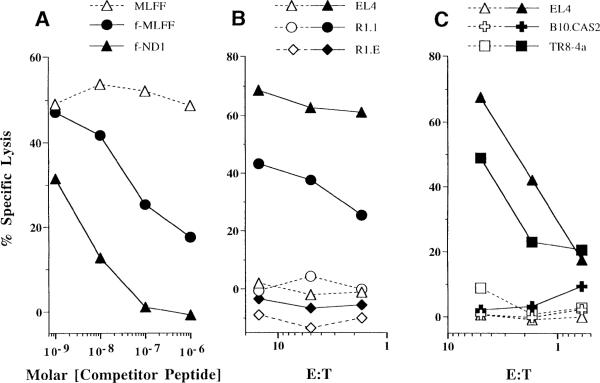
Three lines of evidence indicate that M3 presents the Fr40 epitope to Cb10 CTL effectors. First, competitor peptides initiating with N-formylmethionine (fMet peptides) competitively inhibit lysis of Fr40-coated target cells by Cb10 CTL effectors (Figure 1A). Competition of Fr40 presentation by the corresponding nonformylated competitor requires at least 100-fold higher peptide concentration (Figure 1A), which is consistent with the preferential presentation of fMet peptides by M3 (Shawar et al., 1990; Smith et al., 1994). Second, LmSN-coated allogeneic R1.1 (H-2k) target cells are lysed by Cb10 CTL effectors (Figure 1B), indicating that antigen presentation to Cb10 does not require conventional MHC class Ia or MHC class II molecules. However, R1.E target cells, β2-microglobulin (β2m)-negative mutants of R1.1, do not present LmSN to Cb10 CTL effectors (Figure 1B). Thus, presentation of the Fr40 epitope to Cb10 CTLs requires expression of a β2m-dependent MHC class Ib molecule. The third line of evidence that H2-M3 restricts Cb10 CTL responses is presented in Figure 1C. LmSN-coated B10.CAS2 fibroblasts, which express the (null) H2-M3cas allele, are not lysed by Cb10 CTL effectors, but presentation of the Fr40 epitope is restored in an H2-M3wt transfectant of B10.CAS2 fibroblasts (TR8-4a; Wang et al., 1991). These data provide direct proof that M3 is responsible for presentation of Fr40 to Cb10 CTLs.
Figure 1. Cb10 CTLs Are Restricted by H2-M3wt.
(A) Competition of fMet peptides for presentation to Cb10 CTL effectors. Shown is specific lysis of EL4 target cells coated with HPLC-purified Fr40 plus the indicated concentrations of fMet (closed symbols) or nonformylated (open symbol) competitor peptides. The E:T ratio used was 5:1. Lysis of Fr40-coated targets in the absence of competitors was 47.5%.
(B) Effects of target cell MHC and β2m expression on presentation to Cb10 CTL effectors. Target cells differing in MHC haplotype and β2m expression were coated with uninoculated bacterial culture media (TSB; open symbols) or LmSN (closed symbols) and assayed for lysis by Cb10 CTLs at the indicated E:T. EL4 (H-2b) and R1.1 (H-2k) cells express β2m. R1.E is a β2m-negative mutant of R1.1.
(C) Effects of H2-M3wt expression on presentation to Cb10 CTL effectors. Shown is lysis of TSB- (open symbols) or LmSN- (closed symbols) coated B10.CAS2, TR8-4a, and control EL4 targets at the indicated E:T ratios. B10.CAS2 fibroblasts express a null H2-M3 allele (H2-M3cas2), whereas TR8-4a transfectants and EL4 express H2-M3wt.
Presentation of Exogenous HKLm Antigen to Cb10 CTLs
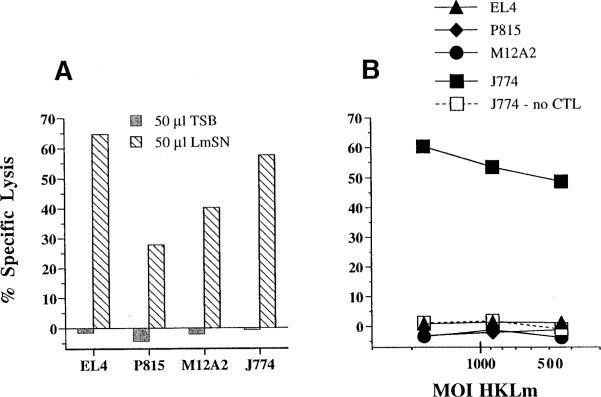
As mentioned above, Cb10 CTLs are restimulated in vitro with HKLm as a source of bacterial antigen. Yet, HKLm preparations do not contain viable bacteria and thus have no obvious route of access to the conventional (cytosolic) MHC class Ia antigen presentation pathway. To investigate more directly the presentation of HKLm to H2-M3-restricted CTLs, we compared the ability of LmSN and washed preparations of HKLm to elicit lysis of target cells by Cb10 CTL effectors (Figures 2A and 2B). Predictably, the LmSN antigen elicited CTL lysis of both J774 macrophages and nonphagocytic EL4 thymoma cells, P815 mastocytoma cells, and M12A2 B cells (Figure 2A). Thus, all three cell types clearly express M3 molecules that load with Listeria peptides present in LmSN. However, Cb10 CTL effectors fail to kill EL4, P815, or M12A2 targets that are incubated with HKLm, despite the observation that macrophage targets readily present the exogenous HKLm antigen in a dose-dependent manner (Figure 2B). Furthermore, target cells incubated with HKLm are not lysed in the absence of CTLs (Figure 2B), demonstrating that HKLm is not toxic to the phagocytic cells. Together, these data suggest that HKLm preparations do not contain high concentrations of soluble targeting activity, and that macrophages can process HKLm to obtain Fr40-like determinants for presentation to H2-M3-restricted CTLs.
Figure 2. Phagocytic Cells Present Exogenous HKLm Antigen to Cb10 CTLs.
51Cr-labeled target cells were precoated for 2 hr with (A) 50 μl of LmSN or (B) an equal volume of HKLm washed and diluted in PBS. Cb10 CTL effectors were added at an E:T of 5:1 and lysis was measured in a 4 hr CTL assay.
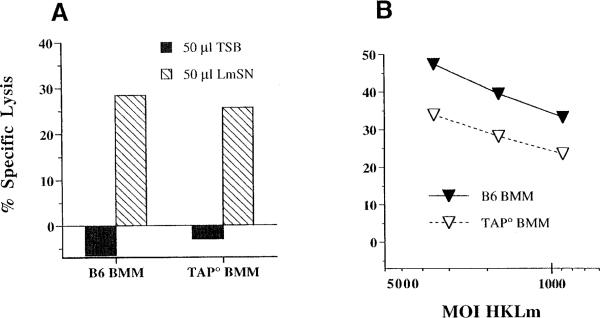
To evaluate whether processing and presentation of HKLm by macrophages utilizes components of the conventional MHC class Ia antigen presentation pathway, we assayed for presentation of LmSN and HKLm by bone marrow–derived macrophages (BMMs) cultured from wild-type C57BL/6 (B6), or Tap1-deficient (TAP°) mice (Figures 3A and 3B). B6 BMMs present both LmSN and HKLm antigens to Cb10 CTLs (Figures 3A and 3B), confirming that primary phagocytes process and present exogenous bacterial antigen to H2-M3-restricted CTLs. Furthermore, presentation of HKLm occurs independent of functional TAP heterodimers, since HKLm-coated TAP° BMMs were also lysed by Cb10 CTLs (Figure 3B). It is clear, however, that antigen-coated TAP° BMMs are less-efficiently lysed than are B6 BMMs. This reduced lysis may reflect reduced surface expression of peptide-receptive MHC class I molecules in the absence of TAP (Day et al., 1995), or it may indicate that HKLm presentation occurs via a TAP-dependent pathway as well. Interestingly, the proliferative responses of Cb10 CTLs in response to HKLm-coated B6 and TAP° splenocytes are nearly equivalent (Lenz and Bevan, 1996), which may indicate delayed release or loading of Fr40 in the absence of TAP. Thus, identification of bacterial antigens presented to H2-M3-restricted CTLs may shed light on the mechanics of this TAP-independent pathway of HKLm presentation.
Figure 3. TAP-Independent Presentation of HKLm to Cb10 CTLs.
Target cells were labeled with 51Cr and precoated with (A) 50 μl of LmSN or (B) dilutions of HKLm and assayed for lysis in a 4 hr CTL assay with Cb10 CTL effectors at an E:T of 5:1. BMMs were cultured from wild-type C57BL/6 mice (B6 BMM) or from Tap-1° mice (TAP° BMM) in L cell–conditioned medium. Lysis of target cells in the absence of Cb10 effectors was less than 5% at all moi.
A Library for Expression Cloning of Listeria Antigens
To gain insight into the nature of bacterial antigens presented by M3, we sought to use an expression cloning technique to identify bacterial antigens presented to H2-M3-restricted CTLs. We screened for expression of Fr40 activity by E. coli transformed with a Listeria genomic library. The library was prepared by cloning Listeria genomic DNA fragments into a shuttle vector, pAT29 (Trieu et al., 1990). This vector was chosen because it permits shuttling of the library between gram-negative and gram-positive hosts, if required for appropriate expression of Listeria gene products.
The library is comprised of 3084 individual E. coli transformants, which have an average insert size of 5.7 ± 1.3 kb, as estimated from 31 randomly selected clones. Thus, the sum of Listeria DNA in the library represents approximately 6 genomes, assuming 3,150 kb/genome (Michel and Cossart, 1992). The completeness of the library was further evaluated by determining the representation of two unlinked single copy Listeria genes: the LLO-encoding gene hly and the p60-encoding gene iap. Using a polymerase chain reaction (PCR)-based assay the library was screened in pools of 96 clones. Based on amplification of PCR products of the appropriate sizes, 11 of 33 pools scored positive for hly and 2 of 33 pools were positive for iap. Assuming that each positive pool contains a single positive transformant, the frequency of hly-positive clones is estimated at 3.6 × 10–3, while iap appears at a frequency of 6.5 × 10–4. These data indicate that both test genes are present in this Listeria expression library, although some individual Listeria genes may be over or underrepresented.
Identification of Transformants that Produce Fr40 Activity
To screen for E. coli transformants potentially expressing the Fr40 epitope, the specific lysis of J774 macrophages coated with lysates of E. coli transformants was measured in CTL assays with Cb10 CTL effectors. J774 macrophage targets were used to facilitate any required processing of Listeria-encoded antigens in the E. coli lysates. These lysates were first screened in pools representing 8 transformant clones per pool, and Cb10 CTL effectors were added to each pool. Of the 386 pools tested, 18 active pools elicited specific lysis of J774 cells which was ≥10% above the level of background lysis seen in the presence of control E. coli lysates (data not shown). These active pools represented 144 individual clones. A screen of lysates prepared from individual clones in these pools revealed 20 active clones, which are listed in Table 1. Seven of these clones (Table 1, group 1) caused target cell lysis in the absence of Cb10 CTLs; presumably these clones produce a Listeria protein that is toxic for J774 macrophages. However, the remainder of the active clones (Table 1, group 2) appear to express a Listeria gene or genes that mark the target cells for destruction by Cb10 CTL effectors. This suggests that the latter group includes clones that express a Listeria gene or genes that give rise to the Fr40 epitope.
Table 1.
Identification of Transformant Clones that Produce CTL Targeting Activity
| Percent Specific Lysis |
||
|---|---|---|
| Transformant | Cb10 CTL | No CTL |
| pAT29a | 12 | 8 |
| Group 1 | ||
| Bb5 | 91 | 100 |
| Ld10 | 81 | 86 |
| Ne5 | 95 | 95 |
| Nh12 | 100 | 94 |
| Ph8 | 100 | 95 |
| Pg12 | 98 | 98 |
| 7Cf2 | 85 | 89 |
| Group2 | ||
| Aa12 | 84 | 24 |
| Lg10 | 66 | 8 |
| Pb12 | 74 | 14 |
| Qe11 | 83 | 8 |
| Rc5 | 80 | 12 |
| Vf8 | 70 | 16 |
| 7Ca10 | 77 | 9 |
| 7Cg3 | 82 | 9 |
| 7Da9 | 79 | 14 |
| 7Eg1 | 71 | 0 |
| 7Fe5 | 71 | 0 |
| 7Gb9 | 66 | 9 |
| 7He3 | 77 | 9 |
E. coli transformed with the parental pAT29 plasmid.
The library was screened for targeting activity and 18 active pools were identified. The transformant clones comprising these active pools were assayed individually for Fr40 production by screening for target cell lysis with or without addition of Cb10 CTL effectors at an E:T of 10:1. Those clones whose lysates elicited > 20% lysis of J774 targets are shown. Transformants in group 1 likely express toxic Listeria-encoded products; those in group 2 express Fr40-like activity (see text).
Subcloning and Sequencing of a Candidate Gene
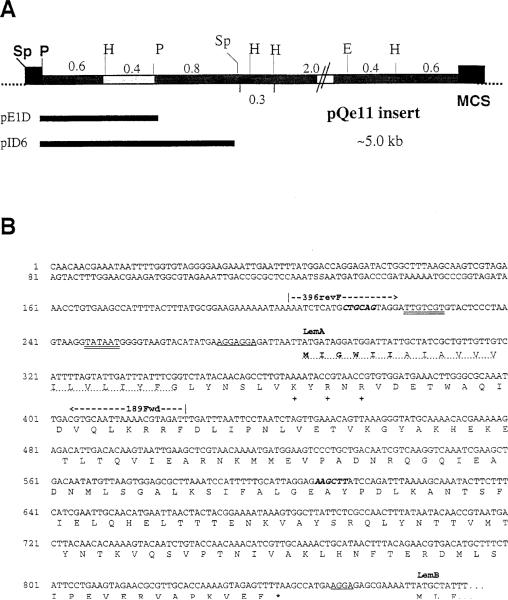
To focus our search for Listeria genes responsible for Fr40 production in the E. coli transformants identified in the library screen, we isolated plasmid DNA from one such transformant (Table 1, Qe11) and generated a restriction map of the insert. The insert in this plasmid (pQe11) was approximately 5 kb long and contained seven restriction sites that could be unambiguously mapped (Figure 4A). By multiple digestions of pQe11 we subcloned six fragments into pUC19. Two additional subclones were generated by excising fragments from pQe11 and reannealing the plasmid. Thus, eight distinct restriction fragments were subcloned. For each subclone, lysates of several insert-bearing transformant colonies were tested for targeting activity using J774 targets and Cb10 CTL effectors. Fr40-like activity was detected in lysates from five subclones, whereas no activity was detected in lysates of the remaining subclones. Furthermore, all Fr40-positive subclones shared a 0.4 kb PstI–HindIII (P–H) fragment, which was absent in the inserts of subclones that did not produce Fr40 activity. These data suggested that this 0.4 kb P–H fragment contained a Listeria gene or gene fragment that permitted expression of Fr40 activity in E. coli transformants.
Figure 4. Restriction Map of pQe11 and Partial Nucleotide Sequence of the Putative Lem Operon.
(A) Insert DNA from the pQe11 plasmid was mapped with the indicated enzymes: Sp, SphI; P, PstI; H, HindIII; E, EcoRI. Individual restriction fragments were subcloned into pUC19 and subclones were tested for production of Fr40-like activity. Activity segregated with the highlighted 0.4 kb H–P fragment. Numbers indicate sizes (in kilobases) of the indicated restriction fragments. The pAT29 multiple cloning site (MCS) is represented by closed boxes at each end of the insert. Indicated below the map are the positions of the ~1.2 kb P–P and ~1.8 kb Sp–Sp inserts of subclones pE1D and pID6.
(B) Partial nucleotide sequence from the Listeria insert DNA of pE1D and pID6. Double-underlined bases represent nucleotides that fit consensus sequences for binding of RNA polymerase (–35 and –10). Single underlines designate ribosome-binding sites (rbs). Bold italics indicate the PstI (CTGCAG) and HindIII (AAGCTT) restriction sites, which flank the 0.4 kb fragment to which the Fr40 epitope was mapped (see text). The nucleotide sequence predicts a polycistronic transcript encoding at least two polypeptides (LemA and LemB). Translations for the LemA coding region and the N terminus of LemB are given using the single letter amino acid code. The predicted LemA membrane-spanning domain is indicated by a dotted underline. Positively charged residues used to predict the membrane orientation of LemA are indicated at positions 27, 29, and 31 with a plus symbol. Amino acid residues in bold type represent the sequence of the LemA hexapeptides used in Figure 6, and an asterisk indicates the stop codon for the LemA protein. The coding sequence for LemB continues at least 103 residues and runs into the multiple cloning site (data not shown). The positions of the 5′ (396revF) and 3′ (189Fwd) oligonucleotide primers used in Figure 5 are also shown.
To identify Listeria genes present in the 0.4 kb P–H fragment, we sequenced two active subclones. These plasmids, pE1D and pID6, carried pQe11-derived inserts of ~1.2 kb and ~1.8 kb, respectively, which overlapped the active P–H fragment (Figure 4A). Analysis of the 0.4 kb sequence revealed five open reading frames (ORFs) with potential polypeptide products of greater than 15 aa. We synthesized decameric fMet peptides corresponding to the N termini of these ORFs and tested each peptide for Fr40-like activity in CTL assays using Cb10 CTL effectors. Remarkably, the fMet peptide corresponding to one Listeria-encoded ORF (329Fwd ORF) triggered dose-dependent CTL lysis of EL4 target cells at nanomolar concentrations, whereas even 10 μM concentrations of the other four peptides had no activity (data not shown). This Listeria-encoded peptide thus demonstrated Fr40-like activity. The DNA 5′ of the ORF encoding this peptide contained appropriately spaced bacterial RNA polymerase (–10 and –35 sites) and ribosome (rbs)-binding sites, consistent with its expression in Listeria. However, database searches with the translated 329Fwd ORF sequence failed to identify homologous gene products, suggesting that this ORF represents a previously unidentified Listeria gene. Together, these data identify this novel gene as a good candidate for the Fr40-encoding gene, and we hereafter refer to it as lemA, for Listeria epitope with M3.
LemA and the Putative lem Operon
To gain insight into the subcellular localization of the LemA protein, we completed sequencing of the lemA gene and surrounding DNA. Analysis of nucleotide sequence obtained from two subclones (Figure 4A, pE1D and pID6) suggests that lemA is transcribed as the first gene of a polycistronic transcript (see Figure 4B). Using the BLAST server at the National Center for Biotechnology Information, we failed to identify any protein sequences in the nonredundant protein sequence database that share extensive homology with LemA. However, analysis of the predicted LemA amino acid sequence reveals a single N-terminal hydrophobic region of 20–26 hydrophobic residues. Cleaved signal peptides of secreted prokaryotic proteins typically have an N-terminal basic region followed by a stretch of hydrophobic residues, a polar region, and a consensus cleavage site for signal peptidase (Simonen and Palva, 1993; Pugsley, 1993). The LemA N terminus lacks basic residues before the hydrophobic stretch as well as a consensus signal peptidase cleavage site, suggesting that its N terminus remains intact. Instead, the protein is predicted to have a single membrane-spanning domain, which orients in the membrane with an Nout–Cin topology and retains its N terminus. This prediction is based on the presence of an N-terminal hydrophobic domain (the N-terminal 20–26 residues), which is followed by several positively charged residues (positions 27, 29, and 31). In accordance with the positive–inside rule (von Heijne, 1986), the charged residues following position 26 and the remainder of the LemA polypeptide should remain within the bacterial cytosol. This topology places the N-terminal formylmethionine of LemA at the exterior (extracytosolic) face of the bacterial plasma membrane.
Detection of lemA in All Fr40-Producing Library Transformants
As noted above, screening of the Listeria library with Cb10 CTLs identified 13 E. coli transformant clones that expressed Fr40 activity (Table 1). Partial mapping of the recombinant plasmids from these clones revealed at least 9 distinct species (data not shown). To determine whether the candidate Fr40-encoding gene was present in each of these heterogeneous plasmids, we assayed for amplification of a diagnostic PCR product using lemA-specific oligonucleotide primers. The PCR primers used (396revF and 189Fwd; see Figure 4B) amplify 119 bp at the 5′ end of the lemA coding sequence plus an additional 64 bp of upstream DNA to yield a 223 bp product. These primers clearly amplify a single product of the predicted size from a Listeria genomic DNA template (Figure 5, lane 15). Conversely, identical PCR conditions failed to amplify this product from pAT29 plasmid control DNA (Figure 5, lane 14), as expected for specific amplification of the Listeria lemA gene. Detection of this lemA-specific PCR product using plasmid DNA templates isolated from all 13 Fr40-producing clones indicates that the 5′ coding region of lemA is present in each clone (Figure 5, lanes 1–13). These data support the notion that only one region of the Listeria genome (lemA) produces targeting activity for Cb10 CTLs.
Figure 5. The N-Terminal Coding Region of lemA Is Present in All Transformants that Produce Fr40 Activity.
lemA-specific PCR primers (lanes 1–15) were used to amplify products from the following template DNAs: plasmid DNA isolated from each Fr40-producing transformant listed in group 2 of Table 1 (lanes 1–13), pAT29 plasmid (lane 14), and Listeria genomic DNA (lane 15). The 396revF and 189Fwd primers were used to detect the 5′ end of the lemA gene anneal as indicated in Figure 4 and amplify a 223 bp product. Lane 16 contains molecular mass standards.
Presentation of LemA Peptides to Cb10 CTLs
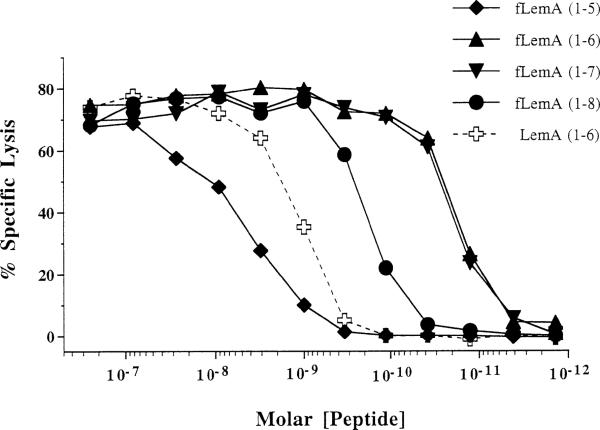
The natural ligands for conventional MHC class Ia molecules are generally 8–10 residue peptides that elicit target cell lysis when present in picomolar concentrations (Germain and Margulies, 1993). The optimal peptide size for presentation by M3 is unknown, although peptides as short as 6 residues target some ND1-specific CTLs (Shawar et al., 1990), and 3- or 4-mer chemotactic fMet peptides block H2-M3-restricted presentation of Listeria peptides (see Figure 1; Pamer et al., 1992). To investigate the effect of peptide length on the efficiency of presentation to the H2-M3-restricted Cb10 CTLs, we assayed CTL lysis of EL4 targets coated with titrated concentrations of 5, 6, 7, and 8 residue peptides corresponding to the LemA N terminus (Figure 6). The peptide concentration required to elicit half-maximal lysis of targets coated with the N-formylated pentapeptide is ~3–10 nM, which is ~30 times that required for the N-formylated 8-mer peptide (Figure 6). The N-formylated 6- and 7-mer peptides were the most active species, were nearly identical in their ability to bind M3 and be recognized by Cb10 CTLs, and targeted lysis at concentrations of 10 pM. The presentation of a nonformylated 6-mer peptide corresponding to the first 6 residues of LemA was also evaluated; the nonformylated peptide was approximately 100-fold less active than the formylated version. These data indicate that the N-terminal 6 and 7 residues of the formylated LemA protein are presented to Cb10 CTLs at 10- to 300-fold lower concentrations than are longer or shorter peptides, suggesting that one or both of these peptides comprises the Fr40 epitope.
Figure 6. fMet Peptides Corresponding to the N Terminus of LemA Efficiently Target Lysis by Cb10 CTLs.
Stocks of synthetic peptides corresponding to the N-terminal 5, 6, 7, or 8 residues of LemA were diluted in PBS and coated onto EL4 targets at the indicated final concentrations. All peptides were synthesized with an N-formylmethionine (fM), except for LemA (1–6), which was synthesized with an unformylated methionine. Peptide-coated targets were assayed for lysis in a 4 hr CTL assay with Cb10 CTL effectors at an E:T of 5:1. Specific lysis of peptide-coated targets in the absence of CTLs was less than 5%.
Discussion
Taking advantage of the fact that L. monocytogenes releases targeting activity for H2-M3-restricted CTLs into bacterial culture supernatants (Figure 1; Pamer et al., 1992) and that phagocytes such as J774 can process heat-killed organisms for presentation to such CTLs, we devised an expression cloning strategy to identify one of these Listeria epitopes. Pools of E. coli transformed with a Listeria genomic library were grown, freeze-thawed and “fed” to 51Cr-labeled J774 targets in the presence of Cb10 CTLs. Transformant clones producing CTL-dependent targeting activity all encoded the N terminus (and presumably the promoter region) of the novel protein LemA. The N-formylated hexamer fMIGWII and heptamer fMIGWIIA synthetic peptides serve as efficient epitopes for these CTLs, causing significant lysis of targets at 10–11 M concentrations. The pentameric peptide is 100- to 1000-fold less active, and the octameric peptide is 10-fold less active. In line with previous work on another epitope for H2-M3-restricted CTLs, the formylated N terminus of the ND1 subunit of mitochrondrial NADH, the deformylated hexamer LemA peptide, is about 100-fold less active. Thus, the formyl group contributes much of the energy for anchoring of these hydrophobic peptides to the M3 groove (Vyas et al., 1995; Wang et al., 1995). The nature of this anchoring was recently elucidated in the crystal structure of M3 complexed with a nonameric mitochondrial peptide (Wang et al., 1995). In this crystal structure, peptide residues 8 and 9 were seen to extend out of the M3 groove, which is consistent with our functional data showing that a 6- or 7-mer is most effective for targeting CTLs. We conclude that the formylated 6- or 7-mer LemA peptides represent the natural Fr40 epitope, which is presented to H2-M3-restricted CTLs during Listeria infection.
During or rapidly following synthesis of most bacterial proteins, the formyl group is removed from the nascent polypeptide by a cytosolic deformylase (Meinnel et al., 1993). The predicted topology of LemA explains how the N terminus of this protein avoids such deformylation. The N terminus of the protein is predicted to orient towards the outside of the bacterium (Nout), while the C terminus should remain within the bacterial cytosol (Cin). Thus, when LemA is assembled in the membrane, its extracytosolic N terminus should be protected from the deformylase activity of the bacterial cytosol. Consistent with this prediction, the N-formylmethionine residue is retained in other bacterial membrane proteins with extracytosolic N termini (Wolfe et al., 1983; Vianney et al., 1994). It was also shown that genetic alteration of a model E. coli membrane protein from an Nout to an Nin topology resulted in removal of the formylmethionine by cytosolic enzymes (von Heijne, 1989). Thus, the pool of bacterial proteins that provide epitopes for presentation by M3 may largely be composed of membrane proteins with extracytosolic N-terminal methionine residues. This pool would include membrane proteins that assume a simple Nout–Cin topology (i.e., a single membrane-spanning domain), as well as those assuming more complex Nout topologies (i.e., multiple membrane-spanning domains). In this light, it is worth noting that the hydrophobic N terminus of the mitochondrial ND1 protein is not thought to be cleaved from the protein (Fischer Lindahl et al., 1991) and, like that of LemA, does not resemble a cleaved signal sequence. Furthermore, the presence of positively charged residues at positions 25 and 26 of the ND1 protein sequence suggests that this protein assumes an Nout topology, possibly contributing to H2-M3-restricted presentation the N-terminal ND1 epitope.
In addition to protection of the formyl group, the topology of LemA may also contribute to the generation and presentation of the Fr40 epitope during growth of Listeria in vitro and in vivo. Thus, the Fr40 activity detected in bacterial cultures may arise from clipping the LemA N terminus by extracellular proteases. Such cleavage may be a rather inefficient and imprecise process, as we observe multiple distinct peaks of Fr40-like activity in HPLC-fractionated LmSN (data not shown). In this case, each peak may represent a differentially processed form of the LemA N terminus. Conversely, the Fr40 activity that elutes from natural peptide extracts of infected cells is more homogeneous (data not shown), suggesting that cellular proteases can also play a role in processing the LemA N terminus. It is unclear whether such host enzymes can process the Fr40 epitope directly from the intact LemA molecule, or whether initial cleavage by bacterial proteases is required.
Data presented in Figures 2 and 3 show that a TAP-independent mechanism exists whereby phagocytic cells process and present heat-killed bacteria to H2-M3-restricted CTLs. Other reports have also described TAP-independent presentation of phagocytosed protein antigens or peptide-coated beads by MHC class Ia molecules (Bachmann et al., 1995; De Bruijn et al., 1995). Akin to the model proposed by De Bruijn et al., we have previously argued that the presentation of HKLm to H2-M3-restricted CTLs may occur via loading of peptide-receptive M3 molecules within phagocytic vacuoles (Lenz and Bevan, 1996). If this is the case, we envision two ways in which the Nout topology of LemA may facilitate such loading. The first mechanism assumes that presentation of HKLm requires proteolytic processing of intact LemA molecules that remain associated with the heat-killed particles. In this scenario, the Nout topology of LemA may facilitate access of phagosomal proteases to the LemA N terminus, promoting excision of the Fr40 epitope. The excised epitope might then readily load peptide-receptive M3 molecules and be shuttled to the macrophage surface. The second mechanism assumes that the Fr40 activity in HKLm preparations is comprised of bacterially processed LemA N-terminal fragments, which remain tightly associated with the Listeria membrane throughout the heat-killing process. In this case, the protruding LemA N terminus may directly bind peptide-receptive M3 molecules, causing their release from the bacterial membrane. While both mechanisms may contribute to processing and presentation of HKLm by phagocytes, unrelated results may support the former mechanism. The data of Brown et al. (1992) suggest that presentation of HKLm to MHC-unrestricted CTLs is pepstatin A sensitive and requires endosomal acidification. However, despite the TAP-independent presentation of HKLm, TAP may still transport cytosolic Fr40 to the ER during infection of cells with viable LLO-positive Listeria, particularly since it has been shown that presentation of the mitochondrial ND1 epitope by M3 is TAP dependent (Attaya et al., 1992). Thus, in addition to its broad tissue distribution and selective binding of fMet peptides, M3 may have evolved the ability to acquire peptides in both cytosolic (TAP-dependent) and vacuolar (TAP-independent) compartments. Such loading may enable H2-M3-restricted CTLs to recognize Listeria-infected cells both prior to and following escape of the bacterium into the host cell cytosol, and may thus suggest that these CTLs are an early component of the host response to Listeria. Additionally, the ability to acquire peptides from phagocytic vacuoles suggests that M3 may play a role in presentation of antigens from both cytosolic and vacuolar bacterial pathogens.
It is interesting to note that Nataraj et al. (1996) have recently reported a biochemical analysis of another hydrophobic Listeria antigen that is recognized by H2-M3-restricted CTLs. This antigen can be extracted from bacterial membranes with detergent, and appears to resist degradation by a number of proteases. However, bioactivity is lost in extracts treated with sodium periodate, leading the authors to conclude that the antigen may be a glycolipid. It has recently become clear that a human class I–like molecule, CD1b, presents hydrophobic lipid-containing moieties (mycolic acid and lipoarabinomannan) to T cells (Beckman et al., 1994; Sieling et al., 1995). These antigens, which are associated with the cell walls of Mycobacterium tuberculosis and Mycobacterium leprae, appear to be processed and presented in a TAP-independent pathway that requires endosomal acidification (Porcelli et al., 1992; Sieling et al., 1995). Together, these results may indicate that in mice (which lack a homolog to human CD1b) M3 evolved to fill a niche similar to that occupied by CD1b in humans. Thus, inasmuch as M3 may present both hydrophobic fMet peptides and glycolipids to the immune system, CD1b may present both glycolipids and hydrophobic peptides. However, proof of this hypothesis awaits the identification of CD1b-restricted peptide ligands and H2-M3-restricted glycolipids.
Experimental Procedures
Reagents and Materials
Chemical reagents were purchased from Sigma Biochemicals (St. Louis, Missouri) unless otherwise noted. Restriction endonucleases were purchased from GIBCO–BRL (Grand Island, New York) and NEB (Beverly, Massachusetts). Oligonucleotide primers for PCR and sequencing were synthesized by Genset Corporation (La Jolla, California). The chemotactic peptide, formyl–Met–Leu–Phe–Phe (fMLFF), was purchased from Sigma Biochemicals. Synthesis of MLFF and the ND1b peptide (residues 1–11) is described elsewhere (Pamer et al., 1992). Peptides representing the N terminus of LemA were synthesized on a model 432A Synergy peptide synthesizer from the Applied Biosystems Division of Perkin Elmer. For synthesis of fMet peptides, empty cartridges were purchased and loaded with 75 μM formyl–Met–OH (Bachem Bioscience, Incorporated, King of Prussia, Pennsylvania). Aliquots of LemA peptide variants were stored at –20°Cas 50 μM stocks in 50% acetonitrile and diluted in phosphate-buffered saline (PBS) prior to use. The Micro BCA Protein Assay Reagent Kit (Pierce, Rockford, Illinois) was used to determine peptide concentrations.
Bacteria and Bacterial Antigens
Virulent Listeria monocytogenes strain 43251 was obtained from the American Type Culture Collection (Rockville, Maryland), and L. monocytogenes 10403S was provided by M. Strom (Northwest Fisheries Science Center, Seattle, Washington). E. coli strain DH10B was purchased from GIBCO–BRL (Grand Island, New York).
Bacteria were cultured at 37°C in trypticase soy broth (TSB; Difco Laboratories, Detroit, Michigan) for antigen preparation and in Luria–Bertani broth (LB) for DNA manipulation. L. monocytogenes strain 10403S is streptomycin resistant and was cultured in 100 μg/ml streptomycin. E. coli transformants were grown in 100 μg/ml spectinomycin (for pAT29-based vectors) or ampicillin (for pUC19-based vectors).
HKLm, was prepared from bacteria that were pelleted from a stationary (48 hr) culture of L. monocytogenes at 6,750 × g. Listeria pellets were washed twice in PBS, then incubated at 67°C for 3 hr. Aliquots at ~5 × 108 HKLm per ml were stored at –80°C. Low molecular mass L. monocytogenes culture supernatants (LmSN) were prepared by filtration through size-exclusion membranes with a 10 kDa cutoff as described (Pamer et al., 1992). Natural peptide extracts were prepared from Listeria-infected J774 macrophages and fractionated on RP-HPLC using a C18 matrix and trifluoroactetic acid/acetonitrile gradients as described (Pamer et al., 1992). Experiments using both Cb10 and JI1.4 CTL effectors (provided by E. G. Pamer, Yale University, New Haven, Connecticut) showed that the epitope recognized by Cb10 CTLs (Fr40) did not coelute with that seen by JI1.4 CTLs (Fr38).
Cell Lines and CTLs
EL4 (H-2b) thymoma, P815 mastocytoma (H-2d), J774 macrophage (H-2d), R1.1 thymoma (H-2k), and R1.E (H-2k) all express H2-M3wt. B10.CAS2 fibroblasts (H-2cas2) express a null allele of H2-M3. TR8-4a is an H2-M3wt transfectant of B10.CAS2 (Wang et al., 1991). M12A2–I-Ab (labeled M12A2 in Figure 2) is an I-Ab transfectant of an H-2d B cell line and was provided by A. R. Rudensky (University of Washington, Seattle, Washington). Cell lines were grown in RP10 medium (RPMI 1640 supplemented with 10% fetal bovine serum, 50 μM 2-mercaptoethanol, l-glutamate, penicillin, gentamicin, and streptomycin). Transfectant cell lines were cultured in RP10 supplemented with 400 μg/ml G418. BMMs were cultured from the bone marrow of C57BL/6 or Tap-1° mice as described (Brunt et al., 1990), using L cell–conditioned media as a source of M-CSF. Breeding pairs of Tap-1° mice were obtained from A. Berns (The Netherlands Cancer Institute, Amsterdam, The Netherlands) and maintained in our colony at the University of Washington.
CTLs were cultured from CD8+-enriched splenocytes of C57BL/6 mice 7 days after intravenous infection with 5000 cfu L. monocytogenes 43251. Enrichment of CD8+ cells and in vitro stimulation with HKLm were as described (Brown et al., 1992). In brief, Listeria-immune splenocytes were panned on plates coated with an anti-CD8 monoclonal antibody (3.168; Sarmiento et al., 1980). Adherent cells were stimulated in upright T-25 Falcon flasks (Beckton Dickinson Labware, Franklin Lakes, New Jersey) containing 1 ml HKLm, and 3 × 107 irradiated syngeneic splenocyte stimulators. CTLs were cloned by limiting dilution; the Cb10 CTL clone was maintained by restimulation with HKLm antigen and irradiated syngeneic splenocytes at 7–10 day intervals. CTLs were cultured in RP10 supplemented with 5% supernatant from 1 day concanavalin A–stimulated rat splenocytes and 50 mM α-methyl mannoside.
CTL Assays
Target cells were labeled with 51Cr for use in standard 4 hr CTL assays as described (Pamer et al., 1992). In brief, 104 labeled target cells (in RP10 media) were added per well of 96-well round-bottomed plates and preincubated 1–2 hr at 37°C with 50 μl of the respective antigen preparation. Following this preincubation, CTL effectors were added at the effector:target (E:T) ratios indicated in individual figures. Following a 4 hr incubation at 37°C, cells were pelleted by centrifugation of plates at 1,500 × g for 5 min, and the cpm 51Cr was determined in 100 μl of supernatant from each well. The peptide competition assay was performed as above, except that competitor peptides were added during the preincubation and remained throughout the assay. LmSN and TSB were used undiluted; LemA peptide variants and competitor peptides were diluted from stocks into PBS. When used in CTL assays, frozen aliquots of HKLm were thawed and organisms were washed twice and resuspended in PBS. The multiplicity of infection (moi) of HKLm was calculated from the absorbance of washed HKLm by assuming 0.1 A600 represented 2 × 108 heat-killed organisms per ml.
Listeria Genomic Library
Listeria 10403S genomic DNA was prepared from 14 hr cultures as described (Flamm et al., 1984), partially digested with Sau3AI, and electrophoresed on 0.6% agarose gels. DNA fragments of 5.0–10.0 kb were isolated from gel slices using the QIAEX DNA extraction kit (QIAGEN, Incorporated, Chatsworth, California) and incubated with Klenow enzyme plus dATP and dGTP to partially fill 3′ overhangs as described (Sambrook et al., 1989). These inserts were ligated to SalI-digested pAT29 made complementary by partial filling of 5′ overhangs with dTTP and dCTP. Recombinant plasmids were electroporated into E. coli DH10B and plated on LB agar containing spectinomycin, IPTG, and X-gal. White transformants were picked and individually seeded in 96-well flat-bottomed plates and expanded overnight at 37°C without shaking. Transformants were stored at –80°C as 15% glycerol stocks in 384-well plates (Nunc, Incorporated, Naperville, Illinois).
PCR Screening
hly and iap were detected in E. coli library transformants by PCR amplification of hly-specific or iap-specific bands from lysates of bacteria pools. Primers used to detect hly were 5′-GGCCCCCTCCTTTGATTA and 5′-CGTGTGTGTTAAGCGGTT, and they amplify a 1.8 kb band. Primers for iap were 5′-CATCCTCCATACCTTCTA and 5′-CTTCATCATACACCGTCT and they amplify a 2.0 kb band. Both primer sets amplify across upstream regions and through coding sequence. Individual positive clones were identified by screening subpools of positive pools with the same primer pairs. Diagnostic restriction enzyme digests of the amplified hly PCR product resulted in the predicted fragment sizes in 4 of 4 clones. Western blots with antisera specific for p60 (provided by E. G. Pamer, Yale University, New Haven, Connecticut; Villanueva et al., 1994) confirmed p60 expression by both iap-positive clones.
To detect lemA in Fr40-positive clones, the primer pair 396revF (5′-AAATCTCATGCTGCAGTAGG) and 189Fwd (5′-AATCTACGTTTAATTGCACG) was used, which specifically amplifies a 223 bp product. Amplification of sequence from the spectinomycin resistance gene (spc) of pAT29 served as a positive control for the PCR. Size markers in Figure 5 were purchased from GIBCO–BRL (φX174 RF DNA–HaeIII fragments).
Screening of the Library for Fr40-Producing Transformants
E. coli transformants were cultured 24 hr with the appropriate antibiotic in 96-well flat-bottomed plates without shaking, transferred to 96-well round-bottomed plates (50 μl/well), then lysed by three cycles of freeze-thawing. 51Cr-labeled J774 target cells were added to thawed lysates and incubated 2 hr at 37°C prior to addition of Cb10 CTL effectors at an E:T of 10:1. A standard 4 hr CTL assay was performed and specific lysis was calculated using a published formula (Moore et al., 1988). The library was originally screened in pools of eight transformants. Lysates prepared from individual transformants were then assayed to identify positive E. coli clones within these pools, which demonstrated ≥20% specific lysis. Individual clones from active subpools were screened with and without addition of Cb10 CTL effectors to CTL assays.
Subcloning of Listeria DNA Encoding Fr40 Activity
Plasmid DNA was prepared from library clone Qe11 using a QIAGEN Plasmid Mini Kit (QIAGEN) and mapped according to standard procedures (Bloch, 1987). Selected restriction fragments were subcloned into pUC19 and lysates of these E. coli subclones were tested for production of Fr40 activity in CTL assays as described above.
DNA Sequencing and Analysis
Plasmid DNA was isolated from subclones pE1D and pID6 and sequenced using the Sequenase Version 2.0 DNA sequencing kit (United States Biochemical Corporation, Cleveland, Ohio). Additional sequencing of pID6 was performed at the Murdock Molecular Biology Facility by Dr. J. Strange (University of Montana, Missoula, Montana). Primers for sequencing included the M13/pUC primers (GIBCO–BRL) and custom oligonucleotides corresponding to internal sequences. Insert DNA was sequenced at least twice and lemA sequence was confirmed in both directions. Predictions of putative LemA membrane spanning regions and topology are based in part on results from sequence analysis using the ISREC (Swiss Institute for Experimental Cancer Research) bioinformatics server (Epalinges, Switzerland).
Acknowledgments
We thank B. T. Cookson, P. J. Fink, and A. M. Pullen for critical reading of the manuscript, B. T. Cookson and M. N. Starnbach for helpful discussions, and E. G. Pamer for providing reagents. This work was supported by the Howard Hughes Medical Institute, and National Institutes of Health grants AI-19335 and CA-09537.
References
- Attaya M, Jameson S, Martinez CK, Hermel E, Aldrich C, Forman J, Fischer Lindahl K, Bevan MJ, Monaco JJ. Ham-2 corrects the class I antigen-processing defect in RMA-S cells. Nature. 1992;355:647–649. doi: 10.1038/355647a0. [DOI] [PubMed] [Google Scholar]
- Bachmann MF, Oxenius A, Pircher H, Hengartner H, Ashton-Richardt PA, Tonegawa S, Zinkernagel RF. TAP1-independent loading of class I molecules by exogenous viral proteins. Eur. J. Immunol. 1995;25:1739–1743. doi: 10.1002/eji.1830250637. [DOI] [PubMed] [Google Scholar]
- Beckman EM, Porcelli SA, Morita CT, Behar SM, Furlong ST, Brenner MB. Recognition of a lipid antigen by CD1-restricted alpha beta+ T cells. Nature. 1994;372:691–694. doi: 10.1038/372691a0. [DOI] [PubMed] [Google Scholar]
- Bloch K. Mapping by multiple endonuclease digestions. In: Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K, editors. Current Protocols in Molecular Biology. Current Protocols; New York: 1987. Unit 3.2. [Google Scholar]
- Brown ML, Fields PE, Kurlander RJ. Metabolic requirements for macrophage presentation of Listeria monocytogenes to immune CD8 cells. J. Immunol. 1992;148:555–561. [PubMed] [Google Scholar]
- Brunt LM, Portnoy DA, Unanue ER. Presentation of Listeria monocytogenes to CD8+ T cells requires secretion of hemolysin and intracellular bacterial growth. J. Immunol. 1990;145:3540–3546. [PubMed] [Google Scholar]
- Day PM, Esquivel F, Lukszo J, Bennink JR, Yewdell JW. Effect of TAP on the generation and intracellular trafficking of peptide-receptive major histocompatibility complex class I molecules. Immunity. 1995;2:137–147. doi: 10.1016/s1074-7613(95)80014-x. [DOI] [PubMed] [Google Scholar]
- De Bruijn MLH, Jackson MR, Peterson PA. Phagocyte-induced antigen-specific activation of unprimed CD8+ T cells in vitro. Eur. J. Immunol. 1995;25:1274–1285. doi: 10.1002/eji.1830250522. [DOI] [PubMed] [Google Scholar]
- De Libero G, Kaufmann SH. Antigen-specific Lyt-2+ cytolytic T lymphocytes from mice infected with the intracellular bacterium Listeria monocytogenes. J. Immunol. 1986;137:2688–2694. [PubMed] [Google Scholar]
- Fischer Lindahl K, Hermel E, Loveland BE, Wang CR. Maternally transmitted antigen of mice: a model transplantation antigen. Annu. Rev. Immunol. 1991;9:351–372. doi: 10.1146/annurev.iy.09.040191.002031. [DOI] [PubMed] [Google Scholar]
- Flamm RK, Hinrichs DJ, Thomashow MF. Introduction of pAM beta 1 into Listeria monocytogenes by conjugation and homology between native L. monocytogenes plasmids. Infect. Immun. 1984;44:157–161. doi: 10.1128/iai.44.1.157-161.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germain RN, Margulies DH. The biochemistry and cell biology of antigen processing and presentation. Annu. Rev. Immunol. 1993;11:403–450. doi: 10.1146/annurev.iy.11.040193.002155. [DOI] [PubMed] [Google Scholar]
- Harty JT, Bevan MJ. CD8+ T cells specific for a single nonamer epitope of Listeria monocytogenes are protective in vivo. J. Exp. Med. 1992;175:1531–1538. doi: 10.1084/jem.175.6.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harty JT, Pamer EG. CD8 T lymphocytes specific for the secreted p60 antigen protect against Listeria monocytogenes infection. J. Immunol. 1995;154:4642–4650. [PubMed] [Google Scholar]
- Kaufmann SH, Rodewald HR, Hug E, De Libero G. Cloned Listeria monocytogenes specific non-MHC-restricted Lyt-2+ T cells with cytolytic and protective activity. J. Immunol. 1988;140:3173–3179. [PubMed] [Google Scholar]
- Kocks C, Gouin E, Tabouret M, Berche P, Ohayon H, Cossart P. L. monocytogenes-induced actin assembly requires the actA gene product, a surface protein. Cell. 1992;68:521–531. doi: 10.1016/0092-8674(92)90188-i. [DOI] [PubMed] [Google Scholar]
- Kurlander RJ, Shawar SM, Brown ML, Rich RR. Specialized role for a murine class I-b MHC molecule in prokaryotic host defenses. Science. 1992;257:678–679. doi: 10.1126/science.1496381. [DOI] [PubMed] [Google Scholar]
- Lenz LL, Bevan MJ. H2-M3 restricted presentation of Listeria monocytogenes antigens. Immunol. Rev. 1996;151 doi: 10.1111/j.1600-065x.1996.tb00705.x. in press. [DOI] [PubMed] [Google Scholar]
- Loveland B, Wang CR, Yonekawa H, Hermel E, Fischer Lindahl K. Maternally transmitted histocompatibility antigen of mice: a hydrophobic peptide of a mitochondrially encoded protein. Cell. 1990;60:971–980. doi: 10.1016/0092-8674(90)90345-f. [DOI] [PubMed] [Google Scholar]
- Lukacs K, Kurlander RJ. MHC-unrestricted transfer of antilisterial immunity by freshly isolated immune CD8 spleen cells. J. Immunol. 1989;143:3731–3736. [PubMed] [Google Scholar]
- Meinnel T, Mechulam Y, Blanquet S. Methionine as translation start signal: a review of the enzymes of the pathway in Escherichia coli. Biochimie. 1993;75:1061–1075. doi: 10.1016/0300-9084(93)90005-d. [DOI] [PubMed] [Google Scholar]
- Michel E, Cossart P. Physical map of the Listeria monocytogenes chromosome. J. Bacteriol. 1992;174:7098–7103. doi: 10.1128/jb.174.22.7098-7103.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore MW, Carbone FR, Bevan MJ. Introduction of soluble protein into the class I pathway of antigen processing and presentation. Cell. 1988;54:777–785. doi: 10.1016/s0092-8674(88)91043-4. [DOI] [PubMed] [Google Scholar]
- Nataraj C, Brown ML, Poston RM, Shawar SM, Rich RR, Fischer Lindahl K, Kurlander RJ. H2-M3wt-restricted, Listeria monocytogenes-specific CD8 T cells recognize a novel, hydrophobic, protease-resistant, periodate-sensitive antigen. Int. Immunol. 1996;8:367–378. doi: 10.1093/intimm/8.3.367. [DOI] [PubMed] [Google Scholar]
- Pamer EG. Direct sequence identification and kinetic analysis of an MHC class I–restricted Listeria monocytogenes CTL epitope. J. Immunol. 1994;152:686–694. [PubMed] [Google Scholar]
- Pamer EG, Harty JT, Bevan MJ. Precise prediction of a dominant class I MHC-restricted epitope of Listeria monocytogenes. Nature. 1991;353:852–855. doi: 10.1038/353852a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pamer EG, Wang CR, Flaherty L, Fischer Lindahl K, Bevan MJ. H-2M3 presents a Listeria monocytogenes peptide to cytotoxic T lymphocytes. Cell. 1992;70:215–223. doi: 10.1016/0092-8674(92)90097-v. [DOI] [PubMed] [Google Scholar]
- Pamer EG, Bevan MJ, Fischer Lindahl K. Do nonclassical, class Ib MHC molecules present bacterial antigens to T cells? Trends Microbiol. 1993;1:35–38. doi: 10.1016/0966-842x(93)90023-k. [DOI] [PubMed] [Google Scholar]
- Porcelli S, Morita CT, Brenner MB. CD1b restricts the response of human CD4–8– T lymphocytes to a microbial antigen. Nature. 1992;360:593–597. doi: 10.1038/360593a0. [DOI] [PubMed] [Google Scholar]
- Pugsley AP. The complete general secretory pathway in gram-negative bacteria. Microbiol. Rev. 1993;57:50–108. doi: 10.1128/mr.57.1.50-108.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Second Edition Cold Spring Harbor Laboratory Press; Plainview, New York: 1989. [Google Scholar]
- Sarmiento M, Glasebrook AL, Fitch FW. IgG or IgM monoclonal antibodies reactive with different determinants on the molecular complex bearing Lyt 2 antigen block T cell–mediated cytolysis in the absences of complement. J. Immunol. 1980;125:2665–2672. [PubMed] [Google Scholar]
- Shawar SM, Cook RG, Rodgers JR, Rich RR. Specialized functions of MHC class I molecules. I. An N-formyl peptide receptor is required for construction of the class I antigen Mta. J. Exp. Med. 1990;171:897–912. doi: 10.1084/jem.171.3.897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shawar SM, Vyas JM, Rodgers JR, Cook RG, Rich RR. Specialized functions of major histocompatibility complex class I molecules. II. Hmt binds N-formylated peptides of mitochondrial and prokaryotic origin. J. Exp. Med. 1991;174:941–944. doi: 10.1084/jem.174.4.941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shawar SM, Vyas JM, Rodgers JR, Rich RR. Antigen presentation by major histocompatibility complex class I-B molecules. Annu. Rev. Immunol. 1994;12:839–880. doi: 10.1146/annurev.iy.12.040194.004203. [DOI] [PubMed] [Google Scholar]
- Sieling PA, Chatterjee D, Porcelli SA, Prigozy TI, Mazzaccaro RJ, Soriano T, Bloom BR, Brenner MB, Kronenberg M, Brennan PJ, Modlin RL. CD1-restricted T cell recognition of microbial lipoglycan antigens. Science. 1995;269:227–230. doi: 10.1126/science.7542404. [DOI] [PubMed] [Google Scholar]
- Sijts AJ, Neisig A, Neefjes J, Pamer EG. Two Listeria monocytogenes CTL epitopes are processed from the same antigen with different efficiencies. J. Immunol. 1996;156:683–692. [PubMed] [Google Scholar]
- Simonen M, Palva I. Protein secretion in Bacillus species. Microbiol. Rev. 1993;57:109–137. doi: 10.1128/mr.57.1.109-137.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith GP, Dabhi VM, Pamer EG, Fischer Lindahl K. Peptide presentation by the MHC class Ib molecule, H2-M3. Int. Immunol. 1994;6:1917–1926. doi: 10.1093/intimm/6.12.1917. [DOI] [PubMed] [Google Scholar]
- Tilney LG, Portnoy DA. Actin filaments and the growth, movement, and spread of the intracellular bacterial parasite, Listeria monocytogenes. J. Cell. Biol. 1989;109:1597–1608. doi: 10.1083/jcb.109.4.1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trieu CP, Carlier C, Poyart SC, Courvalin P. A pair of mobilizable shuttle vectors conferring resistance to spectinomycin for molecular cloning in Escherichia coli and in gram-positive bacteria. Nucl. Acids Res. 1990;18:4296. doi: 10.1093/nar/18.14.4296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez BJA, Kocks C, Dramsi S, Ohayon H, Geoffroy C, Mengaud J, Cossart P. Nucleotide sequence of the lecithinase operon of Listeria monocytogenes and possible role of lecithinase in cell-to-cell spread. Infect. Immun. 1992;60:219–230. doi: 10.1128/iai.60.1.219-230.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vianney A, Lewin TM, Beyer WFJ, Lazzaroni JC, Portalier R, Webster RE. Membrane topology and mutational analysis of the TolQ protein of Escherichia coli required for the uptake of macromolecules and cell envelope integrity. J. Bacteriol. 1994;176:822–829. doi: 10.1128/jb.176.3.822-829.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villanueva MS, Fischer P, Feen K, Pamer EG. Efficiency of MHC class I antigen processing: a quantitative analysis. Immunity. 1994;1:479–489. doi: 10.1016/1074-7613(94)90090-6. [DOI] [PubMed] [Google Scholar]
- von Heijne G. The distribution of positively charged residues in bacterial inner membrane proteins correlates with the trans-membrane topology. EMBO J. 1986;5:3021–3027. doi: 10.1002/j.1460-2075.1986.tb04601.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Heijne G. Control of topology and mode of assembly of a polytopic membrane protein by positively charged residues. Nature. 1989;341:4560–4568. doi: 10.1038/341456a0. [DOI] [PubMed] [Google Scholar]
- Vyas JM, Shawar SM, Rodgers JR, Cook RG, Rich RR. Biochemical specificity of H-2M3a: stereospecificity and space-filling requirements at position 1 maintain N-formyl peptide binding. J. Immunol. 1992;149:3605–3611. [PubMed] [Google Scholar]
- Vyas JM, Rodgers JR, Rich RR. H-2M3a violates the paradigm for major histocompatibility complex class I peptide binding. J. Exp. Med. 1995;181:1817–1825. doi: 10.1084/jem.181.5.1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang CR, Loveland BE, Fischer Lindahl K. H-2M3 encodes the MHC class I molecule presenting the maternally transmitted antigen of the mouse. Cell. 1991;66:335–345. doi: 10.1016/0092-8674(91)90623-7. [DOI] [PubMed] [Google Scholar]
- Wang CR, Castano AR, Peterson PA, Slaughter C, Fischer Lindahl K, Deisenhofer J. Nonclassical binding of formylated peptide in crystal structure of the MHC class Ib molecule H2-M3. Cell. 1995;82:655–664. doi: 10.1016/0092-8674(95)90037-3. [DOI] [PubMed] [Google Scholar]
- Wolf PR, Ploegh HL. How MHC class II molecules acquire peptide cargo: biosynthesis and trafficking through the endocytic pathway. Annu. Rev. Cell Biol. 1995;11:267–306. doi: 10.1146/annurev.cb.11.110195.001411. [DOI] [PubMed] [Google Scholar]
- Wolfe PB, Wickner W, Goodman JM. Sequence of the leader peptidase gene of Escherichia coli and the orientation of leader peptidase in the bacterial envelope. J. Biol. Chem. 1983;258:12073–12080. [PubMed] [Google Scholar]