Abstract
Cytochrome P450 (CYP) 2C8 is responsible for the oxidative metabolism of many clinically available drugs from a diverse number of drug classes (e.g., thiazolidinediones, meglitinides, NSAIDs, antimalarials and chemotherapeutic taxanes). The CYP2C8 enzyme is encoded by the CYP2C8 gene, and several common nonsynonymous polymorphisms (e.g., CYP2C8*2 and CYP2C8*3) exist in this gene. The CYP2C8*2 and *3 alleles have been associated in vitro with decreased metabolism of paclitaxel and arachidonic acid. Recently, the influence of CYP2C8 polymorphisms on substrate disposition in humans has been investigated in a number of clinical pharmacogenetic studies. Contrary to in vitro data, clinical data suggest that the CYP2C8*3 allele is associated with increased metabolism of the CYP2C8 substrates, rosiglitazone, pioglitazone and repaglinide. However, the CYP2C8*3 allele has not been associated with paclitaxel pharmacokinetics in most clinical studies. Furthermore, clinical data regarding the impact of the CYP2C8*3 allele on the disposition of NSAIDs are conflicting and no definitive conclusions can be made at this time. The purpose of this review is to highlight these clinical studies that have investigated the association between CYP2C8 polymorphisms and CYP2C8 substrate pharmacokinetics and/or pharmacodynamics in humans. In this review, CYP2C8 clinical pharmacogenetic data are provided by drug class, followed by a discussion of the future of CYP2C8 clinical pharmacogenetic research.
Keywords: amodiaquine, CYP2C8, cytochrome P450 2C8, human, ibuprofen, NSAID, paclitaxel, repaglinide, thiazolidinedione
The field of pharmacogenetics is aimed at understanding how genetic variation contributes to interindividual variability in drug disposition (i.e., pharmacokinetics) and drug response (i.e., pharmacodynamics). In terms of drug disposition, the cytochrome P450 (CYP) metabolizing enzymes are a major area of study, as they catalyze the oxidative metabolism of numerous drugs and endogenous compounds. Enzymes in the CYP2C family (e.g., CYP2C8, CYP2C9, CYP2C18 and CYP2C19) are significant contributors to drug disposition and metabolize 20% of clinically available drugs [1,2]. Recently, the role of the CYP2C8 isoenzyme has garnered considerable interest in the fields of drug metabolism and pharmacogenetics. This is largely a result of the elucidation of the structure of the CYP2C8 enzyme, characterization of polymorphisms within the CYP2C8 gene and identification of clinically relevant CYP2C8 substrates, such as the thiazolidinediones and repaglinide [1].
In 2005, Totah and colleagues published a comprehensive review describing CYP2C8 substrates, inhibitors, inducers and pharmacogenetics [1]. Since that time, the field of CYP2C8 pharmacogenetics has expanded dramatically, and numerous clinical studies have been published regarding the association between CYP2C8 polymorphisms and the disposition of various substrates. As such, the purpose of this review is to discuss clinical studies that have examined the influence of CYP2C8 polymorphisms on the pharmacokinetics and/or pharmacodynamics of CYP2C8 substrates in humans. First, we present a brief discussion of the CYP2C8 enzyme and CYP2C8 polymorphisms (for a comprehensive background discussion of CYP2C8 and in vitro data, the reader is referred to the review by Totah and Rettie) [1]. Next, we place major focus on the relevant clinical CYP2C8 pharmacogenetic data by drug class, followed by a discussion of the future of CYP2C8 clinical pharmacogenetic research.
CYP2C8 enzyme
CYP2C8 comprises 7% of the total hepatic CYP content and plays an important role in the metabolism of a diverse number of exogenous and endogenous compounds [1]. It is highly expressed in liver, but is also found in extrahepatic sites such as the kidney, heart, adrenal gland, brain, uterus, mammary gland, ovary and duodenum [3,4]. CYP2C8 plays a major role in the metabolism of many clinically available drugs [5,6]. These drugs are summarized in Table 1 and include: thiazolidinediones (i.e., rosiglitazone, pioglitazone and troglitazone), meglitinides (i.e., repaglinide), 3-hydroxy-3-methylglutaryl-coenzyme A (HMG-CoA) reductase inhibitors (i.e., cerivastatin), chemotherapeutic agents (i.e., paclitaxel and all-trans-retinoic acid), antimalarials (i.e., amodiaquine and chloroquine), antiarrhythmics (i.e., amiodarone) and retinoid derivatives (i.e., tazarotenic acid) [1]. Other data suggest that CYP2C8 plays only an intermediate or minor role in the metabolism of other agents such as NSAIDs (i.e., ibuprofen, diclofenac and tenoxicam), fluvastatin, simvastatin acid, carbamazepine, cyclophosphamide, dapsone, diltiazem, ifosfamide, loperamide, methadone, morphine, torsemide, verapamil and zopiclone [1]. CYP2C8 also converts endogenous compounds such as arachidonic acid to biologically active epoxide metabolites, which are important regulators of vascular tone and blood pressure [1,7].
Table 1.
Summary of CYP2C8 substrates.
| Specific drugs | Drug class | Treatment indication | Other CYP enzymes that play a role in metabolism |
|---|---|---|---|
| Major contribution of CYP2C8 to metabolism | |||
| All-trans-retinoic acid | Vitamin A derivative | Promyelocytic leukemia | CYP2C9 |
| Amiodarone | Class III antiarrhythmic | Supraventricular and ventricular arrhythmias | CYP3A4, CYP1A2, CYP2C19, CYP2D6 |
| Amodiaquine | 4-aminoquinoline | Malaria | CYP3A4, CYP1A1, CYP1B1 |
| Cerivastatin* | HMG-CoA reductase inhibitor | Hyperlipidemia | CYP3A4 |
| Chloroquine | 4-aminoquinoline | Malaria | CYP3A4, CYP2D6 |
| Paclitaxel | Taxane | Solid malignant tumors (e.g., breast, ovarian, lung) | CYP3A4, CYP3A5 |
| Repaglinide | Meglitinide | Type 2 diabetes | CYP3A4 |
| Rosiglitazone Pioglitazone | Thiazolidinedione | Type 2 diabetes | CYP2C9, CYP3A4 CYP3A4, CYP1A1 |
| Tazarotenic acid | Retinoid derivative | Acne and psoriasis | – |
| Troglitazone* | Thiazolidinedione | Type 2 diabetes | CYP3A4 |
| Intermediate or minor contribution of CYP2C8 to metabolism | |||
| Carbamazepine | Anticonvulsant | Epilepsy | CYP3A4 (major) |
| Cyclophosphamide | Nitrogen mustard alkylating agent | Lymphoma, leukemia, solid tumors | CYP2A6, CYP2B6, CYP2C9, CYP2C19, CYP3A |
| Dapsone | Antibacterial sulfone | Leprosy and pneumocystis pneumonia | CYP2C9 (major) |
| Diclofenac | NSAID | Pain and inflammation | CYP2C9 (major), CYP3A4, and CYP2C19 |
| Diltiazem | Calcium channel blocker | Hypertension, arrhythmias, angina | CYP3A4, CYP2C9 |
| Fluvastatin | HMG-CoA reductase inhibitor | Hyperlipidemia | CYP2C9 (major), CYP3A4 |
| Ibuprofen | NSAID | Pain and inflammation | CYP2C9 |
| Ifosfamide | Nitrogen mustard alkylating agent | Various cancers | CYP2A6, CYP2B6, CYP2C9, CYP2C19, CYP3A |
| Loperamide | μ-opioid receptor agonist | Antidiarrheal | CYP2B6, CYP2D6, CYP3A4 |
| Methadone | Opioid antagonist | Pain and addiction to opioids | CYP3A4, CYP2D6, CYP2B6 |
| Morphine | Opioid agonist | Pain | CYP3A4 |
| Simvastatin acid | HMG-CoA reductase inhibitor | Hyperlipidemia | CYP3A4 (major) |
| Tenoxicam | NSAID | Pain and inflammation | CYP2C9 (major) |
| Torsemide | Loop diuretic | Edema | CYP2C9 (major) |
| Verapamil | Calcium channel blocker | Hypertension, arrhythmias, angina | CYP3A4 (major), CYP3A5 |
| Zopiclone | Nonbenzodiazepine sedative hypnotic | Insomnia | CYP3A4 (major) |
Withdrawn from the market.
This table was adapted and updated from that previously published [1].
CYP2C8 gene & polymorphisms
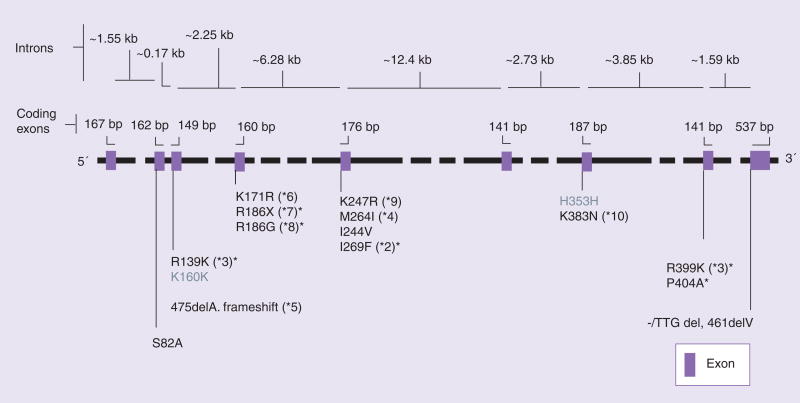
The CYP2C8 enzyme is encoded by the CYP2C8 gene, which spans 31 kb and is located on chromosome 10q24 [3]. CYP2C8 is located in close proximity to CYP2C9, CYP2C19 and CYP2C18, which are also located on chromosome 10q24. In total, the CYP2C gene family spans approximately 400 kb and there is linkage between genes [3,8]. The CYP2C8 gene shares 74% sequence homology with CYP2C9 and is transcriptionally regulated by nuclear receptors such as androstane receptor, pregnane X receptor, glucocorticoid receptor and hepatic nuclear factor-4α [9–11]. The wild-type gene is referred to as CYP2C8*1 (or *1A). A number of common nonsynonymous SNPs have been identified in CYP2C8 including: CYP2C8*2 (Ile269Phe), CYP2C8*3 (linked polymorphisms Arg139Lys and Lys399Arg) and CYP2C8*4 (Ile264Met) (Figure 1). Of note, the CYP2C8*3 allele is in partial linkage disequilibrium with the CYP2C9*2 allele, which is associated with impaired metabolism of many CYP2C9 substrates [8]. Common polymorphisms in the CYP2C8 promoter, -271C>A and -370T>G, have also been identified [12,13]. Additionally, data from the Human Cytochrome P450 Allele Nomenclature Committee and dbSNP show that a number of other rare nonsynonymous polymorphic CYP2C8 alleles exist, including CYP2C8*5 through CYP2C8*14, and several that have not yet been assigned a ‘star’ designation [201,202]. The description, location and frequencies of nonsynonymous and promoter CYP2C8 polymorphic alleles are summarized in Table 2.
Figure 1. CYP2C8 gene structure and coding region polymorphisms.
The CYP2C8 gene, located on chromosome 10q24, spans 31 kb and consists of nine exons. Coding region polymorphisms are shown in the figure. Gray text signifies synonymous polymorphisms, while the black text signifies nonsynonymous polymorphism.
*Alleles coding for documented or predicted reduced enzyme activity.
Figure reproduced from [115].
Table 2.
Putative functional CYP2C8 polymorphisms and allele frequencies.
| Allele | dbSNP number or Human Genome Variation Society number | Nucleotide or amino acid change | Location | Allele frequency: Caucasian (%) | Allele frequency: Chinese (%) | Allele frequency: Japanese (%) | Allele frequency: African (%) |
|---|---|---|---|---|---|---|---|
| CYP2C8*1 (also called *1A) | Wild-type | ||||||
| CYP2C8*1B | rs7909236 | -271C>A | Promoter | 23.3 | 10 | 8.9 | 0 |
| CYP2C8*1C | rs17110453 | -370T>G | Promoter | 11.7 | 27.8 | 34.4 | 0 |
| CYP2C8*2 | rs11572103 | Ile269Phe | Exon 5 | 0 | 0 | 0 | 19 |
| CYP2C8*3 | rs11572080 | Arg139Lys | Exon 3 | 10.8 | 0 | 0 | 0 |
| rs10509681 | Lys399Arg | Exon 8 | 11.7 | 0 | 0 | 0 | |
| CYP2C8*4 | rs1058930 | Ile264Met | Exon 5 | 5.8 | 0 | 0 | 0 |
| CYP2C8*5 | rs72558196 | Thr159X | Exon 3 | – | – | 0.2‡ | – |
| CYP2C8*6 | NT_030059.12:g.15573214G>A | Gly171Ser | Exon 4 | – | – | 0.2‡ | – |
| CYP2C8*7 | rs72558195 | Arg186X | Exon 4 | – | – | 0.1‡ | – |
| CYP2C8*8 | rs72558195 | Arg186Gly | Exon 4 | – | – | 0.1‡ | – |
| CYP2C8*9 | NT_030059.12:g.15566697A>G | Lys247Arg | Exon 5 | – | – | 0.1‡ | – |
| CYP2C8*10 | NT_030059.12:g.15551173G>T | Lys383Asn | Exon 7 | – | – | 0.1‡ | – |
| CYP2C8*12 | rs3832694 | 461delV | Exon 9 | – | – | 0.1‡ | – |
| CYP2C8*13 | NT_030059.12:g.15566768T>G | Ile223Met | Exon 5 | – | – | 0.1‡ | – |
| CYP2C8*14 | NT_030059.12:g.15566725G>C | Ala238Pro | Exon 5 | – | – | 0.1‡ | – |
| Not assigned | rs17851796 | Ala82Ser | Exon 2 | – | – | – | – |
| Not assigned | rs41286886 | Val181Ile | Exon 4 | – | – | – | – |
| Not assigned | rs11572102 | Ile244Val | Exon 5 | 0 | 0 | 0 | 1.7 |
| Not assigned | rs45438799 | Leu361Phe | Exon 7 | – | – | – | – |
| Not assigned | Pro404Ala | Exon 8 | – | – | 0.7‡ | 2.2 |
Polymorphisms are presented based on data summarized in dbSNP [201], the Human Cytochrome P450 Allele Nomenclature Committee [202] and previously published literature [1,7,14,115]. Allele frequencies for CYP2C8*1B, *1C, *2, *3, *4, Ile244Val and Pro404Ala are based on HapMap data available in dbSNP.
If polymorphism allele frequencies were not available in dbSNP, relevant published literature was used [14].
–: Allele frequency data were not available; Caucasian: Centre d’Etude du Polymorphisme Humain (Utah residents with ancestry from northern and western Europe); Chinese: Han Chinese in Beijing, China; Japanese: Japanese in Tokyo, Japan; African: Yoruba in Ibadan, Nigeria
Recently, several groups have characterized CYP2C8 haplotypes in different racial groups. Using data from the International HapMap Project, Rodriguez-Antona and colleagues characterized CYP2C8 haplotypes in 54 unrelated Caucasian individuals [13]. They inferred haplotypes from 12 HapMap tag SNPs and SNPs with a putative functional significance. Seven common haplotypes (frequency greater than 2%) were identified and named (A, B, C1, C2, C3, D and E). The SNPs included in these haplotypes are shown in Table 3.
Table 3.
Common CYP2C8 haplotypes in Caucasians‡.
| SNPs | Hap A | Hap B | Hap C1 | Hap C2 | Hap C3 | Hap D | Hap E |
|---|---|---|---|---|---|---|---|
| rs11188172 G>A (-2048, promoter) | G | A | G | G | G | G | G |
| rs17110453 T>G (-370, promoter, CYP2C8*1C) | T | T | G | G | T | T | T |
| rs7909236 C>A (-271, promoter, CYP2C8*1B) | C | A | C | C | C | C | C |
| rs1934956 G>A (intron 1) | G | G | G | G | G | G | A |
| rs3216029 or rs11572078 (intron 2) | – | – | T | T | T | – | – |
| rs2275622 G>A (intron 2) | G | A | G | G | G | G | A |
| rs11572080 G>A (exon 3, Arg139Lys, CYP2C8*3) | G | G | G | G | G | A | G |
| rs2185571 G>A (intron 3) | A | G | G | G | G | G | G |
| rs1113129 C>G (intron 5) | C | C | G | G | G | C | C |
| rs1058930 C>G (exon 5, Ile264Met, CYP2C8*4) | C | C | C | G | C | C | C |
| rs2275620 A>T (intron 7) | A | T | A | A | A | A | T |
| rs10509681 A>G (exon 8, Lys399Arg, CYP2C8*3) | A | A | A | A | A | G | A |
As reported by [13].
SNPs included in the haplotypes were HapMap tagging SNPs and CYP2C8 SNPs with putative functional significance.
The haplotypes were inferred using the CYP2C8 reference sequence (AL359672). Haplotype frequencies were 30.5% (Hap A), 19.6% (Hap B), 7.4% (Hap C1), 6.5% (Hap C2), 5.4% (Hap C3), 15.3% (Hap D) and 6.9% (Hap E).
This table was adapted from [13].
Considering that there is a large degree of racial diversity in CYP2C8 haplotype structure, Saito and colleagues set out to characterize CYP2C8 haplotypes using 40 polymorphisms in 437 Japanese individuals [14]. A total of 40 haplotypes without amino acid changes and nine haplotypes with amino acid changes were inferred. The most common haplotypes were *1d (36.6%), *1e (28.9%), *1f (11.3%), *1B (8.5%). Using network analysis, haplotypes that were found in more than two patients were further classified into six groups, *IA (2.1%), *IB (10.3%), *ID (38.1%), *IE (43.5%), *IG (3%) and *IJ (1.3%).
Given the close proximity of CYP2C8 and CYP2C9 on chromosome 10, and the linkage that exists between these two genes [8], Speed and colleagues recently characterized haplotypes containing CYP2C8 and CYP2C9 polymorphisms in 2500 individuals from 45 populations around the world [15]. Ten SNPs were studied across a 132 kb region containing CYP2C8 and CYP2C9. These SNPs included: rs4918758 (CYP2C9), rs1799853 (CYP2C9, Arg144Cys), rs1934953 (CYP2C8), rs10509681 (CYP2C8, Lys399Arg), rs11572103 (CYP2C8, Ile269Phe), rs1058930 (CYP2C8, Ile264Met), rs11572093 (CYP2C8), rs11572080 (CYP2C8, Arg139Lys), rs11572076 (CYP2C8) and rs17110453 (CYP2C8). From these SNPs, 17 common haplotypes were inferred (haplotypes A through Q). Haplotype G contained three coding region SNPs, CYP2C9 Arg144Cys, CYP2C8 Arg139Lys and CYP2C8 Lys399Arg and occurred at a frequency of 10% in Europeans. The investigators also found that 89% of chromosomes with either the CYP2C8 Arg139Lys or CYP2C8 Lys399Arg variants also carried the CYP2C9 Arg144Cys variant allele. The investigators went on to hypothesize that due to the strong association of these three variants, it may be difficult in clinical studies to differentiate if observed associations are a result of CYP2C8 or CYP2C9 polymorphisms [15]. Of note, haplotypes B, L and M, which contain the CYP2C8 Ile269Phe (*2) polymorphism, were common in Africans (frequency of 6–28%), but were absent in other races studied. The investigators were unable to compare their haplotype results to those of Rodriguez-Antona and colleagues because only four polymorphisms were studied in common between the two studies. Future investigations elucidating the optimal panel of polymorphisms to include in CYP2C8 haplotype analysis are needed.
CYP2C8 clinical pharmacogenetic research is a direct result of in vitro work, which identified some CYP2C8 polymorphisms to have putative functional significance. Specifically, CYP2C8*3 is associated with reduced CYP2C8 metabolic activity for the substrates, paclitaxel and arachidonic acid, compared with wild-type enzyme [12,16,17]. Decreased CYP2C8 metabolic activity has also been demonstrated for CYP2C8*2 [16]. Although not statistically significant, data suggest that median CYP2C8*4 metabolic activity for paclitaxel is lower than wild-type enzyme [12]. More recently, in vitro analysis of promoter polymorphisms showed that the CYP2C8*1B allele (-271C>A) is associated with higher rates of transcription compared with wild-type [13]. In regards to less common polymorphisms, the CYP2C8*8 allele (Arg186Gly) is associated with reduced enzymatic activity compared with wild-type, and the CYP2C8*7 allele (Arg186X) results in a prematurely terminated protein. However, these polymorphisms are rare and appear to be race specific, occurring primarily in Asian populations [18].
While in vitro data suggest that variant CYP2C8*3 and CYP2C8*2 alleles result in decreased metabolism of CYP2C8 substrates, clinical pharmacogenetic data have yielded sometimes contradictory results. Hereafter, we present CYP2C8 clinical pharmacogenetic data for clinically available drugs, by drug class. This is followed by a discussion of the future of CYP2C8 clinical pharmacogenetic research. For a review of the influence of CYP2C8 polymorphisms on arachidonic acid metabolism and clinical outcomes, the reader is referred to a recent review by Theken and Lee [7].
Antidiabetic agents
Thiazolidinediones
The thiazolidinediones, rosiglitazone and pioglitazone, are peroxisome-proliferator-activated receptor-γ agonists that modulate the transcription of numerous genes involved in glucose metabolism, lipid metabolism, adipocyte differentiation and insulin sensitivity [19]. Rosiglitazone and pioglitazone are indicated for the treatment of Type 2 diabetes [20]. Both rosiglitazone and pioglitazone are extensively metabolized in the liver by CYP2C8 [3,21,22]. Troglitazone, a thiazolidinedi-one that was withdrawn from the market due to idiosyncratic hepatotoxicity, is also a CYP2C8 substrate [23]. Other CYP enzymes that contribute a lesser degree to rosiglitazone and pioglitazone metabolism include CYP2C9 (rosiglitazone), CYP3A4 (pioglitazone) and CYP1A1 (pioglitazone) [24,25]. Rosiglitazone undergoes CYP2C8-mediated N-demethylation and hydroxylation, followed by sulfate and glucuronic acid conjugation [21,24]. N-desmethylrosiglitazone and para-O-sulfate rosiglitazone are the primary metabolites, and both are considered less potent than the parent drug. Pioglitazone undergoes CYP2C8-mediated hydroxylation and oxidation, followed by sulfate and glucuronide conjugation. M-IV (hydroxy-derivative of pioglitazone) and M-III (keto-derivative) are the principal metabolites found in vivo [25].
Recent clinical data suggest that CYP2C8 polymorphisms contribute to interindividual variability in thiazolidinedione pharmacokinetics (Table 4). Kirchheiner and colleagues considered the influence of the CYP2C8*3 polymorphism on single dose and multiple-dose rosiglitazone (8 mg) pharmacokinetics in German healthy volunteers [26]. Following a single-dose, mean rosiglitazone area under the plasma concentration–time curve (AUC) was 36% lower and weight-adjusted oral clearance was 39% higher in CYP2C8*3 homozygotes compared with wild-type homozygotes (p = 0.03, both comparisons). Rosiglitazone AUC was 7% lower and weight-adjusted oral clearance was 15% higher in heterozygotes compared with wild-type homozygotes (not statistically significant, both comparisons). Similar pharmacokinetic results by genotype were found after 14 days of rosiglitazone administration. In multivariate regression analysis, significant predictors of rosiglitazone AUC were CYP2C8*3 genotype and body weight, which accounted for 48% of the variability in rosiglitazone AUC. The investigators also conducted a population pharmacokinetic analysis of their data, which revealed a 23 and 46% higher rosiglitazone oral clearance in heterozygotes and CYP2C8*3 homozygotes, respectively, compared with wild-type homozygotes.
Table 4.
Summary of thiazolidinedione CYP2C8 clinical pharmacogenetic–pharmacokinetic studies.
| Drug | Study description | Sample size and genotype distributions | PK results in relation to CYP2C8 genotype | Covariates | Ref. |
|---|---|---|---|---|---|
| Rosiglitazone | 8 mg single dose and multiple-dose PK |
CYP2C8*1/*1, n = 14 CYP2C8*1/*3, n = 13 CYP2C8*3/*3, n = 4 |
↓AUC and ↑CL in *3/*3 and *1/*3 vs *1/*1 | Linear regression with inclusion of sex, age and weight | [26] |
| Rosiglitazone | 4 mg single dose, PK |
CYP2C8*1/*1, n = 19 CYP2C8*1/*3, n = 7 |
↓AUC and ↑CL in *1/*3 vs *1/*1 | Linear regression with inclusion of sex, age, weight, and SLCO1B1 diplotype | [27] |
| Rosiglitazone | 8 mg single-dose PK (cross-over trimethoprim drug interaction study) |
CYP2C8*1/*1, n = 4 CYP2C8*1/*3, n = 3 CYP2C8*1/*2, n = 1 |
No association between CYP2C8*2 or CYP2C8*3 and PK | None reported | [29] |
| Rosiglitazone | 4 mg single-dose PK (cross-over fluvoxamine drug interaction study) |
CYP2C8*1/*1, n = 10 CYP2C8*1/*3, n = 10 CYP2C8*3/*3, n = 3 |
No association between CYP2C8*3 allele and PK | None reported | [30] |
| Pioglitazone | 15 mg single-dose PK (cross-over trimethoprim drug interaction study) |
CYP2C8*1/*1, n = 8 CYP2C8*1/*3, n = 5 CYP2C8*3/*3, n = 3 |
↓AUC and ↓ t1/2 in *3/*3 and *1/*3 vs *1/*1 | Weight | [28] |
AUC: Area under the plasma concentration–time curve; CL: Oral clearance; PK: Pharmacokinetics; t1/2: Half life.
The association between the CYP2C8*3 polymorphism and rosiglitazone pharmacokinetics in healthy Caucasian volunteers was recently evaluated in our laboratory [27]. No CYP2C8*3 homozygotes were identified in our population. Following a single dose of rosiglitazone 4 mg, mean AUC was 29% lower and weight-adjusted oral clearance was 39% higher in heterozygotes compared with wild-type homozygotes (p = 0.002 and p = 0.03, respectively). The half-life of rosiglitazone was also shorter in heterozygotes compared with wild-type homozygotes (3.5 h versus 4 h), although this did not reach statistical significance (p = 0.09). In multivariate regression analysis, significant predictors of rosiglitazone AUC were CYP2C8*3 genotype and body weight, which accounted for 42% of the variability in rosiglitazone AUC.
Tornio and colleagues evaluated the effects of the CYP2C8*3 allele on single-dose pioglitazone (15 mg) pharmacokinetics in healthy volunteers [28]. The weight-adjusted pioglitazone AUC was 34% lower in CYP2C8*3 homozygotes and 26% lower in heterozygotes compared with wild-type homozygotes (p < 0.05, both comparisons). The half-life of pioglitazone was significantly shorter in heterozygotes (3.4 h) and CYP2C8*3 homozygotes (3.3 h) compared with wild-type homozygotes (4.5 h). The investigators also evaluated the effects of trimethoprim (a CYP2C8 inhibitor) on pioglitazone pharmacokinetics, but found that CYP2C8 genotype did not significantly influence the extent to which trimethoprim increased the plasma concentrations of pioglitazone.
While these three studies demonstrated lower rosiglitazone and pioglitazone plasma exposure in carriers of the CYP2C8*3 allele, two other studies reported no association between CYP2C8 polymorphisms and thiazolidinedione pharmacokinetics. As part of a healthy volunteer drug–drug interaction study between trimethoprim and rosiglitazone, Hruska and colleagues found that rosiglitazone AUC did not differ significantly between wild-type homozygotes and those carrying a CYP2C8*3 or *2 allele [29]. In a different healthy volunteer study, Pederson and colleagues found that the pharmacokinetics of a single dose of rosiglitazone 4 mg did not differ significantly by CYP2C8 genotype, nor did genotype influence the magnitude of the drug–drug interaction between rosiglitazone and fluvoxamine (a CYP2C8 inhibitor) [30].
Although the data are not entirely consistent, three of five studies suggest that the CYP2C8*3 allele is associated with increased rosiglitazone and pioglitazone metabolism. In studies that included CYP2C8*3 homozygotes, a gene–dose effect appears to be present, both for rosiglitazone and pioglitazone. Of note, variability in the magnitude of the differences in AUC and oral clearance between heterozygotes and wild-type homozygotes was evident upon comparison of some of the studies. This may be explained by differences in body weight and gender distribution between genotype groups. Additionally, all studies did not genotype for the CYP2C9*2 and CYP2C9*3 alleles, which may have confounded the study findings, particularly for rosiglitazone, which undergoes some CYP2C9 metabolism. In terms of the studies that did not find an association between CYP2C8 polymorphisms and rosiglitazone pharmacokinetics, this is most likely to be due to small sample sizes and the fact that these studies were not originally powered to detect differences between genotype groups (i.e., both were drug–drug interaction studies).
In terms of clinical relevance, the issue that remains to be determined is whether these modest thiazolidinedione pharmacokinetic differences by CYP2C8 genotype will translate into pharmacodynamic differences in patients with Type 2 diabetes. Both rosiglitazone and pioglitazone have wide therapeutic indices. However, changes in plasma drug exposure, as a result of CYP2C8 polymorphisms, may influence the risk of concentration-dependent adverse effects (e.g., weight gain, edema) or the magnitude of glucose reductions or insulin sensitization. In light of recent cardiovascular risk findings surrounding rosiglitazone, it seems prudent at this point in time to focus attention on the potential influence of CYP2C8 polymorphisms on pioglitazone pharmacokinetics and pharmacodynamics in long-term studies of patients with Type 2 diabetes. Additionally, recent data suggest that polymorphisms in drug receptor and effector protein genes (e.g., PPAR-γ, adiponectin) influence response to thiazolidinediones [31]. Thus, consideration of both drug–metabolism and drug–target genetics must be given to comprehensively elucidate interindividual variability in thiazolidinedione response.
Repaglinide
Repaglinide, an agent in the meglitinide class, is a nonsulfonylurea insulin secretagogue that is used to lower postprandial glucose levels in patients with Type 2 diabetes. Repaglinide is metabolized through oxidative biotransformation and glucuronidation to several metabolites, M1, M2 and M4 [32,33]. Both CYP2C8 and CYP3A4 have been implicated in the metabolism of repaglinide; however, in vivo it appears that CYP2C8 may contribute to a greater extent than CYP3A4 [33,34]. This is supported by drug–drug interaction data that showed that gemfibrozil (a CYP2C8 inhibitor) increased repaglinide plasma concentrations to a greater extent than the CYP3A4 inhibitors, itraconazole and clarithromycin [35,36]. Given the important role of CYP2C8 in repaglinide metabolism, investigations have been conducted examining the influence of CYP2C8 polymorphisms on repaglinide pharmacokinetics (Table 5).
Table 5.
Summary of repaglinide CYP2C8 clinical pharmacogenetic–pharmacokinetic studies.
| Drug | Study description | Sample size and genotype distributions | PK results in relation to CYP2C8 genotype | Covariates | Ref. |
|---|---|---|---|---|---|
| Repaglinide | 0.25 mg single-dose PK |
CYP2C8*1/*1, n = 19 CYP2C8*1/*3, n = 6 CYP2C8*1/*4, n = 3 |
↓AUC and ↓ Cmax in *1/*3 vs *1/*1 ↓AUC in *1/*4 vs *1/*1, although not significant | None reported | [37] |
| Repaglinide | 0.25 mg single-dose PK |
CYP2C8*1/*1, n = 41 CYP2C8*1/*3, n = 10 CYP2C8*1/*4, n = 5 |
↓AUC and ↓ Cmax in *1/*3 vs *1/*1 | Linear regression analysis with inclusion of SLCO1B1, ABCB1 and CYP3A5 genotype | [38] |
| Repaglinide | 0.25 mg single-dose PK | Not described | ↓AUC in haplotype B vs noncarriers; ↓AUC in haplotype D vs noncarriers; ↑AUC in haplotype C vs noncarriers | Data were stratified for the SLCO1B1 521T>C polymorphism | [13] |
| Repaglinide | 0.25 mg single-dose PK (cross-over gemfibrozil drug interaction study) |
CYP2C8*1/*1, n = 7 CYP2C8*1/*3, n = 3 |
↑ratio of M2 and M4 to parent in *1/*3 vs *1/*1 | None reported SLCO1B1 genotype was also determined for each subject | [40] |
| Repaglinide | 2 mg single-dose PK (cross-over grapefruit juice drug interaction study) |
CYP2C8*1/*1, n = 24 CYP2C8*1/*3, n = 11 CYP2C8*3/*3, n = 1 |
No association between CYP2C8*3 and PK | None reported | [41] |
AUC: Area under the plasma concentration–time curve; Cmax: Maximum plasma concentrations; PK: Pharmacokinetics.
Niemi and colleagues set out to determine whether CYP2C8 genotype affects the pharmacokinetics of single-dose repaglinide 0.25 mg in healthy Caucasian volunteers [37]. Repaglinide mean AUC and maximium plasma concentration (Cmax) were 45 and 39% lower, respectively, in subjects with the CYP2C8*1/*3 genotype compared with wild-type homozygotes. Repaglinide AUC was also 13% lower in subjects with the CYP2C8*1/*4 genotype compared with wild-type homozygotes, although this was not statistically significant. Niemi and colleagues then went on to expand their study of repaglinide pharmacogenetics in a larger analysis of their completed or ongoing repaglinide healthy volunteer cohorts [38]. In this analysis, they evaluated the effects of polymorphisms in the CYP2C8 and CYP3A5 genes, and the drug transporter genes, ABCB1 and SLCO1B1, on the pharmacokinetics of repaglinide following a single 0.25 mg dose. Consistent with their previous data, repaglinide were significantly 48 and mean AUC and Cmax 44% lower, respectively, in subjects with the CYP2C8*1/*3 genotype compared with wild-type homozygotes. Significant predictors of repaglinide AUC in multiple regression analysis were CYP2C8*1/*3 genotype and SLCO1B1 genotype. Taken together, these two investigations were the first to suggest that the CYP2C8*3 allele is associated with increased metabolism in vivo for certain substrates, namely repaglinide.
Given the association between CYP2C8 genotype and repaglinide pharmacokinetics, these investigators went on to conduct a CYP2C8 haplotype–phenotype study of repaglinide [13]. First, CYP2C8 haplotypes were characterized in 54 Caucasian liver samples and CYP2C8 activity was assessed by measuring paclitaxel 6α-hydroxylation. Of 12 tagging polymorphisms, 7 common haplotypes were inferred (i.e., A, B, C1, C2, C3, D and E). In vitro, haplotype B was associated with increased paclitaxel metabolism, while haplotype C was associated with decreased paclitaxel metabolism. Of note, the CYP2C8*3 and CYP2C8*4 alleles were not significantly associated with paclitaxel metabolism in the liver samples. Next, the investigators studied the influence of these haplotypes on single-dose repaglinide 0.25 mg pharmacokinetics in 68 Caucasian healthy volunteers. Carriers of haplotype B had a 34% reduction in repaglinide AUC compared with noncarriers (p = 0.036), and carriers of haplotype C had a 34% increase in repaglinide AUC compared with noncarriers (p = 0.046). Haplotype D, which contains the CYP2C8*3 allele, had 50% lower repaglinide AUC compared with noncarriers (p = 0.035). Of note, these findings were observed in SLCO1B1 c.521 T/C heterozygotes, and the CYP2C8 haplotype findings did not reach statistical significance in SLCO1B1 c.521 wild-type homozygotes. In further functional analyses, the investigators characterized CYP2C8 -271C>A as the putative polymorphism causing increased activity of haplotype B. These data suggest that, in humans, haplotypes B and D are associated with increased repaglinide metabolism, while haplotype C is associated with decreased repaglinide metabolism.
CYP2C8 genotyping has been conducted in a few repaglinide drug–drug interaction studies. In a drug–drug interaction study of ciclosporin (inhibitor of organic anion-transporting polypeptide 1B1, which is encoded by the SLCO1B1 gene) and repaglinide, CYP2C8 polymorphisms did not affect the extent of the interaction or the baseline pharmacokinetics of repaglinide [39]. However, there was only one subject with the CYP2C8*1/*3 genotype and one subject with the CYP2C8*1/*4 genotype in this study. Thus definitive CYP2C8 genotype conclusions cannot be drawn due to an inadequate sample size for comparisons. Another study evaluated the interaction between gemfibrozil and repaglinide in healthy volunteers. In the study population, the ratios of M2 and M4 to parent repaglinide were highest in the 3 subjects possessing the CYP2C8*1/*3 genotype [40]. Although the number of heterozygotes was small, these data are consistent with previous findings of increased repaglinide metabolism associated with the CYP2C8*3 allele. In a drug–drug interaction study of grapefruit juice and repaglinide, the CYP2C8*3 allele was not significantly associated with repaglinide pharmacokinetics when repaglinide was administered alone [41].
Most studies have found the CYP2C8*3 allele to be associated with increased repaglinide metabolism, while one study found no association. A possible explanation for this discrepancy is that a higher dose of repaglinide (2 mg) was used in the drug–drug interaction study that observed no CYP2C8 genotype association. The authors hypothesized that the effects of the CYP2C8*3 allele on repaglinide pharmacokinetics may be relevant at lower doses (e.g., 0.25 mg used in most studies), but at higher clinically relevant doses other factors, such as CYP3A4 metabolism, may come into play diluting the CYP2C8 genotype effect. Additionally, it is well-recognized that SLCO1B1 polymorphisms are important predictors of repaglinide pharmacokinetics [38]. SLCO1B1 polymorphisms were evaluated in most, but not all, repaglinide studies. Thus, imbalances in SLCO1B1 genotypes between CYP2C8 genotype groups may have confounded the results. In future repaglinide CYP2C8 pharmacogenetic studies, consideration should be given to stratification by SLCO1B1 genotype and evaluation of CYP2C8 haplotypes should be conducted across a range of clinically applicable doses. Furthermore, the clinical implication of CYP2C8 and SLCO1B1-mediated variability in repaglinide pharmacokinetics on repaglinide pharmacodynamics merits investigation in patients with Type 2 diabetes.
HMG-CoA reductase inhibitors
The HMG-CoA reductase inhibitors, ‘statins’, are used extensively in the treatment of hyperlipidemia. CYP2C8 is thought to play a major role in the metabolism of cerivastatin and a minor role in the metabolism of fluvastatin and simvastatin acid.
Cerivastatin
Cerivastatin is a lipophilic statin that undergoes CYP2C8-mediated metabolism to the active metabolites M-1 and M-23 [42]. In vitro, gemfibrozil markedly inhibited the formation of the M-23 metabolite, and to a lesser extent the M-1 metabolite, presumably through CYP2C8 inhibition [43]. CYP3A4 contributes to a minor extent in the formation of M-1 [42]. In 2001, cerivastatin was withdrawn from the worldwide market due to a higher incidence of rhabdomyolysis compared with other statins [44,45]. Rhabdomyolysis is the consequence of excessive muscle breakdown that results in acute renal failure, cardiac complications and compression of the nerves and blood vessels [46]. Although the mechanisms of statin-induced rhabdomyolysis remain uncertain, one possible risk factor for rhabdomyolysis is elevated concentrations of statins in the plasma [46]. This may be a result of high statin doses, drug–drug interactions that increase statin plasma exposure or dysfunctional CYP metabolizing enzymes [46]. An important risk factor for cerivastatin-associated rhabdomyolysis was concomitant treatment with gemfibrozil, which increased cerivastatin concentrations several-fold through inhibition of CYP2C8-mediated metabolism [47].
The association between CYP2C8 polymorphisms and cerivastatin-associated side effects, such as rhabdomyolysis, has not been formally assessed. However, a case report identified three CYP2C8 polymorphisms, 475delA (exon 3), G874T (exon 6) and T1551C (exon 9), in a Japanese woman who developed rhabdomyolysis after 22 days of cerivastatin therapy [48]. The authors concluded that the 475delA polymorphism, which results in a frameshift and premature protein truncation, was the putative polymorphism causing decreased CYP2C8 metabolizing enzyme activity. In a subsequent analysis, the authors evaluated the cerivastatin metabolite ratios in this patient’s serum, and found the ratio of M-23:M-1 to be lower than in control patients [49]. This finding suggests that decreased metabolism of cerivastatin was evident in this patient, and this may be due to CYP2C8 gene polymorphisms, as previously discussed. While the potential associations between CYP2C8 polymorphisms and decreased cerivastatin metabolism are intriguing, the clinical relevance of the 475delA polymorphism is unclear, as homozygozity for this allele is extremely rare [48].
Fluvastatin
Fluvastatin is a racemic mixture of an active (+)-3R, 5S form and an inactive (−)-3S, 5R form. Both enantiomers are principally metabolized by CYP2C9 (75%) and to a lesser extent by CYP3A4 (~20%) and CYP2C8 (~5%) [50,51]. Given the small contribution of CYP2C8 to fluvastatin’s overall metabolism, it would not be expected that CYP2C8 polymorphisms would contribute in a major way to the disposition of the drug. Furthermore, gemfibrozil, a CYP2C8 inhibitor, was shown to have no effect on the pharmacokinetics of fluvastatin [52]. Kirchheiner and colleagues evaluated the influence of CYP2C9 and CYP2C8 polymorphisms on fluvastatin pharmacokinetics in 26 healthy German volunteers [53]. Neither CYP2C8*3, nor CYP2C9*2, significantly influenced the pharmacokinetics of fluvastatin 40 mg following 14 days of therapy. Of note, the mean AUC of the active (+)-3R, 5S enantiomer was threefold higher and the mean AUC of the inactive (−)-3S, 5R enantiomer was fourfold higher in CYP2C9*3 homozygotes (n = 3) compared with wild-type homozygotes (n = 5). In summary, although CYP2C9 polymorphisms influence the pharmacokinetics of fluvastatin, genetic variation in CYP2C8 does not contribute to a significant extent.
Other statins
Most of the other statins (e.g., atorvastatin and lovastatin) do not undergo CYP2C8-mediated metabolism [54]. In vitro data from human liver microsomes showed that CYP2C8 contributed only a minor extent to the metabolism of simvastatin acid [55]. Rosuvastatin, the newest statin, does not undergo extensive metabolism; however, approximately 10% of the parent drug is metabolized by CYP2C9 [56]. Thus, it is unlikely that the CYP2C9*2 polymorphism, or the partially linked CYP2C8*3 polymorphism, would significantly influence the disposition or response of rosuvastatin. This hypothesis is supported by drug–drug interaction data, which showed that CYP2C9 inhibition with fluconazole only modestly increased the AUC of rosuvastatin by 14% [57]. Furthermore, genome-wide studies have not identified CYP2C9 or CYP2C8 polymorphisms as key determinants of statin response. Instead, it appears that polymorphisms in other CYP enzyme genes (e.g., CYP3A5), drug-transporter genes (e.g., SLCO1B1) and drug-target genes (e.g., apolipoprotein E and HMGCR) may be more likely contributors to interpatient variability in statin response and adverse events [58–61].
NSAIDs
Drugs in the nonsteroidal anti-inflammatory class are some of the most widely prescribed and over-the-counter medications. While CYP2C9 contributes to the metabolism of most NSAIDS, recent data show that CYP2C8 polymorphisms may influence interindividual variability in the pharmacokinetics of some NSAIDs, namely ibuprofen and diclofenac [62].
Ibuprofen
Ibuprofen, a racemic mixture of S-(+) and R-(−) enantiomers, is one of the most commonly used NSAIDs. The S-enantiomer is mainly responsible for the anti-inflammatory pharmacologic activity of ibuprofen, and the R-enantiomer is capable of conversion to the S-enantiomer in vivo [63,64]. CYP2C9 contributes to the metabolism of both the S- and R-enantiomers, and CYP2C8 plays a role in the metabolism of the R-enantiomer [65]. However, recent data suggest controversy in the extent to which CYP2C8 is involved in R-ibuprofen metabolism. For example, in vitro data suggest that CYP2C8 contributes less than 10% to the clearance of R-ibuprofen [66]. However, in vivo, administration of gemfibrozil, a CYP2C8 inhibitor, increased the AUC of R-ibuprofen to a greater extent than S-ibuprofen (34 vs 7%, respectively) [67]. Studies have been conducted that examined the effects of CYP2C8 polymorphisms on the disposition of ibuprofen in humans (Table 6).
Table 6.
Summary of NSAID CYP2C8 clinical pharmacogenetic–pharmacokinetic studies.
| Drug | Study description | Sample size and genotype distributions | PK results in relation to CYP2C8 genotype | Covariates | Ref. |
|---|---|---|---|---|---|
| Ibuprofen | 400 mg single-dose PK |
CYP2C8*1/*1, n = 9 CYP2C8*1/*3, n = 10 CYP2C8*3/*3, n = 6 |
↓R-ibuprofen CL ↑ in *1/*3 and *3/*3 vs *1/*1 | CYP2C9 genotype was also determined for each subject | [68] |
| Ibuprofen | 400 mg single-dose PK |
CYP2C8*1/*1, n = 93 CYP2C8*1/*3, n = 35 CYP2C8*3/*3, n = 2 |
↓Ibuprofen CL in *1/*3 and *3/*3 vs *1/*1 ↓R- and S-ibuprofen CL with *3 allele vs *1/*1 | CYP2C9 genotype was also determined for each subject | [69] |
| Ibuprofen | 600 mg single-dose PK (cross-over bioequivalence study |
CYP2C8*1/*1, n = 43 CYP2C8*1/*2, n = 3 CYP2C8*1/*3, n = 11 CYP2C8*1/*4, n = 10 CYP2C8*3/*3, n = 2 |
↑R-ibuprofen CL in *1/*3 vs *1/*1 | CYP2C9 genotype was also determined for each subject Weight-adjusted clearance was also assessed | [70] |
| Diclofenac | 50 mg single-dose PK |
CYP2C8*1/*1, n = 71 CYP2C8*1/*3, n = 19 CYP2C8*1/*4, n = 7 CYP2C8*3/*3, n = 5 |
↑Urinary ratio of parent to metabolite in *3 and *4 carriers vs *1/*1 | None reported | [73] |
| Tenoxicam | 20 mg single-dose PK (cross-over bioequivalence study) |
CYP2C8*1/*1, n = 10 CYP2C8*1/*3, n = 5 CYP2C8*1/*4, n = 2 CYP2C8*3/*3, n = 1 |
No association between CYP2C8 polymorphisms and PK | Analysis of variance which included confounders such as age, sex, menstrual cycle phase, body weight and formulation of drug CYP2C9 genotype was also determined for all subjects | [77] |
CL: Oral clearance; PK: Pharmacokinetics.
In a healthy volunteer study, Spanish Caucasians were administered a single dose of ibuprofen 400 mg [68]. In order to minimize the potential confounding of CYP2C9 genotype on ibuprofen pharmacokinetics, subjects with the CYP2C9*3/*3 genotype were excluded from the study. R-ibuprofen clearance was significantly 1.7-fold and 1.6-fold lower in CYP2C8*3 homozygotes and heterozygotes, respectively, compared with wild-type homozygotes (p = 0.03). R-ibuprofen half-life was also significantly longer in CYP2C8*3 homozygotes (9 h) and heterozygotes (4.2 h) compared with CYP2C8*1 homozygotes (2 h, p < 0.025). These data suggest that the CYP2C8*3 genotype is associated with decreased R-ibuprofen metabolism. However, 13 of the 16 individuals who carried a CYP2C8*3 allele were also carriers of at least one copy of the CYP2C9*2 allele, therefore, it is difficult to draw definitive conclusions regarding the relative contributions of CYP2C8*3 versus CYP2C9*2 on interindividual variability in R-ibuprofen metabolism.
In another healthy volunteer study, Spanish Caucasian individuals were administered a single dose of ibuprofen 400 mg [69]. CYP2C8*3 homozygotes had approximately ninefold lower R-ibuprofen clearance and S-ibuprofen clearance than wild-type homozygotes. Of note, the CYP2C9*2 allele was associated with ibuprofen pharmacokinetics only in individuals who also carried the CYP2C8*3 allele. In regards to the enantiospecific clearance of ibuprofen, a modest genotype effect was observed, with the CYP2C8*3 allele being associated with reduced R-ibuprofen clearance, and both the CYP2C8*3 and CYP2C9*3 alleles being associated with reduced S-ibuprofen clearance. In contrast to these findings, a healthy volunteer bioequivalence study of single-dose ibuprofen 600 mg showed 24% higher clearance of R-ibuprofen in subjects with the CYP2C8*1/*3 genotype compared with wild-type homozygotes [70]. Given these conflicting findings, controversy exists regarding the role of CYP2C8 and CYP2C9 polymorphisms in ibuprofen metabolism. Potential explanations for the discrepancies between studies include differences in sample size, differences in CYP2C8 and CYP2C9 genotype combinations, linkage between the CYP2C8*3 and CYP2C9*2 polymorphisms, small number of CYP2C8*3 homozygotes evaluated in most studies, lack of control for body weight in some studies, and differences in the extent of conversion of R-ibuprofen to S-ibuprofen in vivo.
Diclofenac
Diclofenac is a phenylacetic acid NSAID that is metabolized by CYP2C9 to the major metabolite 4-hydroxydiclofenac, and by CYP2C8, CYP3A4 and CYP2C19 to the minor metabolite, 5-hydroxydiclofenac [62,71,72]. Diclofenac is also glucuronidated by UGT2B7 to diclofenac acyl glucuronide, which is subsequently metabolized by CYP2C8 to 4-hydroxydiclofenac acyl glucuronide [62,71,72]. Dorado and colleagues sought to determine the impact of CYP2C8 polymorphisms on diclofenac metabolism, with a particular focus on the 5-hydroxylation pathway [73]. The investigators analyzed data from healthy Spanish volunteers who had previously participated in a diclofenac CYP2C9 pharmacogenetic study. The subjects had received a single dose of enteric-coated diclofenac 50 mg followed by an 8 h urine collection for evaluation of diclofenac and its metabolites. In this analysis, the study population was genotyped for the CYP2C8*2, *3 and *4 alleles. The mean urinary ratio of diclofenac:5-hydroxydiclofenac was significantly higher in carriers of the CYP2C8*3 or CYP2C8*4 alleles compared with wild-type homozygotes. However, there was substantial overlap in the urinary ratios between genotype groups and plasma concentrations of diclofenac and its metabolites were not reported. As such, definitive conclusions regarding the impact of CYP2C8 polymorphisms on diclofenac metabolism cannot be drawn from this study. Considering that only 65% of diclofenac and its metabolites are renally eliminated, with biliary excretion accounting for the other 35%, it is likely that other polymorphisms, or non-genetic factors, may contribute to diclofenac metabolism and excretion [74].
Diclofenac has been associated with rare, but serious, hepatotoxicity [75]. It is hypothesized that the 5-hydroxydiclofenac metabolic pathway or the UGT2B7 glucuronidation pathway may play a role in diclofenac adduct formation, which may subsequently contribute to hepatotoxicity [72]. Aithal and colleagues previously demonstrated that the CYP2C9 genotype was not associated with the risk of diclofenac-induced hepatitis [76]. The investigators then went on to assess the role of CYP2C8 polymorphisms on the risk of diclofenac hepatotoxicity [72]. The study included 24 case patients who had experienced diclofenac hepatotoxicity, and 160 control patients (i.e., those who had received diclofenac but who had not developed hepatotoxicity). This control group was comprised of 48 patients from a hospital setting and 112 patients from the community setting. All patients were genotyped for the CYP2C8*3 and *4 alleles and haplotypes were assigned. The overall frequency of CYP2C8 haplotypes differed significantly between cases and hospital controls (p = 0.04). Case patients who carried the CYP2C8*4 allele had a non-significant increased odds of hepatotoxicity compared with the hospital control group. In contrast, genes involved in the metabolism and biliary excretion (i.e., UGT2B7 and ABCC2, respectively) of diclofenac were associated more strongly with diclofenac hepatotoxicity. Thus, it appears that CYP2C8 polymorphisms are not the strongest predictors of diclofenac hepatic adverse events in patients.
Tenoxicam
Tenoxicam is an NSAID that is available in Europe and Canada. It is extensively metabolized by CYP2C9 and possibly to a lesser extent by CYP2C8 [62,77]. Peiro and colleagues sought to determine if polymorphic CYP2C9 and CYP2C8 alleles influence variability in tenoxicam pharmacokinetics [77]. In this cohort of Spanish healthy volunteers, CYP2C8 polymorphisms (*2, *3 and *4) were not significantly associated with the pharmacokinetics of a single 20 mg dose of tenoxicam. However, significant increases in tenoxicam AUC and elimination half-life were observed for CYP2C9*3 carriers compared with noncarriers. Although the sample size of this study was small, these data suggest that CYP2C9, rather than CYP2C8, is the principal determinant of tenoxicam pharmacokinetics in humans.
Effects of genotype on NSAID-related bleeding
Studies have also been conducted evaluating the association between CYP2C8 genotype and NSAID-induced adverse effects (e.g., gastrointestinal bleeding) and NSAID-induced protective effects (e.g., decreased colorectal cancer risk). Acute gastrointestinal bleeding is one of the most common adverse effects of NSAID therapy. Previously, CYP2C9 genetic variation, namely CYP2C9*2, was associated with an increased risk of gastrointestinal bleeding in patients receiving NSAID therapy [78]. Given that CYP2C9*2 is in partial linkage disequilibrium with CYP2C8*3, and many NSAIDs are substrates for both enzymes, Blanco and colleagues set out to determine whether CYP2C9 and CYP2C8 polymorphisms were associated with gastrointestinal bleeding in a cross- sectional study of NSAID users (n = 134 bleeding cases, n = 177 nonbleeding controls) [79]. The use of a number of different NSAIDs was observed in this study, some of which have not been documented to be CYP2C8 substrates. Nonetheless, the frequencies of the CYP2C8*3 and CYP2C9*2 alleles were higher in NSAID users who experienced a bleed versus those that did not experience a bleed (CYP2C8*3, odds ratio: 3.4, p < 0.002; CYP2C9*2, odds ratio: 2.7, p = 0.013). In multivariable regression analysis, significant predictors of NSAID-induced bleeding were CYP2C8*3 genotype and drinking habits. Further analysis of the data revealed that the highest bleeding risk was in patients who possessed both the variant CYP2C8*3 and CYP2C9*2 alleles. The authors hypothesized that these polymorphisms conferred an increased risk of gastrointestinal bleeding due to decreased metabolic clearance, and increased plasma concentrations of NSAIDs. Given the limitations of this study (e.g., case–control design, small sample size, multiple NSAIDs included – some of which are metabolized by multiple CYP isoforms, small number of CYP2C8*3 homozygotes, and lack of pharmacokinetic correlates), these findings merit replication in additional populations. In regards to protective effects, NSAIDs (i.e., aspirin and ibuprofen) have been associated with a reduction in the risk of colorectal cancer. McGreavey and colleagues hypothesized that individuals who possessed variant CYP2C8 or CYP2C9 alleles would have decreased NSAID metabolism, which may lead to an enhanced protective effect from these agents [80]. They evaluated this hypothesis in a case–control study of 478 patients with colorectal cancer and 733 controls. NSAID use was associated with a significant reduction in the risk of colorectal cancer. However, the CYP2C8*3, CYP2C9*2 and CYP2C9*3 alleles did not modify the protective effects of NSAIDs in this population.
In summary, available data suggest that CYP2C8 polymorphisms may contribute to interindividual variability in the pharmacokinetics and risk of adverse events of certain NSAIDs, namely ibuprofen and possibly diclofenac. However, given the conflicting genetic associations observed in these studies, particularly for ibuprofen, definitive conclusions cannot be made at this time. Futhermore, future NSAID CYP2C8 pharmacogenetic studies should include well-characterized groups in regards to the CYP2C8 and CYP2C9 alleles. Additionally, studies are needed to determine if the observed decreased metabolism of ibuprofen and diclofenac in carriers of polymorphic CYP2C8 alleles is associated with differences in clinical response to these agents. Lastly, while many NSAIDs are known to be CYP2C9 substrates, the contribution of CYP2C8 metabolism has not been elucidated for many agents (e.g., naproxen, piroxicam and celecoxib) [62]. Given the high sequence homology between the two enzymes, it is possible that CYP2C8 contributes to the metabolism of some of these substrates. Additional in vitro studies are needed to assess the contribution of CYP2C8 versus CYP2C9 to the metabolism of clinically available NSAIDs.
Cancer drugs
Paclitaxel is a chemotherapeutic agent that is used in the treatment of solid tumors, particularly breast, lung and ovarian cancer. CYP2C8 is the principal enzyme that catalyzes the formation of the major metabolite, 6α-hydroxypaclitaxel [81]. CYP3A4 and CYP3A5 have also been shown to contribute to the metabolism of paclitaxel to a lesser extent [82]. Given paclitaxel’s affinity for CYP2C8, it has been used as the primary probe in studies examining the effects of CYP2C8 polymorphisms on substrate disposition in vitro. These studies have demonstrated a significant reduction in paclitaxel metabolism associated with the CYP2C8*3 allele, and a modest reduction in paclitaxel metabolism with the CYP2C8*4 allele [12,16,17]. It is well recognized that a high degree of inter-individual variability exists in paclitaxel clearance [83]. Consequently, a number of clinical pharmacogenomic studies have been conducted to determine if CYP2C8 polymorphisms, namely CYP2C8*3, explain interindividual variability in paclitaxel disposition in cancer patients (Table 7).
Table 7.
Summary of paclitaxel CYP2C8 clinical pharmacogenetic–pharmacokinetic studies
| Drug | Study description | Sample size and genotype distributions | PK results in relation to CYP2C8 genotype | Covariates | Ref. |
|---|---|---|---|---|---|
| Paclitaxel | 80–225 mg/m2, breast, ovarian, esophageal cancer (n = 97) |
CYP2C8*1/*2, n = 1 CYP2C8*1/*3, n = 11 CYP2C8*1/*4, n = 4 CYP2C8*3/*3, n = 3 |
No association between CYP2C8 genotype and PK | Population pharmacokinetic analysis was performed Additional genetic variables were CYP3A4*3, CYP3A5*3C and ABCB1 3435C>T | [84] |
| Paclitaxel | 575–775 mg/m2, breast cancer (n = 93) |
CYP2C8*1/*3, n = 11 CYP2C8*1/*4, n = 6 CYP2C8*3/*3, n = 1 CYP2C8*4/*4, n = 1 |
No association between CYP2C8 genotype and PK | Additional polymorphisms evaluated were ABCB1 1236C>T, ABCB1 2677G>T/A; ABCG2 421C>A; CYP1B1*3; CYP3A4*1B; and CYP3A5*3C | [85] |
| Paclitaxel | 175–210 mg/m2, lung, thymic, breast cancer (n = 199) |
CYP2C8*IA, 2.1% CYP2C8*IB, 10.3% CYP2C8*ID, 38.1% CYP2C8*IE, 43.5% CYP2C8*IG, 3% CYP2C8*IJ, 1.3% |
↑3-p-hydroxypaclitaxel AUC in haplotype *IG group carriers vs noncarriers | CYP3A4 genotypes were also determined | [14] |
| Paclitaxel | 135–175 mg/m2, ovarian cancer (n = 38) |
CYP2C8*1/*3, n = 6 CYP2C8*1/*4, n = 4 |
↓CL in patients with the CYP2C8*1/*3 and ABCB1 2677G/T vs CYP2C8 wild-type homozygotes and ABCB1 2677G/T | Data were stratified according to the ABCB1 2677G>T/A polymorphism. The ABCB1 1236C>T, ABCB1 3435C>T, and CYP3A4*1B polymorphisms were also evaluated | [90] |
Three other groups set out to determine the influence of polymorphic CYP2C8 alleles on paclitaxel disposition and outcomes in Japanese cancer patients. However, no polymorphic CYP2C8 alleles were found in their respective populations [87–89].
AUC: Area under the plasma concentration–time curve; CL: Clearance; PK: Pharmacokinetics.
Henningsson and colleagues considered the influence of CYP2C8 polymorphisms on high-dose paclitaxel pharmacokinetics in a retrospective study of 97 Caucasian patients with breast, ovarian or esophageal cancer [84]. The CYP2C8*3 allele was not significantly associated with paclitaxel clearance. Additional genetic analysis revealed that polymorphisms in CYP3A4, CYP3A5 and the ABCB1 drug transporter gene were also not associated with paclitaxel pharmacokinetics. In another study, Marsh and colleagues evaluated the association between CYP2C8 polymorphisms and paclitaxel pharmacokinetics and progression-free survival in 93 patients with breast cancer [85]. CYP2C8*3 and CYP2C8*4 alleles were not significantly associated with paclitaxel clearance or treatment outcomes. Additionally, polymorphisms in the CYP3A4, CYP3A5, CYP1B1, ABCB1 and ABCG2 genes were not significantly associated with paclitaxel clearance. Marsh and colleagues then went on to study the influence of 27 polymorphisms in 16 candidate genes on paclitaxel and docetaxel (both in combination with carboplatin) treatment outcomes and toxicity in a retrospective analysis of 914 patients in the Scottish Randomized Trial in Ovarian Cancer [86]. CYP2C8 polymorphisms were not significantly associated with paclitaxel or docetaxel progression-free survival or toxicity in this retrospective analysis. Other groups set out to determine the influence of polymorphic CYP2C8 alleles on paclitaxel disposition and outcomes in Japanese cancer patients. However, no polymorphic CYP2C8*2 or *3 alleles were found in their respective populations [87–89]. Thus, these polymorphisms are unlikely to be the causative factors to explain interindividual variability in paclitaxel pharmacokinetics or response in Japanese patients.
Along these lines, Saito and colleagues set out to determine CYP2C8 haplotype structure in a Japanese population (n = 437), and to determine the association between CYP2C8 haplotypes and paclitaxel pharmacokinetics in 199 Japanese cancer patients [14]. The investigators found nonsynonymous CYP2C8 polymorphisms to be rare, and focused their analysis on 40 haplotypes that contained only synonymous polymorphisms. Haplotypes were classified into six groups (*IA, *IB, *ID, *IE, *IG and *IJ). The pharmacokinetics of paclitaxel and paclitaxel metabolites (major, 6α-hydroxypaclitaxel; minor, 3′-p-hydroxypaclitaxel and 3′-p-dihydroxypaclitaxel) were then compared between haplotype groups. The 3-p-hydroxypaclitaxel median AUC was 2.5-fold higher, and the 3-p-hydroxypaclitaxel to paclitaxel AUC ratio was 64% higher, in carriers of the *IG group haplotype compared with noncarriers of the *IG group haplotype. CYP3A4 is the primary enzyme responsible for the metabolism of paclitaxel to 3-p-hydroxypaclitaxel, and CYP2C8 is responsible for the subsequent conversion of 3-p-hydroxypaclitaxel to 3′-p-dihydroxypaclitaxel. The investigators reported that the observed pharmacokinetic differences of 3′-p-hydroxypaclitaxel were not due to differences in polymorphic CYP3A4 alleles between the CYP2C8 diplotype groups. The authors hypothesized that the *IG group haplotype, which is comprised of intronic polymorphisms (mainly IVS3–21T>A and IVS4+151G>A), confers reduced CYP2C8 activity. However, no significant associations were observed between the CYP2C8 *IG group haplotype and paclitaxel or 6-hydroxypaclitaxel pharmacokinetics. It is unclear why the *IG haplotype was not associated with 6-hydroxypaclitaxel pharmacokinetics given that CYP2C8 is the principal enzyme responsible for the formation of this metabolite. Nonetheless, the role of the CYP2C8*IG haplotype in paclitaxel pharmacokinetics, clinical response and toxicity merits further analysis in additional studies, particularly in Asian populations.
More recently, Green and colleagues evaluated the role of CYP2C8, CYP3A4 and ABCB1 polymorphisms on paclitaxel pharmacokinetics and toxicity in 38 Caucasian ovarian cancer patients [90]. CYP2C8*3 heterozygotes (n = 2) had a 55% lower clearance than CYP2C8 wild-type homozygotes, but these results were observed only in patients who were ABCB1 2677G/T heterozygotes. In multivariable regression analysis, both CYP2C8*3 and ABCB1 2677G/T were modest predictors of paclitaxel clearance (p = 0.076 for both factors). In terms of toxicity, patients with the CYP2C8*1/*3 genotype had a higher risk of motor neuropathy and hematological toxicities.
When evaluated together, most studies did not find an association between the CYP2C8*3 allele and paclitaxel pharmacokinetics. Discrepancies between the data could be due to inadequate study sample sizes to assess these genetic effects, different paclitaxel doses and infusion times, and the infrequency of the CYP2C8*3 allele in Asian patients. Furthermore, paclitaxel is a substrate for P-glycoprotein; therefore, differences in the frequencies of polymorphic ABCB1 alleles between CYP2C8 genotype groups may have potentially confounded the results between studies. Recent in vitro data suggest that common CYP2C8 haplotypes are associated with altered paclitaxel 6α-hydroxylation [13]. Specifically, haplotype B was associated with increased paclitaxel metabolism, while haplotype C was associated with decreased paclitaxel metabolism in vitro. Whether these haplotypes explain inter-individual variability in paclitaxel pharmacokinetics in humans has yet to be determined. In the future, pharmacokinetic studies that include patient groups that are well-characterized for pertinent population-specific CYP2C8 and ABCB1 haplotypes may help elucidate the role of genetic polymorphisms on interindividual variability in paclitaxel pharmacokinetics and clinical response.
Antimalarial drugs
Amodiaquine
Amodiaquine is an antimalarial drug that is used as monotherapy, or in combination therapy, in many countries, particularly Africa. Amodiaquine undergoes CYP2C8-mediated metabolism to N-desethylamodiaquine, which has a longer half-life than the parent drug (9–18 days versus 5.2 h) [91]. Other enzymes that have been suggested to contribute a small extent to amodiaquine metabolism include CYP3A4 and extrahepatic CYP1A1 and CYP2B1 [91,92]. One study evaluated the effects of CYP2C8 polymorphisms on the efficacy and toxicity of amodiaquine monotherapy in a group of patients (n = 275) infected with malaria in West Africa [93]. The CYP2C8*2 allele was common in this population (11.5%) but was not associated with malaria treatment outcomes compared with wild-type homozygotes. In terms of adverse events, an increased incidence of self-reported abdominal pain was observed in CYP2C8*2 carriers compared with wild-type homozygotes (52 vs 30%, respectively). In this same report, amodiaquine metabolism was evaluated in recombinant CYP2C8 proteins and showed threefold and sixfold lower intrinsic clearance higher Km in CYP2C8*2 compared with CYP2C8*1. The CYP2C8*3 enzyme also demonstrated decreased activity compared with CYP2C8*1. Another study evaluated the pharmacokinetics of amodiaquine, in combination with artesunate, in 103 Ghanian children [94]. In this population pharmacokinetic analysis, the CYP2C8*2 allele was associated with lower plasma concentrations of N-desethylamodiaquine compared with wild-type homozygotes; however, this difference was not statistically significant. CYP2C8 genotype was not associated with efficacy or toxicity in this study. Together, these two clinical studies suggest that CYP2C8*2 is not a major determinant of amodiaquine efficacy or toxicity in adults or children. This is hypothesized to be due to the fact that both the parent drug and metabolite are pharmacologically active. In vitro data suggest that the CYP2C8*3 allele is associated with marked reductions in amodiaquine metabolism [93]. Given that CYP2C8*3 is more prevalent in Caucasians, and CYP2C8*2 is more prevalent in Africans, the in vitro finding of marked reduction in amodiaquine metabolism with the CYP2C8*3 allele is most relevant for Caucasian populations. Additional studies are needed to more comprehensively elucidate the effects of CYP2C8*2 and CYP2C8*3 alleles on pharmacokinetics, efficacy and toxicity of amodiaquine in both African and Caucasian populations.
Chloroquine
Chloroquine is another agent used in the prevention and treatment of malaria. Chloroquine is metabolized to desethylchloroquine and bisdesethylchloroquine, both of which are pharmacologically active metabolites. In vitro, CYP2C8, CYP3A4 and CYP3A5 were significant contributors to chloroquine metabolism [95]. The influence of CYP2C8 polymorphisms on chloroquine pharmacokinetics and pharmacodynamics in humans has yet to be determined.
Other CYP2C8 substrates
A diverse number of other drugs have been implicated as CYP2C8 substrates through in vitro studies. However, to our knowledge, clinical studies evaluating the effects of polymorphic CYP2C8 alleles on drug disposition or response of these agents have not been published. Brief descriptions of these drugs and relevant available in vitro CYP2C8 pharmacogenetic data are provided below.
All-trans-retinoic acid
All-trans-retinoic acid (ATRA), or tretinoin, is the acid form of vitamin A and is used in the treatment of acute promyelocytic leukemia. It is also used topically in the treatment of acne. In vitro data suggest that CYP2C8 is a major contributor to the formation of 4-hydroxy-ATRA, which is the first step in the termination of ATRA action. Significant variability exists in the pharmacokinetics of ATRA in cancer patients [96]. To our knowledge, the extent to which CYP2C8 polymorphisms influence ATRA metabolism and therapeutic efficacy or toxicity has yet to be determined in clinical studies.
Loperamide
Loperamide is a μ-opioid receptor agonist that is used in the treatment of diarrhea. In human liver microsomes, loperamide was shown to be metabolized by CYP2C8, CYP3A4, CYP2B6 and CYP2D6 [97]. In this study, CYP2C8 and CYP3A4 played the most important roles in loperamide N-demethylation [97]. In a healthy volunteer drug–drug interaction study, gemfibrozil increased loperamide AUC and Cmax 2.2-fold and 1.6-fold, respectively [98]. To our knowledge, the impact of CYP2C8 polymorphisms on loperamide pharmacokinetic has not yet been published.
Methadone
Methadone is an opioid receptor antagonist that is used in the treatment of chronic pain and opioid dependence. In vitro, CYP2C8 was shown to metabolize both the R- and S-enantiomers of methadone, with a greater effect on the R-enantiomer [99]. Considering that the R-enantiomer is the more pharmacologically active form in vivo, the potential influence of CYP2C8 polymorphisms on the metabolism of the R-enantiomer may be clinically significant and warrants further study.
Morphine
Morphine is an opioid analgesic that is glucuronidated to morphine-3-glucuronide and morphine-6-glucuronide. In addition, morphine undergoes N-demethylation to normorphine. In human liver microsomes, CYP3A4 was found to contribute a major extent, and CYP2C8 a minor extent, to normorphine formation [100]. Given that the N-demethylation pathway is a minor contributor to morphine metabolism, and CYP2C8 contributes only a small extent, it is unlikely that polymorphisms in CYP2C8 would explain a significant amount of the variability in morphine pharmacokinetics or pharmacodynamics.
Amiodarone
Amiodarone is an antiarrhythmic agent used in the treatment of supraventricular and ventricular arrhythmias. Amiodarone is metabolized by CYP2C8 to desethylamiodarone, which is a pharmacologically active metabolite [101]. An in vitro study examining the effects of the CYP2C8*1, CYP2C8*3 and CYP2C8 P404A alleles on amiodarone metabolism was carried out in Hep G2 cells [102]. CYP2C8*3 was not associated with a reduction in amiodarone metabolism in this study. Instead, the study found that CYP2C8 404A was associated with 48% lower amiodarone intrinsic clearance compared with the wild-type allele. This was hypothesized to be due to decreased expression of the protein compared with wild-type. The implications of this finding on the pharmacokinetics and pharmacodynamics of amiodarone in patients remains to be determined. However, considering that the 404A allele has only been identified at low frequency in Japanese individuals, it is unlikely that it will have broad clinical effects in patients treated with amiodarone [17].
Zopiclone
Zopiclone is a nonbenzodiazepine sedative hypnotic that is used in the treatment of insomnia. Zopiclone is extensively hepatically metabolized to N-oxide-zopiclone, which has low pharmacologic activity, and N-desmethyl-zopiclone, which is pharmacologically inactive. An in vitro study showed that CYP3A4 was the major enzyme involved in the metabolism of zopiclone to both metabolites, and that CYP2C8 contributed to the metabolism of N-desmethyl-zopiclone [103]. However, in a healthy volunteer drug–drug interaction study, gemfibrozil did not significantly increase the plasma concentrations of zopiclone [104]. Thus, in vivo, it appears that CYP2C8 contributes a minor extent to zopiclone metabolism and CYP2C8 polymorphisms are unlikely to have clinically significant effects on zopiclone pharmacokinetics.
Tazarotenic acid
Tazarotenic acid (tazarotene) is used in the topical treatment of psoriasis and acne. It is a retinoid prodrug that is converted to its active metabolite tazarotenic acid, which is subsequently converted to an inactive sulfoxide metabolite. In human liver microsomes, the metabolism of tazarotenic acid was mediated, in part by CYP2C8 [105]. Topical administration results in low plasma concentrations of tazarotenic acid. Thus it is unlikely that CYP2C8 polymorphisms would contribute a significant extent to variability in the pharmacokinetics of tazarotenic acid when it is used in topical administration [106]. However, CYP2C8 polymorphisms may be relevant to the pharmacokinetics of an oral formulation of tazarotenic acid, which is under development for the treatment of psoriasis.
Verapamil
Verapamil is an L-type calcium channel blocker used in the treatment of cardiovascular conditions such as hypertension, angina and arrhythmias. Verapamil is administered as a racemic mixture of R- and S-enantiomers, with the S-enantiomer being the more pharmacologically active form [107]. Additionally, the S-enantiomer undergoes a quicker rate of hepatic first pass metabolism than the R-enantiomer [108]. In vitro data have demonstrated that CYP2C8 contributes to the metabolism of both R-verapamil and S-verapamil, and further contributes to the metabolism of norverpamil, one of the primary metabolites [109]. The extent to which CYP2C8 polymorphisms influence the pharmacokinetics and pharmacodynamics of verapamil in humans has not been assessed.
Other agents
In vitro studies have also demonstrated that CYP2C8 has a minor role in the metabolism of other substrates such as: diltiazem, carbamazepine, cyclophosphamide, dapsone, ifosfamide and torsemide [110–114]. To our knowledge the influence of CYP2C8 polymorphisms has not been studied for these drugs. However, given that CYP2C8 plays only a minor role in the metabolism of these substrates, it is unlikely that CYP2C8 polymorphisms would have clinically relevant effects for these particular drugs.
Conclusion
CYP2C8 has emerged as an important enzyme that is responsible for the metabolism of a diverse group of substrates in humans. Available data suggest that genetic variation in CYP2C8 alters the in vivo disposition of many commonly used drugs. In clinical studies, the polymorphic CYP2C8*3 allele is associated with increased metabolism of thiazolidinediones and repaglinide. By contrast, all but one study showed no effect of the CYP2C8*3 allele on paclitaxel pharmacokinetics. At this time, the data regarding the impact of the CYP2C8*3 allele on ibuprofen metabolism are conflicting and inconclusive. The CYP2C8*2 allele, which is common in Africans, has not been associated with altered pharmacokinetics of the substrate amodiaquine in humans. Unfortunately, data regarding the effects of CYP2C8*4 on substrate pharmacokinetics are unclear, as too few subjects with this variant genotype were included in clinical studies. More recently, novel CYP2C8 haplotypes in Caucasians and Japanese patients have been identified, and these have been associated with altered pharmacokinetics of repaglinide and paclitaxel.
Future perspective
While the CYP2C8 clinical pharmacogenetic data are intriguing, there are several issues that merit consideration in future clinical studies. First, future CYP2C8 pharmacogenetic–pharmacokinetic studies should conduct a priori sample size calculations based on the differences that would like to be detected between genotype groups. In turn, subjects should be prospectively stratified and enrolled based on these CYP2C8 genotype numbers. This will ensure suitable power to detect potential pharmacokinetic differences between genotype groups. While some investigators have done this, in most situations, this has not been the case. This is likely due to the fact that prospectively-stratified genotype panel studies are difficult to conduct because it is often challenging to find a sufficient number of subjects who possess CYP2C8 homozygous variant genotypes. Second, population-specific studies are needed to assess the effects of certain CYP2C8 alleles on substrate pharmacokinetics. A good example of this is CYP2C8*2. To date, few studies, with the exception of amodiaquine, have attempted to elucidate the role of this polymorphism on substrate pharmacokinetics in individuals of African descent. Along these lines, the enrollment of patients with the CYP2C8*4 allele has been too few in most studies to make meaningful conclusions regarding the role of this polymorphism on substrate pharmacokinetics in humans. Third, future studies should consider characterizing CYP2C8 haplotypes rather than focusing on single polymorphisms, as has been the case in most studies to date. Fourth, for drugs that are metabolized by both CYP2C8 and CYP2C9, polymorphisms within both of these genes should be evaluated in order to provide a more comprehensive assessment of their relative contribution to drug disposition and response. Fifth, human CYP2C8 is transcriptionally regulated by nuclear receptors. The extent to which polymorphisms in nuclear receptor genes influence the downstream transcriptional regulation of CYP2C8 has not been assessed. Future clinical studies examining polymorphisms in these regulator genes and their association with CYP2C8 substrate pharmacokinetics are needed. In addition, many of the CYP2C8 substrates discussed in this review are also substrates for drug transporters. In many cases, drug-transporter polymorphisms influence the pharmacokinetics of the substrate (e.g., SLCO1B1–repaglinde; ABCB1–paclitaxel). As such, future pharmacokinetic studies should carefully consider the confounding influence of these drug transporter polymorphisms when evaluating CYP2C8 genetics. In addition, future analysis of studies should include important nongenetic covariates, such as body weight, to improve the statistical power of genotype comparisons. Lastly, for drugs in which the pharmacokinetics are affected by CYP2C8 genotype (e.g., repaglinide, rosiglitazone and pioglitazone), pharmacodynamic studies are needed in relevant patient populations to determine if CYP2C8-mediated differences in pharmacokinetics translate into meaningful inter-individual differences in pharmacodynamics.
Executive summary
CYP2C8 enzyme
CYP2C8 plays an important role in the metabolism of a diverse number of exogenous and endogenous compounds.
CYP2C8 gene & polymorphisms
Common nonsynonymous polymorphisms exist in the CYP2C8 gene and the CYP2C8*3 and CYP2C8*2 polymorphisms have been studied most often in clinical investigations.
The frequency of polymorphic CYP2C8 alleles differs by race and ethnicity. CYP2C8*2 is common in Blacks, but is rare in Caucasians and Asians. In contrast, CYP2C8*3 is common in Caucasians, but is rare in Blacks or Asians.
In vitro, the CYP2C8*2 and *3 alleles have been associated with decreased metabolism of paclitaxel and arachidonic acid.
Clinical data suggest that the CYP2C8*3 allele is associated with increased metabolism of rosiglitazone, pioglitazone and repaglinide; however, for other substrates (e.g., ibuprofen and paclitaxel), the data regarding the association between CYP2C8 polymorphisms and substrate pharmacokinetics are conflicting.
Antidiabetic agents
In clinical studies, the CYP2C8*3 allele is associated with increased metabolism, and decreased plasma concentrations, of the oral antidiabetic drugs rosiglitazone, pioglitazone and repaglinide.
HMG-CoA reductase inhibitors
CYP2C8 plays a major role in the metabolism of cerivastatin, and a minor role in the metabolism of fluvastatin and simvastatin acid.
To date, data suggest that CYP2C8 polymorphisms do not influence fluvastatin pharmacokinetics in healthy volunteers.
NSAIDs
The CYP2C8*3 allele has been associated with decreased ibuprofen metabolism in two studies and increased ibuprofen metabolism in one study. Thus, the data regarding the influence of the CYP2C8*3 allele on the metabolism of ibuprofen are inconclusive at this time.
One pharmacokinetic study and one case–control study suggest CYP2C8 polymorphisms may influence diclofenac metabolism and risk of hepatotoxicity, respectively. However, these data merit replication in additional populations.
Cancer drugs
Paclitaxel is an in vitro probe drug for CYP2C8 activity; however, most clinical studies have reported no association between CYP2C8 polymorphisms and paclitaxel pharmacokinetics or pharmacodynamics in cancer patients.
Antimalarial drugs
The CYP2C8*2 allele is not a significant predictor of the pharmacokinetics or clinical response of the antimalarial drug, amodiaquine, in African patients.
Footnotes
For reprint orders, please contact: reprints@futuremedicine.com
Financial & competing interests disclosure
Dr Aquilante currently holds investigator-initiated research grants from the NIH (K23 DK073197), American College of Clinical Pharmacy and Tibotec Therapeutics. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Bibliography
Papers of special note have been highlighted as:
▪ of interest
▪▪ of considerable interest
- 1 ▪▪.Totah RA, Rettie AE. Cytochrome P450 2C8: substrates, inhibitors, pharmacogenetics, and clinical relevance. Clin Pharmacol Ther. 2005;77:341–352. doi: 10.1016/j.clpt.2004.12.267. Comprehensive review of CYP2C8 substrates, inhibitors and pharmacogenetics. [DOI] [PubMed] [Google Scholar]
- 2.Evans WE, Relling MV. Moving towards individualized medicine with pharmacogenomics. Nature. 2004;429:464–468. doi: 10.1038/nature02626. [DOI] [PubMed] [Google Scholar]
- 3.Klose TS, Blaisdell JA, Goldstein JA. Gene structure of CYP2C8 and extrahepatic distribution of the human CYP2Cs. J Biochem Mol Toxicol. 1999;13:289–295. doi: 10.1002/(sici)1099-0461(1999)13:6<289::aid-jbt1>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 4.Delozier TC, Kissling GE, Coulter SJ, et al. Detection of human CYP2C8, CYP2C9, and CYP2J2 in cardiovascular tissues. Drug Metab Dispos. 2007;35:682–688. doi: 10.1124/dmd.106.012823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schoch GA, Yano JK, Wester MR, Griffin KJ, Stout CD, Johnson EF. Structure of human microsomal cytochrome P450 2C8. Evidence for a peripheral fatty acid binding site. J Biol Chem. 2004;279:9497–9503. doi: 10.1074/jbc.M312516200. [DOI] [PubMed] [Google Scholar]
- 6.Schoch GA, Yano JK, Sansen S, Dansette PM, Stout CD, Johnson EF. Determinants of cytochrome P450 2C8 substrate binding: structures of complexes with montelukast, troglitazone, felodipine, and 9-cis-retinoic acid. J Biol Chem. 2008;283:17227–17237. doi: 10.1074/jbc.M802180200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7 ▪.Theken KN, Lee CR. Genetic variation in the cytochrome P450 epoxygenase pathway and cardiovascular disease risk. Pharmacogenomics. 2007;8:1369–1383. doi: 10.2217/14622416.8.10.1369. Comprehensive review of the influence of CYP2C8 polymorphisms on arachidonic acid metabolism and clinical outcomes. [DOI] [PubMed] [Google Scholar]
- 8.Yasar U, Lundgren S, Eliasson E, et al. Linkage between the CYP2C8 and CYP2C9 genetic polymorphisms. Biochem Biophys Res Commun. 2002;299:25–28. doi: 10.1016/s0006-291x(02)02592-5. [DOI] [PubMed] [Google Scholar]
- 9.Johnson EF, Stout CD. Structural diversity of human xenobiotic-metabolizing cytochrome P450 monooxygenases. Biochem Biophys Res Commun. 2005;338:331–336. doi: 10.1016/j.bbrc.2005.08.190. [DOI] [PubMed] [Google Scholar]
- 10.Ferguson SS, Chen Y, LeCluyse EL, Negishi M, Goldstein JA. Human CYP2C8 is transcriptionally regulated by the nuclear receptors constitutive androstane receptor, pregnane X receptor, glucocorticoid receptor, and hepatic nuclear factor 4α. Mol Pharmacol. 2005;68:747–757. doi: 10.1124/mol.105.013169. [DOI] [PubMed] [Google Scholar]
- 11.Kojima K, Nagata K, Matsubara T, Yamazoe Y. Broad but distinct role of pregnane X receptor on the expression of individual cytochrome p450s in human hepatocytes. Drug Metab Pharmacokinet. 2007;22:276–286. doi: 10.2133/dmpk.22.276. [DOI] [PubMed] [Google Scholar]
- 12.Bahadur N, Leathart JB, Mutch E, et al. CYP2C8 polymorphisms in Caucasians and their relationship with paclitaxel 6α-hydroxylase activity in human liver microsomes. Biochem Pharmacol. 2002;64:1579–1589. doi: 10.1016/s0006-2952(02)01354-0. [DOI] [PubMed] [Google Scholar]
- 13 ▪.Rodriguez-Antona C, Niemi M, Backman JT, et al. Characterization of novel CYP2C8 haplotypes and their contribution to paclitaxel and repaglinide metabolism. Pharmacogenomics J. 2008;8:268–277. doi: 10.1038/sj.tpj.6500482. Important study that characterized CYP2C8 haplotypes and their functional significance in Caucasian individuals. [DOI] [PubMed] [Google Scholar]
- 14 ▪.Saito Y, Katori N, Soyama A, et al. CYP2C8 haplotype structures and their influence on pharmacokinetics of paclitaxel in a Japanese population. Pharmacogenet. Genomics. 2007;17:461–471. doi: 10.1097/FPC.0b013e32805b72c1. Important study that characterized CYP2C8 haplotypes and their association with paclitaxel metabolism in Japanese cancer patients. [DOI] [PubMed] [Google Scholar]
- 15 ▪.Speed WC, Kang SP, Tuck DP, Harris LN, Kidd KK. Global variation in CYP2C8–CYP2C9 functional haplotypes. Pharmacogenomics J. 2009;9(4):283–290. doi: 10.1038/tpj.2009.10. Characterized CYP2C8 and CYP2C9 haplotypes in a large population of individuals from around the world. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dai D, Zeldin DC, Blaisdell JA, et al. Polymorphisms in human CYP2C8 decrease metabolism of the anticancer drug paclitaxel and arachidonic acid. Pharmacogenetics. 2001;11:597–607. doi: 10.1097/00008571-200110000-00006. [DOI] [PubMed] [Google Scholar]
- 17.Soyama A, Saito Y, Hanioka N, et al. Non-synonymous single nucleotide alterations found in the CYP2C8 gene result in reduced in vitro paclitaxel metabolism. Biol Pharm Bull. 2001;24:1427–1430. doi: 10.1248/bpb.24.1427. [DOI] [PubMed] [Google Scholar]
- 18.Hichiya H, Tanaka-Kagawa T, Soyama A, et al. Functional characterization of five novel CYP2C8 variants, G171S, R186X, R186G, K247R, and K383N, found in a Japanese population. Drug Metab Dispos. 2005;33:630–636. doi: 10.1124/dmd.105.003830. [DOI] [PubMed] [Google Scholar]
- 19.Yki-Jarvinen H. Thiazolidinediones. N Engl J Med. 2004;351:1106–1118. doi: 10.1056/NEJMra041001. [DOI] [PubMed] [Google Scholar]
- 20.American Diabetes Association. Standards of medical care in diabetes – 2008. Diabetes Care. 2008;31:S12–S54. doi: 10.2337/dc08-S012. [DOI] [PubMed] [Google Scholar]
- 21.Baldwin SJ, Clarke SE, Chenery RJ. Characterization of the cytochrome P450 enzymes involved in the in vitro metabolism of rosiglitazone. Br J Clin Pharmacol. 1999;48:424–432. doi: 10.1046/j.1365-2125.1999.00030.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jaakkola T, Laitila J, Neuvonen PJ, Backman JT. Pioglitazone is metabolised by CYP2C8 and CYP3A4 in vitro: potential for interactions with CYP2C8 inhibitors. Basic Clin Pharmacol Toxicol. 2006;99:44–51. doi: 10.1111/j.1742-7843.2006.pto_437.x. [DOI] [PubMed] [Google Scholar]
- 23.Yamazaki H, Shibata A, Suzuki M, et al. Oxidation of troglitazone to a quinone-type metabolite catalyzed by cytochrome P-450 2C8 and P-450 3A4 in human liver microsomes. Drug Metab Dispos. 1999;27:1260–1266. [PubMed] [Google Scholar]
- 24.Avandia Prescribing Information. GlaxoSmithKline (2007).
- 25.Actos Prescribing Information. Takeda Pharmaceuticals America, Inc. (2007)
- 26 ▪.Kirchheiner J, Thomas S, Bauer S, et al. Pharmacokinetics and pharmacodynamics of rosiglitazone in relation to CYP2C8 genotype. Clin Pharmacol Ther. 2006;80:657–667. doi: 10.1016/j.clpt.2006.09.008. First study to show a significant association between CYP2C8 polymorphisms and rosiglitazone pharmacokinetics. [DOI] [PubMed] [Google Scholar]
- 27.Aquilante CL, Bushman LR, Knutsen SD, Burt LE, Rome LC, Kosmiski LA. Influence of SLCO1B1 haplotype on rosiglitazone pharmacokinetics in healthy volunteers. Hum Genomics. 2008;3(1):7–16. doi: 10.1186/1479-7364-3-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28 ▪.Tornio A, Niemi M, Neuvonen PJ, Backman JT. Trimethoprim and the CYP2C8*3 allele have opposite effects on the pharmacokinetics of pioglitazone. Drug Metab Dispos. 2008;36:73–80. doi: 10.1124/dmd.107.018010. First study to show an association between CYP2C8 polymorphisms and pioglitazone pharmacokinetics. [DOI] [PubMed] [Google Scholar]
- 29.Hruska MW, Amico JA, Langaee TY, Ferrell RE, Fitzgerald SM, Frye RF. The effect of trimethoprim on CYP2C8 mediated rosiglitazone metabolism in human liver microsomes and healthy subjects. Br J Clin Pharmacol. 2005;59:70–79. doi: 10.1111/j.1365-2125.2005.02263.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pedersen RS, Damkier P, Brosen K. The effects of human CYP2C8 genotype and fluvoxamine on the pharmacokinetics of rosiglitazone in healthy subjects. Br J Clin Pharmacol. 2006;62:682–689. doi: 10.1111/j.1365-2125.2006.02706.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aquilante CL. Pharmacogenetics of thiazolidinedione therapy. Pharmacogenomics. 2007;8:917–931. doi: 10.2217/14622416.8.8.917. [DOI] [PubMed] [Google Scholar]
- 32.Prandin Prescribing Information. Novo Nordisk, Inc. (2006)
- 33.Bidstrup TB, Bjornsdottir I, Sidelmann UG, Thomsen MS, Hansen KT. CYP2C8 and CYP3A4 are the principal enzymes involved in the human in vitro biotransformation of the insulin secretagogue repaglinide. Br J Clin Pharmacol. 2003;56:305–314. doi: 10.1046/j.0306-5251.2003.01862.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kajosaari LI, Laitila J, Neuvonen PJ, Backman JT. Metabolism of repaglinide by CYP2C8 and CYP3A4 in vitro: effect of fibrates and rifampicin. Basic Clin Pharmacol Toxicol. 2005;97:249–256. doi: 10.1111/j.1742-7843.2005.pto_157.x. [DOI] [PubMed] [Google Scholar]
- 35.Niemi M, Backman JT, Neuvonen M, Neuvonen PJ. Effects of gemfibrozil, itraconazole, and their combination on the pharmacokinetics and pharmacodynamics of repaglinide: potentially hazardous interaction between gemfibrozil and repaglinide. Diabetologia. 2003;46:347–351. doi: 10.1007/s00125-003-1034-7. [DOI] [PubMed] [Google Scholar]
- 36.Niemi M, Neuvonen PJ, Kivisto KT. The cytochrome P4503A4 inhibitor clarithromycin increases the plasma concentrations and effects of repaglinide. Clin Pharmacol Ther. 2001;70:58–65. doi: 10.1067/mcp.2001.116511. [DOI] [PubMed] [Google Scholar]
- 37 ▪.Niemi M, Leathart JB, Neuvonen M, Backman JT, Daly AK, Neuvonen PJ. Polymorphism in CYP2C8 is associated with reduced plasma concentrations of repaglinide. Clin Pharmacol Ther. 2003;74:380–387. doi: 10.1016/S0009-9236(03)00228-5. First study to show an association between CYP2C8 polymorphisms and repaglinide pharmacokinetics. [DOI] [PubMed] [Google Scholar]
- 38.Niemi M, Backman JT, Kajosaari LI, et al. Polymorphic organic anion transporting polypeptide 1B1 is a major determinant of repaglinide pharmacokinetics. Clin Pharmacol Ther. 2005;77:468–478. doi: 10.1016/j.clpt.2005.01.018. [DOI] [PubMed] [Google Scholar]
- 39.Kajosaari LI, Niemi M, Neuvonen M, Laitila J, Neuvonen PJ, Backman JT. Cyclosporine markedly raises the plasma concentrations of repaglinide. Clin Pharmacol Ther. 2005;78:388–399. doi: 10.1016/j.clpt.2005.07.005. [DOI] [PubMed] [Google Scholar]
- 40.Tornio A, Niemi M, Neuvonen M, et al. The effect of gemfibrozil on repaglinide pharmacokinetics persists for at least 12 h after the dose: evidence for mechanism-based inhibition of CYP2C8 in vivo. Clin Pharmacol Ther. 2008;84:403–411. doi: 10.1038/clpt.2008.34. [DOI] [PubMed] [Google Scholar]
- 41.Bidstrup TB, Damkier P, Olsen AK, Ekblom M, Karlsson A, Brosen K. The impact of CYP2C8 polymorphism and grapefruit juice on the pharmacokinetics of repaglinide. Br J Clin Pharmacol. 2006;61:49–57. doi: 10.1111/j.1365-2125.2005.02516.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Muck W. Clinical pharmacokinetics of cerivastatin. Clin Pharmacokinet. 2000;39:99–116. doi: 10.2165/00003088-200039020-00002. [DOI] [PubMed] [Google Scholar]
- 43.Wang JS, Neuvonen M, Wen X, Backman JT, Neuvonen PJ. Gemfibrozil inhibits CYP2C8-mediated cerivastatin metabolism in human liver microsomes. Drug Metab Dispos. 2002;30:1352–1356. doi: 10.1124/dmd.30.12.1352. [DOI] [PubMed] [Google Scholar]
- 44.Lucas RA, Weathersby BB, Rocco VK, Pepper JM, Butler KL. Rhabdomyolysis associated with cerivastatin: six cases within 3 months at one hospital. Pharmacotherapy. 2002;22:771–774. doi: 10.1592/phco.22.9.771.34071. [DOI] [PubMed] [Google Scholar]
- 45.Law M, Rudnicka AR. Statin safety: a systematic review. Am J Cardiol. 2006;97:C52–C60. doi: 10.1016/j.amjcard.2005.12.010. [DOI] [PubMed] [Google Scholar]
- 46.Omar MA, Wilson JP, Cox TS. Rhabdomyolysis and HMG-CoA reductase inhibitors. Ann Pharmacother. 2001;35:1096–1107. doi: 10.1345/aph.10228. [DOI] [PubMed] [Google Scholar]
- 47.Backman JT, Kyrklund C, Neuvonen M, Neuvonen PJ. Gemfibrozil greatly increases plasma concentrations of cerivastatin. Clin Pharmacol Ther. 2002;72:685–691. doi: 10.1067/mcp.2002.128469. [DOI] [PubMed] [Google Scholar]
- 48.Ishikawa C, Ozaki H, Nakajima T, et al. A frameshift variant of CYP2C8 was identified in a patient who suffered from rhabdomyolysis after administration of cerivastatin. J Hum Genet. 2004;49:582–585. doi: 10.1007/s10038-004-0188-6. [DOI] [PubMed] [Google Scholar]
- 49.Ozaki H, Ishikawa CT, Ishii T, et al. Clearance rates of cerivastatin metabolites in a patient with cerivastatin-induced rhabdomyolysis. J Clin Pharm Ther. 2005;30:189–192. doi: 10.1111/j.1365-2710.2005.00633_1.x. [DOI] [PubMed] [Google Scholar]
- 50.Scripture CD, Pieper JA. Clinical pharmacokinetics of fluvastatin. Clin Pharmacokinet. 2001;40:263–281. doi: 10.2165/00003088-200140040-00003. [DOI] [PubMed] [Google Scholar]
- 51.Lescol Prescribing Information. Novartis Pharmaceuticals Corporation. (2006)
- 52.Spence JD, Munoz CE, Hendricks L, Latchinian L, Khouri HE. Pharmacokinetics of the combination of fluvastatin and gemfibrozil. Am J Cardiol. 1995;76:A80–A83. doi: 10.1016/s0002-9149(05)80024-4. [DOI] [PubMed] [Google Scholar]
- 53.Kirchheiner J, Kudlicz D, Meisel C, et al. Influence of CYP2C9 polymorphisms on the pharmacokinetics and cholesterol-lowering activity of (−)-3S,5R-fluvastatin and (+)-3R,5S-fluvastatin in healthy volunteers. Clin Pharmacol Ther. 2003;74:186–194. doi: 10.1016/S0009-9236(03)00121-8. [DOI] [PubMed] [Google Scholar]
- 54.Shitara Y, Sugiyama Y. Pharmacokinetic and pharmacodynamic alterations of 3-hydroxy-3-methylglutaryl coenzyme A (HMG-CoA) reductase inhibitors: drug–drug interactions and interindividual differences in transporter and metabolic enzyme functions. Pharmacol Ther. 2006;112:71–105. doi: 10.1016/j.pharmthera.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 55.Prueksaritanont T, Ma B, Yu N. The human hepatic metabolism of simvastatin hydroxy acid is mediated primarily by CYP3A, and not CYP2D6. Br J Clin Pharmacol. 2003;56:120–124. doi: 10.1046/j.1365-2125.2003.01833.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Crestor Prescribing Information. AstraZeneca Pharmaceticals (2009).
- 57.Cooper KJ, Martin PD, Dane AL, Warwick MJ, Schneck DW, Cantarini MV. The effect of fluconazole on the pharmacokinetics of rosuvastatin. Eur J Clin Pharmacol. 2002;58:527–531. doi: 10.1007/s00228-002-0508-8. [DOI] [PubMed] [Google Scholar]
- 58.Link E, Parish S, Armitage J, et al. SLCO1B1 variants and statin-induced myopathy – a genomewide study. N Engl J Med. 2008;359:789–799. doi: 10.1056/NEJMoa0801936. [DOI] [PubMed] [Google Scholar]
- 59.Chasman DI, Posada D, Subrahmanyan L, Cook NR, Stanton VP, Jr, Ridker PM. Pharmacogenetic study of statin therapy and cholesterol reduction. JAMA. 2004;291:2821–2827. doi: 10.1001/jama.291.23.2821. [DOI] [PubMed] [Google Scholar]
- 60.Thompson JF, Man M, Johnson KJ, et al. An association study of 43 SNPs in 16 candidate genes with atorvastatin response. Pharmacogenomics J. 2005;5:352–358. doi: 10.1038/sj.tpj.6500328. [DOI] [PubMed] [Google Scholar]
- 61.Wilke RA, Moore JH, Burmester JK. Relative impact of CYP3A genotype and concomitant medication on the severity of atorvastatin-induced muscle damage. Pharmacogenet Genomics. 2005;15:415–421. doi: 10.1097/01213011-200506000-00007. [DOI] [PubMed] [Google Scholar]
- 62.Rodrigues AD. Impact of CYP2C9 genotype on pharmacokinetics: are all cyclooxygenase inhibitors the same? Drug Metab Dispos. 2005;33:1567–1575. doi: 10.1124/dmd.105.006452. [DOI] [PubMed] [Google Scholar]
- 63.Davies NM. Clinical pharmacokinetics of ibuprofen. The first 30 years. Clin Pharmacokinet. 1998;34:101–154. doi: 10.2165/00003088-199834020-00002. [DOI] [PubMed] [Google Scholar]
- 64.Evans AM. Enantioselective pharmacodynamics and pharmacokinetics of chiral non-steroidal anti-inflammatory drugs. Eur J Clin Pharmacol. 1992;42:237–256. doi: 10.1007/BF00266343. [DOI] [PubMed] [Google Scholar]
- 65.Hamman MA, Thompson GA, Hall SD. Regioselective and stereoselective metabolism of ibuprofen by human cytochrome P450 2C. Biochem Pharmacol. 1997;54:33–41. doi: 10.1016/s0006-2952(97)00143-3. [DOI] [PubMed] [Google Scholar]
- 66.Chang SY, Li W, Traeger SC, et al. Confirmation that cytochrome P450 2C8 (CYP2C8) plays a minor role in (S)-(+)- and (R)-(−)-ibuprofen hydroxylation in vitro. Drug Metab Dispos. 2008;36:2513–2522. doi: 10.1124/dmd.108.022970. [DOI] [PubMed] [Google Scholar]
- 67.Tornio A, Niemi M, Neuvonen PJ, Backman JT. Stereoselective interaction between the CYP2C8 inhibitor gemfibrozil and racemic ibuprofen. Eur J Clin Pharmacol. 2007;63:463–469. doi: 10.1007/s00228-007-0273-9. [DOI] [PubMed] [Google Scholar]
- 68.Martinez C, Garcia-Martin E, Blanco G, Gamito FJ, Ladero JM, Agundez JA. The effect of the cytochrome P450 CYP2C8 polymorphism on the disposition of (R)-ibuprofen enantiomer in healthy subjects. Br J Clin Pharmacol. 2005;59:62–69. doi: 10.1111/j.1365-2125.2004.02183.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Garcia-Martin E, Martinez C, Tabares B, Frias J, Agundez JA. Interindividual variability in ibuprofen pharmacokinetics is related to interaction of cytochrome P450 2C8 and 2C9 amino acid polymorphisms. Clin Pharmacol Ther. 2004;76:119–127. doi: 10.1016/j.clpt.2004.04.006. [DOI] [PubMed] [Google Scholar]
- 70.Lopez-Rodriguez R, Novalbos J, Gallego-Sandin S, et al. Influence of CYP2C8 and CYP2C9 polymorphisms on pharmacokinetic and pharmacodynamic parameters of racemic and enantiomeric forms of ibuprofen in healthy volunteers. Pharmacol Res. 2008;58:77–84. doi: 10.1016/j.phrs.2008.07.004. [DOI] [PubMed] [Google Scholar]
- 71.Tang W. The metabolism of diclofenac – enzymology and toxicology perspectives. Curr Drug Metab. 2003;4:319–329. doi: 10.2174/1389200033489398. [DOI] [PubMed] [Google Scholar]
- 72.Daly AK, Aithal GP, Leathart JB, Swainsbury RA, Dang TS, Day CP. Genetic susceptibility to diclofenac-induced hepatotoxicity: contribution of UGT2B7, CYP2C8, and ABCC2 genotypes. Gastroenterology. 2007;132:272–281. doi: 10.1053/j.gastro.2006.11.023. [DOI] [PubMed] [Google Scholar]
- 73.Dorado P, Cavaco I, Caceres MC, Piedade R, Ribeiro V, Llerena A. Relationship between CYP2C8 genotypes and diclofenac 5-hydroxylation in healthy Spanish volunteers. Eur J Clin Pharmacol. 2008;64:967–970. doi: 10.1007/s00228-008-0508-4. [DOI] [PubMed] [Google Scholar]
- 74.Voltaren Prescribing Information. Novartis Pharmaceuticals Corporation (2009).
- 75.de Abajo FJ, Montero D, Madurga M, Garcia Rodriguez LA. Acute and clinically relevant drug-induced liver injury: a population based case–control study. Br J Clin Pharmacol. 2004;58:71–80. doi: 10.1111/j.1365-2125.2004.02133.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Aithal GP, Day CP, Leathart JB, Daly AK. Relationship of polymorphism in CYP2C9 to genetic susceptibility to diclofenac-induced hepatitis. Pharmacogenetics. 2000;10:511–518. doi: 10.1097/00008571-200008000-00004. [DOI] [PubMed] [Google Scholar]
- 77.Peiro AM, Novalbos J, Zapater P, et al. Pharmacogenetic relevance of the CYP2C9*3 allele in a tenoxicam bioequivalence study performed on Spaniards. Pharmacol Res. 2009;59:62–68. doi: 10.1016/j.phrs.2008.09.018. [DOI] [PubMed] [Google Scholar]
- 78.Martinez C, Blanco G, Ladero JM, et al. Genetic predisposition to acute gastrointestinal bleeding after NSAIDs use. Br J Pharmacol. 2004;141:205–208. doi: 10.1038/sj.bjp.0705623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Blanco G, Martinez C, Ladero JM, et al. Interaction of CYP2C8 and CYP2C9 genotypes modifies the risk for nonsteroidal anti-inflammatory drugs-related acute gastrointestinal bleeding. Pharmacogenet Genomics. 2008;18:37–43. doi: 10.1097/FPC.0b013e3282f305a9. [DOI] [PubMed] [Google Scholar]
- 80.McGreavey LE, Turner F, Smith G, et al. No evidence that polymorphisms in CYP2C8, CYP2C9, UGT1A6, PPARδ and PPARγ act as modifiers of the protective effect of regular NSAID use on the risk of colorectal carcinoma. Pharmacogenet Genomics. 2005;15:713–721. doi: 10.1097/01.fpc.0000174786.85238.63. [DOI] [PubMed] [Google Scholar]
- 81.Rahman A, Korzekwa KR, Grogan J, Gonzalez FJ, Harris JW. Selective biotransformation of taxol to 6 α-hydroxytaxol by human cytochrome P450 2C8. Cancer Res. 1994;54:5543–5546. [PubMed] [Google Scholar]
- 82.Harris JW, Rahman A, Kim BR, Guengerich FP, Collins JM. Metabolism of taxol by human hepatic microsomes and liver slices: participation of cytochrome P450 3A4 and an unknown P450 enzyme. Cancer Res. 1994;54:4026–4035. [PubMed] [Google Scholar]
- 83.Somlo G, Doroshow JH, Synold T, et al. High-dose paclitaxel in combination with doxorubicin, cyclophosphamide and peripheral blood progenitor cell rescue in patients with high-risk primary and responding metastatic breast carcinoma: toxicity profile, relationship to paclitaxel pharmacokinetics and short-term outcome. Br J Cancer. 2001;84:1591–1598. doi: 10.1054/bjoc.2001.1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Henningsson A, Marsh S, Loos WJ, et al. Association of CYP2C8, CYP3A4, CYP3A5, and ABCB1 polymorphisms with the pharmacokinetics of paclitaxel. Clin Cancer Res. 2005;11:8097–8104. doi: 10.1158/1078-0432.CCR-05-1152. [DOI] [PubMed] [Google Scholar]
- 85.Marsh S, Somlo G, Li X, et al. Pharmacogenetic analysis of paclitaxel transport and metabolism genes in breast cancer. Pharmacogenomics J. 2007;7:362–365. doi: 10.1038/sj.tpj.6500434. [DOI] [PubMed] [Google Scholar]
- 86.Marsh S, Paul J, King CR, Gifford G, McLeod HL, Brown R. Pharmacogenetic assessment of toxicity and outcome after platinum plus taxane chemotherapy in ovarian cancer: the Scottish Randomised Trial in Ovarian Cancer. J Clin Oncol. 2007;25:4528–4535. doi: 10.1200/JCO.2006.10.4752. [DOI] [PubMed] [Google Scholar]
- 87.Yamaguchi H, Hishinuma T, Endo N, et al. Genetic variation in ABCB1 influences paclitaxel pharmacokinetics in Japanese patients with ovarian cancer. Int J Gynecol Cancer. 2006;16:979–985. doi: 10.1111/j.1525-1438.2006.00593.x. [DOI] [PubMed] [Google Scholar]
- 88.Jiko M, Yano I, Sato E, et al. Pharmacokinetics and pharmacodynamics of paclitaxel with carboplatin or gemcitabine, and effects of CYP3A5 and MDR1 polymorphisms in patients with urogenital cancers. Int J Clin Oncol. 2007;12:284–290. doi: 10.1007/s10147-007-0681-y. [DOI] [PubMed] [Google Scholar]
- 89.Nakajima M, Fujiki Y, Kyo S, et al. Pharmacokinetics of paclitaxel in ovarian cancer patients and genetic polymorphisms of CYP2C8, CYP3A4, and MDR1. J Clin Pharmacol. 2005;45:674–682. doi: 10.1177/0091270005276204. [DOI] [PubMed] [Google Scholar]
- 90.Green H, Soderkvist P, Rosenberg P, et al. Pharmacogenetic studies of paclitaxel in the treatment of ovarian cancer. Basic Clin Pharmacol Toxicol. 2009;104:130–137. doi: 10.1111/j.1742-7843.2008.00351.x. [DOI] [PubMed] [Google Scholar]
- 91.Li XQ, Bjorkman A, Andersson TB, Ridderstrom M, Masimirembwa CM. Amodiaquine clearance and its metabolism to N-desethylamodiaquine is mediated by CYP2C8: a new high affinity and turnover enzyme-specific probe substrate. J Pharmacol Exp Ther. 2002;300:399–407. doi: 10.1124/jpet.300.2.399. [DOI] [PubMed] [Google Scholar]
- 92.Jewell H, Maggs JL, Harrison AC, O’Neill PM, Ruscoe JE, Park BK. Role of hepatic metabolism in the bioactivation and detoxication of amodiaquine. Xenobiotica. 1995;25:199–217. doi: 10.3109/00498259509061845. [DOI] [PubMed] [Google Scholar]
- 93.Parikh S, Ouedraogo JB, Goldstein JA, Rosenthal PJ, Kroetz DL. Amodiaquine metabolism is impaired by common polymorphisms in CYP2C8: implications for malaria treatment in Africa. Clin Pharmacol Ther. 2007;82:197–203. doi: 10.1038/sj.clpt.6100122. [DOI] [PubMed] [Google Scholar]
- 94.Adjei GO, Kristensen K, Goka BQ, et al. Effect of concomitant artesunate administration and cytochrome P4502C8 polymorphisms on the pharmacokinetics of amodiaquine in Ghanaian children with uncomplicated malaria. Antimicrob Agents Chemother. 2008;52:4400–4406. doi: 10.1128/AAC.00673-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kim KA, Park JY, Lee JS, Lim S. Cytochrome P450 2C8 and CYP3A4/5 are involved in chloroquine metabolism in human liver microsomes. Arch Pharm Res. 2003;26:631–637. doi: 10.1007/BF02976712. [DOI] [PubMed] [Google Scholar]
- 96.Rigas JR, Francis PA, Muindi JR, et al. Constitutive variability in the pharmacokinetics of the natural retinoid, all-trans-retinoic acid, and its modulation by ketoconazole. J Natl Cancer Inst. 1993;85:1921–1926. doi: 10.1093/jnci/85.23.1921. [DOI] [PubMed] [Google Scholar]
- 97.Kim KA, Chung J, Jung DH, Park JY. Identification of cytochrome P450 isoforms involved in the metabolism of loperamide in human liver microsomes. Eur J Clin Pharmacol. 2004;60:575–581. doi: 10.1007/s00228-004-0815-3. [DOI] [PubMed] [Google Scholar]
- 98.Niemi M, Tornio A, Pasanen MK, Fredrikson H, Neuvonen PJ, Backman JT. Itraconazole, gemfibrozil and their combination markedly raise the plasma concentrations of loperamide. Eur J Clin Pharmacol. 2006;62:463–472. doi: 10.1007/s00228-006-0133-z. [DOI] [PubMed] [Google Scholar]
- 99.Wang JS, DeVane CL. Involvement of CYP3A4, CYP2C8, and CYP2D6 in the metabolism of (R)- and (S)-methadone in vitro. Drug Metab Dispos. 2003;31:742–747. doi: 10.1124/dmd.31.6.742. [DOI] [PubMed] [Google Scholar]
- 100.Projean D, Morin PE, Tu TM, Ducharme J. Identification of CYP3A4 and CYP2C8 as the major cytochrome P450 s responsible for morphine N-demethylation in human liver microsomes. Xenobiotica. 2003;33:841–854. doi: 10.1080/0049825031000121608. [DOI] [PubMed] [Google Scholar]
- 101.Ohyama K, Nakajima M, Nakamura S, Shimada N, Yamazaki H, Yokoi T. A significant role of human cytochrome P450 2C8 in amiodarone N-deethylation: an approach to predict the contribution with relative activity factor. Drug Metab Dispos. 2000;28:1303–1310. [PubMed] [Google Scholar]
- 102.Soyama A, Hanioka N, Saito Y, et al. Amiodarone N-deethylation by CYP2C8 and its variants, CYP2C8*3 and CYP2C8 P404A. Pharmacol Toxicol. 2002;91:174–178. doi: 10.1034/j.1600-0773.2002.910404.x. [DOI] [PubMed] [Google Scholar]
- 103.Becquemont L, Mouajjah S, Escaffre O, Beaune P, Funck-Brentano C, Jaillon P. Cytochrome P-450 3A4 and 2C8 are involved in zopiclone metabolism. Drug Metab Dispos. 1999;27:1068–1073. [PubMed] [Google Scholar]
- 104.Tornio A, Neuvonen PJ, Backman JT. The CYP2C8 inhibitor gemfibrozil does not increase the plasma concentrations of zopiclone. Eur J Clin Pharmacol. 2006;62:645–651. doi: 10.1007/s00228-006-0155-6. [DOI] [PubMed] [Google Scholar]
- 105.Attar M, Dong D, Ling KH, Tang-Liu DD. Cytochrome P450 2C8 and flavin-containing monooxygenases are involved in the metabolism of tazarotenic acid in humans. Drug Metab Dispos. 2003;31:476–481. doi: 10.1124/dmd.31.4.476. [DOI] [PubMed] [Google Scholar]
- 106.Tang-Liu DD, Matsumoto RM, Usansky JI. Clinical pharmacokinetics and drug metabolism of tazarotene: a novel topical treatment for acne and psoriasis. Clin Pharmacokinet. 1999;37:273–287. doi: 10.2165/00003088-199937040-00001. [DOI] [PubMed] [Google Scholar]
- 107.Covera-HS Prescribing Information. Pfizer (2006).
- 108.Vogelgesang B, Echizen H, Schmidt E, Eichelbaum M. Stereoselective first-pass metabolism of highly cleared drugs: studies of the bioavailability of L- and D-verapamil examined with a stable isotope technique. Br J Clin Pharmacol. 1984;58:S796–S803. doi: 10.1111/j.1365-2125.2004.02299.x. discussion S804–S796 (2004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Tracy TS, Korzekwa KR, Gonzalez FJ, Wainer IW. Cytochrome P450 isoforms involved in metabolism of the enantiomers of verapamil and norverapamil. Br J Clin Pharmacol. 1999;47:545–552. doi: 10.1046/j.1365-2125.1999.00923.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kerr BM, Thummel KE, Wurden CJ, et al. Human liver carbamazepine metabolism. Role of CYP3A4 and CYP2C8 in 10,11-epoxide formation. Biochem Pharmacol. 1994;47:1969–1979. doi: 10.1016/0006-2952(94)90071-x. [DOI] [PubMed] [Google Scholar]
- 111.Chang TK, Weber GF, Crespi CL, Waxman DJ. Differential activation of cyclophosphamide and ifosphamide by cytochromes P-450 2B and 3A in human liver microsomes. Cancer Res. 1993;53:5629–5637. [PubMed] [Google Scholar]
- 112.Winter HR, Wang Y, Unadkat JD. CYP2C8/9 mediate dapsone N-hydroxylation at clinical concentrations of dapsone. Drug Metab Dispos. 2000;28:865–868. [PubMed] [Google Scholar]
- 113.Miners JO, Coulter S, Birkett DJ, Goldstein JA. Torsemide metabolism by CYP2C9 variants and other human CYP2C subfamily enzymes. Pharmacogenetics. 2000;10:267–270. doi: 10.1097/00008571-200004000-00008. [DOI] [PubMed] [Google Scholar]
- 114.Sutton D, Butler AM, Nadin L, Murray M. Role of CYP3A4 in human hepatic diltiazem N-demethylation: inhibition of CYP3A4 activity by oxidized diltiazem metabolites. J Pharmacol Exp Ther. 1997;282:294–300. [PubMed] [Google Scholar]
- 115 ▪.Gil JP, Gil Berglund E. CYP2C8 and antimalaria drug efficacy. Pharmacogenomics. 2007;8:187–198. doi: 10.2217/14622416.8.2.187. Comprehensive review of the association between CYP2C8 polymorphisms and antimalaria drug efficacy. [DOI] [PMC free article] [PubMed] [Google Scholar]
Websites
- 201.CYP2C8 allele nomenclature. www.cypalleles.ki.se/cyp2c8.htm.
- 202.dbSNP Home Page. www.ncbi.nlm.nih.gov/SNP.