Abstract
Objective
Our aim was to determine if the expression pattern of CLP-1 in developing heart is consistent with its role in controlling RNA transcript elongation by transcriptional elongation factor b (P-TEFb) and if the inhibitory control exerted over P-TEFb by CLP-1 is released under hypertrophic conditions.
Methods
We performed immunoblot and immunofluorescence analysis of CLP-1 and the P-TEFb components cdk9 and cyclin T in fetal mouse heart and 2 day post-natal mouse cardiomyocytes to determine if they are co-localized. We induced hypertrophy in rat cardiomyocytes either by mechanical stretch or treatment with hypertrophic agents such as endothelin-1 and phenylephrine to determine if CLP-1 is released from P-TEFb in response to hypertrophic stimuli. The involvement of the Jak/STAT signal transduction pathway in this process was studied by blocking this pathway with the Jak2 kinase inhibitor, AG490, and assessing the association of CLP-1 with P-TEFb complexes.
Results
We found that CLP-1 is expressed along with P-TEFb components in developing heart during the period in which knockout mice lacking the CLP-1 gene develop cardiac hypertrophy and die. Under conditions of hypertrophy induced by mechanical stretch or agonist treatment, CLP-1 dissociates from the P-TEFb complex, a finding consistent with the de-repression of P-TEFb kinase activity seen in hypertrophic cardiomyocytes. Blockage of Jak/STAT signaling by AG490 prevented release of CLP-1 from P-TEFb despite the ongoing presence of hypertrophic stimulation by mechanical stretch.
Conclusions
CLP-1 expression in developing heart and isolated post-natal cardiomyocytes colocalizes with P-TEFb expression and therefore has the potential to regulate RNA transcript elongation by controlling P-TEFb cdk9 kinase activity in heart. We further conclude that the dissociation of CLP-1 from P-TEFb is responsive to hypertrophic stimuli transduced by cellular signal transduction pathways. This process may be part of the genomic stress response resulting in increased RNA transcript synthesis in hypertrophic cardiomyocytes.
1. INTRODUCTION
Myocardial hypertrophy is a major risk factor for the development of congestive heart failure. When confronted by hypertrophic stimuli, the heart myocardium mounts an adaptive response to increase hemodynamic output by increasing cardiac muscle cell contractility (1). This involves transmission of the hypertrophic stimuli via signal transduction pathways that together instigate a rapid increase in the expression of early response genes as well as a fetal program of contractile and non-contractile protein genes (2–5). Concurrent with this is a global increase in RNA and protein synthesis, a phenomenon typical of hypertrophic cells (6–9). More recent studies using microarray analysis have identified an even wider spectrum of genes that exhibit increased expression with hypertrophy (10,11).
The immediate, wide-scale genomic response to hypertrophic stimuli is likely to be achieved not through the specific activation of genes, but rather through a generalized activation of the transcriptional apparatus or of some common part of that apparatus critical to the synthesis of a wide spectrum of RNA transcripts. Along these lines, recent studies have implicated Transcription Elongation Factor b, or P-TEFb, a complex of cyclin dependent kinase (cdk) 9 with cyclin T1, in the hypertrophic response (12–14). A major substrate of P-TEFb is RNA polymerase II which upon phosphorylation elongates nascent transcripts to form full-length mRNAs (15, 16). In HeLa cells, P-TEFb appears to be dynamically partitioned between an inactive versus an active kinase state suggesting a possible influence on transcriptional processes that could be subject to regulatory control (17, 18). Maintenance of the inactive state was initially attributed to a small nuclear RNA called 7SK that associates with cyclin T1 to repress cdk9 kinase activity (19, 20). The linkage of 7SK RNA, P-TEFb kinase activity, and RNA pol II phosphorylation with the adaptive response of cardiac muscle cells to hypertrophic stimuli was first discovered by Sano et al. (12) who showed that various hypertrophic stimuli could de-repress P-TEFb kinase activity by causing the dissociation of the 7SK RNA inhibitor. More recent studies in HeLa cells have shown that the actual inhibitor of P-TEFb cdk9 kinase activity is not 7SK RNA but a protein called HEXIM1 that binds to P-TEFb using 7SK RNA as a support (18, 21).
HEXIM1 exhibits all the properties previously attributed to 7SK RNA such as its binding to P-TEFb to inhibit cdk9 kinase activity and its release resulting in the de-repression of the cdk9 kinase.(18, 21). These findings have provided greater insight into how P-TEFb is regulated generally and suggest that an early and critical step in the adaptive response of cardiac muscle cells to hypertrophic stimuli may be the dissociation of HEXIM1 from the P-TEFb complex. We have cloned the chicken and mouse cDNA homologs of HEXIM1, called CLP-1, and have knocked out the CLP-1 gene in mice (22–24). CLP-1 knockout mice die during fetal development with hearts that exhibit the physical and genetic hallmarks of cardiac hypertrophy. In CLP-1 knockout mice, the complete absence of CLP-1 could be likened to a “chronic” state of dissociation of this inhibitor from P-TEFb leading to a persistent state of P-TEFb deregulation and phosphorylation and activation of RNA pol II and its elongation activity. This, in turn, could lead to the global increase in gene transcription and RNA production seen in hypertrophic cardiomyocytes. In this report, we show the presence of cdk9, cyclin T1 and CLP-1 in heart during the period in which CLP-1 knockout fetuses develop hypertrophy and demonstrate experimentally the involvement of CLP-1 in cardiac hypertrophy by establishing its co-localization in cardiac cells with cdk9 and cyclin T and its dissociation from P-TEFb complexes in cardiac cells rendered hypertrophic by mechanical stretch or treatment with hypertrophic agents in vitro. We also demonstrate involvement of the Jak/STAT signaling pathway in this process by showing that CLP-1 is not released from P-TEFb under hypetrophic conditions when Jak2 kinase signaling is inhibited.
2. METHODS
This investigation conforms with the Guide for the Care and Use of Laboratory Animals published by the US National Institutes of Health (NIH Publication No. 85-23, revised 1996).
2.1. Abbreviations used
CLP-1, cardiac lineage protein-1; cdk9, cyclin dependent kinase 9; P-TEFb, positive transcription elongation factor b; RNA polymerase II, RNA pol II; STAT3, Signal Transducer and Activator of Transcription 3; MAPK, mitogen-activated protein kinase; GAPDH, glyceraldehyde-3-phosphate-dehydrogenase; ANF, atrial natriuretic factor.
2.2. Analysis of P-TEFb components in developing tissues
Protein lysates from heart, liver, and kidney of mice at different stages of development and post-natal ages were resolved by gel electrophoresis and transferred to nitrocellulose membranes. The membranes were probed with anti-CLP-1 antibody (rabbit anti-HEXIM1 antiserum, a generous gift from Dr. Q. Zhou, University of California, Berkeley) or anti-GAPDH antibody (ab9485, Abcam, Cambridge, Massachusetts, USA) and then incubated with horseradish peroxidase-conjugated secondary antibody (Santa Cruz Biotechnology, Santa Cruz, California, USA). The same membranes were stripped and re-probed with anti-Cdk9 antibody (sc-13130, Santa Cruz Biotechnology) or anti Cyclin T1 antibody (sc-10750, Santa Cruz Biotechnology). Antibody reactivity was visualized with ECL detection systems (Amersham Biosciences, Pittsburgh, Pennsylvania, USA).
2.3. Immunohistochemistry and immunofluoresence of CLP-1, cdk9, cyclin T, and MF20
Serial sections from paraffin-embedded fetal hearts were mounted and incubated with either rabbit anti-CLP-1 antibody (1:250 dilution; affinity purified CLP-1 rabbit antiserum made to the peptide HRQQERAPLSKFGD, Proteintech Group Inc., Chicago, Illinois, USA) or rabbit anti-cdk9 antibody (1:100 dilution, Santa Cruz Biotechnology). Slides were washed and incubated with horseradish peroxidase-conjugated secondary antibody followed by development with diaminobenzidene (ABC reagent system, Vector Labs, Burlingham, California, USA) and counterstaining with eosin. For immunofluoresence studies, 2-day post-natal rat cardiomyocytes were plated on collagen-coated glass coverslips and fixed in cold 4% paraformaldehyde. Cells were incubated with primary antibody (rabbit anti- CLP-1 antibody (Proteintech Group Inc., 1:50 dilution); goat anti-cyclin T antiserum (Santa Cruz Biotechnology, 1:100 dilution); goat anti-cdk9 antiserum (Santa Cruz biotechnology; 1:250 dilution); MF20 mouse hybridoma supernatant (Developmental Studies Hybridoma Bank; 1:25 dilution)) followed by incubation with biotin-conjugated secondary antibody and fluorescein or Texas Red-conjugated avidin. Incubation with MF20 was followed by incubation with anti-rabbit IgG antibody (AlexaFluor 647 (Far Red), Molecular Probes/Invitrogen, Carlsbad, California, USA, 1:200–300 dilutions). Confocal imaging was carried out using a BioRad Radiance microscope with LaserSharp 2000 software.
2.4. Mechanical stretch of cardiomyocytes, treatment with hypertrophic agents, and analysis of P-TEFb components
Cardiac cells of 2–4-day-old WKY rats were isolated using a neonatal cardiomyocyte isolation system (Worthington Biochemical Corp, Lakewood, New Jersey, USA). After pre-plating to reduce contaminating non-muscle cells, cardiomyocytes were plated on Bioflex collagen 6-well plates (Flexcell, McKeesport, Pennsylvania, USA) and cultured in DMEM/F-12 (1:1) media supplemented with 10% fetal bovine serum for approximately 1.5 days prior to switching to serum-free medium. Mechanical stretch was applied after 24 h in serum-free medium by applying a computer-controlled vacuum suction under the flexible-bottomed Bioflex collagen plates with Flexercell Strain Unit FX-4000 (Flexcell) (25). The vacuum varied in one-second cycles sufficient to promote 10–15% elongation of the cardiomyocytes at the point of maximal distension of the culture surface. For experiments requiring inhibition of the Jak 2 kinase, AG490, dissolved in DMSO, was added to a final concentration of 500 nM. Control cells received DMSO. For agonist treatment, isolated cardiomyocytes were cultured for 36 hours, switched to serum-free medium and cultured for another 24 hours at which time agonists were added (Endothelin-1 (Sigma), 200 nM final concentration; phenylephrine (Sigma), 100 uM final concentration) and the cells cultured for another 24 hours and harvested for analysis. For analysis of all control and experimental cultures, cell lysates were clarified by centrifugation and then incubated with goat anti-cyclin T1 antibody (Santa Cruz Biotechnology) followed by precipitation with protein G agarose. Blots of immunoprecipitated proteins were incubated with primary antibody followed by detection using horseradish peroxide-coupled secondary antibody and the ECL enhanced chemiluminescence assay (Amersham Biosciences). For STAT3 analysis, cardiomyocyte lysates were fractionated directly by SDS-PAGE, transferred to nitrocellulose and probed with anti-STAT3 antibody (Santa Cruz Biotechnology).
2.5. ANF analysis
Total RNA was isolated from cardiomyocytes using Trizol (Invitrogen). Northern blot analysis of atrial natriuretic factor mRNA levels was carried out as described (23).
3. RESULTS
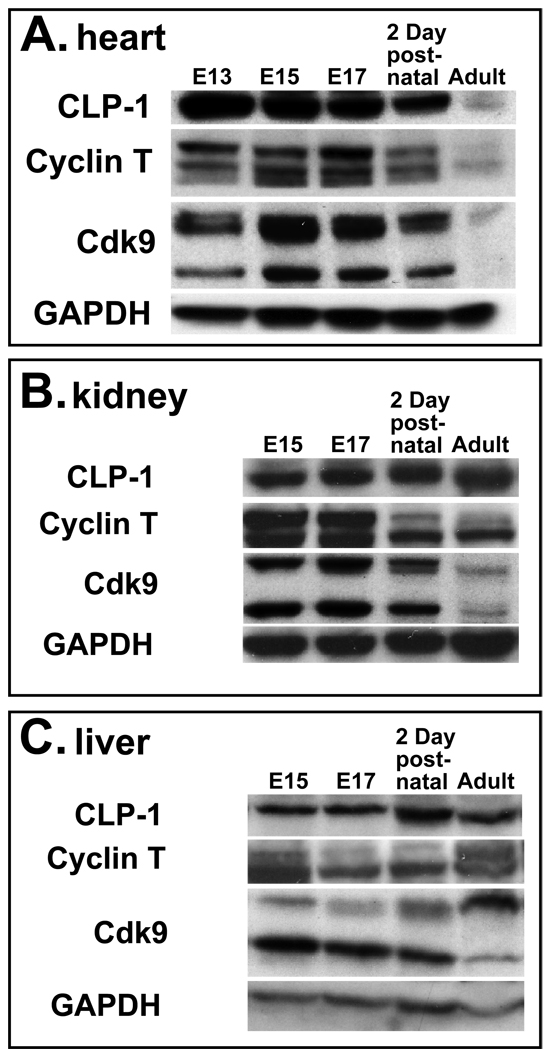
We sought to determine if the P-TEFb components regulating transcriptional elongation are expressed in cardiomyocytes between embryonic day (E)15 to E 19 when CLP-1 knockout fetuses develop cardiac hypertrophy and in 2 day old rat cardiomyocytes rendered hypertrophic by mechanical stretch. CLP-1 is expressed at high levels early in heart development then decrease slightly as the heart grows attaining their lowest levels in adult heart (Figure 1A). In kidney and liver, CLP-1 levels are relatively constant throughout development and maintained at similar levels in the adult organs (Figure 1B and C). Cyclin T is expressed in three isoforms, cyclin T1, cyclin T2a, and cyclin T2b, all of which can bind and activate cdk9 (26). In pre-natal heart, cyclin T levels are high then diminish to lower levels in the adult heart (Figure 1A), while kidney and liver exhibit differential expression patterns among the three isoforms (Figure 1B and C). Cdk9 is expressed in two isoforms, 55Kd and 42Kd (27, 28) both of which appear to be maximally expressed between E15 and E19 in heart (Figure 1A). In liver, the 55 Kd and 42 Kd cdk9 isoforms were expressed in an inverse fashion that cumulatively resulted in a constant level of cdk9 protein, while kidney expressed relatively unchanged levels during development with lowered levels in adult. A third immunoreactive band is present in heart and kidney migrating slightly slower than the 55Kd cdk9 isoform. This is likely to be the phosphorylated form of cdk9 that is required for kinase activity and the recruitment of P-TEFb to the HEXIM1/7SK RNA complex (29, 30). A subsequent incubation of the stripped heart immunoblot with less primary antibody to cdk9 and for shorter incubation times made more apparent the differential expression of the lower molecular weight cdk9 isoform in heart between E15 to E17. This peak of cdk9 isoform expression coincides with the period in which CLP-1 knockout fetuses develop cardiac hypertrophy (24).
Figure 1.
Expression of CLP-1, cyclin T and cdk9 proteins in developing and post-natal heart (A), kidney (B), and liver (C). One hundred micrograms of protein lysate was loaded per lane and expression of CLP-1, cyclin T, cdk9, and GAPDH proteins detected by immunoblot analysis. GAPDH expression served as loading control. The E15 to E17 pre-natal period encompasses the time period in which CLP-1 knockout fetuses develop hypertrophy and die (24). Definitive liver and kidney tissues were not identifiable in E13 embryos and thus not harvested.
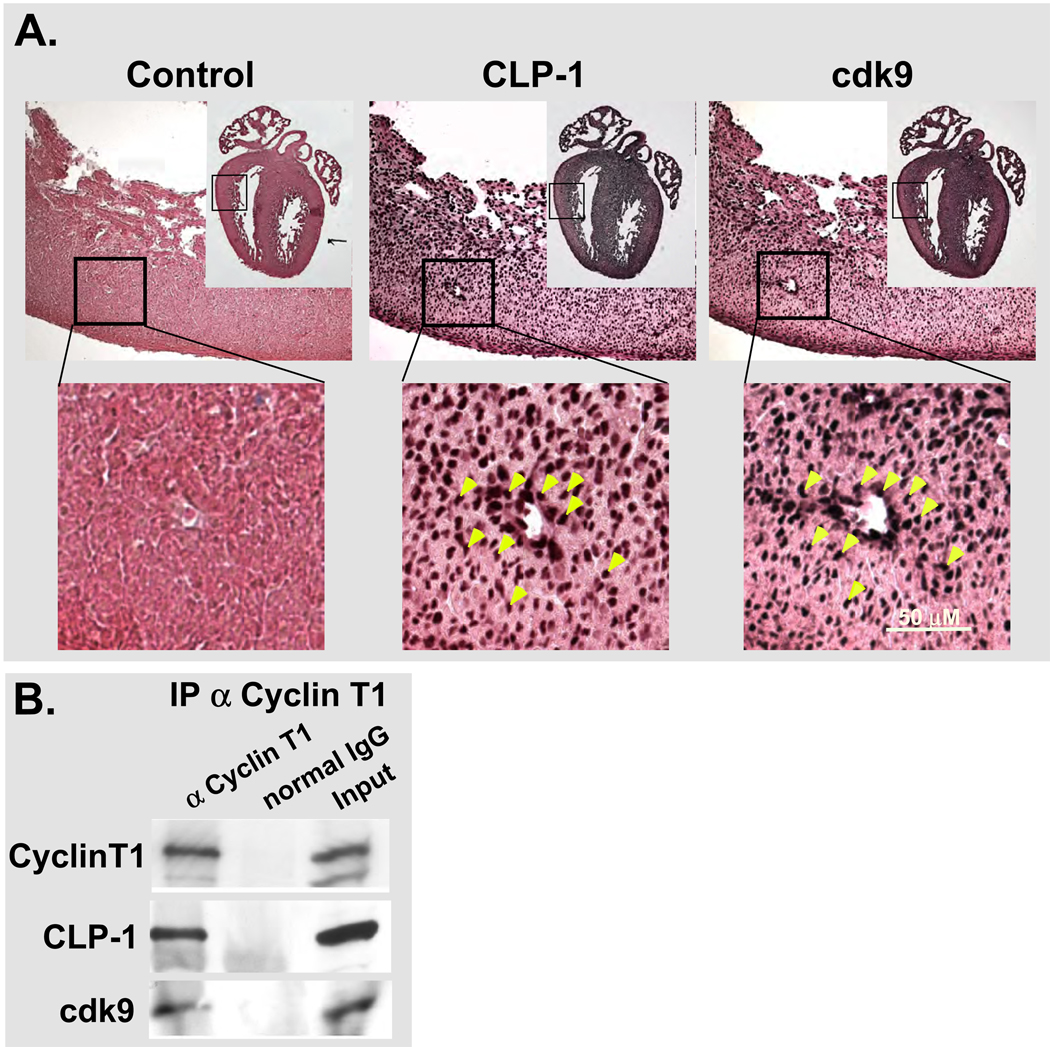
If CLP-1 regulates cdk9 kinase activity in vivo and under hypertrophic conditions, then both proteins should be expressed in the E16-18 heart myocardium since this is when CLP-1 knockout fetuses develop hypertrophy (24). To confirm this, we sought to localize CLP-1 and cdk9 proteins in the myocardium of the E17 heart by staining alternate serial sections of E17 heart with antibody to CLP-1 and cdk9 (Figure 2A). The majority of nuclei expressed CLP-1 or cdk9 and a random measurement of selected nuclei clearly showed co-localization (see yellow arrows, Figure 2A). Such co-localization is indicative of the association of CLP-1 with p-TEFb components at the molecular level. To confirm this, we performed co-immunoprecipitations of fetal heart lysates with cyclin T1 antibody followed by analysis of immunoprecipitates for CLP-1 and cdk9 (Figure 2B). Cyclin T1 immunoprecipitates showed the presence of CLP-1 and cdk9 confirming our co-localization findings and establishing CLP-1’s association with P-TEFb in fetal heart cells during the same time period in which CLP-1 knockout fetuses develop hypertrophy.
Figure 2.
Expression of cdk9 and CLP-1 in E17 and E19 heart. A. Serial paraffin sections from E17 mouse heart showing co-localization of cdk9 and CLP-1 in heart cells. Boxed regions of hearts show area depicted in images (a transected vein (white area in image) was used to co-align images for comparing overlap in nuclear stainings). B. Immunoprecipitation of E19 heart lysates showing co-immunoprecipitation of CLP-1 with P-TEFb components. Input refers to the non-precipitated lysate.
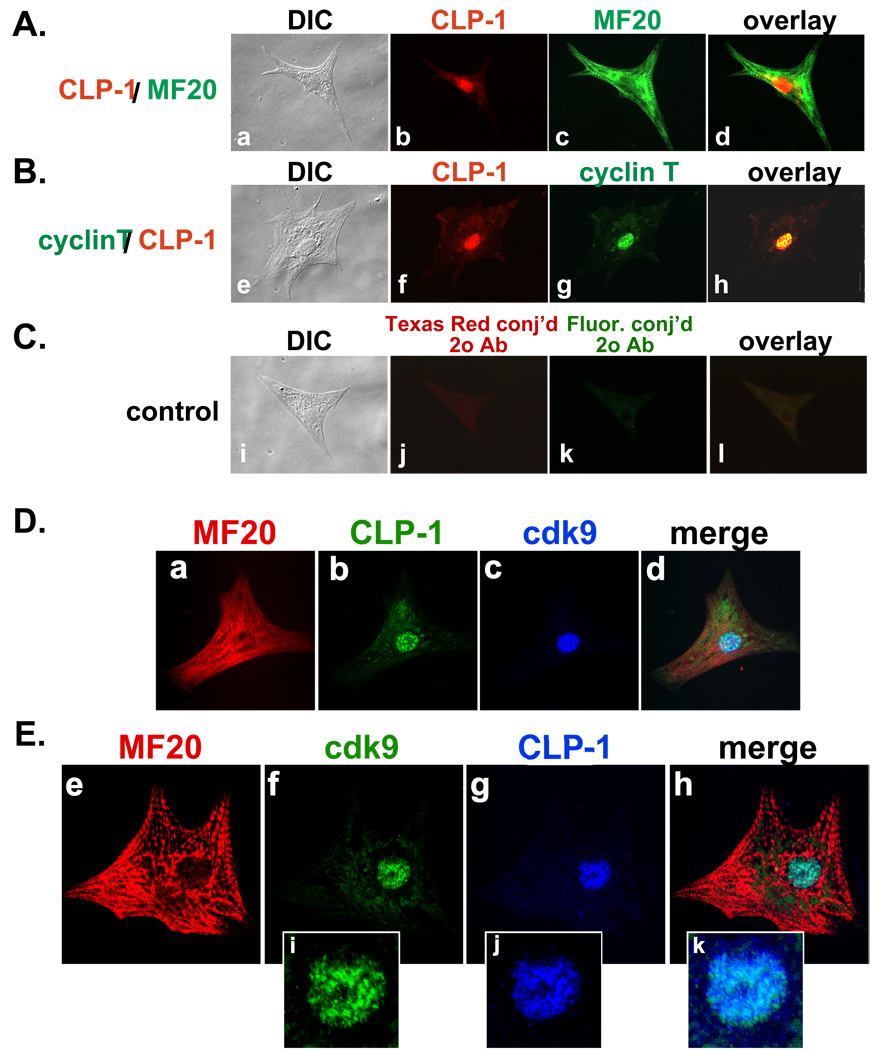
To further demonstrate that CLP-1 is co-expressed with cdk9 and cyclin T in cardiomyocytes and thus capable of regulating P-TEFb activity, we used double-labeled fluorescent immunocytochemistry on isolated cardiomyocytes obtained from 2 day post-natal rat hearts and plated in short-term cultures. These are the same cell type used in the mechanical stretch experiments of this study (see below). Cardiomyocytes were distinguished by their expression of myosin filaments as revealed by staining with the MF20 antibody (Figure 3A, D, and E). Double staining with antisera to MF20 and CLP-1 showed CLP-1 expression in the nuclei of these MF20-positive cardiomyocytes (Figure 3A, D, and E). Cyclin T and CLP-1 staining showed co-localization to cell nuclei (Figure 3B). Confocal microscopy of cardiomyocytes using triple staining for cardiac myosin, CLP-1 and cdk9 showed that CLP-1 is co-localized with cdk9 in cardiomyocytes (Figure 3D, E). Interestingly, higher magnification showed co-localization of cdk9 with CLP-1 in a non-uniform or “speckled” staining pattern (Figure 3E; panels i, j, and k). Such a “speckled” pattern has been seen for cdk9 and cyclin T expression in HeLa cell nuclei (31).
Figure 3.
Immunocytochemical co-localization of CLP-1, cyclin T and cdk9 in isolated 2 day post natal rat cardiomyocytes. A–C: (A) CLP-1 is in the nucleus of MF20-positive cardiomyocytes; (B) nuclear co-localization of CLP-1 and cyclin T; (C) control (no 1° antibody). (DIC, Differential Interference Contrast microscopy). D,E: Confocal microscopy of cardiomyocytes. D and E: CLP-1 and cdk9 are co-localized to the nucleus of MF20-positive cardiomyocytes. Panels i-k, magnified images of nuclei showing speckled type nuclear staining of cdk9 and CLP-1.
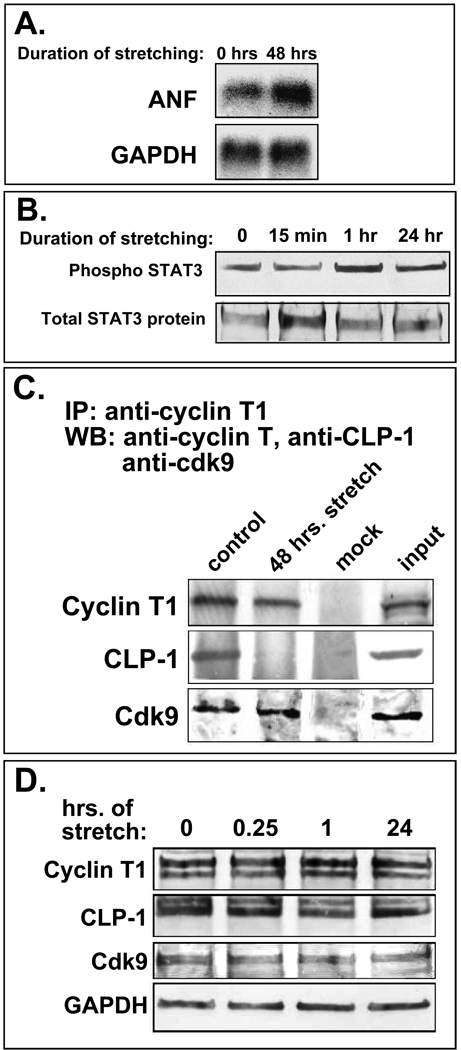
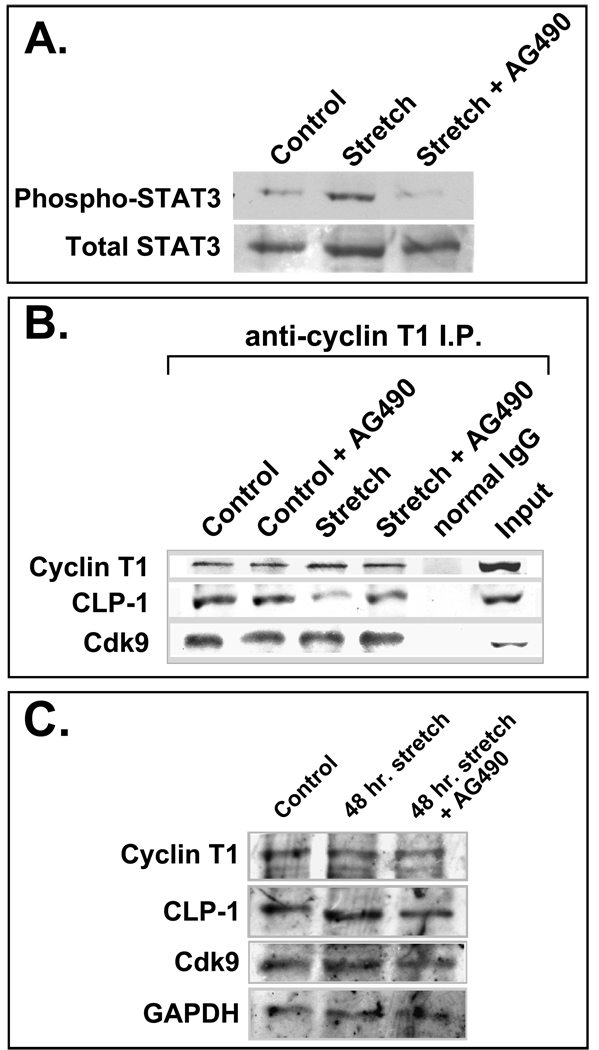
The co-localization of CLP-1 with cdk9 and cyclin T proteins in cardiomyocte nuclei suggests that CLP-1 is in a position to regulate P-TEFb activity under normal and hypertrophic conditions. While the dissociation of 7SK RNA from p-TEFb under hypertrophic conditions has already been experimentally shown (12), the dissociation of CLP-1, the actual kinase inhibitor, has yet to be demonstrated. Suggestive evidence for this derives from our interpretation of the cardiac hypertrophy evident in CLP-1 knockout mice: absence of CLP-1 may lead to de-regulation of cdk9, constitutive phosphorylation and activation of RNA pol II, and increased global RNA transcript output. To demonstrate experimentally that hypertrophic stimuli can trigger the release of CLP-1 from P-TEFb, we have resorted to the in vitro system of cultured cardiomyocytes rendered hypertrophic by mechanical stretch. This system exhibits the same signaling and growth-related events seen in hypertrophic cardiomyocytes in vivo (32) and provides a useful experimental system with which to study the molecular events underlying the adaptive hypertrophic response. Two-day post-natal rat cardiomyocytes were stretched for 48 hours (see Methods) and then analysed for the induction of hypertrophy and the presence of CLP-1 in P-TEFb complexes. The attainment of hypertrophy was confirmed by the increase in expression of atrial natriuretic factor (ANF) mRNA (33) and activation of STAT3 (34) (Figure 4A and B). After one hour of mechanical stretch, STAT3 was activated as indicated by its increased phosphorylation. Together, these data indicate that mechanical stretch rendered these cardiomyocytes hypertrophic.
Figure 4.
A. Stretched cardiomyocytes express markers of hypertrophy. Northern blot showing increased ANF (atrial natriuretic factor) mRNA levels in stretched cardiomyocytes. GAPDH is loading control. B. Phosphorylation of STAT 3 increases with continued stretch of cardiomyocytes (1 hr and 24 hr); overall STAT3 protein levels remain constant. C. CLP-1 is released from p-TEFb complexes in stretch-induced hypertrophic cardiomyocytes. Immunoblot of p-TEFb components in stretched and non-stretched cardiomyocytes showing markedly reduced levels of CLP-1 in P-TEFb complexes from cardiomyocytes stretched for 48 hours; cyclin T1 and cdk9 levels remain unchanged. D. Total levels of p-TEFb components (free and complexed) remain unchanged with stretch. GAPDH is loading control.
We next assessed the status of cdk9 and CLP-1 in P-TEFb complexes in stretched versus non-stretched cardiomyocytes by immunoprecipitating P-TEFb with anti-cyclin T1 antibody and assaying for the presence of cdk9 and CLP-1 by immunoblot analysis (Figure 4C). CLP-1 was present in P-TEFb complexes isolated from unstretched cardiomyocytes while it was markedly decreased in P-TEFb complexes from cardiomyocytes subjected to 48 hours of mechanical stretch. The relative amounts of cyclin T1 and cdk9 in P-TEFb appeared unchanged between unstretched and stretched cardiomyocytes. The absence of CLP-1 in P-TEFb complexes isolated from mechanically stretched cardiomyocytes was not due to an absence of CLP-1 protein as there was no change in the level of CLP-1 protein in cardiomyocytes subjected to mechanical stretch compared to unstretched control cells (Figure 4D). (See also Figure 5C for 48 hour stretching regimen). Together, these data show that hypertrophic stimuli in the form of mechanical stretch can induce the dissociation of CLP-1 from P-TEFb.
Figure 5.
Inhibition of the Jak2 kinase results in decreased dissociation of CLP-1 under hypertrophic conditions. A. Mechanical stretch induces phosphorylation of STAT3; addition of AG490 prevents STAT3 phosphorylation. B. AG490 blocks the dissociation of CLP-1 from the p-TEFb complex. CLP-1 levels are reduced in cyclin T1 immunoprecipitates from stretched cardiomyocytes relative to non-stretched controls, but remain unchanged in stretched cardiomyocytes treated with AG490. Cardiomyocytes not receiving AG490 received vehicle alone. C. Total levels of P-TEFb (free and complexed) remain unchanged in 48 hour stretched and non-stretched cardiomyocytes.
Among the several signaling pathways activated by mechanical stretch, two major pathways are prominantly involved in the transduction of the hypertrophic mechanical stretch signal, the Janus-associated kinase/signal transducers and activators of transcription, or Jak/STAT, pathway and the mitogen-activated protein kinase, or MAPK, pathway (35, 36). It is well documented that the Jak/STAT signal transduction pathway is activated in cardiomyocytes rendered hypertrophic by mechanical stretch, resulting in phosphorylation of STAT3 (34), as well as by hypertrophic agonists such as cardiotrophin-1 (37), leukemia inhibitory factor (LIF) (38), and Angiotensin II (39, 40). AG490 is a potent blocker of the Jak2 kinase that phosphorylates STAT3 and has been shown to inhibit STAT3 phosphorylation (41) and to block hypertrophic growth (42, 43). We sought to investigate if CLP-1 dissociation from P-TEFb complexes was responsive to the Jak/STAT signaling pathway by inhibiting the Jak2 kinase during mechanical stretch. Two-day post-natal rat cardiomyocytes stretched for 48 hours showed activation of the Jak/STAT pathway as evidenced by increased STAT3 phosphorylation (Figure 5A, lane 2, “Stretch”) that was reduced to control levels with AG490 treatment (Figure 5A, lane 3, “Stretch + AG490”). To determine if blockage of the Jak/STAT pathway also blocks CLP-1’s dissociation from p-TEFb, AG490 was added to control and stretched cardiomyocytes during the 48-hour stretching regimen after which P-TEFb complexes were isolated and assayed for the presence of CLP-1. Stretched cardiomyocytes showed a decrease in the amount of CLP-1 associated with P-TEFb complexes (Figure 5B, compare lane 3 (“Stretch”) with lane 1 (“Control”)). In AG490-treated stretched cells, the amount of CLP-1 remaining associated with P-TEFb complexes was similar to AG490-treated non-stretched or control cells indicating that little or no CLP-1 dissociated from P-TEFb complexes with AG490 treatment (Figure 5B, compare lane 4 (“Stretch + AG490”) with lane 2 (“Control + AG490”)). There was no change in overall protein levels of P-TEFb components or CLP-1 with stretching or AG490 treatment (Figure 5C). These results indicate that the release of CLP-1 from P-TEFb is likely to be an integral and requisite step in the response of cardiomyoyctes to hypertrophic stimuli and that this process is directly responsive to hypertrophic signal transducers such as the Jak/STAT pathway.
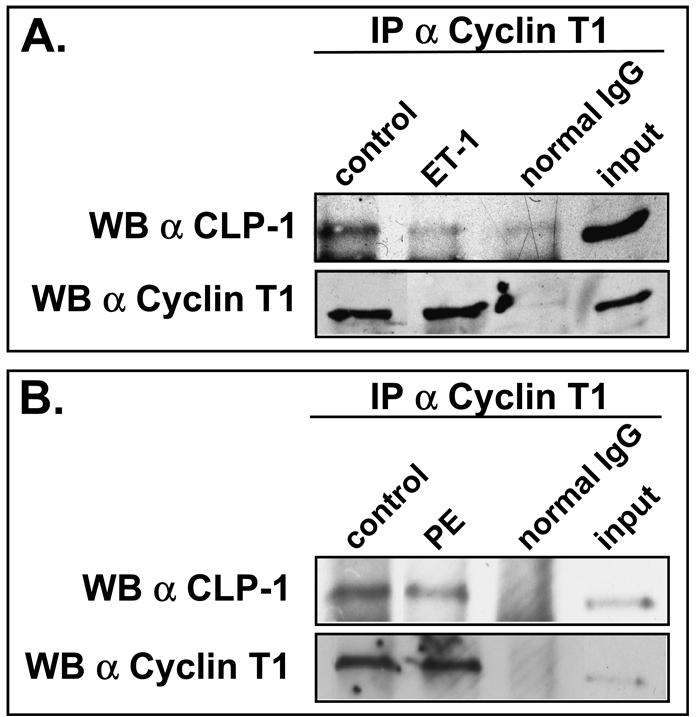
De-repression of P-TEFb is also seen in cardiomyocytes rendered hypertrophic by other means, for example, endothelin-treated cardiomyocytes that progress to hypertrophy and in transgenic or experimentally induced mouse models of hypertrophy (12). We therefore used endothelin-1 treatment as well as another hypertrophic agent, phenylephrine (44), to determine if CLP-1 dissociates from P-TEFb in cardiomyocytes treated with these hypertrophic agents. Immunoprecipitated P-TEFb from treated cardiomyocytes showed a decreased level of CLP-1 when compared to untreated cardiomyocytes indicating that as with mechanical stretch, CLP-1 responded to these hypertrophic agents by dissociating from P-TEFb (Figure 6A and B).
Figure 6.
CLP-1 dissociates from P-TEFb in cardiomyocytes treated with hypertrophic agents. A. Immunoprecipitated cyclin T1 shows less associated CLP-1 in endothelin-1-treated cardiomyocytes than untreated cells (control). B. Immunoprecipitated cyclin T1 shows less associated CLP-1 in phenylephrine-treated cardiomyocytes than in untreated cells. Input is non-immunoprecipitated cell lysates.
4. DISCUSSION
Our studies show that in cardiomyocytes rendered hypertrophic by mechanical stretch or treatment with hypertrophic agents, CLP-1 is dissociated from P-TEFb complexes. This process could be a common feature of the hypertrophic response to most hypertrophic stimuli since de-repression of P-TEFb has been shown to occur in cardiomyoyctes rendered hypertrophic by a variety of stimuli (12). Of the multiple signal transduction pathways activated by mechanical stretch, two appear to play a prominent role in stretch-induced hypertrophy: the Jak/STAT pathway and the MAPK pathway (35). The observation that CLP-1 does not dissociate from P-TEFb in the presence of the Jak2 kinase inhibitor AG490 suggests that dissociation of CLP-1 from P-TEFb is responsive to transduction of the hypertrophic signal by the Jak/STAT pathway. Inhibition of the Jak2 kinase prevents the phosphorylation and translocation of STATs to the nucleus. Blockage of STAT translocation and CLP-1 dissociation by AG490 treatment suggests that they might be linked, either directly via interaction between STATs and the P-TEFb complex itself (45) or indirectly, by the absence of a requisite STAT-dependent gene product that promotes CLP-1 dissociation from P-TEFb. Treatment of cardiomyocytes with the hypertrophic agents endothelin-1 and phenylephrine also results in the dissociation of CLP-1 from P-TEFb complexes. Since both endothelin-1 and phenylephrine signal primarily through the MAPK pathway (44, 46), it appears that multiple signaling pathways are involved in regulating CLP-1’s dissociation from P-TEFb. The convergence of the Jak/STAT and MAPK pathways at the CLP-1-P-TEFb interface suggests that this transcriptional regulatory complex may be a nexus point for the convergence of these and perhaps other hypertrophic pathways that reprogram cardiac gene expression to respond to hypertrophic stress (47).
Our studies show that cdk9 and CLP-1 are expressed in E13-E19 fetal hearts and that in rat neonatal cardiomyocytes, the cells used in our stretch studies, cdk9, cyclin T, and CLP-1 are co-localized in cell nuclei. These data, along with the demonstration of CLP-1 dissociation from P-TEFb complexes in response to hypertrophic stimuli, provide some insight into why our CLP-1 knockout mice die of cardiac hypertrophy (24). One possibility is that in the absence of the negative regulator CLP-1, elevated levels of constitutively active P-TEFb will phosphorylate RNA pol II leading to increased elongation activity and transcription from competent gene loci that increase cellular mRNA levels. While it is unclear whether this is sufficient to trigger hypertrophy, studies by Sano et al. (12) in which 7SK RNA was eliminated in cultured cardiomyocytes by antisense RNA showed increased cdk9 activity and increased RNA synthesis, two results consistent with the onset of hypertrophy. Constitutively active RNA pol II could foster a state of increased gene responsiveness that promotes a more rapid and robust response to hypertrophic stimuli. The period between E16 and E19 coincides with the fetal heart beginning to take on the increased hemodynamic demands imposed upon it by the maturing fetal circulatory system. This obviously does not pose a risk to wild-type fetuses expressing CLP-1, but in CLP-1 knockout fetuses the absence of CLP-1 may make the genetic component of the adaptive response to increased hemodynamic demand that much more responsive. In accord with this are recent studies showing that mice with elevated cdk9 kinase activity in heart rapidly develop hypertrophy and heart failure (48) and our own unpublished data showing that CLP-1 (+/−) mice are more susceptible than wild-type CLP-1 (+/+) mice to hypertrophic insults such as trans-aortic constriction. Perhaps the high levels of CLP-1 in fetal heart allow for normal cardiovascular development by precluding a “hyper-response” to the hemodynamic load imposed by the expanding fetal circulatory system.
In summary, our studies suggest that regulation of P-TEFb activity is likely to be an important control point in the mounting of the hypertrophic response. Curtailing or preventing CLP-1’s ability to bind P-TEFb and inhibit cdk9 kinase activity may lead to a P-TEFb population sufficiently biased to the de-repressed, kinase active state as to impart a state of cellular hyper-responsiveness to hypertrophic stimuli.
Acknowledgments
This work was supported by a grant from the National Institutes of Health (HL 07339, M.A.Q. Siddiqui). We thank Mr. Weimin Liu for his expertise and help with confocal microscopy.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Schaub MC, Hefti MA, Zuellig RA, Morano I. Modulation of contractility in human cardiac hypertrophy by myosin essential light chain isoforms. Cardiovasc Res. 1998;37:381–404. doi: 10.1016/s0008-6363(97)00258-7. [DOI] [PubMed] [Google Scholar]
- 2.Izumo S, Nadal-Ginard B, Mahdavi V. Protooncogene induction and reprogramming of cardiac gene expression produced by pressure overload. Proc Natl Acad Sci USA. 1988;85:339–343. doi: 10.1073/pnas.85.2.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lompre A, Mercadier J, Schwartz K. Changes in Gene Expression during Cardiac Growth. Int Rev Cyto. 1991;124:137–185. doi: 10.1016/s0074-7696(08)61526-0. [DOI] [PubMed] [Google Scholar]
- 4.Molkentin JD, Dorn II GW., 2nd Cytoplasmic signaling pathways that regulate cardiac hypertrophy. Annu Rev Physiol. 2001;63:391–426. doi: 10.1146/annurev.physiol.63.1.391. [DOI] [PubMed] [Google Scholar]
- 5.Olson EN, Schneider MD. Sizing up the heart: development redux in disease. Genes Dev. 2003;17:1937–1956. doi: 10.1101/gad.1110103. [DOI] [PubMed] [Google Scholar]
- 6.Kurnick NB, Lindsay PA. Nucleic acids in compensatory renal hypertrophy. Lab Invest. 1968;18:700–708. [PubMed] [Google Scholar]
- 7.Koide T, Rabinowitz M. Biochemical correlates of cardiac hypertrophy. II. Increased rate of RNA synthesis in experimental cardiac hypertrophy in the rat. Circ Res. 1969;24:9–18. doi: 10.1161/01.res.24.1.9. [DOI] [PubMed] [Google Scholar]
- 8.Nair KG, Cutilletta AF, Zak R, Koide T, Rabinowitz M. Biochemical correlates of cardiac hypertrophy. I. Experimental model; changes in heart weight, RNA content, and nuclear RNA polymerase activity. Circ Res. 1968;23:451–462. doi: 10.1161/01.res.23.3.451. [DOI] [PubMed] [Google Scholar]
- 9.Norman JT, Bohman RE, Fischmann G, Bowen JW, McDonough A, Slamon D, et al. Patterns of mRNA expression during early cell growth differ in kidney epithelial cells destined to undergo compensatory hypertrophy versus regenerative hyperplasia. Proc Natl Acad Sci U S A. 1988;85:6768–6772. doi: 10.1073/pnas.85.18.6768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Johnatty SE, Dyck JRB, Michael LH, Olson EN, Abdellatif M. Identification of Genes Regulated During Mechanical Load-induced Cardiac Hypertrophy. J Mol Cell Cardiol. 2000;32:805–815. doi: 10.1006/jmcc.2000.1122. [DOI] [PubMed] [Google Scholar]
- 11.Aronow BJ, Toyokawa T, Canning A, Haghighi K, Delling U, Kranias E, et al. Divergent transcriptional responses to independent genetic causes of cardiac hypertrophy. Physiological Genomics. 2001;6:19–28. doi: 10.1152/physiolgenomics.2001.6.1.19. [DOI] [PubMed] [Google Scholar]
- 12.Sano M, Abdellatif M, Oh H, Xie M, Bagella L, Giordano A, et al. Activation and function of cyclin T-Cdk9 (positive transcription elongation factor-b) in cardiac muscle-cell hypertrophy. Nat Med. 2002;8:1310–1317. doi: 10.1038/nm778. [DOI] [PubMed] [Google Scholar]
- 13.Sano M, Schneider MD. Cyclins that don't cycle--cyclin T/cyclin-dependent kinase-9 determines cardiac muscle cell size. Cell Cycle. 2003;2:99–104. [PubMed] [Google Scholar]
- 14.Sano M, Schneider MD. Cyclin-dependent kinase-9: an RNAPII kinase at the nexus of cardiac growth and death cascades. Circ Res. 2004;95:867–876. doi: 10.1161/01.RES.0000146675.88354.04. [DOI] [PubMed] [Google Scholar]
- 15.Marshall NF, Peng J, Xie Z, Price DH. Control of RNA polymerase II elongation potential by a novel carboxyl-terminal domain kinase. J Biol Chem. 1996;271:27176–27183. doi: 10.1074/jbc.271.43.27176. [DOI] [PubMed] [Google Scholar]
- 16.Price DH. P-TEFb, a cyclin-dependent kinase controlling elongation by RNA polymerase II. Mol Cell Biol. 2000;20:2629–2634. doi: 10.1128/mcb.20.8.2629-2634.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chiu YL, Cao H, Jacque JM, Stevenson M, Rana TM. Inhibition of human immunodeficiency virus type 1 replication by RNA interference directed against human transcription elongation factor P-TEFb (CDK9/CyclinT1) J Virol. 2004;78:2517–2529. doi: 10.1128/JVI.78.5.2517-2529.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yik JH, Chen R, Nishimura R, Jennings JL, Link AJ, Zhou Q. Inhibition of P-TEFb (CDK9/Cyclin T) kinase and RNA polymerase II transcription by the coordinated actions of HEXIM1 and 7SK snRNA. Mol Cell. 2003;12:971–982. doi: 10.1016/s1097-2765(03)00388-5. [DOI] [PubMed] [Google Scholar]
- 19.Nguyen VT, Kiss T, Michels AA, Bensaude O. 7SK small nuclear RNA binds to and inhibits the activity of CDK9/cyclin T complexes. Nature. 2001;414:322–325. doi: 10.1038/35104581. [DOI] [PubMed] [Google Scholar]
- 20.Yang Z, Zhu Q, Luo K, Zhou Q. The 7SK small nuclear RNA inhibits the CDK9/cyclin T1 kinase to control transcription. Nature. 2001;414:317–322. doi: 10.1038/35104575. [DOI] [PubMed] [Google Scholar]
- 21.Michels AA, Fraldi A, Li Q, Adamson TE, Bonnet F, Nguyen VT, et al. Binding of the 7SK snRNA turns the HEXIM1 protein into a P-TEFb (CDK9/cyclin T) inhibitor. EMBO J. 2004;23:2608–2619. doi: 10.1038/sj.emboj.7600275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ghatpande S, Goswami S, Mathew S, Rong G, Cai L, Shafiq S, et al. Identification of a novel cardiac lineage-associated protein (cCLP-1): A candidate regulator of cardiogenesis. Dev Biol. 1999;208:210–221. doi: 10.1006/dbio.1998.9180. [DOI] [PubMed] [Google Scholar]
- 23.Huang F, Wagner M, Siddiqui MAQ. Structure, expression and functional characterization of the mouse CLP-1 gene. Gene. 2002;292:245–259. doi: 10.1016/s0378-1119(02)00596-6. [DOI] [PubMed] [Google Scholar]
- 24.Huang F, Wagner M, Siddiqui MAQ. Ablation of the CLP-1 gene leads to down-regulation of the HAND1 gene and abnormality of the left ventricle of the heart and fetal death. Mech Dev. 2004;121:559–572. doi: 10.1016/j.mod.2004.04.012. [DOI] [PubMed] [Google Scholar]
- 25.Pikkarainen S, Tokola H, Majalahti-Palviainen T, Kerkela R, Hautala N, Bhalla SS, Charron F, Nemer M, Vuolteenaho O, Ruskoaho H. GATA-4 is a nuclear mediator of mechanical stretch-activated hypertrophic program. J Biol Chem. 2003;278:23807–23816. doi: 10.1074/jbc.M302719200. [DOI] [PubMed] [Google Scholar]
- 26.Peng J, Zhu Y, Milton JT, Price DH. Identification of multiple cyclin subunits of human P-TEFb. Genes Dev. 1998;12:755–762. doi: 10.1101/gad.12.5.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu H, Herrmann CH. Differential localization and expression of the Cdk9 42k and 55k isoforms. J Cell Physiol. 2005;203:251–260. doi: 10.1002/jcp.20224. [DOI] [PubMed] [Google Scholar]
- 28.Shore SM, Byers SA, Dent P, Price DH. Characterization of Cdk9(55) and differential regulation of two Cdk9 isoforms. Gene. 2005;350:51–58. doi: 10.1016/j.gene.2005.01.015. [DOI] [PubMed] [Google Scholar]
- 29.Chen R, Yang Z, Zhou Q. Phosphorylated positive transcription elongation factor b (P-TEFb) is tagged for inhibition through association with 7SK snRNA. J Biol Chem. 2004;279:4153–4160. doi: 10.1074/jbc.M310044200. [DOI] [PubMed] [Google Scholar]
- 30.Li Q, Price JP, Byers SA, Cheng D, Peng J, Price DH. Analysis of the large inactive P-TEFb complex indicates that it contains one 7SK molecule, a dimer of HEXIM1 or HEXIM2, and two P-TEFb molecules containing Cdk9 phosphorylated at threonine 186. J Biol Chem. 2005;280:28819–28826. doi: 10.1074/jbc.M502712200. [DOI] [PubMed] [Google Scholar]
- 31.Herrmann CH, Mancini MA. The Cdk9 and cyclin T subunits of TAK/P-TEFb localize to splicing factor-rich nuclear speckle regions. J Cell Sci. 2001;114:1491–1503. doi: 10.1242/jcs.114.8.1491. [DOI] [PubMed] [Google Scholar]
- 32.Sadoshima J, Izumo S. The cellular and molecular response of cardiac myocytes to mechanical stress. Annu Rev Physiol. 1997;59:551–571. doi: 10.1146/annurev.physiol.59.1.551. [DOI] [PubMed] [Google Scholar]
- 33.Sadoshima J, Jahn L, Takahashi T, Kulik TJ, Izumo S. Molecular characterization of the stretch-induced adaptation of cultured cardiac cells. An in vitro model of load-induced cardiac hypertrophy. J Biol Chem. 1992;267:10551–10560. [PubMed] [Google Scholar]
- 34.Pan J, Fukuda K, Saito M, Matsuzaki J, Kodama H, Sano M, et al. Mechanical stretch activates the JAK/STAT pathway in rat cardiomyocytes. Circ Res. 1999;84:1127–1136. doi: 10.1161/01.res.84.10.1127. [DOI] [PubMed] [Google Scholar]
- 35.Ruwhof C, van der Laarse A. Mechanical stress-induced cardiac hypertrophy: mechanisms and signal transduction pathways. Cardiovasc Res. 2000;47:23–37. doi: 10.1016/s0008-6363(00)00076-6. [DOI] [PubMed] [Google Scholar]
- 36.Lammerding J, Kamm RD, Lee RT. Mechanotransduction in cardiac myocytes. Ann NY Acad Sci. 2004;1015:53–70. doi: 10.1196/annals.1302.005. [DOI] [PubMed] [Google Scholar]
- 37.Freed DH, Borowiec AM, Angelovska T, Dixon IM. Induction of protein synthesis in cardiac fibroblasts by cardiotrophin-1: integration of multiple signaling pathways. Cardiovasc Res. 2003;60:365–375. doi: 10.1016/s0008-6363(03)00534-0. [DOI] [PubMed] [Google Scholar]
- 38.Kodama H, Fukuda K, Pan J, Makino S, Baba A, Hori S, Ogawa S. Leukemia inhibitory factor, a potent cardiac hypertrophic cytokine, activates the JAK/STAT pathway in rat cardiomyocytes. Circ Res. 1997;81:656–663. doi: 10.1161/01.res.81.5.656. [DOI] [PubMed] [Google Scholar]
- 39.Pan J, Fukuda K, Kodama H, Makino S, Takahashi T, Sano M, Hori S, Ogawa S. Role of angiotensin II in activation of the JAK/STAT pathway induced by acute pressure overload in the rat heart. Circ Res. 1997;81:611–617. doi: 10.1161/01.res.81.4.611. [DOI] [PubMed] [Google Scholar]
- 40.Mascareno E, Dhar M, Siddiqui MAQ. Signal transduction and activator of transcription (STAT) protein-dependent activation of angiotensinogen promoter: a cellular signal for hypertrophy in cardiac muscle. Proc Natl Acad Sci U S A. 1998;95:5590–5594. doi: 10.1073/pnas.95.10.5590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guo Y, Mascareno E, Siddiqui MAQ. Distinct components of Janus kinase/signal transducer and activator of transcription signaling pathway mediate the regulation of systemic and tissue localized rennin-angiotensin system. Mol Endocrinol. 2004;18:1033–1041. doi: 10.1210/me.2003-0231. [DOI] [PubMed] [Google Scholar]
- 42.Fukuzawa J, Booz GW, Hunt RA, Shimizu N, Karoor V, Baker KM, et al. Cardiotrophin-1 increases angiotensinogen mRNA in rat cardiac myocytes through STAT3 : an autocrine loop for hypertrophy. Hypertension. 2000;35:1191–1196. doi: 10.1161/01.hyp.35.6.1191. [DOI] [PubMed] [Google Scholar]
- 43.Kodama H, Fukuda K, Pan J, Sano M, Takahashi T, Kato T, et al. Significance of ERK cascade compared with JAK/STAT and PI3-K pathway in gp130-mediated cardiac hypertrophy. Am J Physiol Heart Circ Physiol. 2000;279:H1635–H1644. doi: 10.1152/ajpheart.2000.279.4.H1635. [DOI] [PubMed] [Google Scholar]
- 44.Bogoyevitch MA, Glennon PE, Sugden PH. Endothelin-1, phorbol esters and phenylephrine stimulate MAP kinase activities in ventricular cardiomyocytes. FEBS Lett. 1993;317:271–275. doi: 10.1016/0014-5793(93)81291-7. [DOI] [PubMed] [Google Scholar]
- 45.Giraud S, Hurlstone A, Avril S, Coqueret O. Implication of BRG1 and cdk9 in the STAT3-mediated activation of the p21waf1 gene. Oncogene. 2004;23:7391–7398. doi: 10.1038/sj.onc.1207972. [DOI] [PubMed] [Google Scholar]
- 46.Sugden PH. An overview of endothelin signaling in the cardiac myocyte. J Mol Cell Cardiol. 2003;35:871–886. doi: 10.1016/s0022-2828(03)00153-6. [DOI] [PubMed] [Google Scholar]
- 47.Backs J, Olson EN. Control of cardiac growth by histone acetylation/deacetylation. Circ Res. 2006;98:15–24. doi: 10.1161/01.RES.0000197782.21444.8f. [DOI] [PubMed] [Google Scholar]
- 48.Sano M, Wang SC, Shirai M, Scaglia F, Xie M, Sakai S, et al. Activation of cardiac Cdk9 represses PGC-1 and confers a predisposition to heart failure. EMBO J. 2004;23:3559–3569. doi: 10.1038/sj.emboj.7600351. [DOI] [PMC free article] [PubMed] [Google Scholar]