Abstract
The accurate prediction of the secondary and tertiary structure of an RNA with different folding algorithms are dependent on several factors, including the energy functions. However, an RNA higher-order structure cannot be accurately predicted from its sequence based on a limited set of energy parameters. The inter- and intra-molecular forces between this RNA and other small molecules and macromolecules, in addition to other factors in the cell such as pH, ionic strength, and temperature influence the complex dynamics associated with a single stranded RNA's transitioning to its secondary and tertiary structure. Since all of the factors that affect the formation of an RNAs three-dimensional structure cannot be determined experimentally, statistically derived potential energy has been used in the prediction of protein structure. In the current work, we evaluate the statistical free energy of various secondary structure motifs, including base-pair stacks, hairpin loops, and internal loops, using their statistical frequencies obtained from the comparative analysis of more than 50 000 RNA sequences stored in the RNA Comparative Analysis Database (rCAD) at the Comparative RNA Web (CRW) Site. Statistical energies were computed from the structural statistics for several datasets. While the statistical energies for base-pair stacks correlate with experimentally derived free energy values, suggesting a Boltzmann-like distribution, variation is observed between different molecules and their location on the phylogenetic tree of life. Our statistical energies for several structural elements were utilized in the Mfold RNA folding algorithm. The combined statistical energies for base-pair stacks, hairpins and internal loop flanks results in a significant improvement in the accuracy of secondary structure prediction; however, the hairpin flanks contribute the most.
Keywords: statistical potentials, RNA folding, thermodynamic stability, comparative analysis
Introduction
It has been appreciated since canonical base-pairs were first identified and arranged anti-parallel and consecutive with one another to form regular helices, that G:C, A:U, and G:U base pairs are stabilized by hydrogen bonding and base stacking. And with experimental calorimetric measurements of simple oligonucleotides that base-pair with one another, the majority of the known free energy values were determined for consecutive ‘neighbor joining’ base-pairs by Turner and his collaborators.1 While it is appreciated that base-pair stacks (Figure 1A) contribute a significant extent to the stability of an RNA structure, the relative contribution of base stacking and the hydrogen bonding that forms base-pairs and other interactions in RNA structure to the overall stability of an RNA structure is not well understood.2; 3 While the full extent of the types of RNA structural elements and helices have not been identified and characterized, the energetic contribution for only a small percentage of the characterized RNA structural elements have been determined. It is known that approximately 66% of the nucleotides in larger RNAs like the 16S and 23S rRNA form regular secondary structure helix.4 The remaining third of the nucleotides are involved in more complex secondary and tertiary structures.5 A partial list of these includes: U-turns,6 lone pair tri loops,7 a very high percentage of unpaired A's in the secondary structure8 that are involved in several motifs, including - A minor motifs,9; 10, E and E-like motifs,8 UAA/GAN internal loop motif,11 GNRA tetraloops,11; 12 and a high percentage of A:A and A:G juxtapositions at the ends of regular helices,13. The majority of the base pairs in these structural motifs form unusual non-canonical base pair types and their base pair conformations.14; 15; 16
Figure 1.
Illustrations of 4 secondary structures. (a) An example of a base-pair stack that is denoted UA/CG. (b) An example of a base-pair stack that is denoted CG/UA. (c) An example of a hairpin flank that is denoted GC/CA. (d) An example of an internal loop that is denoted GC/CG/AG/AA.
With an incomplete knowledge about all of the possible RNA structural motifs and their energetic stabilities in different structural environments, an alternative approach to the simplified energy model dominated by base-pair stacks in regular secondary structure helices is needed. An analysis of the population of 750 optimal and sub-optimal structures for two different 16S rRNA sequences revealed that the accuracy of the most stable (or optimal) predicted structures with Mfold is higher when the majority of the predicted structures with a minimal variation among themselves have a minimal difference in their total ΔG.17 These results are consistent with the subsequent work of Ding et al. who performed cluster analysis on a distribution of folded structures to improve the accuracy of prediction.18
While experimental approaches have been essential to our understanding of macromolecular structure and their energetic stabilities, it's not feasible to determine the energetic stabilities for all possible structural motifs. In contrast, an analysis of high resolution crystal structures in parallel with the statistical analysis of different sets of comparative macromolecular sequences that form identical or very similar secondary and tertiary structure has been utilized to determine knowledge-based potentials or scoring functions. This approach has been used frequently in protein structure prediction19; 20 following the work by Scheraga.21 An important assumption involved in the conversion of structural statistics into (pseudo) free energy is the Boltzmann-like distribution of the structure, which has been substantiated for proteins.22; 23
Stochastic grammar-based models24; 25; 26; 27 have been used as a non-physical-based method to model RNA. Recently Do28 et al developed, a method that maximizes the expectation of the objective function related to the accuracy of the prediction. Although this knowledge-based approach has only recently been applied to RNA, there has been an increased effort to apply statistically derived potentials to the prediction of RNA structure. A few years ago, Dima and coworkers29 extracted RNA structural statistics from experimentally determined high-resolution structures from the Protein Data Bank to attain PDB-derived potentials for consecutive base-pairs in helices.30 Statistics of the tertiary interactions in the three-dimensional structures of RNAs in PDB were not determined due to the complexity and uncertainty of the sets of nucleotides to study. They determined, to a first approximation, that the structural potentials derived from the base pairings in the secondary structure of experimentally determined 3D structures are similar to those energy values determined experimentally by Turner and collaborators.
Recently, Das et al. developed a Rosetta-like scheme to predict tertiary structure of small RNA sequences of ∼30 nucleotides.31 In their scheme, a statistical potential is inferred from the distance and angle distributions of base-pairs in the ribosome crystal structure, following the method by Sykes and Levitt.32 Parisien33 et al. predicts the secondary and tertiary structures of short RNA molecules with the statistics of nucleotide cyclic motifs using the dynamic programming Waterman-Byers algorithm. Recently, Jonikas et al developed a coarse-grain nucleotide based model to effectively predict structure34 It has been appreciated since the first few tRNA sequences were determined, that different RNA sequences with similar function can form a similar secondary and tertiary structure4; 35 Comparative analysis of tRNA sequences revealed the classic cloverleaf secondary structure36 and parts of the three-dimensional structure.37 Comparative analysis has also revealed other RNA secondary structures that are each similar to different sets of RNA sequences with similar functions. This list includes 5S rRNA,38 16S rRNA,39; 40; 41 23S rRNA,42; 43; 44 RNase P,45 and group I and II introns.46; 47; 48 Covariation and other comparative analysis have the potential to be extremely accurate when there are a sufficient number and diversity of properly aligned sequences. For the rRNA, 97-98% of the base-pairs predicted with covariation analysis were present in the high-resolution crystal structure of the ribosome.5
Due to the emerging technologies that rapidly determine nucleic acid sequences for entire genomes, we are obtaining nucleotide sequences for an increasing number of RNA families with significant increases in the number of sequences per family. To facilitate the analysis of these increasing comparative RNA datasets and to enhance the analysis of these datasets, we have collaborated with Microsoft Research to develop an RNA Comparative Analysis Database (rCAD) that cross-indexes sequence, structure, and phylogenetic information (Ozer, Doshi, Cannone, Xu, and Gutell, manuscript in preparation). In this work, we have utilized rCAD to obtain structural statistics from the comparative analysis of these sequences, and derived statistical energies that can be used for RNA structure prediction. We demonstrate that structural motifs beyond base-pair stacking in helices are also important in determining the RNA structure. The statistical energies derived from sequence information has the potential to significantly improve the accuracy of RNA secondary structure prediction.
Results
Correlation of Base-Pair Stack Statistical Energies and Experimental Thermodynamic Stability
With comparative structural information, we have examined the base-pair stack statistics and their correlation with thermodynamic stability extracted from RNA duplex melting experiments.49 Since statistically derived energies require a reference value to set the absolute scale, we normalized the lowest base-pair stack (BP-ST) statistical energies (SE) with the lowest BP-ST experimental value (See Material and Methods). We have computed molecule-specific BP-ST SE for tRNA (for amino acids A, D, E, G, I, L, M, F), Eukaryotic 5S rRNA, Bacterial 5S rRNA, and Bacterial 16S rRNA. A BP-ST SE was also determined by combining all the sequences from the four datasets (Supplemental Tables). In Figure 2, we plot the statistical energy against experimental data. The correlation coefficient and standard deviation of difference between SE and experimental values for each BP-ST are reported in Table 1. The highest correlation coefficient (0.870) between the statistical and experimental free energies is found for Bacterial 16S rRNA, followed closely by Eukaryotic 5S rRNA (0.857) and tRNA (0.833). The statistical energies for Bacterial 5S rRNA is the least correlated with experimental stability (0.477). More than 80% of the nucleotides in the all-sequence dataset are from Bacterial 16S rRNA, thus the calculated BP-ST SE is biased by the Bacterial 16S rRNA. (Table 1). As a result, the correlation of the all-sequence base-pair SE with experimental stability is also high (0.86). The standard deviations for all-sequence base-pair stack SE, Bacterial 16S rRNA, Bacterial 5S rRNA, Eukaryotic 5S rRNA, and tRNA are 0.525, 0.507, 1.194, 0.677, and 0.988, respectively. In comparison, the PDB-derived base-pair stack energies have a correlation coefficient of 0.7764 with experimental stability (Figure 2F) and a standard deviation of 0.8138.29 All structural statistics extracted from each molecular dataset and free energy data used in Figure 2 are included in the Supplemental Tables S.1 – 10.
Figure 2.
(a) Base-pair stack statistical energy (SE) derived from all-sequence dataset, (b) Bacterial 16S rRNA, (c) Bacterial 5S rRNA, (d) Eukaryotic 5S rRNA, and (e) tRNA versus free energy obtained experimentally for a given base-pair stack. (f) PDB-derived statistical potentials versus free energy obtained experimentally. Experimental values are in kcal/mol.
Table 1.
For statistical energies derived from sequences of each specified molecule, the number of nucleotides in base-pair stacks, correlation coefficient and standard deviation compared with experimental base-pair stack energiesa are presented. The last row lists equivalent information for PDB-derived statistical potentials.
For comparison with experimental base-pair stack free energy we have used symmetric SE for correlation calculations. Hence, 5′ and 3′ directionality are equivalent, e.g. GC/CG and CG/GC are considered to be the same structure. However, asymmetric SE, in which 5′ and 3′ directions are differentiated, will be used to study the folding applications of BP-ST SE. Overall the statistics of base-pair stack frequency show a good correlation with experiment. However a few, base-pair stacks differ more significantly. One of them involves consecutive UG base pairs. The UG/GU base-pair stack statistical energy (SE) from the Bacterial 16S rRNA (-1.0 kcal/mol) and all molecules (-0.82 kcal/mol)(Supplemental Tables S.4 and S5) datasets are much lower than that used by Mfold (-0.5 kcal/mol). However, the value for that base-pair stack in Mfold has been adjusted from the experimental value of 0.47 kcal/mol.50 Additionally, the experimental stability of UG/GU has the largest error (0.96 kcal.mol) among all the base-pair stacks since only one duplex containing the UG/GU was measured which was insufficient for linear regression.50 The statistical potentials for the UG/GU base-pair stack derived from PDB structures also varied significantly from the experimental energy values.29
The largest discrepancy between the all-sequence SE and experimental stability is for the UG/UG (-1.23 kcal/mol) and GU/GU (-0.12 kcal/mol) base-pair stacks (Supplemental Table S.5). The corresponding experimental energy for UG/UG is 0.30 kcal/mol and GU/GU is 1.30 kcal/mol (Supplemental Table S.12). As noted earlier, the statistical energy for these are significantly different from the experimental data.50 The UA/AU base-pair stack statistical energies (Supplemental Tables S.1 – S.5) are all consistently below -2.3 kcal/mol and differ from the experimental value by more than 1 kcal/mol. This indicates that UA/AU base-pair stacks occur more frequently in these RNA molecules than suggested by the stability derived from duplex melting experiment. The GC/GC and GC/CG base-pair stacks determined from calorimetric experiments are the most stable structures with the stability of the former being slightly lower than the latter by 0.1 kcal/mol. However, the lowest (i.e. most stable) base-pair stack SE is GC/CG for the all-sequence, Bacterial 16S rRNA, and Bacterial 5S rRNA datasets. The difference between the SE of the GC/CG base-pair stack for each aforementioned datasets (Supplemental Tables S.3 – S.5) and the corresponding experimental value (Supplemental Table S.12) is 0.9 kcal/mol on average. The GC/GC BP-ST SEs do not immediately follow GC/CG in terms of stability. The PDB-derived potential29 GC/CG base-pair stack minimum agrees with that of our BP-ST SE.
Overall, base-pair stack SE derived from comparative sequences analysis show a better correlation with experimental free energy1 than the PDB-derived potentials. However, noticeable differences exist between SEs of different phylogenetic domains, which concur with experimental observations. For example, although the Escherichia coli loop E of Bacterial 5S rRNA and Spinacia oleracia of Eukaryotic 5S rRNA are found to be isosteric and are able to bind to the same L25 protein, they show substantial differences of stability in various ionic conditions.51 In addition, sequences of RNA molecules have been confirmed by Kiparisov et al to be highly specific and optimized through evolution as only 7 alleles in Saccharomyces cerevisiae 5S rRNA are found to be viable.52 The high correlation indicates that the distribution of base-pair stacks in three out of the four molecule specific datasets, or when combined, is Boltzmann-like. The agreement is rather remarkable especially given that the experiments were performed on isolated duplexes while the statistics were collected from biological sequences, which also represents tertiary contacts and many other factors in the cell. For example, structural studies have supported that hairpin loops can be stabilized by tertiary interactions that are not considered in the oligomer experiments. 13;53;54
Decomposition of Pairing and Stacking
With the statistical frequency from comparative structures, we also modeled the individual contributions of the base pairing (BP) and stacking (ST) from the base-pair stack (BP-ST) energy by fitting the decomposed energies to experimental base-pair stack energies. As described earlier, statistical energies (SE) for individual (Watson-Crick, G-U) base-pairs and stacks are computed from statistical frequency with equations (4) and (5), respectively. A total of 16 different types of stacks can form from the 6 acceptable base pair types. The total energy is then expressed as a function of the individual pair and stack contributions in equation (6). Two assumptions are made in this decomposition model: (1) the contributions of the base-pair and stack are independent and can be separated and (2) the total energy of a base pair stack is obtained from the sum of the decomposed base-pair and stacks. Although the assumptions may be an oversimplification, this examination reveals information on the relationship of the two interactions. Coefficients α and β characterize the relative contribution of each term. By fitting to the 21 base-pair stacks simultaneously using linear least-square regression, we determined 2.3893 for α, 0.6189 for β, and 0.4088 for C. With this decomposition model, we “predict” the energy for each of the 16 canonical base-pair stacks. As shown in Table 2, the values calculated are in good agreement with experimental energies, with a correlation coefficient of 0.882 and RMS error of 0.51 kcal/mol. Since we fit for C, the decomposed base-pair and stack SE only reflect their relative contributions. Accordingly, positive energetic values for stacks only indicate that the base-pair stack containing that stack has a higher energy than the undecomposed value, C. The decomposed statistics and corresponding energy for each base-pair and stacks, total energy computed from the decomposition model are available in Supplemental Tables S.13 and S.14. Although almost all adjacent nucleotides that are involved in base-pairing are stacked, many adjacent nucleotides in other structures, such as hairpin loops and non-consecutive nucleotides that flank coaxial stacked helices, are stacked as well. We also attempted to count all adjacent nucleotides in a sequence as stacks, which yielded similar results.
Table 2.
For each base-pair stack, the decomposed base-pair SE, decomposed stacking SE, base-pair stack energy computed from the decomposition model (equation (6)), base-pair stack SE, experimental base-pair stack energy (kcal/mol), and the difference between base-pair stack SE and experimental energy are presented. SE are in units of kcal/mol and are derived from statistics using all sequences.
| Base-pair 1 | Base-pair 2 | Decomp Base-pair SE | Decomp Stacking SE | Decomp. Base-pair + Stack SE | BP-ST SE | Exp BP-ST energy | Difference of Decomp. BP-ST SE and Exp BP-ST Energy |
|---|---|---|---|---|---|---|---|
| CG | CG | -3.1814 | -0.2416 | -3.01 | -2.43 | -2.40 | 0.61 |
| GC | CG | -3.2819 | -0.2444 | -3.12 | -3.40 | -3.30 | -0.18 |
| GC | GC | -3.3824 | -0.0578 | -3.03 | -2.70 | -3.40 | -0.37 |
| GU | CG | -1.7603 | -0.1206 | -1.47 | -2.32 | -2.10 | -0.63 |
| GU | GC | -1.8608 | -0.1169 | -1.57 | -2.35 | -2.50 | -0.93 |
| GU | GU | -0.3392 | -0.1761 | -0.11 | 0.14 | 1.30 | 1.41 |
| UG | CG | -1.724 | -0.2009 | -1.52 | -1.76 | -1.40 | 0.12 |
| UG | GC | -1.8245 | -0.2202 | -1.64 | -1.93 | -1.50 | 0.14 |
| UG | GU | -0.3029 | -0.0461 | 0.06 | -0.83 | -0.50 | -0.56 |
| UG | UG | -0.2667 | -0.1602 | -0.02 | -1.22 | 0.30 | 0.32 |
| AU | CG | -2.6577 | 0.1496 | -2.10 | -3.19 | -2.10 | 0.00 |
| AU | GC | -2.7583 | -0.0203 | -2.37 | -3.09 | -2.20 | 0.17 |
| AU | GU | -1.2367 | 0.1435 | -0.68 | -2.03 | -1.40 | -0.72 |
| AU | UG | -1.2004 | 0.2242 | -0.57 | -1.33 | -0.60 | -0.03 |
| AU | AU | -2.1341 | 0.4631 | -1.26 | -1.65 | -1.10 | 0.16 |
| UA | CG | -2.8042 | 0.0226 | -2.37 | -3.19 | -2.10 | 0.27 |
| UA | GC | -2.9048 | -0.0084 | -2.50 | -2.91 | -2.40 | 0.10 |
| UA | GU | -1.3832 | 0.1658 | -0.81 | -1.62 | -1.30 | -0.49 |
| UA | UG | -1.3469 | 0.1261 | -0.81 | -1.07 | -1.00 | -0.19 |
| UA | AU | -2.2806 | 0.4795 | -1.39 | -2.53 | -0.90 | 0.49 |
| UA | UA | -2.4271 | 0.4124 | -1.61 | -2.04 | -1.30 | 0.31 |
Since no direct experimental data has been determined for the individual energy contribution for base-pairs or base stacks in isolation from one another, no reference values are available to establish the absolute scale. Thus, the constant C, which is intended to be the intrinsic or sequence-independent stability factor of a base pair stack was added to the decomposition model. Hence, the energy value of every base-pair stack includes the value of C in addition to the sequence specific term. Although no conclusion is drawn from the absolute contributions of pairing or stacking, our model elucidates the relative stability between different base-pair stacks. By comparing the individual contributions from pairing and stacking with the base-pair stack SE, it is evident that the relative stability of base-pair stacks is dominated by the sequences in pairing. Figure 3 reveals a strong BP SE correlation (0.86) with the Watson-Crick/GU base-pairs SE determined using all sequences. By contrast, the stacking SE shows almost no correlation with base-pair stack SE (correlation coefficient of 0.18). The statistics collected in the current study clearly shows that pairing is responsible for the variation of stabilities of base-pair stacking. It is worth emphasizing that this finding does not dispute the importance of the base stack and that it provides more stability as compared to the base-pair. 2; 55; 56 Specifically, suppose that ΔGa and ΔGb are two base-pair stacks. It is possible for the absolute energy of each structure to be primarily stabilized by base stacking. However, ΔΔGba, the relative stability of the base-pair stacks could be largely due to the difference between the base-pair energy. Hence, it is likely that stacking is the driving force for helix formation, yet such interactions are not as specific as the complementarities from hydrogen bonding.
Figure 3.
(a) Base-pair statistical energy versus base-pair stack statistical energy with a correlation coefficient of 0.8743. (b) Stacking statistical energy versus base-pair stack statistical energy with a correlation coefficient of 0.0071.
Application of Statistical Energies to Folding
We investigated the ability of the statistically derived energies of BP-ST, hairpin flanks, and internal loops to improve the prediction of an RNA's secondary structure. We incorporated our statistical-energies in place of those energy parameters determined experimentally in the Mfold program. Statistical energy (SE) derived from the molecule-specific and the all-sequence datasets are utilized within Mfold to predict the secondary structure for tRNA, Eukaryotic rRNA, Bacterial 5S rRNA, and Bacterial 16S rRNA.
Similar to previous studies,50 the folding accuracy is evaluated by comparing base-pairs that are accurately predicted by Mfold with base-pairs determined by comparative sequence analysis5 (see details in Material and Methods). Although Mfold determines the optimal (most stable) structure and a set of sub-optimal secondary structure models, the structure with the lowest energy was used in our analysis for folding accuracy. Furthermore, the experimental energy values were obtained from oligonucleotide duplexes where base-pair stacks have no directionality. However, the anisotropic nature of a base-pair stack is apparent when, for example, it is adjacent to a hairpin loop. Thus, equation (1) and its corresponding modifications were used to evaluate symmetric and asymmetric BP-ST energies for folding accuracy. As shown in Supplemental Tables S.6 – S.10, asymmetric SE offered slightly better folding results overall and will be used in the folding evaluation.
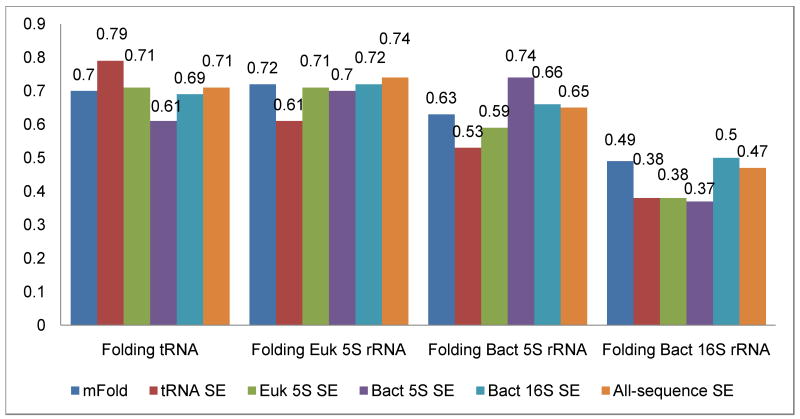
Figure 4 provides the accuracy of folding tRNA, Eukaryotic 5S rRNA, Bacterial 5S rRNA, Bacterial 16S rRNA, using BP-ST SE derived from each molecular and phylogenetic dataset and the all-sequence dataset. The accuracy in secondary structure prediction is either improved or remains the same when the SE derived from each dataset is applied to the sequences within the same dataset. For example, when tRNA-specific SEs are used in place of the experimental BP-ST energies to fold tRNA sequences, the accuracy improved from 0.70 to 0.79. The improvement in the prediction accuracy is more significant for Bacterial 5S rRNA; 0.74 vs. 0.63 for SE and experimentally derived energy values. However, for other datasets (e.g., Bacterial 16S rRNA and Eukaryotic 5S rRNA) the prediction accuracy was similar between the molecule-specific SEs and the experimentally derived energy values. The prediction accuracy usually decreases when the BP-ST SE determined from one molecule/phylogenetic dataset is utilized to predict the secondary structure for another dataset. And last, prediction accuracy for the SE derived from all-sequence dataset or the Bacterial 16S rRNA is similar with the experimentally determined energy values. These results indicated that, the statistics of base-pair stacking in these two datasets is indeed Boltzmann like, and the statistically derived energy is as reliable as the experimental measurements.
Figure 4.
Each group of bars represents folding accuracy of (from left to right) tRNA, Eukaryotic 5S rRNA, Bacterial 5S rRNA, and Bacterial 16S rRNA. Within each group, each bar represents (from left to right) unmodified Mfold, base-pair stack SE derived using tRNA, Eukaryotic 5S rRNA, Bacterial 5S rRNA, Bacterial 16S rRNA, and all-sequence dataset.
Extension of Statistical Energies to Hairpin Flanks and Internal Loops
Our analysis has shown some improvement in the prediction of RNA secondary structure helices from the structural statistics of consecutive base pairs within a helix. About 66% of the nucleotides in an RNA structure predicted with covariation analysis form a base pair. And the vast majority of these are G:C, A:U, and G:U pairings that occur within a regular helix. The remaining third of the RNA secondary structure form hairpin, internal, and multistem loops. However, an analysis of the three-dimensional structure and our growing knowledge about the variety of structural motifs that occur in RNA structure provides a foundation to improve the accuracy in the prediction of an RNAs secondary and even tertiary structure.
The majority of these unpaired nucleotides in the secondary structure are base-paired with non-canonical pairing types and conformation.14 And nearly all of the nucleotides in the rRNA high-resolution structure are stacked onto another nucleotide. While the majority of these stackings are formed between adjacent nucleotides that are base-paired, a significant number of nucleotides that are stacked are not base-paired or consecutive.
Towards that end, Lee, Gardner, and Gutell (manuscript in preparation) have identified and characterized a set of different types of base stackings at the ends of helices that add stability to the helix and potentially protect the ends of the helix while bridging the ends of regular helices with different structural motifs that occur in the hairpin, internal, and multistem loops in an RNA secondary structure. Many of these helix cappings are associated with numerous structural motifs that have been identified and characterized for their chemical structure and energetic properties. One example, the UAA/GAN motif57 has several non-canonical base pairs and unpaired nucleotides that form a longer co-axial stack that bridges the two flanking helices. Another example, the E loop and the E-like loop have been well characterized8; 58 and also contain several non-canonical base pairs that form a contiguous stack onto the regular secondary structure helix.
While the current thermodynamic based folding algorithms consider these unpaired regions of the secondary structure to be destabilizing, as stated earlier, it is known that base stacking contributes more to the overall stability of the RNA structure. While our longer term objectives are to determine the structural potentials associated with all of the structural motifs in the unpaired regions of the secondary structure, for this paper we only determine those structural potentials that can be utilized within the Mfold program.
The statistical energies for hairpin flanks (HF) and internal loops (IL) have been evaluated and utilized in the prediction of an RNA secondary structure with Mfold. HF and IL SE are computed according to equations (7), (10), (12) and (13). The statistical energies for HF, IL (1×1, 1×2, 2×2, and flanks) are available in Supplementary Tables S.16 – 20, respectively. The minimum and maximum SE values were calibrated with those determined in experiments.50 Statistical energies are derived with comparative structures from several RNA datasets: tRNA, Eukaryotic 5S rRNA, Bacterial 5S rRNA, and Bacterial 16S rRNA. All-sequence SE is obtained from the Boltzmann-average of the individual datasets since the occurrences of HF and IL are sparse in structures. We then replaced the energy values for HF-IL in Mfold with the corresponding SE, and set the rest to maximum experimental energy values.
We compare the SE of those motifs from the all-sequence dataset with the corresponding free energy obtained from experiment. Hairpin flanks yield a low correlation with experiment (0.5132) which is computed from SE in Supplemental Table S.16 and experimental values from Mathews work.50 We analyze the differences in SE values between each of the datasets. SE values are only shown for the Base-pair/hairpin flanks that occur within each dataset. The arrangement of the nucleotides are described in the following example. The hairpin flank denoted GC-AC, as illustrated in Figure 1C, is composed of a GC base-pair at the end of a helix and the A is an unpaired nucleotide flanking the paired G and the C is an unpaired nucleotide flanking the paired C. These nucleotides correspond to the first two - columns of the Supplemental Table S.16. With six types of helix ending base pairs (GC, CG, etc.) and 16 possible unpaired flanking pairs (AA, AC, AG, AU, etc.), 96 possible BP/HF flanks are possible. Of the 55 observed, 27 occur in only one dataset (excluding the all-sequence dataset), 16 occur in two datasets, nine occur in three datasets, and three occur in all four datasets.
The most pronounced patterns are:
The hairpin flanking nucleotides that are associated with the most helix ending base pair types is GA, occurring with six different base pairs, followed by CA, UC and UU that both have four different helix ending base pairs.
The vast majority of the most stable helix flanking nucleotides in hairpin loops are GA, UC, UU, and CA with most helix ending base pair types.
The other very stable BP/HF nucleotide sets that do not have a GA, UC, UU, or CA hairpin flank are: CG/AA; CG/AC; GC/AC; GC/AG; UA/UA; CG/UG
Several of these BP/HF sets are associated with known tetraloops, although not exclusively. For example, CG/UG is associated with one of the most stable statistical energies occurs frequently in numerous RNA molecules with the sequence C(UUCG)G; This motif has been determined to be very stable.11; 59 Many but not all of the family of BP/HF nucleotide sets with a GA hairpin flank are associated with the GNRA tetraloop.11; 12 The closing base pairs of these tetraloops were investigated experimentally and generally consistent with our statistical energies for the BP/HF.60 Some of the other GA BP/HP are associated with hairpin loops with six nucleotides. Two experimental studies revealed that the G and A in the hairpin flank form a base pair with a sheared conformation.61; 62
A previous study revealed that many helices in the ribosomal RNAs have an AA or AG flanking the end of the helix in the unpaired region of the secondary structure.13 This analysis revealed that all of the GA flanks associated with a hairpin loop form a base pair. Two of the three most stable BP/HF statistical energies have a helix ending CG base pair and a AA (-2.81) and GA (-2.8) flank (the statistical energy for the most stable BP/HF - AU/UC is -2.87). An experimental study revealed that the hairpin flank GA is more stable than AA for a hairpin loop with the same intervening four nucleotides.63 We anticipate that the accuracy of the prediction of an RNA secondary structure will be further enhanced once we associate the BP/HF statistical energies with the size and sequence of the hairpin loop.
The 1×1 IL-SE has a low correlation with experiment (0.18) as computed from Supplemental Table S.17. Two of the most stable BP/IL/BP 1×1 internal loops has a UU juxtaposition. The next most stable BP/IL/BP 1×1 internal loop has a GA juxtaposition. The UU juxtaposition in the 1×1 internal loops occurs with eight different sets of base pair types that flank both sides of the non-canonical set of nucleotides. Of these, five are considered more stable with statistical energy values > -1.0. The GA and AG juxtapositions in the 1×1 internal loops are both associated with seven different sets of flanking basepairs. However, while six of the seven GA's have a stability greater than -1.0, only two of the AG's have an energy value greater than -1.0. Five of the six UC juxtapositions have an energy value greater than -1.0 and all three of the CU juxtapositions have energy values greater than -1.0. While some of our statistical energies are similar to those determined with experiments, others are very different. For example the UG/U-U/AU (BP/IL/BP) loop statistical energy is underestimated (-1.59 kcal/mol) compared to the 1.7 kcal/mol obtained from experiment50. The AU/G-G/GC loop SE (-0.45 kcal/mol) is also underestimated, compared with the -1.4 kcal/mol of experiment. No 1×1 IL (Supplemental Tables S.17), were observed in the tRNA datasets we analyzed. The Eukaryotic 5S rRNA contains six and the Bacterial 5S rRNA has eight. All of these are considered stable (energy value greater than -1.0 and all have a different set of BP/IL/BP sequences).
The number of occurrences of internal loops with one nucleotide on one side of the helix and two on the other is low. None were present in the tRNA and two 5S rRNA datasets analyzed. Seven were present in the Bacterial 16S rRNA. Of the nine 1×2 internal loops, 24 of the 27 nucleotides are a purine. The correlation between the 1×2 IL-SE (0.11) with experiment is low; The largest underestimation of SE is for the GC/A-AA/AU loop (0.31 kcal/mol) compared with experiment (3.2 kcal/mol).
Twenty five different 2×2 internal loops with unique sets of nucleotides within the internal loop and the two base pairs that flank the internal loop occur in the Bacterial 16S rRNA dataset. These 2×2 internal loops are not in the tRNA and 5S rRNA datasets analyzed here. Of the 25 different 2×2 internal loops, nine of them have tandem GA/AG internal loop, four have the tandem AA/AG IL, four more have the GA/AA IL, and two have the AA/AA IL. In total, 19 have one of the 2×2 internal loops that form a unique structural motif family.64 The G in the GA juxtaposition can be replaced with an A and still maintain the same sheared conformation in many of the tandem GA motifs. Experimental studies revealed an association between the base pairs that flank the tandem GA 2×2 internal loop65 The 2×2 internal loop in three of the 27 BP/2×2 IL/BP is UU/UU. Tandem UU mismatches have been studied experimentally and are stable in some structural environments.66 The 2×2 IL-SE has a low correlation with experiment as well (0.08). The AU/AA-AG/GC loop is underestimated with -4.70 kcal/mol while experiment has 1.00 kcal/mol.
While the SE values determined from the nucleotide frequencies of consecutive base pairs are similar to and sometimes slightly better than the experimentally determined energy values for the same consecutive base pairs, the incorporation of hairpin flank and internal loop SE terms significantly increase the accuracy of the prediction (Figure 5). Using all statistical energies (base-pair stack, hairpin flank, and internal loop), we attain an increase in folding accuracy from 0.70 to 0.89 in tRNA, from 0.72 to 0.84 in Eukaryote 5S rRNA, from 0.63 to 0.88 in Bacterial 5S rRNA, and from 0.49 to 0.56 in Bacterial 16S rRNA. The substantial improvement suggests that the structural elements, by themselves and/or coordinated with the ends of the secondary structure helix stabilizes the higher-order structure of the RNA molecule. The statistical energies for the hairpin flank contributed more to the improvement in the prediction of an RNA secondary structure than for the internal loops and base-pair stacks. In particular, while the folding accuracy increased from 0.70 (for experimental energy values) to 0.89 (for all statististical energies), statistical energies for only internal loops increased the accuracy to 0.78, while statistical energies for only hairpin flanks increased the accuracy to 0.85. Accordingly, the folding accuracy of Eukaryotic 5S rRNA when only hairpin and only internal loop statistical energies are used are 0.79 and 0.74, respectively, in constrast with 0.72 (experimental) and 0.84 (for all statistical energies). However, statistical energies for hairpin flanks do not always contribute more than internal loop flanks to the accuracy of the prediction of an RNA secondary structure. The internal loop flank statistical energy increases the accuracy of RNA folding more than hairpin flanks for Bacterial 5S rRNA. This may be due to the small sampling of the molecule. While the improvement in the folding accuracy with HF SE is not as significant for Bacterial 16S rRNA, the overall improvement does occur. This observation emphasizes the significance of hairpin flanks to RNA structures.
Figure 5.
Each group of bars represents the folding accuracy of (from left to right) tRNA, Eukaryotic 5S rRNA, Bacterial 5S rRNA, and Bacterial 16S rRNA. Within each group, each bar represents (from left to right) unmodified Mfold, base-pair stack SE, hairpin flank SE, internal loop SE, and all available SE (base-pair stack, hairpin flank, and internal loops).
While internal loops within 5S rRNA ranges from 1 – 8 nucleotides in length and 16S rRNA range from 1 – 12 nucleotides in length, only energies for internal loops of lengths with less than four nucleotides can be utilized by Mfold. As a result, the statistical energy contributions for the longer internal loops are not used. Similarly, Mfold has energy functions based on the length and nucleotide composition of hairpins and yet cannot assign an energy contribution for specific hairpin loops with more than 4 nucleotides. This is particularly important since tRNA are mainly composed of loops of lengths 7 and 8, while 5S rRNA are composed of hairpin loops that are longer than 4 nucleotides as well. Hence, the inclusion of energy parameters for larger hairpin loops in to the folding algorithm should increase the prediction accuracy. Although bulge loops are known to be important to the stability of RNA, the simple model that only accounts for the length of the loop to be used as input to evaluate energy does not lend itself to much improvement when statistical methods are applied. Additional development of Mfold would be needed to incorporate statistical energies of bulge loops. Furthermore, many unpaired nucleotides in an RNA's secondary structure participate in tertiary interactions to further stabilize the structure.67,68,69,70 Information on such interactions may be needed to determine loop energies. In addition, a special bonus is used by Mfold when empirical results deviate from the general rules. For example, hairpin loops are checked for a special GU closure and given a bonus.50 Due to these limitations, a new framework beyond the current Mfold is necessary to utilize the statistical energies that can be evaluated from comparative structures.
While the statistical energies derived from one molecular/phylogenetic dataset have the potential to significantly improve the prediction accuracy for sequences in the same dataset (i.e. self-folding), we expect the same set of statistical energies derived from numerous molecular RNA sequences that span the phylogenetic tree of life can be determined and utilized by an RNA folding algorithm to accurately predict the secondary structure for any RNA molecule.
The accuracy of folding each molecule using statistical energies derived from molecule-specific and all-sequence datasets is shown in Figure 6. The statistical energies for BP-ST-HF-IF structural parameters for the all-sequence dataset did predict the secondary structures more accurately (∼0.1) with Mfold than the experimentally derived energy values for tRNA, Eukaryotic 5S rRNA, and Bacterial 5S rRNA (tables or figures that substantiate this claim). The improved accuracy for the Bacterial 16S rRNA dataset was moderate (∼0.05). However, as noted earlier, Bacterial 16S rRNA has a larger number of HF and IL structural elements that cannot be utilized by the current Mfold program.
Figure 6.
Each group of bars represents folding accuracy of (from left to right) tRNA, Eukaryotic 5S rRNA, Bacterial 5S rRNA, and Bacterial 16S rRNA. Within each group, each bar represents (from left to right) unmodified Mfold, base-pair stack, hairpin flank, and internal loop SEs derived using tRNA, Eukaryotic 5S rRNA, Bacterial 5S rRNA, Bacterial 16S rRNA, and all-sequence dataset.
The use of hairpin flank and internal loop energies with base-pair stack energies will enhance the prediction accuracy for some self and cross folding, and decrease the prediction accuracy for other folding. For example, tRNA-specific BP-ST-HF-IL statistical energies increase the accuracy to 89% of the base-pairs in tRNA. However, the statistical energies derived from Eukaryotic and Bacterial 5S rRNA decrease the accuracy for sequences in the tRNA dataset to 0.62 and 0.65, respectively. This corresponds to a decrease by almost 0.3 when the SE values for a different molecule/phylogenetic dataset are used. However, when only BP-ST SE is examined (Figure 5), the difference in folding accuracy is about 0.1 between the tRNA-specific SE and SE from other molecules. Similar trends are observed for other molecules. This is again due to the infrequent occurrences of secondary structures such as hairpin flanks and internal loops as opposed to BP-ST. It is possible that additional comparative structures from other molecules in the future would be helpful. Currently, it appears that Boltzmann average between molecular contributions is an effective approach to retain the “signals” and to derive a general SE for these motifs.
Discussion
Structural statistics from comparative sequence analysis can be used to generate statistical energies that agree with experimental measurements. For base-pair stack, a correlation coefficient of ∼0.9 has been achieved between the energies derived structural statistics of Bacterial 16S rRNA and the all-sequence dataset and the free energy values extracted from experiments. Statistics from individual molecules, such as Bacterial 5S rRNA and tRNA, express specificity, and to an extent, rigidity as Smith et al57 have found nucleotides in Loop E, Helix I, and Helix IV that are lethal to Saccharomyces cerevisiae. However, statistics from a single molecule may not sufficiently represent the complete Boltzmann distribution of base-pair stacks due to biologically-driven biases as well as the small sample size of each small molecule as shown by the number of nucleotides in Table 1, although the sampling issue will improve as more sequences are aligned. Conversely, the dataset for Bacterial 16S rRNA has a vast sample size (slightly less than 1.5 million nucleotides) and, subsequently, represents a Boltzmann-like distribution.
Utilizing the structural statistics, the base-pair stack energy was decomposed into its corresponding base-pair and stacking contributions. We find that base-pairing is the main factor that determines the relative stability of that particular motif. Base-pairing stability is highly correlated to base-pair stacking stability with a correlation coefficient of 0.87 while no correlation is observed between the base-pair stacking and decomposed stacking statistical energy. The analysis suggests that the stacking likely contributes to the intrinsic stability in a non-specific way while the base-pairing, driven by the hydrogen bonding, renders the sequence specificity in the BP-ST.
We further evaluated the SE for base-pair stack, internal loop and hairpin-flank by applying them in Mfold to predict the secondary structures. The statistical energies have been derived from sequences of individual molecules (molecule-specific) and all combined (all-sequence). Molecule-specific base-pair stack energies improve folding accuracy for some molecules and have little effect on others, as compared to the experimental values used by original Mfold. Using all-sequence base-pair stack SE, we observe the accuracy of folding prediction to be comparable to that of free energy obtained experimentally. When SE for hairpin loops and internal loops are included, we see dramatic improvements in the folding accuracy to 0.80, 0.79, and 0.77, and 0.53 for tRNA, Eukaryotic 5S rRNA, Bacterial 5S rRNA, and Bacterial 16S rRNA, respectively. Much of the improvement in accuracy is due to the application of hairpin flank statistical energies. Since Mfold does not utilize energy parameters for structures such as internal loops that have more than 3 nucleotides, not all statistical energies can be employed.
Overall, the prediction accuracy when utilizing the SE values from one dataset to the sequences in another dataset is worse than the prediction accuracy for the experimentally determined energy values. Consistent with our previous analysis, the results suggest that individual RNA molecules have insufficient HF and IL statistics for a Boltzmann-like distribution and cares need to be taken when combining the statistics from different molecules into general statistical energies. More importantly, we have demonstrated here that motifs beyond BP-ST are critical to a more complete understanding of RNA folding and to the refinement of folding algorithms.
Material and Methods
RNA Comparative Structure
The RNA molecules - 5S rRNA, 16S rRNA, and 23S rRNA are available for different phylogenetic groups from the rCAD database. The rCAD database is implemented in the Microsoft SQL Server, a relational database management system. Analysis performed on rCAD can be accessed online at the Comparative RNA Web Site CRW) at http://www.rna.ccbb.utexas.edu. The rCAD system stores over 50 000 RNA sequences, 1746521 nucleotides, and comparative structures. We have rich information on base-pair stack statistics of 319 Bacterial 16S rRNA sequences, 650 tRNA sequences, 263 Eukaryotic 5S rRNA sequences, and 96 Bacterial 5S rRNA. Sequences of the above molecules are available from all phylogenetic domains including Eukaryote, Bacteria, and Archaea. Sequences with similarity of greater than 97% have been pruned to eliminate duplicates. We assume an ensemble of good distribution.71 The ribosomal RNA is studied for its rich structural diversity, its functional abilities and for its well-conserved qualities within phylogenetic domains. The secondary structures of all sequences are evaluated from comparative analysis.4; 42; 72; 73 Structural motifs such as base-pair stacks, hairpin flanks, hairpin loops, internal loops, multi-stem loops, and bulges are stored in tables within the SQL Server.
Base-pair Stack Statistical Energy
Statistics of base-pair stacks found via comparative analysis have been collected to calculate statistical energies. A base-pair stack is denoted AB/CD where AB and CD are base-pairs, and A and D are on the 5′ ends of the stack. For example, the UA/GC base-pair stack is identified in Figure 1A. The sample size of base-pair stacks obtained from sequence analysis is three orders of magnitude greater that those obtained from crystal structures.29 In addition to containing a larger sample size than the set of crystal structures, sequence data contains a greater diversity spanning a larger portion from the phylogenetic tree of life and, hence, a more complete ensemble. Statistics were obtained by counting base-pair stacks composed of canonical Watson-Crick (CG and AU), and GU base-pairs, which are available in the Supplemental Materials.
Statistical energies are calculated from base-pair stack statistics (see Supplemental Figures and Tables) and are evaluated as:
| (1) |
where
| (2) |
and
| (3) |
The indices i, j, k, l represent any of the nucleotides A, C, G, or U. We define NST(ij,kl) as the number of base-pair stacks composed of ij and kl. The total number of base-pair stacks is NST. The delta function δij,kl is equal to 1 if ij=kl and 0 otherwise. Pi is the probability of occurrence of nucleotide i. The scaling factor, λ, is determined by setting , where the numerator is the minimum experimental base-pair stack energy50 and the denominator is the minimum base-pair stack statistical energy. Any statistical energies that are higher than the corresponding maximum experimental value are set equal to the maximum experimental value.
Equations (1)–(3) are used as a common treatment of base-pair stacks where symmetric base-pair stacks such as UA/CG (Figure 1A) and CG/UA (Figure 1B) are considered to be equivalent. For example, the base-pair stack indicated in Figure 1A and Figure 1B are degenerate since the same nucleotides are on the 5′ end of the strands (namely U and C) while A and G are on the 3′ end of the strands in both figures. These two rotated configurations are commonly considered to be equivalent and their statistics would be the average of the two conformations. However, such treatment may not accurately represent the distribution of base-pair stack configurations and asymmetry of directionality of RNA structures, such as the consideration of individual nucleotides that are immediately 5′ and 3′ to a hairpin loop. Hence, we will investigate the asymmetric statistical energy as well. Only slight modifications are needed to equation (2) by eliminating the sum and equation (3) by eliminating the delta function. The statistical energy is normalized by the reference state, P(rand), calculated from the probability of finding each individual nucleotide in the sequences of a given molecule.
We have developed molecule-specific energies by sampling sequences that are specific to particular molecules. For example, to evaluate tRNA-specific energies, we sample from the set of tRNA sequences.
Extracting Components of Base-Pair Stacking Free Energy
We attempt to separate the base-pair stacking energy into pairing and stacking components to elucidate the contributions of the two interactions. In order to evaluate the energy of base-paring, we have
| (4) |
where i′ and j′ are the nucleotides in consideration, PBP(i′ j′) is the probability of finding the base-pair consisting of i′ and j′, and (i′ j′) is the reference state computed as the probability of finding individual nucleotides i′ and j′. The energy of stacking is evaluated as
| (5) |
where i″ and j″ are the nucleotides in consideration, PST (i″ j″) is the probability of finding the stack consisting of i″ and j″, and essentially is same reference state as that in eq (4). With energies for base-pairs, stacks, and base-pair stacks, we can use a linear fit to estimate the relative contribution of base-pairs and stacks using the following form:
| (6) |
In the equation above, nucleotides ij and kl are base-pairs and ik and jl are stacked.
Additional Statistical Energy Terms
In addition to base-pair stacks, the energetics of other secondary structural motifs, although limited in Mfold, may be modified to further refine our free energy calculations and potentially improve folding results. Statistical potentials can be developed by identifying nucleotides at the ends of hairpin loops and nucleotides that flank those loops as shown in Figure 1C. We let i be the nucleotide in a base-pair that surround the 5′ end of the hairpin loop, j be the nucleotide on the 5′ end of the loop, k be the nucleotide on the 3′ end of the loop, and l be nucleotide in a base-pair (with nucleotide i) that surround the 3′ end of the loop. Then the free energy of hairpin flanks can be evaluated as follows:
| (7) |
where
| (8) |
and
| (9) |
The indices i and j represent any of the nucleotides A, C, G, or U. We define NHF(i, j) as the number of hairpins composed of nucleotides, i and j, surrounding the motifs. The total number of hairpin flanks is NHF. Pi is the probability of occurrence of nucleotide i. The scaling factor, λ, is estimated in a similar manner to the previous scaling factors by comparing the minimum value of experimental data50 with the minimum value of ΔGHF(i, j). Furthermore, hairpin flank statistical potentials are limited to the highest energy found in experimental results.
Internal loops are unpaired nucleotides surrounded by helices as shown in Figure 1D. Energy terms for internal loops are derived using nucleotides of the loops on the 5′ and 3′ strands as well as the base-pairs surrounding those nucleotides. Loops of different lengths on each strand use a different energy function. For the example of internal loops with 2 nucleotides on the 5′ and 3′ ends, we evaluate the free energy of internal loops as:
| (10) |
where
| (11) |
and
| (12) |
The indices i1 and i4 are the base-pairs around the internal loop on the 5′ end of the molecule. Indices i2, and i3 are the nucleotides consisting of the internal loop on the 5′ end of the molecule. Indices j1, j2, j3, and j4 are the corresponding nucleotides on the 3′ end of the molecule. We define NIL(i1, i2, i3, i4, j1, j2, j3, j4) as the number of internal loops composed of nucleotides, i1, i2, i3, i4, j1, j2, j3, and j4. Similarly, internal loops with 1 nucleotide on the 5′ and 3′ ends are evaluated as:
| (12) |
In this case, i1, i3, j1, and j3 are the surrounding base-pairs while i2 and j2 are the nucleotides of the internal loops. Finally, internal loops with 1 nucleotide on one strand and 2 nucleotides on another strand are evaluated as:
| (13) |
The nucleotides i1, i3, j1, and j4 are the surrounding base-pairs and the rest are the internal loops. The scaling factor, λ, was estimated similarly to the previous scaling factors by comparing the minimum value of experimental data50 with the minimum value of ΔGIL(i1, i2, i3, j1, j2, j3, j4). Note that λ is uniquely calculated for each equation. Furthermore, hairpin flank statistical potentials are limited to the highest energy found in experimental results. All statistically derived energies can be downloaded and applied to the original Mfold program at http://biomol.bme.utexas.edu/∼wuch/statistical energies.
Statistical Energy Derived from All-Sequence Dataset
In addition to molecule-specific potentials, sequences from all molecules are combined to derive the all-sequence statistical energy that can potentially be applied to prediction. For base-pair stacks, we have directly combined all sequences from all molecules to derive the statistical energy. For hairpin flanks and internal loops, however, due to the limited occurrences in smaller molecules such as the 5S rRNA and tRNA, a different approach was used to combine the statistics from individual molecules. Hence, the all-sequence hairpin flank statistical energy and all-sequence internal loop statistical energy are derived by averaging the molecule-specific statistical energies from all molecules using Boltzmann weights:
| (14) |
For each structural element P, such as the base-pair stack and internal loops discussed above, the all-sequence statistical energy is a weighted sum of energies from each molecule i.
Evaluation of Statistical Potentials
We applied different combinations of base-pair stack (BP-ST), hairpin flank (HF), and internal loop (IL) statistical energies (SE) to the Mfold program50 developed by Zuker et al to test its ability to predict RNA folding. The Mfold program was also modified to utilize asymmetric BP-ST SE in energy calculations. Five sets of asymmetric SE were derived from tRNA, Eukaryotic 5S rRNA, Bacterial 5S rRNA, Bacterial 16S rRNA, and all sequences combined. Each set of SE utilizing BP-ST and BP-ST-HF-IL terms were used to fold each of the four molecules.
In order to make performance comparisons of energy values, the number of base-pairs of a structure predicted by Mfold that are in agreement with comparative sequence analysis5 is divided by all base-pairs determined by comparative sequence analysis. Only canonical (Watson-Crick G-C, A-U, and G-U) base-pairs are counted. Although this performance measurement does not count erroneously predicted base-pairs, false positives affect the predicted structure and may preclude correct base-pairs from forming. This metric is simple and sufficient in evaluating the accuracy of folding.
Supplementary Material
Acknowledgments
The authors acknowledge the Robert A. Welch Foundation (grant numbers F-1691 and F-1427), the National Institutes of Health (R01GM079686 and R01GM067317), and Microsoft Research for financial gifts and a TCI grant. Kishore Doshi is acknowledged for his contribution to the development of the rCAD system. The authors are also grateful to resources provided by the Texas Advanced Computing Center.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Zuker M, Jaeger JA, Turner DH. A comparison of optimal and suboptimal RNA secondary structures predicted by free energy minimization with structures determined by phylogenetic comparison. Nucleic Acids Res. 1991;19:2707–14. doi: 10.1093/nar/19.10.2707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yakovchuk P, Protozanova E, Frank-Kamenetskii MD. Base-stacking and base-pairing contributions into thermal stability of the DNA double helix. Nucleic Acids Research. 2006;34:564–574. doi: 10.1093/nar/gkj454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guckian KM, Schweitzer BA, Ren RXF, Sheils CJ, Tahmassebi DC, Kool ET. Factors contributing to aromatic stacking in water: Evaluation in the context of DNA. Journal of the American Chemical Society. 2000;122:2213–2222. doi: 10.1021/ja9934854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gutell RR, Weiser B, Woese CR, Noller HF. Comparative anatomy of 16-S-like ribosomal-RNA. Progress in Nucleic Acid Research and Molecular Biology. 1985;32:155–216. doi: 10.1016/s0079-6603(08)60348-7. [DOI] [PubMed] [Google Scholar]
- 5.Gutell RR, Lee JC, Cannone JJ. The accuracy of ribosomal RNA comparative structure models. Current Opinion in Structural Biology. 2002;12:301–310. doi: 10.1016/s0959-440x(02)00339-1. [DOI] [PubMed] [Google Scholar]
- 6.Gutell RR, Cannone JJ, Konings D, Gautheret D. Predicting U-turns in ribosomal RNA with comparative sequence analysis. Journal of Molecular Biology. 2000;300:791–803. doi: 10.1006/jmbi.2000.3900. [DOI] [PubMed] [Google Scholar]
- 7.Lee JC, Cannone JJ, Gutell RR. The lonepair triloop: A new motif in RNA structure. Journal of Molecular Biology. 2003;325:65–83. doi: 10.1016/s0022-2836(02)01106-3. [DOI] [PubMed] [Google Scholar]
- 8.Gutell RR, Cannone JJ, Shang Z, Du Y, Serra MJ. A story: Unpaired adenosine bases in ribosomal RNAs. Journal of Molecular Biology. 2000;304:335–354. doi: 10.1006/jmbi.2000.4172. [DOI] [PubMed] [Google Scholar]
- 9.Nissen P, Ippolito JA, Ban N, Moore PB, Steitz TA. RNA tertiary interactions in the large ribosomal subunit: The A-minor motif. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:4899–4903. doi: 10.1073/pnas.081082398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Battle DJ, Doudna JA. Specificity of RNA-RNA helix recognition. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:11676–11681. doi: 10.1073/pnas.182221799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Woese CR, Winker S, Gutell RR. Architecture of Ribosomal-Rna -Constraints on the Sequence of Tetra-Loops. Proceedings of the National Academy of Sciences of the United States of America. 1990;87:8467–8471. doi: 10.1073/pnas.87.21.8467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Michel F, Westhof E. Modeling of the 3-Dimensional Architecture of Group-I Catalytic Introns Based on Comparative Sequence-Analysis. Journal of Molecular Biology. 1990;216:585–610. doi: 10.1016/0022-2836(90)90386-Z. [DOI] [PubMed] [Google Scholar]
- 13.Elgavish T, Cannone JJ, Lee JC, Harvey SC, Gutell RR. AA.AG@Helix.Ends: A: A and A: G base-pairs at the ends of 16 S and 23 S rRNA helices. Journal of Molecular Biology. 2001;310:735–753. doi: 10.1006/jmbi.2001.4807. [DOI] [PubMed] [Google Scholar]
- 14.Lee JC, Gutell RR. Diversity of base-pair conformations and their occurrence in rRNA structure and RNA structural motifs. Journal of Molecular Biology. 2004;344:1225–1249. doi: 10.1016/j.jmb.2004.09.072. [DOI] [PubMed] [Google Scholar]
- 15.Leontis NB, Stombaugh J, Westhof E. The non-Watson-Crick base pairs and their associated isostericity matrices. Nucleic Acids Research. 2002;30:3497–3531. doi: 10.1093/nar/gkf481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xin YR, Olson WK. BPS: a database of RNA base-pair structures. Nucleic Acids Research. 2009;37:D83–D88. doi: 10.1093/nar/gkn676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Doshi KJ, Cannone JJ, Cobaugh CW, Gutell RR. Evaluation of the suitability of free-energy minimization using nearest-neighbor energy parameters for RNA secondary structure prediction. Bmc Bioinformatics. 2004;5 doi: 10.1186/1471-2105-5-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ding Y, Chan CY, Lawrence CE. RNA secondary structure prediction by centroids in a Boltzmann weighted ensemble. Rna-a Publication of the Rna Society. 2005;11:1157–1166. doi: 10.1261/rna.2500605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Floudas CA, Fung HK, McAllister SR, Monnigmann M, Rajgaria R. Advances in protein structure prediction and de novo protein design: A review. Chemical Engineering Science. 2006;61:966–988. [Google Scholar]
- 20.Shen MY, Sali A. Statistical potential for assessment and prediction of protein structures. Protein Science. 2006;15:2507–2524. doi: 10.1110/ps.062416606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tanaka S, Scheraga HA. Medium- and long-range interaction parameters between amino acids for predicting three-dimensional structures of proteins. Macromolecules. 1976;9:945–50. doi: 10.1021/ma60054a013. [DOI] [PubMed] [Google Scholar]
- 22.Bryant SH, Lawrence CE. The Frequency Of Ion-Pair Substructures In Proteins Is Quantitatively Related To Electrostatic Potential - A Statistical-Model For Nonbonded Interactions. Proteins-Structure Function and Genetics. 1991;9:108–119. doi: 10.1002/prot.340090205. [DOI] [PubMed] [Google Scholar]
- 23.Finkelstein AV, Badretdinov AY, Gutin AM. Why Do Protein Architectures Have Boltzmann-Like Statistics. Proteins-Structure Function and Genetics. 1995;23:142–150. doi: 10.1002/prot.340230204. [DOI] [PubMed] [Google Scholar]
- 24.Sakakibara Y, Brown M, Hughey R, Mian IS, Sjolander K, Underwood RC, Haussler D. Stochastic Context-Free Grammars for Transfer-RNA Modeling. Nucleic Acids Research. 1994;22:5112–5120. doi: 10.1093/nar/22.23.5112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Durbin R, Eddy SR, Krogh A, Mitchison G. Biological Sequence Analysis: Probabilistic Models of Proteins and Nucleic Acids. Cambridge University Press; 1998. [Google Scholar]
- 26.Knudsen B, Hein J. RNA secondary structure prediction using stochastic context-free grammars and evolutionary history. Bioinformatics. 1999;15:446–454. doi: 10.1093/bioinformatics/15.6.446. [DOI] [PubMed] [Google Scholar]
- 27.Knudsen B, Hein J. Pfold: RNA secondary structure prediction using stochastic context-free grammars. Nucleic Acids Research. 2003;31:3423–3428. doi: 10.1093/nar/gkg614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Do CB, Woods DA, Batzoglou S. CONTRAfold: RNA secondary structure prediction without physics-based models. Bioinformatics. 2006;22:E90–E98. doi: 10.1093/bioinformatics/btl246. [DOI] [PubMed] [Google Scholar]
- 29.Dima RI, Hyeon C, Thirumalai D. Extracting stacking interaction parameters for RNA from the data set of native structures. Journal of Molecular Biology. 2005;347:53–69. doi: 10.1016/j.jmb.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 30.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Research. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Das R, Baker D. Automated de novo prediction of native-like RNA tertiary structures. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:14664–14669. doi: 10.1073/pnas.0703836104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sykes MT, Levitt M. Describing RNA structure by libraries of clustered nucleotide doublets. Journal of Molecular Biology. 2005;351:26–38. doi: 10.1016/j.jmb.2005.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Parisien M, Major F. The MC-Fold and MC-Sym pipeline infers RNA structure from sequence data. Nature. 2008;452:51–55. doi: 10.1038/nature06684. [DOI] [PubMed] [Google Scholar]
- 34.Jonikas MA, Radmer RJ, Laederach A, Das R, Pearlman S, Herschlag D, Altman RB. Coarse-grained modeling of large RNA molecules with knowledge-based potentials and structural filters. RNA-a Publication of the RNA Society. 2009;15:189–199. doi: 10.1261/rna.1270809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Woese CR, Gutell R, Gupta R, Noller HF. Detailed Analysis of the Higher-Order Structure of 16s-Like Ribosomal Ribonucleic-Acids. Microbiological Reviews. 1983;47:621. doi: 10.1128/mr.47.4.621-669.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Holley RW, Apgar J, Everett GA, Madison JT, Marquise M, Merrill SH, Penswick JR, Zamir A. Structure Of A Ribonucleic Acid. Science. 1965;147:1462. doi: 10.1126/science.147.3664.1462. [DOI] [PubMed] [Google Scholar]
- 37.Levitt M. Detailed Molecular Model For Transfer Ribonucleic Acid. Nature. 1969;224:759. doi: 10.1038/224759a0. [DOI] [PubMed] [Google Scholar]
- 38.Fox GE, Woese CR. 5S-RNA Secondary Structure. Nature. 1975;256:505–507. doi: 10.1038/256505a0. [DOI] [PubMed] [Google Scholar]
- 39.Woese CR, Magrum LJ, Gupta R, Siegel RB, Stahl DA, Kop J, Crawford N, Brosius J, Gutell R, Hogan JJ, Noller HF. Secondary Structure Model For Bacterial 16S Ribosomal-RNA - Phylogenetic, Enzymatic And Chemical Evidence. Nucleic Acids Research. 1980;8:2275–2293. doi: 10.1093/nar/8.10.2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zwieb C, Glotz C, Brimacombe R. Secondary Structure Comparisons Between Small Subunit Ribosomal-RNA Molecules From 6 Different Species. Nucleic Acids Research. 1981;9:3621–3640. doi: 10.1093/nar/9.15.3621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stiegler P, Carbon P, Zuker M, Ebel JP, Ehresmann C. Secondary Structure And Topography Of 16S-Ribosomal RNA From Escherichia-Coli. Comptes Rendus Hebdomadaires Des Seances De L Academie Des Sciences Serie D. 1980;291:937–940. [PubMed] [Google Scholar]
- 42.Noller HF, Kop J, Wheaton V, Brosius J, Gutell RR, Kopylov AM, Dohme F, Herr W, Stahl DA, Gupta R, Woese CR. Secondary Structure Model for 23s Ribosomal-Rna. Nucleic Acids Research. 1981;9:6167–6189. doi: 10.1093/nar/9.22.6167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Glotz C, Zwieb C, Brimacombe R, Edwards K, Kossel H. Secondary Structure of the Large Subunit Ribosomal-Rna from Escherichia-Coli, Zea-Mays Chloroplast, and Human and Mouse Mitochondrial Ribosomes. Nucleic Acids Research. 1981;9:3287–3306. doi: 10.1093/nar/9.14.3287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Branlant C, Krol A, Machatt MA, Pouyet J, Ebel JP, Edwards K, Kossel H. Primary and Secondary Structures of Escherichia-Coli Mre-600-23s Ribosomal-Rna - Comparison with Models of Secondary Structure for Maize Chloroplast 23s Ribosomal-Rna and for Large Portions of Mouse and Human 16s Mitochondrial Ribosomal-Rnas. Nucleic Acids Research. 1981;9:4303–4324. doi: 10.1093/nar/9.17.4303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.James BD, Olsen GJ, Liu JS, Pace NR. The Secondary Structure of Ribonuclease-P Rna, the Catalytic Element of a Ribonucleoprotein Enzyme. Cell. 1988;52:19–26. doi: 10.1016/0092-8674(88)90527-2. [DOI] [PubMed] [Google Scholar]
- 46.Michel F, Jacquier A, Dujon B. Comparison of Fungal Mitochondrial Introns Reveals Extensive Homologies in Rna Secondary Structure. Biochimie. 1982;64:867–881. doi: 10.1016/s0300-9084(82)80349-0. [DOI] [PubMed] [Google Scholar]
- 47.Cech TR. Conserved Sequences and Structures of Group-I Introns - Building an Active-Site for Rna Catalysis - a Review. Gene. 1988;73:259–271. doi: 10.1016/0378-1119(88)90492-1. [DOI] [PubMed] [Google Scholar]
- 48.Michel F, Umesono K, Ozeki H. Comparative and Functional-Anatomy of Group-Ii Catalytic Introns - a Review. Gene. 1989;82:5–30. doi: 10.1016/0378-1119(89)90026-7. [DOI] [PubMed] [Google Scholar]
- 49.Xia TB, SantaLucia J, Burkard ME, Kierzek R, Schroeder SJ, Jiao XQ, Cox C, Turner DH. Thermodynamic parameters for an expanded nearest-neighbor model for formation of RNA duplexes with Watson-Crick base pairs. Biochemistry. 1998;37:14719–14735. doi: 10.1021/bi9809425. [DOI] [PubMed] [Google Scholar]
- 50.Mathews DH, Sabina J, Zuker M, Turner DH. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. Journal of Molecular Biology. 1999;288:911–940. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
- 51.Vallurupalli P, Moore PB. The solution structure of the loop E region of the 5 S rRNA from spinach chloroplasts. Journal of Molecular Biology. 2003;325:843–856. doi: 10.1016/s0022-2836(02)01270-6. [DOI] [PubMed] [Google Scholar]
- 52.Kiparisov S, Petrov A, Meskauskas A, Sergiev PV, Dontsova OA, Dinman JD. Structural and functional analysis of 5S rRNA in Saccharomyces cerevisiae. Molecular Genetics and Genomics. 2005;274:235–247. doi: 10.1007/s00438-005-0020-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gutell RR, Woese CR. Higher-Order Structural Elements in Ribosomal-Rnas - Pseudo-Knots and the Use of Noncanonical Pairs. Proceedings of the National Academy of Sciences of the United States of America. 1990;87:663–667. doi: 10.1073/pnas.87.2.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liang XG, Kuhn H, Frank-Kamenetskii MD. Monitoring single-stranded DNA secondary structure formation by determining the topological state of DNA catenanes. Biophysical Journal. 2006;90:2877–2889. doi: 10.1529/biophysj.105.074104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kool ET, Morales JC, Guckian KM. Mimicking the structure and function of DNA: Insights into DNA stability and replication. Angewandte Chemie-International Edition. 2000;39:990–1009. doi: 10.1002/(sici)1521-3773(20000317)39:6<990::aid-anie990>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- 56.Protozanova E, Yakovchuk P, Frank-Kamenetskii MD. Stacked-unstacked equilibrium at the nick site of DNA. Journal of Molecular Biology. 2004;342:775–785. doi: 10.1016/j.jmb.2004.07.075. [DOI] [PubMed] [Google Scholar]
- 57.Smith MW, Meskauskas A, Wang P, Sergiev PV, Dinman JD. Saturation Mutagenesis of 5S rRNA in Saccharomyces cerevisiae. Molecular and Cellular Biology. 2001;21:8264–8275. doi: 10.1128/MCB.21.24.8264-8275.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wimberly B, Varani G, Tinoco I. The Conformation Of Loop-E Of Eukaryotic 5S-Ribosomal RNA. Biochemistry. 1993;32:1078–1087. doi: 10.1021/bi00055a013. [DOI] [PubMed] [Google Scholar]
- 59.Tuerk C, Gauss P, Thermes C, Groebe DR, Gayle M, Guild N, Stormo G, Daubentoncarafa Y, Uhlenbeck OC, Tinoco I, Brody EN, Gold L. CUUCGG Hairpins - Extraordinarily Stable RNA Secondary Structures Associated With Various Biochemical Processes. Proceedings of the National Academy of Sciences of the United States of America. 1988;85:1364–1368. doi: 10.1073/pnas.85.5.1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Antao VP, Tinoco I. Thermodynamic Parameters For Loop Formation In RNA And DNA Hairpin Tetraloops. Nucleic Acids Research. 1992;20:819–824. doi: 10.1093/nar/20.4.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Huang SG, Wang YX, Draper DE. Structure of a hexanucleotide RNA hairpin loop conserved in ribosomal RNAs. Journal of Molecular Biology. 1996;258:308–321. doi: 10.1006/jmbi.1996.0252. [DOI] [PubMed] [Google Scholar]
- 62.Fountain MA, Serra MJ, Krugh TR, Turner DH. Structural features of a six-nucleotide RNA hairpin loop found in ribosomal RNA. Biochemistry. 1996;35:6539–6548. doi: 10.1021/bi952697k. [DOI] [PubMed] [Google Scholar]
- 63.Serra MJ, Lyttle MH, Axenson TJ, Schadt CA, Turner DH. RNA Hairpin Loop Stability Depends On Closing Base-Pair. Nucleic Acids Research. 1993;21:3845–3849. doi: 10.1093/nar/21.16.3845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gautheret D, Konings D, Gutell RR. A Major Family Of Motifs Involving G-Center-Dot-A Mismatches In Ribosomal-RNA. Journal of Molecular Biology. 1994;242:1–8. doi: 10.1006/jmbi.1994.1552. [DOI] [PubMed] [Google Scholar]
- 65.Walter AE, Wu M, Turner DH. The Stability and Structure of Tandem Ga Mismatches in Rna Depend on Closing Base-Pairs. Biochemistry. 1994;33:11349–11354. doi: 10.1021/bi00203a033. [DOI] [PubMed] [Google Scholar]
- 66.Santalucia J, Kierzek R, Turner DH. Stabilities Of Consecutive A.C, C.C, G.G, U.C, And U.U Mismatches In RNA Internal Loops - Evidence For Stable Hydrogen-Bonded U.U And C.C+ Pairs. Biochemistry. 1991;30:8242–8251. doi: 10.1021/bi00247a021. [DOI] [PubMed] [Google Scholar]
- 67.Gate JH, Gooding AR, Podell E, Zhou KH, Golden BL, Szewczak AA, Kundrot CE, Cech TR, Doudna JA. RNA tertiary structure mediation by adenosine platforms. Science. 1996;273:1696–1699. doi: 10.1126/science.273.5282.1696. [DOI] [PubMed] [Google Scholar]
- 68.Costa M, Michel F. Frequent Use Of The Same Tertiary Motif By Self-Folding RNAs. Embo Journal. 1995;14:1276–1285. doi: 10.1002/j.1460-2075.1995.tb07111.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jaeger L, Michel F, Westhof E. Involvement Of A GNRA Tetraloop In Long-Range Tertiary Interactions. Journal of Molecular Biology. 1994;236:1271–1276. doi: 10.1016/0022-2836(94)90055-8. [DOI] [PubMed] [Google Scholar]
- 70.Lee JC, Gutell RR, Russell R. The UAA/GAN internal loop motif: A new RNA structural element that forms a cross-strand AAA stack and long-range tertiary interactions. Journal of Molecular Biology. 2006;360:978–988. doi: 10.1016/j.jmb.2006.05.066. [DOI] [PubMed] [Google Scholar]
- 71.Bloch F. Fundamentals of statistical mechanics: manuscript and notes of Felix Bloch. Imperial College Press; World Scientific; London: Singapore: 2000. [Google Scholar]
- 72.Woese CR, Gutell R, Gupta R, Noller HF. Detailed Analysis Of The Higher-Order Structure Of 16S-Like Ribosomal Ribonucleic-Acids. Microbiological Reviews. 1983;47:621–&. doi: 10.1128/mr.47.4.621-669.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gutell RR. Collection Of Small-Subunit (16S- And 16S-Like) Ribosomal-Rna Structures. Nucleic Acids Research. 1994;22:3502–3507. doi: 10.1093/nar/22.17.3502. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.