Abstract
Variants in the engulfment and cell motility 1 (ELMO1) gene are associated with nephropathy due to type 2 diabetes mellitus (T2DM) in a Japanese cohort. We comprehensively evaluated this gene in African American (AA) T2DM patients with end-stage renal disease (ESRD). Three hundred nine HapMap tagging SNPs and 9 reportedly associated SNPs were genotyped in 577 AA T2DM-ESRD patients and 596 AA non-diabetic controls, plus 43 non-diabetic European American controls and 45 Yoruba Nigerian samples for admixture adjustment. Replication analyses were conducted in 558 AAs with T2DM-ESRD and 564 controls without diabetes. Extension analyses included 328 AA with T2DM lacking nephropathy and 326 with non-diabetic ESRD. The original and replication analyses confirmed association with four SNPs in intron 13 (permutation p-values for combined analyses = 0.001-0.003), one in intron 1 (P=0.004) and one in intron 5 (P=0.002) with T2DM-associated ESRD. In a subsequent combined analysis of all 1,135 T2DM-ESRD cases and 1,160 controls, an additional 7 intron 13 SNPs produced evidence of association (P = 3.5×10-5 – P=0.05). No associations were seen with these SNPs in those with T2DM lacking nephropathy or with ESRD due to non-diabetic causes. Variants in intron 13 of the ELMO1 gene appear to confer risk for diabetic nephropathy in AA.
Keywords: Association, Nephropathy, African American, Diabetes
Introduction
African Americans (AA) with diabetes have an increased risk of end-stage renal disease (ESRD) relative to European Americans (EA) (Boone, 2000). T2DM-associated nephropathy is the most frequent cause of ESRD among incident AA dialysis patients in the United States (Brancati et al., 1992, Geiss et al., 1993). The engulfment and cell motility 1 (ELMO1) gene, located on chromosome 7p14.2-14.1, was previously associated with T2DM-associated nephropathy in a Japanese cohort of 640 patients (Shimazaki et al., 2005).
ELMO1 is a soluble cytoplasmic protein that functionally cooperates with CRKII and dedicator of cytokinesis 180 (DOCK180) to mediate cytoskeletal rearrangements during phagocytosis of apoptotic cells and cell motility in mammalian cells (Gumienny et al., 2001). Functional studies of ELMO1 expression reveal that increased levels were observed in COS cells cultured at high glucose concentration, loss of cell adhesion properties and enhanced synthesis of collagen and fibronectin in ELMO1 transfected cells suggest a pathological role in kidney disease (Shimazaki et al., 2005, Shimazaki et al., 2006).
AA are approximately five times more likely than EA with diabetes to develop kidney disease (Bryson et al., 2006, Collins et al., 2005), and AA with T2DM have an 8.1 fold risk (P=0.0005) for developing ESRD in the presence of a family history of ESRD (Freedman et al., 1995). Although a genetic component for T2DM-associated nephropathy has been established, a limited number of genetic variants have shown association in AA.
To investigate the role of ELMO1 in susceptibility to T2DM-associated nephropathy in an AA population, we genotyped 309 single nucleotide polymorphisms (SNPs) selected from HapMap on the basis of linkage disequilibrium (LD), and nine previously-associated SNPs in 577 AA T2DM patients with ESRD and 596 AA controls without known diabetes or kidney disease (Set 1). In addition, we adjusted for the impact of admixture on association results using ancestry informative markers (AIMs). Nominally associated SNPs were then genotyped in independent populations of 558 additional AA subjects with T2DM-ESRD and 564 AA controls (Set 2), as well as in 328 AA with longstanding T2DM lacking nephropathy and 326 AA with non-diabetic forms of ESRD.
Research Design and Methods
This study was conducted under Institutional Review Board approval from Wake Forest University School of Medicine, and adhered to the tenets of the Declaration of Helsinki. Recruitment of AA and EA patients and controls has been described previously (Sale et al., 2007). Two independently recruited groups of unrelated patients with T2DM-ESRD and non-diabetic controls were evaluated in this study. The first consisted of 577 AA with T2DM-ESRD and 596 AA non-diabetic controls (Set 1). Replication studies were performed in an independent population of 558 additional AAT2DM-ESRD subjects and 564 AA non-diabetic controls (Set 2). Briefly, AA patients with T2DM-ESRD, born in North Carolina, South Carolina, Georgia, Tennessee or Virginia, were recruited from dialysis facilities in northwestern North Carolina. A diagnosis of T2DM was based upon participants reporting an initial diagnosis of diabetes mellitus after age 35 years, receiving oral hypoglycemic agents or dietary therapy without insulin for at least one year after initial diagnosis, and active treatment with diabetes medications. Cases with ESRD had T2DM diagnosed at least 5 years prior to initiating renal replacement therapy, background or greater diabetic retinopathy and/or > 3+ proteinuria on urinalysis in the absence of other causes of nephropathy. The AA controls and 39 EA controls for admixture analyses born in North Carolina, South Carolina, Georgia, Tennessee or Virginia, without a current diagnosis of T2DM or renal disease, were recruited from community sources. The majority of controls were not tested for diabetes; however, serum glucose values were obtained for 256 of the 596 AA controls (mean 93.6 mg/dl). Five of these individuals had non-fasting serum glucose levels greater than 126 mg/dl, suggesting the overall misclassification rate of controls is likely well below 2%. Three hundred twenty-eight unrelated AA patients with T2DM for greater than 10 years and lacking nephropathy (defined as serum creatinine concentrations ≤ 1.5 mg/dl, without proteinuria) were recruited from local general internal medicine and endocrinology clinics in northwestern North Carolina. Additionally, 326 AA subjects with non-diabetic forms of ESRD (predominantly hypertension- and chronic glomerular disease-associated ESRD) were recruited from the same dialysis facilities as patients with T2DM-ESRD. Hypertension-associated ESRD was diagnosed in non-diabetic subjects without other diseases known to cause nephropathy, when hypertension preceded dialysis initiation by more than 5 years, and typically in the presence of proteinuria ≤ 30 mg /dl or 500 mg/24 hr. In the absence of a quantified urinary protein measurement, this diagnosis was also applied when provided by the patient’s nephrologist on the CMS 2728 form. Chronic glomerulonephritis (CGN) was diagnosed in non-diabetic patients having proteinuria ≥ 100 mg/dl or 500 mg/24 hr and/or hematuria with or without urinary red blood cell casts. These subjects had idiopathic and secondary forms of glomerular disease (e.g., systemic lupus erythematosus). DNA extraction was performed using the PureGene system (Gentra Systems, Minneapolis, MN). DNA was obtained from 44 Yoruba Nigerians (YRI) from the National Institute of General Medical Sciences Human Variation Collection (Coriell Cell Repositories, Camden, NJ).
SNP selection and genotyping
Genetically, AA populations in the U.S. are mosaic admixtures of ancestral European and African chromosomal segments. In the absence of a comprehensive AA SNP database of allele frequencies, we used the genotypic data of the YRI and CEPH Europeans (CEU) from the International HapMap project (HapMap #20, on NCBI B35 assembly) in order to capture the most common haplotypes in AA. Using the entire ELMO1 transcript plus 5kb upstream and downstream of the gene, we uploaded the YRI genotypic data into Haploview 3.2 (Barrett et al., 2005), selected markers with a minor allele frequency (MAF) ≥ 0.05, and excluded SNPs with a designability score of < 1.0 based on the Illumina platform. The aggressive tagging (2- or 3-haplotype tagging of SNPs) option of Tagger (de Bakker et al., 2005) was used to search for predictors that captured all alleles of interest. Next, we uploaded the CEU genotypic data into Haploview 3.2 (Barrett et al., 2005), forced the inclusion of the already selected YRI tagSNPs and 9 SNPs previously associated SNPs with T2DM nephropathy (Shimazaki et al., 2005), and identified any additional CEU tagSNPs required to capture SNPs with MAF ≥ 0.05. This yielded a total of 318 (3 SNPs in 3′ UTR, 1 coding, 7 promoter, 3 intron boundary and 304 non-coding) SNPs at an average density of 1 SNP every 2 kb, with the largest gap 31.6 kb and the smallest 31 bp. Tagger (de Bakker et al., 2005) results indicated that 2- or 3-SNP haplotypes were not required to capture ungenotyped YRI and CEU HapMap SNPs in this gene.
Genotyping was performed in Set 1 by Illumina Genotyping Services (Illumina Inc., San Diego, CA) (Buetow et al., 2001). The genotyping success rates were >98.5% in cases and >97.1% in controls. For quality control (QC) purposes, each plate contained 2 duplicates and 4 Illumina controls. Concordance rates for 50 replicate pairs in Set 1 were >99.7%.
Genotyping of nominally associated SNPs in subsequent populations was performed using iPlex methodology on a MassARRAY genotyping system (Sequenom Inc., San Diego, CA) (Buetow et al., 2001). Primer sequences are available on request. Fragment sizes were determined by mass spectrometry (Bruker Daltonics, Billerica, MA), and the genotypes were viewed and analyzed using SpectroTyper software (Sequenom Inc., San Diego, CA). Genotyping success rates were >95% in cases and >93% in controls. The blind duplicate reproducibility rate for Set 2 was ≥99.5%.
Genotyping for admixture analyses
Seventy bi-allelic Admixture Informative Markers (AIMs), selected to maximize European and African allele frequency differences and sample all non-acrocentric arms of the autosomal genome, were genotyped by Illumina Inc.’s Custom Genotyping Service (San Diego, CA) in Set 1. Genotyping success rates for AIMs were >97.4% in AA cases and >95.6% in AA controls.
The AIMs were genotyped in the replication sample population (Set 2), and the extension cohorts of T2DM without nephropathy, and non diabetic-ESRD using iPlex methodology on the MassARRAY genotyping system (Sequenom Inc., San Diego, CA) (Buetow et al., 2001). Primer sequences are available on request. Genotyping success rates for AIMs were >96.2% in AA cases and >96.4% in AA controls.
Direct Sequencing
Due to failure to genotype on the MassARRAY genotyping platform (Sequenom Inc., San Diego, CA) (Buetow et al., 2001), two SNPs: rs6462731 and rs17171047 were subjected to bi-directional sequencing using BigDye Terminator v.1.1 Cycle Sequencing Kits (Applied Biosystems, Foster City, CA) and an Applied Biosystems 3730×l DNA Analyzer (Applied Biosystems, Foster City, CA), and data was viewed using Sequencher 4.1.4 software (Gene Codes Corporation, Ann Arbor, MI). Two 350 bp amplification products containing rs6462731 and rs17170147 were bi-directionally sequenced, and genotypes were independently reviewed and scored by 3 individuals. Only genotypes obtained via sequencing were used for analyses presented. For QC purposes, each plate contained two duplicate samples and four inter-plate controls.
Statistical Methods
Each SNP was tested for departures from Hardy Weinberg equilibrium (HWE) expectations via the χ2 square goodness-of-fit test; these tests were computed separately for cases and controls. The pairwise linkage disequilibrium (LD) statistic D’ and the haplotype block definition of the four gamete method (Wang et al., 2002) calculated using Haploview 3.2 (Barrett et al., 2005).
All SNPs were tested for genotypic association in the case-control study under the following models: the three a priori genetic models (dominant, additive and recessive) and 2- and 3-SNP haplotype moving windows using our software program SNPGWA (http://www.phs.wfubmc.edu/web/public_bios/sec_gene/downloads.cfm). Our replication criteria were: (1) P<0.05 for both sets; (2) associations in the same direction, with similar MAF>0.10 for each case-control group. SNPs that showed nominal evidence for replication using these criteria were then testing using a permutation test (1,000 permutations) as implemented in SNPGWA.
Individual ancestral proportions were estimated using an EM algorithm (FRAPPE) (Tang et al., 2005) under a two population model. Logistic regression tests of genetic (dominant, additive and recessive) models of association included adjustments for individual estimates of African ancestry implemented in the program SNPADMIX (http://www.phs.wfubmc.edu/web/public_bios/sec_gene/downloads.cfm).
Results
Population characteristics
Characteristics of the AA case and control populations are shown in Table 1. Controls were significantly younger than T2DM-ESRD cases (P<0.0001), although they were significantly older than the mean age at T2DM diagnosis (P<0.0001) for both Set 1 and Set 2. Age data were unavailable for 25% of controls in Set 1 and 18% of controls in Set 2. BMI data are not presented, since measures reflect weight on dialysis in cases and were unavailable for the majority of controls. Similar proportions of females were present in the T2DM-ESRD cases in Set 1 and Set 2 (61% and 62%, respectively), with the higher female proportion likely due to participation and survival bias.
Table 1.
Demographic data for AA Cases and Controls
| Set 1 T2DM- ESRD Cases Mean±SD (n*) |
Set 2 T2DM- ESRD Cases Mean±SD (n*) |
Set 1 Non-diabetic Controls (Mean±SD (n*) |
Set 2 Non-diabetic Controls (Mean±SD (n*) |
|
|---|---|---|---|---|
| Female (% (n) | 61% (352/577) | 62% (344/558) | 51% (306/596) | 71% (328/564) |
| Age at exam (years) | 62.2±10.3 (541) | 61.1± 9.7 (558) | 49.3± 9.8 (448) | 52.3± 12.1 (462) |
| BMI (kg/m2) | 29.6±7.0 (559) | 29.6±6.8 (544) | 29.8±7.1 (450) | 29.5±7.2 (456) |
|
Age at T2DM diagnosis
(years) |
41.8±11.6 (544) | 40.1±12.2 (558) | - | - |
| Age at ESRD onset (years) | 59.0±10.5 (560) | 57.6±10.1 (558) | - | - |
| T2DM Duration (years) | 20.3±10.5 (511) | 20.9±10.4 (558) | - | - |
n = number with data available. Set 1 and Set 2 cases all were diagnosed with T2DM and ESRD.
Estimates of ancestry
FRAPPE (Tang et al., 2005) analysis indicated that the mean proportion of African ancestry in Set 1 was: 0.817±0.133 in T2DM-ESRD cases and 0.791±0.131 in controls; and in Set 2 was 0.814±0.138 in T2DM-ESRD cases and 0.793±0.126 in controls (Table 1). The mean proportion of African ancestry for controls was significantly less than seen in cases for both Set 1 (P=0.001) and Set 2 (P=0.008).
SNP association analyses
A total of 311 bi-allelic variants in the ELMO1 gene were successfully genotyped in Set 1. Based on a threshold significance of P<0.01, 3 SNPs (0.9%) deviated from HWE in cases and 1 SNP (0.3%) deviated from HWE in controls (Supplementary Table 2). While departure from HWE may indicate genotyping errors, it may also reflect admixture differences. There was initial nominal evidence for association with rs3778713, rs17171024 and rs1647791 and T2DM-ESRD, however these SNPs were not among those that later replicated in Set 2.
Haplotype block structure using the four gamete method (Wang et al., 2002) employed in Haploview 3.2 (Barrett et al., 2005) revealed that 250 SNPs (80.4%) fell into 67 blocks of high LD across the gene in the Set 1 AA controls, with mean block length of 4.3 kb (largest block 21 kb), and an average of 3.7 SNPs per block (ranging from 2 to 9 SNPs). In Set 1, AA T2DM-ESRD cases, 260 (83.6%) SNPs fell into 64 blocks of high LD, with mean block length of 4.7 kb (largest block 34 kb), with an average of 4.1 SNPs per block (ranging from 2 to 9 SNPs).
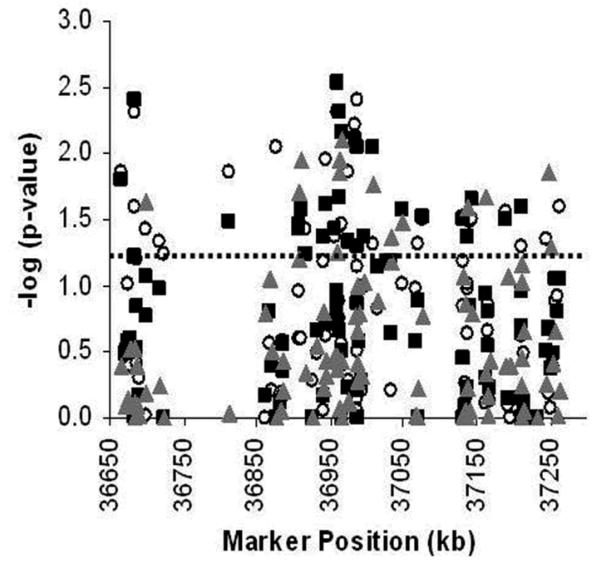
Single-SNP genotypic association results for Set 1 are presented in Figure 1 and Supplementary Table 1. Genotype frequencies and counts for each SNP are shown in Supplementary Table 2. Two- and 3-SNP moving windows analyses were also performed (data not shown). Ninety-eight SNPs (31.5%) showed nominal evidence of association (P<0.05) in one or more tests of association: allelic (N=27), dominant (N=25), additive (N=25), or recessive (N=13) genotypic models, 2- (N=35) or 3-SNP (N=37) haplotypic analyses. The most significant genotypic associations in Set 1 were with 11 SNPs located in intron 13 (Supplementary Table 1).
Figure 1. Single SNP Genotypic association results for 311 SNPs in Set 1.
X-axis: Marker positions in kb; Y-axis: -log10 (global P-value). The P<0.05 nominal level of significance is indicated by the dashed line.
The 98 nominally associated SNPs were subjected to further genotyping in an independent population of 558 additional AA T2DM-ESRD cases and 564 AA non-diabetic controls (Set 2), and extension analyses were performed in 328 AA with T2DM lacking nephropathy and 326 AA with non-diabetic forms of ESRD subjects.
Association analysis of associated SNPs in T2DM-ESRD subjects (Set 2)
A total of 96 SNPs were successfully genotyped in 558 T2DM-ESRD cases and 564 AA controls in the second round of genotyping (Set 2). Single-SNP genotypic association results for Set 2 are presented in Figures 2 and 3, and Supplementary Table 3. Genotype frequencies and counts are shown in Supplementary Table 4. All SNPs were examined for departures from HWE (P<0.01). Four SNPs (4.2%) were inconsistent with HWE in both cases and controls: rs6462731, rs6949576, rs1558688 and rs13225050, however these were not among the replicated SNPs.
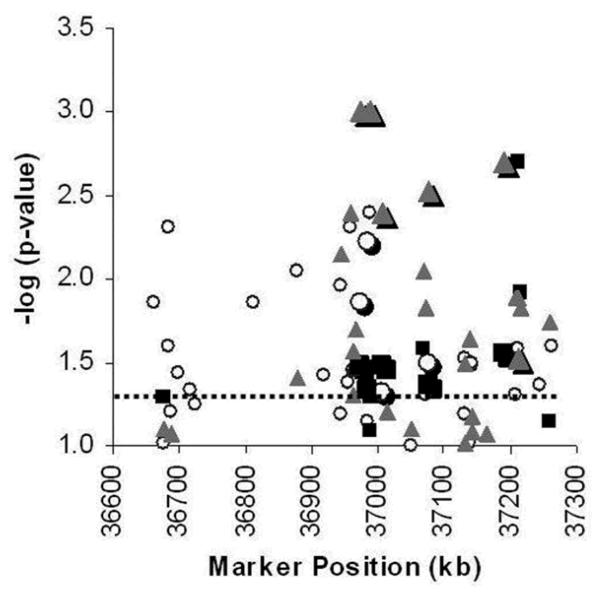
Figure 2. Single-SNP results for the dominant model genotypic test of association with T2DM-ESRD.
Ninety-six SNPs genotyped in Set 1 (N=1,173) (white circles), Set 2 (N=1,122) (gray squares), and Set 1 and Set 2 Combined (N=2,295) (gray triangles). X- axis: Marker positions in kb; Y-axis: -log10 (global P-value). The P<0.05 nominal level of significance is indicated by the dashed line. Replicated SNPs are represented by enlarged data points.
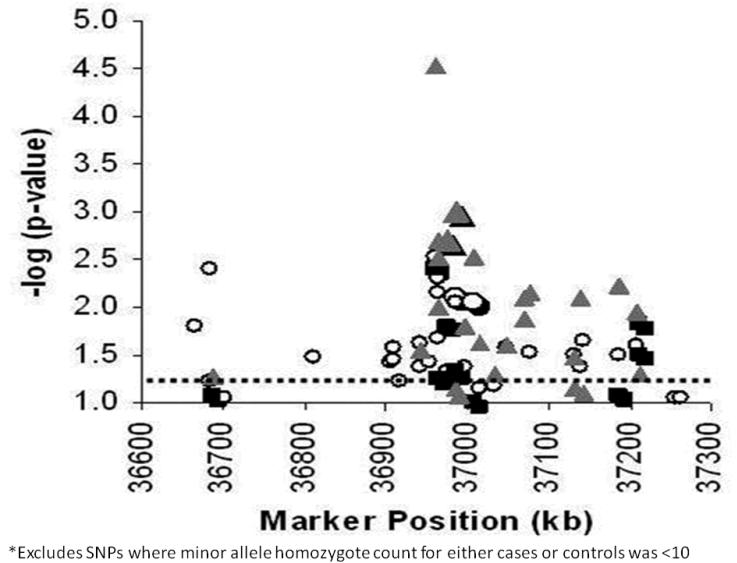
Figure 3. Single-SNP results for the additive model genotypic test of association with T2DM-ESRD.
Ninety-six SNPs genotyped in Set 1 (N=1,173) (white circles), Set 2 (N=1,122) (black squares), and Set 1 and Set 2 Combined (N=2295) (gray triangles). X-axis: Marker positions in kb; Y-axis: -log10 (global P-value). The P<0.05 nominal level of significance is indicated by the dashed line. Replicated SNPs are represented by enlarged data points.
Twenty (20.8%) SNPs showed nominal evidence of association (P<0.05) in one or more tests of association in Set 2: dominant (N=8), additive (N=6), or recessive (N=4) genotypic models, 2- (N=6) and 3-SNP (N=7) haplotypic analyses (Supplementary Table 3).
We compared Set 1 results with Set 2 and considered association confirmed when SNPs met the following criteria: (1) P<0.05 for both sets; (2) associations in the same direction, with similar MAF>0.10 for each case-control group. Based on this approach we observed six SNPs consistently associated with T2DM-ESRD. Results for these SNPs from permutation tests and admixture adjustment are shown in Table 2. Genotypic associations were seen with SNPs: rs1345365 (Set 1: dominant P=0.014; Pa=0.046, Set 2: P=0.037; Pa=0.061); rs1981740 (additive Set 1: P=0.008, Pa=0.041; Set 2: P=0.042;, Pa=0.125); rs2058730 (dominant Set 1: P=0.048, Pa=0.142; Set 2: P=0.033, Pa=0.069); rs10951509 (additive Set 1: P=0.009, Pa=0.055; Set 2: P=0.044, Pa=0.115); rs2717972 (dominant Set 1: P=0.032, Pa=0.068; Set 2: P=0.035, Pa=0.066) and rs9969311 (Set 1: dominant P=0.028, Pa=0.029; Set 2: P=0.026, Pa=0.025) (Table 2). SNPs rs9969311 is located in intron 1, rs2717972 in intron 5 and rs1345365, rs1981740, rs2058730 and rs10951509 in intron 13. All of the replicated SNPs were consistent with HW proportions (P <0.01) in cases (P values ranging 0.47-0.93) and controls (P values ranging 0.02-0.71).
Table 2.
Replicated single-SNP genotypic tests of association with T2DM-ESRD (permutation P<0.05 in both Set 1 and Set 2)
| Marker | Set | MAF | Dominant Model* |
Dominant Admixture Adjusted |
OR (95% CI) | Additive Model |
Additive Admixture Adjusted |
OR (95% CI) |
|---|---|---|---|---|---|---|---|---|
| Cases/ Controls |
P-value | P-value | P-value | P-value | ||||
| rs9969311 | 1 | 0.21/ 0.17 | 0.028 | 0.029 | 1.32 (1.03-1.68) | - | - | - |
| 2 | 0.20/ 0.17 | 0.026 | 0.025 | 1.33 (1.03-1.7) | - | - | - | |
| Combined (Set 1 & 2) |
0.20/ 0.17 | 0.004 | 0.002 | 1.32 (1.11-1.57) | - | - | - | |
| rs2717972 | 1 | 0.45/ 0.50 | 0.032 | 0.068 | 0.75 (0.58-0.98) | - | - | - |
| 2 | 0.47/ 0.50 | 0.035 | 0.066 | 0.75 (0.57-0.99) | - | - | - | |
| Combined (Set 1 & 2) |
0.46/ 0.50 | 0.002 | 0.010 | 0.75 (0.62-0.91) | - | - | - | |
| rs1345365 | 1 | 0.30/ 0.34 | 0.014 | 0.046 | 0.75 (0.59-0.94) | 0.047 | 0.178 | 0.84 (0.70-1.00) |
| 2 | 0.31/ 0.36 | 0.037 | 0.061 | 0.77 (0.61-0.98) | 0.018 | 0.035 | 0.80 (0.67-0.96) | |
| Combined (Set 1 & 2) |
0.31/ 0.35 | 0.001 | 0.006 | 0.76 (0.64-0.90) | 0.002 | 0.014 | 0.82 (0.72-0.93) | |
| rs1981740 | 1 | - | - | - | - | 0.008 | 0.041 | 0.80 (0.67-0.94) |
| 2 | - | - | - | - | 0.042 | 0.125 | 0.84 (0.71-1.00) | |
| Combined (Set 1 & 2) |
- | - | - | - | 0.002 | 0.012 | 0.82 (0.73-0.92) | |
| rs10951509 | 1 | - | - | - | - | 0.009 | 0.055 | 0.79 (0.66-0.94) |
| 2 | - | - | - | - | 0.044 | 0.115 | 0.84 (0.70-1.00) | |
| Combined (Set 1 & 2) |
- | - | - | - | 0.004 | 0.013 | 0.81 (0.72-0.92) | |
| rs2058730 | 1 | 0.30/ 0.34 | 0.048 | 0.142 | 0.79 (0.63-1.00) | - | - | - |
| 2 | 0.31/ 0.34 | 0.033 | 0.069 | 0.77 (0.61-0.98) | - | - | - | |
| Combined (Set 1 & 2) |
0.30/ 0.34 | 0.003 | 0.020 | 0.78 (0.66-0.93) | - | - | - |
Test models refer to the minor allele. Major allele is defined as most common allele in controls. Bold: P-values <0.05.
The four associated intron 13 SNPs fell into two blocks of LD (data not shown), the first block contained SNPs rs1345365, rs1981740, and rs10951509 and the second block contained rs2058730. The three ELMO1 SNPs contained in the single block had pairwise values of D’=0.99 (r2=0.78) between rs1345365 and rs1981740, D’=0.96 (r2=0.93) between rs1345365 and rs10951509, and D’=0.96 (r2=0.78) between rs1981740 and rs10951509.
The MAF for rs1981740, rs10951509 rs2058730 and rs1345365 in AA (AA controls MAF=0.41; 0.36; 0.34; 35 AA cases MAF= 0.37; 0.31; 0.30; 31) are intermediate between the HapMap YRI (0.34; 0.28; 0.30; 0.29) and HapMap CEU (0.81; 0.73; 0.57; 0.68) frequencies.
Analyses of combined association results
Given the modest power to detect association using each set individually, we also performed association analyses on the combined Set 1 + Set 2 genotypic data. A total of 7 (7.3%) and 6 (6.2%) variants were inconsistent with HW proportions (P<0.01) in cases and controls respectively. Two SNPs rs6462731 and rs6949576 deviated from HWE in cases and controls, but were retained for exploratory analyses. Marker rs6462731 deviated from HW proportions in Set 2 T2DM-ESRD cases (P<0.0001) due to an increase of the homozygote ‘AA’ genotype, counterbalanced with an increase of the homozygote ‘GG’ genotype in the Set 2 controls (P=1.6×10-5).
We detected a total of 27 SNPs (28.1%) nominally associated (P < 0.05) with T2DM-ESRD in one or more test of association in the combined Set 1 and Set 2: dominant (N=21), additive (N=20) and recessive (N=8) genotypic models (Figures 2 and 3 and Supplementary Table 5). Eleven of these variants are located in intron 13: rs6462731, rs6967682, rs6462733, rs6979467, rs7782590, rs2160430, rs6462740 and included the four replicated polymorphisms rs1345365, rs1981740, rs2058730 and rs10951509.
Extension analyses in subjects with T2DM lacking nephropathy and non-diabetic kidney disease
To determine whether the associated genetic variants predispose separately to T2DM, or ESRD as a sequela of T2DM, or to ESRD per se (T2DM and non-diabetic disease causes of ESRD), we performed association studies in a population of 328 AA affected with T2DM lacking nephropathy, 326 AA with non-diabetic forms of ESRD and independent controls. The replicated T2DM-ESRD SNPs were not associated with T2DM alone (P values ranging from 0.07 to 0.59) or non-diabetic ESRD (P values ranging from 0.21 to 0.87). Nominal associations with different ELMO1 SNPs were observed with non-diabetic ESRD (two SNPs in intron 13) and T2DM (six SNPs in intron 13) (data not shown).
Discussion
This is the first analysis of the ELMO1 gene in AA affected with T2DM-ESRD. Our interest in ELMO1 gene was due to association between nine intronic SNPs (rs7804092, rs3807163, rs4723596, rs4723593, rs741301, rs11983698, rs1558688, rs7799004, rs1541727) and type 2 diabetic nephropathy in a Japanese population (Shimazaki et al., 2005). Reported associations in the Japanese (Shimazaki et al., 2005) were not detected in our AA population (P-values ranging 0.12 to 0.97 in Set 1), so these SNPs were not investigated in replication analyses.
Replicated associations with T2DM-ESRD were detected with four SNPs located in intron 13 (rs1345365, rs1981740, rs10951509 and rs2058730). For all four SNPs, the minor alleles appear to be protective against T2DM-ESRD, with odds ratios between 0.77-0.84. The frequencies of the replicated intron 13 SNPs (rs1345365, rs2058730, rs10951509 and rs1981740) more closely resemble the frequencies of the HapMap YRI samples, suggesting African influence for disease susceptibly for these markers. There are regions of intron 13 that show a high level of sequence conservation between human, mouse and rhesus monkey. Using HapMap YRI intron 13 genotype data, the four replicated SNPs fall within two blocks of high LD. The first block of 23 kb consisting of 23 HapMap SNPs, and includes rs1345365, rs1981740 and rs10951509. SNP rs2058730 is within a 30kb block containing 40 HapMap SNPs and this SNP is approximately 13.5 kb from the conserved intronic region. It is possible that this region of ELMO1 contains transcription regulatory elements or has a role in regulating splicing events. This hypothesis will need to be explored with functional analysis of the evolutionarily conserved sequences.
The relationship between the associated intron 5 rs2717972 major ‘G’ allele and T2DM-ESRD susceptibility is unknown. However, the minor ‘risk’ A allele appears to be ancestral since it is present in the chimpanzee sequence. This SNP is consistent with HWE (cases: P=0.99; controls P=0.06), but the fact that the ‘A’ allele frequencies are greater in both combined cases (0.46) and controls (0.50) than both pseudo-parental populations, HapMap YRI (MAF 0.53) and HapMap CEU (0.64), suggests this result should be treated with caution.
The more common ‘risk’ G allele of intron 1 SNP rs9969311 appears to be ancestral since it is present in the chimpanzee sequence. The fact that MAF are higher in AA T2DM-ESRD cases (0.20) and controls (0.17) than both the YRI (0.10) and CEU (0.12) pseudo-parental populations, suggests this result should also be treated with caution. However the sample sizes of the AA populations are considerably larger than those of HapMap, and may reflect diverse African ancestry.
In an attempt to avoid false associations due to admixture, individual ancestral proportions were calculated using the program FRAPPE (Tang et al., 2005) and covariate adjustments using these estimates were performed for all SNPs. The replicated SNPs (rs1345365, rs1981740, rs2058730, rs10951509, rs2717972 and rs9969311) all remained significant in at least one model of association in the combined set after adjustment (Table 2). After Bonferroni correction for the three models of genotypic association, one would consider a P<0.0001 and P<0.0002 for Set 1 and Set 2 as significant evidence for association. Although none of the SNPs would meet this threshold, associations were replicated in two independent AA case-control populations, and the effects of these variants appear confined to diabetic nephropathy, consistent with a previous report (Shimazaki et al., 2005).
It is possible that a small number of our reportedly non-diabetic controls could have had undiagnosed T2DM. To evaluate this, we were able to measure a blood sugar in 256 of the controls and 5 had non-fasting serum glucose concentrations greater than 126 mg/dl. This suggests that the overall misclassification rate among controls is likely well below 2%. Although this did not impact our ability to detect positive associations, we cannot rule out the possibility that this may have reduced our power to detect more subtle influences of additional variants, and led to underestimation of effect sizes. The lack of significant effects in non-diabetic ESRD patients and T2DM patients without nephropathy may be attributable to their smaller sample sizes; however trends suggest the replicated associations were specific to diabetic nephropathy. We estimate 37% power to detect an OR of 1.30 based on a MAF > 0.05 at a significance level of 0.05 in the extension cohort (Dupont and Plummer, 1990).
Initial associations observed by Shimazaki et al. (2005), were detected by comparing a diabetic population with nephropathy and diabetic patients with retinopathy in the absence of nephropathy. When combined with our data suggesting that associations are specific to T2DM-ESRD patients, it appears that the effects of these variants may be specific to diabetic kidney disease. Shimazaki et al. (2006) demonstrated that cells stably transfected with ELMO1 manifested diminished adherence to the glomerular basement membrane (GBM) (with the potential for podocyte maladherence to the GBM) and secreted increased amounts of collagen and fibronectin and reduced amounts of matrix metalloproteinases. Therefore, ELMO1 polymorphisms could play a role in the development of glomerulosclerosis and renal fibrosis in the presence of high glucose concentration.
This study represents the first comprehensive evaluation of variation across the entire ELMO1 gene in an AA population, and has also accounted for the impact of admixture on association results. The majority of replicated T2DM-ESRD associations in AA, as well as nominally significant associations in the combined T2DM-ESRD datasets, were located within intron 13 of ELMO1. The results indicate that this region merits further examination to investigate the biological relevance of variations in this region. Investigation of this gene in other populations with diabetic nephropathy is also warranted.
Supplementary Material
Acknowledgments
We thank patients and staff of the Southeastern Kidney Council/ESRD Network 6 and individuals recruited as controls for their participation, Joyce Byers, Mitzie Spainhour, Candace Gordon, Matt Stiegert, and Mark Hansen and colleagues at Illumina Inc. This work was supported by grants DK066358, DK072550, Wake Forest University General Clinical Research Center M01 RR07122, and a Career Development Award from the American Diabetes Association (MMS).
References
- Barrett JC, Fry B, Maller J, Daly MJ. Bioinformatics. 2005;21:263–5. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Boone CA. Nephrol Nurs J. 2000;27:597–600. [PubMed] [Google Scholar]
- Brancati FL, Whittle JC, Whelton PK, Seidler AJ, Klag MJ. Jama. 1992;268:3079–84. [PubMed] [Google Scholar]
- Bryson CL, Ross HJ, Boyko EJ, Young BA. Am J Kidney Dis. 2006;48:720–6. doi: 10.1053/j.ajkd.2006.07.023. [DOI] [PubMed] [Google Scholar]
- Buetow KH, Edmonson M, MacDonald R, Clifford R, Yip P, Kelley J, Little DP, Strausberg R, Koester H, Cantor CR, Braun A. Proc Natl Acad Sci U S A. 2001;98:581–4. doi: 10.1073/pnas.021506298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins AJ, Kasiske B, Herzog C, Chavers B, Foley R, Gilbertson D, Grimm R, Liu J, Louis T, Manning W, Matas A, McBean M, Murray A, St Peter W, Xue J, Fan Q, Guo H, Li S, Roberts T, Snyder J, Solid C, Wang C, Weinhandl E, Arko C, Chen SC, Dalleska F, Daniels F, Dunning S, Ebben J, Frazier E, Johnson R, Sheets D, Forrest B, Berrini D, Constantini E, Everson S, Frederick P, Eggers P, Agodoa L. Am J Kidney Dis. 2005;45:A5–7. doi: 10.1053/j.ajkd.2004.10.009. [DOI] [PubMed] [Google Scholar]
- de Bakker PI, Yelensky R, Pe’er I, Gabriel SB, Daly MJ, Altshuler D. Nat Genet. 2005;37:1217–23. doi: 10.1038/ng1669. [DOI] [PubMed] [Google Scholar]
- Dupont WD, Plummer WD., Jr. Control Clin Trials. 1990;11:116–28. doi: 10.1016/0197-2456(90)90005-m. [DOI] [PubMed] [Google Scholar]
- Freedman BI, Tuttle AB, Spray BJ. Am J Kidney Dis. 1995;25:710–3. doi: 10.1016/0272-6386(95)90546-4. [DOI] [PubMed] [Google Scholar]
- Geiss LS, Herman WH, Goldschmid MG, DeStefano F, Eberhardt MS, Ford ES, German RR, Newman JM, Olson DR, Sepe SJ, et al. MMWR CDC Surveill Summ. 1993;42:1–20. [PubMed] [Google Scholar]
- Gumienny TL, Brugnera E, Tosello-Trampont AC, Kinchen JM, Haney LB, Nishiwaki K, Walk SF, Nemergut ME, Macara IG, Francis R, Schedl T, Qin Y, Van Aelst L, Hengartner MO, Ravichandran KS. Cell. 2001;107:27–41. doi: 10.1016/s0092-8674(01)00520-7. [DOI] [PubMed] [Google Scholar]
- Sale MM, Smith SG, Mychaleckyj JC, Keene KL, Langefeld CD, Leak TS, Hicks PJ, Bowden DW, Rich SS, Freedman BI. Diabetes. 2007;56:2638–42. doi: 10.2337/db07-0012. [DOI] [PubMed] [Google Scholar]
- Shimazaki A, Kawamura Y, Kanazawa A, Sekine A, Saito S, Tsunoda T, Koya D, Babazono T, Tanaka Y, Matsuda M, Kawai K, Iiizumi T, Imanishi M, Shinosaki T, Yanagimoto T, Ikeda M, Omachi S, Kashiwagi A, Kaku K, Iwamoto Y, Kawamori R, Kikkawa R, Nakajima M, Nakamura Y, Maeda S. Diabetes. 2005;54:1171–8. doi: 10.2337/diabetes.54.4.1171. [DOI] [PubMed] [Google Scholar]
- Shimazaki A, Tanaka Y, Shinosaki T, Ikeda M, Watada H, Hirose T, Kawamori R, Maeda S. Kidney Int. 2006;70:1769–76. doi: 10.1038/sj.ki.5001939. [DOI] [PubMed] [Google Scholar]
- Tang H, Peng J, Wang P, Risch NJ. Genet Epidemiol. 2005;28:289–301. doi: 10.1002/gepi.20064. [DOI] [PubMed] [Google Scholar]
- Wang N, Akey JM, Zhang K, Chakraborty R, Jin L. Am J Hum Genet. 2002;71:1227–34. doi: 10.1086/344398. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.