SUMMARY
In proneural groups of cells in the morphogenetic furrow of the developing Drosophila eye phosphorylated mitogen activated protein kinase (MAPK) antigen is held in the cytoplasm for hours. We have developed a reagent to detect nuclear MAPK non-antigenically and report our use of this reagent to confirm that MAPK nuclear translocation is regulated by a second mechanism in addition to phosphorylation. This ‘cytoplasmic hold’ of activated MAPK has not been observed in cell culture systems. We also show that MAPK cytoplasmic hold has an essential function in vivo: if it is overcome, developmental patterning in the furrow is disrupted.
Keywords: MAP kinase, Drosophila, Compound eye
INTRODUCTION
The Drosophila compound eye is composed of about 800 similar facets or ommatidia, each of which contains eight photoreceptor and 12 accessory cells (Ready et al., 1976). Early in life, the presumptive eye is a columnar monolayer epithelium in the eye-antennal imaginal disc, which grows by unpatterned proliferation until the third larval instar, when a wave of cell-cycle synchronization, cell-type specification and patterning moves across the disc from posterior to anterior, called the morphogenetic furrow (Ready et al., 1976). As the furrow moves a new column of future ommatidia is formed roughly every 2 hours (Basler and Hafen, 1989). In the furrow all cells are held in the G1 phase of the cell cycle (Thomas et al., 1994).
The first cell type specified is the R8 photoreceptor (Tomlinson and Ready, 1987) and the central event in the formation of this founding cell is the progressive restriction of the transcription factor Atonal to individual cells (Jarman et al., 1994; White and Jarman, 2000). Atonal expression begins about 10−15 cells anterior to the morphogenetic furrow, initially in all cell nuclei in a rising gradient (Fig. 1). Then expression is lost from some cells and retained in others, to produce the ‘intermediate groups’ (arrow in Fig. 1E) (Jarman et al., 1995). By the next column, only one cell retains Atonal: the presumptive R8 photoreceptor cell (Baker et al., 1996).
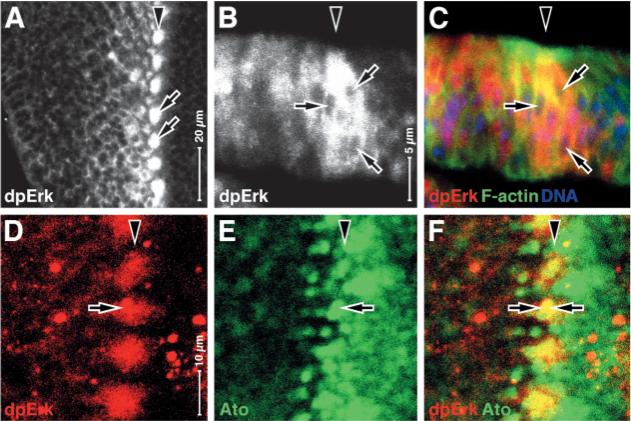
Fig. 1.
MAP kinase phosphorylation in the Atonal-positive intermediate groups in phase 1. Wild-type third instar eye-imaginal discs, anterior rightwards and (B,C) apical upwards. Arrowheads indicate furrow in all panels. (A) Antigen dpErk (which indicates MAPK activation); arrows indicate cell clusters with elevated dpErk. (B,C) dpErk in the large clusters is predominantly cytoplasmic (white in B, red in C). Arrows indicate unstained nuclei. (D-F) Co-localization of dpErk (red) and Atonal (green). Note that dpErk clusters correspond to the Atonal-expressing intermediate groups (arrows).
The remaining cells are recruited after the R8 (Tomlinson, 1988; Wolff and Ready, 1993) and each of these later recruitment steps depends on inductive signaling via the Ras pathway, either via the Drosophila EGF receptor homolog (Egfr) or Sevenless (Sev) (reviewed by Freeman, 1997). This Ras signal is also modulated by Notch and Hedgehog signaling (Dominguez, 1999; Tomlinson and Struhl, 2001; Rebay, 2002; Tsuda et al., 2002). If the Egfr signal is blocked by removing the positive ligand Spitz, then the R8 founder cells begin to form normally, but fail to recruit any subsequent cells (Freeman, 1996; Tio and Moses, 1997). To simplify, cell-type specification and patterning in the developing Drosophila compound eye can be described as occurring in two phases. In phase 1 the ommatidial founder cells (the R8s) are specified at precise positions by a process of lateral inhibition which involves the progressive restriction of Atonal to a single ommatidial founder cell (the future R8 photoreceptor). In phase 2, the founder cell (and later the newly recruited cells that are added) send inductive signals to recruit the remaining cells.
The formation of a precisely spaced pattern of founder cells in phase 1 involves Notch mediated lateral inhibition (Cagan and Ready, 1989; Baker and Zitron, 1995; Baker et al., 1996; Baker and Yu, 1998). The role of the Notch signal is not simple: it initially induces Atonal expression and only later restricts it (Baker and Zitron, 1995; Baker et al., 1996). Furthermore, this Notch signal is genetically upstream of Egfr pathway signaling (Chen and Chien, 1999; White and Jarman, 2000).
It has been suggested that the Egfr/MAPK signal is required for the formation of the founding R8 photoreceptor cells in phase 1, either as an inductive or an inhibitory signal, or both (via positive and/or negative ligands) (Baker and Rubin, 1989; Baker and Rubin, 1992; Zak and Shilo, 1992; Xu and Rubin, 1993; Freeman, 1996; Spencer et al., 1998). Dominant gain-of-function mutations of Egfr (EgfrElp) reduce the number of founder cells (Baker and Rubin, 1989; Baker and Rubin, 1992; Zak and Shilo, 1992). Genetic loss-of-functions tests in mosaic clones were difficult at first, because the Egfr/Ras signal functions in cell cycle regulation (a MAPK signal is normally required for cells to pass the G1/S checkpoint) and also later to regulate apoptosis (Dickson, 1998; Freeman, 1998; Kurada and White, 1998; Assoian and Schwartz, 2001; Baker and Yu, 2001; Jones and Kazlauskas, 2001; Baonza et al., 2002; Howe et al., 2002). To overcome this operational problem, we removed Egfr function by means of a temperature-sensitive mutation and found that the R8 cells form in a nearly normal pattern (Kumar et al., 1998). Others then used an alternative approach to derive genetic loss-of-function mosaic clones for elements of the Egfr pathway (the Minute technique) and they obtained similar results. Loss-of-function genetic tests have been carried out for Egfr itself (Kumar et al., 1998; Baonza et al., 2001; Yang and Baker, 2001), the positive ligands Spitz and Vein (Freeman, 1996; Tio and Moses, 1997; Spencer et al., 1998), the inhibitory ligand Argos (Yang and Baker, 2001), Ras (Halfar et al., 2001; Yang and Baker, 2001), and Raf (Yang and Baker, 2001). In each case, removing Ras pathway signaling during phase 1 eliminates neither the initial upregulation of Atonal nor its subsequent downregulation to (mostly) single founder cells. Indeed, rather than being regulated by the Egfr/Ras pathway, atonal function is genetically upstream of MAPK activation (Chen and Chien, 1999; White and Jarman, 2000). However, in gain-of-function experiments, there is evidence that activation of MAPK signaling in the morphogenetic furrow can inhibit Atonal expression (Kumar et al., 1998; Chen and Chien, 1999). Thus, we conclude that a subset of cells in the furrow can be specified as R8 founder cells without any direct function of the Egfr/Ras pathway, and indeed, that pathway activity may be antagonistic to R8 cell specification at this stage.
We and others have observed highly regulated activation of MAPK at the earliest stage of ommatidial cluster formation (the intermediate groups), by the use of antibodies specific to diphosphorylated MAPK (dpErk antigen) (see Fig. 1) (Gabay et al., 1997; Kumar et al., 1998; Spencer et al., 1998; Chen and Chien, 1999; Yang and Baker, 2003). This regulated activation of MAPK in the furrow is controlled by Egfr (Dominguez et al., 1998; Kumar et al., 1998; Chen and Chien, 1999; Baonza et al., 2001; Halfar et al., 2001; Yang and Baker, 2001). This is apparently paradoxical: how can the Egfr pathway specifically activate MAPK in the nascent ommatidial clusters in the morphogenetic furrow, yet also be dispensable for their foundation? We observed that the activated MAPK (dpErk) antigen is predominantly cytoplasmic at this stage and thus unable to regulate nuclear targets, although it may still affect cytoplasmic proteins (see Fig. 1) (Kumar et al., 1998). Thus, we suggested that while the pathway is activated in a patterned manner, the nuclear MAPK signal may be blocked at the level of MAPK nuclear translocation after its phosphorylation, a mechanism that we call ‘cytoplasmic hold’.
In vertebrates MAPK phosphorylation has been shown to induce a conformational change and homodimerization (Zhang et al., 1994; Wang et al., 1997; Cobb and Goldsmith, 2000). Although some MAPKs such as mammalian Erk3, are constitutively nuclear (Cheng et al., 1996), most (such as Erk1 and Erk2) are activated in the cytoplasm and then move to the nucleus (Khokhlatchev et al., 1998; Cobb and Goldsmith, 2000; Robinson et al., 2002). In cultured cells, MAPK nuclear translocation follows phosphorylation within a few minutes (Chen et al., 1992; Lenormand et al., 1993). Thus, our suggestion that phospho-MAPK may be subject to developmentally regulated cytoplasmic hold (Kumar et al., 1998) was novel but was based only on the observed subcellular distribution of dpErk antigen, which might be subject to a number of artifacts.
The experiments presented here address two questions: does MAPK cytoplasmic hold really occur in the morphogenetic furrow and, if so, what is its function? We adopted an approach that had been used to demonstrate the nuclear translocation of Notch (Struhl and Adachi, 1998). We created and expressed proteins with Drosophila Rolled MAPK fused to an exogenous transcription factor (based on yeast GAL4), which can activate a reporter gene only if it can reach the nucleus. We report that the nuclear translocation of MAPK is indeed regulated by a second means and does not directly follow from phosphorylation during phase 1 (but not in phase 2). Furthermore, we show that if MAPK cytoplasmic hold is overcome in phase 1 Atonal expression is reduced. We conclude that MAPK cytoplasmic hold occurs during phase 1 and is released later to allow Ras pathway signaling to operate during ommatidial assembly (phase 2). We also conclude that this early block is necessary for the patterning events that focus Atonal expression to produce the precise array of R8 photoreceptors.
MATERIALS AND METHODS
DNA constructs and Drosophila transformation
pDMERKA1BS containing rolled (MAPK) was a gift of S. L. Zipursky (Biggs and Zipursky, 1992). Coding sequence (CDS) was excised and the NdeI site removed by PCR to yield fragments 5′ to 3′ as follows: EcoRI, in frame NdeI, rolled CDS (ending ‘QPDNAP’), (SG)4 including BamHI site, HSV epitope tag ‘QPELAPEDPED’, three stop codons, PstI and XbaI. This inserted EcoRI/XbaI in pBLUESCRIPTKSM13+ (Stratagene) yielded ‘pBSRolledCDS’. A 710 bp BamHII fragment encoding GAL4 2−147, then ‘PEFPGIW’, then HSV glycoprotein D 413−490 obtained from N951 (a gift from G. Struhl) was inserted via BamHI into pBSRolledCDS yielding ‘pBSRolledGal4Vp16’. The encoded 540 residue protein was called ‘MG’. Two oligonucleotides were annealed to yield a 62 bp fragment as follows: 5′ EcoRI, SV40 NLS (Kalderon et al., 1984a; Kalderon et al., 1984b), ‘TPPKKKRKVEDPKD’ with N-terminal ‘M’, C-terminal ‘HMLE’, NdeI, XhoI. This was inserted into pBSRolledCDS and pBSRolledGal4Vp16 between the EcoRI and NdeI sites, to produce ‘pBSSV40Rolled’ (encoding a protein called ‘NM’) and ‘pBSSV40RolledGal4Vp16’ (encoding a protein called ‘NMG’). Fragments encoding NM and NMG with a blunt 5′ end and a Pst1 sticky end were then excised from pBSSV40Rolled and pBSSV40RolledGal4Vp16 with SalI, (sticky end rendered blunt by end-repair), then cut with PstI. These fragments were then inserted between the EcoRV and PstI sites in pP{Hsp70-CaSpeR} (Bell et al., 1991), yielding ‘HS:M’, ‘HS:NM’, ‘HS:MG’ and ‘HS:NMG’. Fragments encoding NM and NMG were also excised from pBSSV40Rolled and pBSSV40RolledGal4Vp16 using ClaI and XbaI, and were inserted into pHSX (a derivative of pHSS) (Seifert et al., 1986) with flanking NotI sites (a gift of D. Bowtell). Fragments encoding NM and NMG were then excised using Not1 and inserted into the unique NotI site of pGMR(1N) (a derivative of pGMR; gift of B. Hay) (Hay et al., 1997) yielding ‘GMR:MG’ and ‘GMR:NMG’. All constructs were confirmed by sequencing. Drosophila transformation was as described previously (Rubin and Spradling, 1982), but using pCaWc (Karess and Rubin, 1984). Many independent lines were obtained for most constructs, and five of each were obtained for analysis. Only three lines of HS:MG and two of HS:NMG were obtained, despite extensive efforts. These elements may be toxic when inserted at many sites that permit high level expression.
Drosophila stocks, mosaic clones and temperature-shift regimes
Materials were sourced as follows:
UAS:GFP, UAS:lacZ was a gift from J. Fischer
Wild-type Canton-S was a gift from D. Gailey
GMR:Gal4 was a gift from S. L. Zipursky (Pignoni and Zipursky, 1997)
rl1 (Biggs et al., 1994), rlSem (Brunner et al., 1994) were gifts from the Bloomington center
Egfrtsla (Kumar et al., 1998)
hhts2 (Ma et al., 1993)
Nts is Nl1N-ts1 (Shellenbarger and Mohler, 1975)
ato3, an antigen positive, functional null allele (Jarman et al., 1995)
Clones were obtained as described previously (Xu and Rubin, 1993) using FRT 82B, Ub:GFP and eyFLP (Newsome et al., 2000)
w1118 (Levis et al., 1985)
Nts and hhts shifts were as follows: 1 hour 29°C, 1 hour 37°C, 2 hours 25°C
Egfrtsla shifts were as follows: 22 hours 29°C, 1 hour at 37°C, 2 hours at 29°C
The HS:M and HS:NM time course experiment was carried out as follows: the first larvae were dissected before induction, then kept for 1 hour at 37°C; more larvae were dissected immediately, then kept at 25°C with dissections at 1, 2, 4, 6 and 8 hours.
The HS:MG and HS:NMG experiments were carried out as follows: 1 with hour at 37°C then 2 hours at 25°C.
Immunohistochemistry in situ hybridization and SEM
Primary antibodies: rabbit anti-β-galactosidase (Cortex Biochem), mouse anti-dpErk (Sigma) (Gabay et al., 1997), mouse anti-HSV-Tag (Novagen), rabbit anti-Ato (Jarman et al., 1993), rabbit anti-Spalt (a gift from R. Schuh) (Kuhnlein et al., 1994), mouse anti-BarH1 (a gift from K. Saigo) (Higashijima et al., 1992b), guinea pig anti-Sens (a gift from G. Mardon) (Frankfort et al., 2001), mouse anti-Cut (mAb 2B10) (from Developmental Studies Hybridoma Bank), mouse anti-Pros (mAb MR1A) (from Developmental Studies Hybridoma Bank), rat anti-Elav (from Developmental Studies Hybridoma Bank) and mouse anti-Glass (mAb 9B2.1) (from Developmental Studies Hybridoma Bank). Secondary antibodies were conjugated to FITC or TRITC or Cy5 (Jackson Laboratories). F-actin was visualized with phalloidin (Molecular Probes). DNA was stained with SYTO24 (Molecular Probes). Immunohistochemistry on imaginal discs was as described previously (Tomlinson and Ready, 1987). SEM on adult flies was as described previously (Tio and Moses, 1997).
RESULTS
Expression of MAPK-GAL4/VP16 reveals regulated nuclear translocation in the developing eye
We made four versions of the Drosophila MAPK Rolled (Biggs et al., 1994) for expression in the developing eye (see Fig. 2A, and Materials and Methods): (1) the full-length natural amino acid sequence of Rolled called ‘M’ (for ‘Mapk’); (2) ‘NM’ (‘Nuclear-Mapk’), which adds the strong nuclear localization signal (NLS) from SV40 virus large T antigen (Kalderon et al., 1984a; Kalderon et al., 1984b) fused to the N-terminus of ‘M’; (3) ‘MG’ (‘Mapk-Gal4vp16), which contains the entire sequence of Rolled, followed by the yeast GAL4 DNA binding domain (which is not known to contain a nuclear localization signal) with an acidic activation domain from herpes simplex virus protein 16 (Sadowski et al., 1988); and (4) ‘NMG’ (‘Nuclear-Mapk-Gal4vp16’), which contains a SV40 NLS on the N terminus of MG. Each version was also engineered to carry a C-terminal epitope tag. These proteins were inserted into two Drosophila transformation vectors to drive their expression under heat induction (hsp70 gene promoter or ‘HS’) or the eye-specific Glass transcription factor (‘Glass-Mediated-Response’ or ‘GMR’) to produce six constructs: HS:M, HS:NM, HS:MG, HS:NMG, GMR:MG and GMR:NMG.
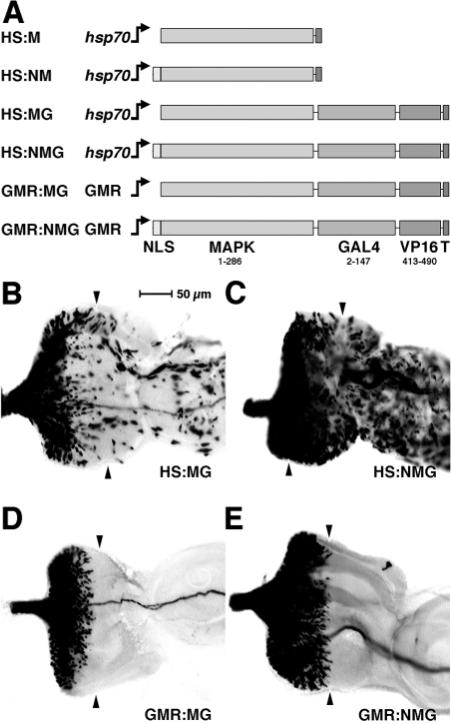
Fig. 2.
MAP kinase-Gal4/Vp16 fusion constructs and their activity in the developing eye. (A) MAP kinase-Gal4/Vp16 constructs. Protein linear maps drawn to scale. NLS, SV40 nuclear localization sequence; MAPK, entire 286 residue natural sequence of the Drosophila rolled MAP kinase gene (Biggs and Zipursky, 1992); GAL4, residues 2−147 of the S. cerevisiae Gal4 (includes DNA-binding domain, but no known NLS sequence); VP16, residues 413−490 of HSV protein 16 (Sadowski et al., 1988); T, 11 residue epitope tag from HSV glycoprotein D (Novagen). (B-E) Eye discs are shown; anterior rightwards, dorsal upwards. In B-E, the fusion construct carried is indicated in the bottom left-hand corner and in all cases activity is detected antigenically using UAS:lacZ. Arrowheads indicate the furrow.
We assessed the transcriptional activating activity of the four Gal4-Vp16 constructs using a UAS:lacZ reporter in third instar eye imaginal discs. In all cases, we tested multiple independently derived transgenic lines for each construct and in each case they gave indistinguishable results. We induced the hsp70 promoter by heat induction (Bonner et al., 1984). and in the case of HS:MG we observed that a subset of cells express β-galactosidase antigen (some in the antennal disc, in the peripodial membrane and in the developing retina; Fig. 2B). In the retina, HS:MG driven reporter gene activity is sporadic on the anterior side, and then is seen in rising numbers of cells posterior to the furrow. By contrast, HS:NMG produces reporter gene expression in many more cells, in all parts of the disc (Fig. 2C). This difference between HS:MG and HS:NMG strongly suggests that a developmentally regulated nuclear localization signal is present in the MAPK part of the MG fusion protein, and that this is over-ridden by strong dominant NLS activity in the NMG protein (associated with the SV40 NLS placed there). The GMR promoter is active only posterior to the morphogenetic furrow (and in the larval photoreceptor of Bolwig's organ) where the Glass transcription factor is expressed (Moses et al., 1989; Moses and Rubin, 1991; Ellis et al., 1993; Hay et al., 1997). We observe reporter gene activity in the GMR:NMG lines immediately posterior to the furrow, but only after a delay in the GMR:MG lines (Fig. 2D,E). These data from the GMR constructs are consistent with the same regulated NLS activity as seen in the HS lines: in the absence of the SV40 NLS, the MG protein can only activate the reporter some distance posterior to the furrow in the developing retina.
Reporter gene activation is not seen in the same cells that show high level dpErk expression in the furrow
In the morphogenetic furrow, large groups of cells (the ‘intermediate’ groups) express high levels of dpErk, but this antigen is predominantly cytoplasmic (Fig. 1) (Kumar et al., 1998). A simple hypothesis might be that in the HS:MG experiment described above, the MG fusion protein requires some time to translocate to the nucleus. This hypothesis suggests that all the cells that express dpErk cytoplasmically early would later go on to show reporter gene activation by MG. However, when we double stained induced HS:MG eye discs for the reporters and dpErk, we found that the prominent column of dpErk expression intermediate groups was not followed in a later column by a prominent column of reporter gene expressing clusters (Fig. 3A,B). We conclude that the majority of the dpErk antigen in the intermediate groups does not later move to the nucleus and activate gene expression, rather we suggest that it is probably dephosphorylated within the next column or two (2−4 hours), in the cytoplasm, without first passing through the nucleus.
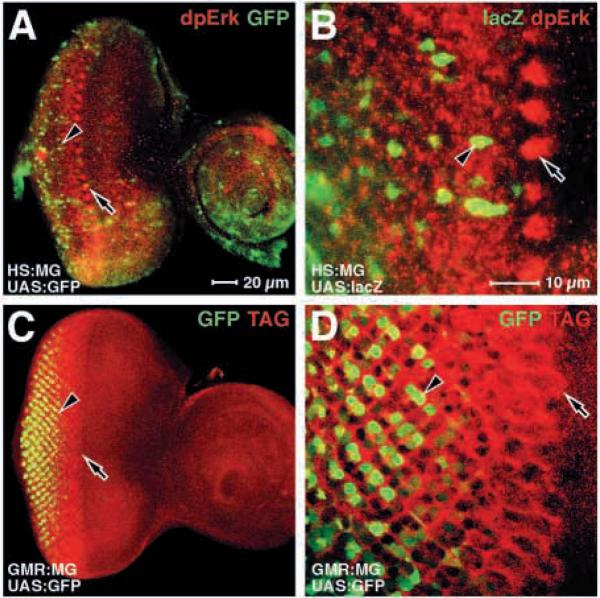
Fig. 3.
Neither dpErk antigen or the MG fusion protein are detected in cell nuclei in the morphogenetic furrow. Eye discs are shown; anterior rightwards. A,C and B,D are pairs at the same magnification. Genotypes are indicated in the bottom left-hand corner; antigens are indicated in the top right-hand corner. (A,B) The activity of the MG fusion protein in activating reporter genes (UAS:GFP and UAS:lacZ as indicated), relative to the endogenous dpErk antigen. Note that not only is reporter gene activity later than the high level dpErk antigen cell clusters in the furrow, but that the pattern of clusters is not reiterated by the reporter gene. (C,D) The same specimen showing the MG fusion protein expressed specifically behind the furrow (GMR:MG). Note that although the MG protein can be detected directly in most or all cells from the furrow (the leading edge of the TAG, arrow), MG activity is detected later and in a more restricted subset of cells (GFP, arrowhead).
It is also possible that the MG protein in these discs is somehow delayed in its expression, so that it does not act early in the furrow. To test this, we stained for the epitope tag that we had placed in the constructs. For HS:MG we observe a general elevation of expression after heat induction, consistent with an unpatterned general induction (not shown). For GMR:MG, we observe elevated expression from the furrow to the back of the disc, consistent with Glass activation. Significantly, we observe reporter gene expression only much later and in a regulated subset of cells (Fig. 3C,D). We are thus confident that our transgenic lines do express MG and NMG fusion proteins as expected, but that they activate the reporter only in a regulated subset of cells: we can see widespread and general expression of MG fusion protein with the tag, but we see activity in only a regulated subset of cells.
In vivo calibration of reporter gene activity
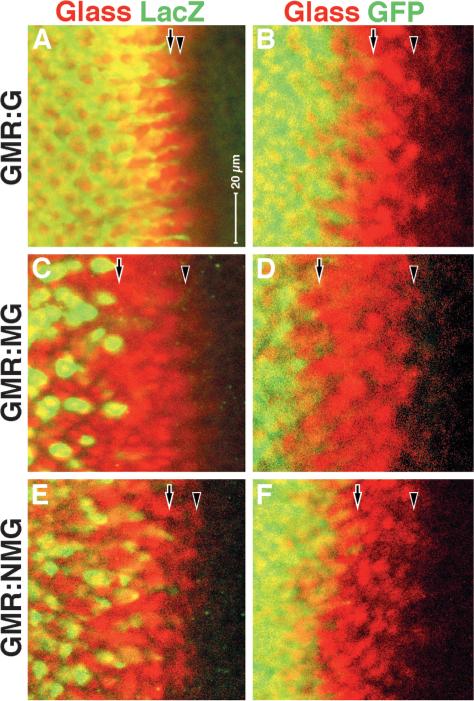
The two reporter genes (UAS:lacZ and UAS:GFP) could themselves be responsible for the long delay in reporter gene activation seen by MG induction following the furrow. Indeed, GFP is known to require time to reveal its expression, because the fluorescent molecule is the product of a series of enzymatic reactions catalyzed by GFP, which requires time (Matz et al., 2002). Although this would seem to be controlled by the much reduced delay seen in the NMG lines, we calibrated the expression of both reporters in third instar eye imaginal discs in vivo. We colocalized both β-galactosidase and GFP reporters in eye discs with Glass protein itself, with the GAL4 activity driven directly by GMR or by the MG or NMG fusions (Fig. 4). As Glass is responsible for the induction in all three cases, the spatial delay between Glass antigen and reporter expression in each case is the sum of the delays imposed by driver expression at the transcriptional level and later and by reporter gene expression. We make two significant observations: (1) GFP reporter is expressed in most or all target cells, but only after a delay of several columns (8−10 hours); (2) β-galactosidase reporter is expressed only in a subset of target cells, but its induction is much faster (seen within one column or 2 hours). Thus, the UAS:lacZ reporter appears to have a higher threshold of activation than UAS:GFP. These operational limitations of the two reporter genes must be borne in mind in analyzing the results reported below.
Fig. 4.
Calibration of the delay between driver and reporter expression. Eye discs are shown; anterior rightwards, dorsal upwards. (A,B) GMR directly driving Gal4 (GMR:G); (C,D) GMR driving the MAP kinase-Gal4/Vp16 fusion (GMR:MG); (E,F) GMR driving the nuclear localized MAP kinase-Gal4/Vp16 fusion (GMR:NMG). Reporters (A,C,E) UAS:lacZ, (B,D,F) UAS:GFP. In all cases, Glass antigen is colocalized with the reporter antigen (β-galactosidase or GFP) as indicated. Leading edges of Glass expression (arrowheads) and of reporter expression (arrows).
MAPK-GAL4/VP16 fusion proteins retain MAPK/rolled function in vivo
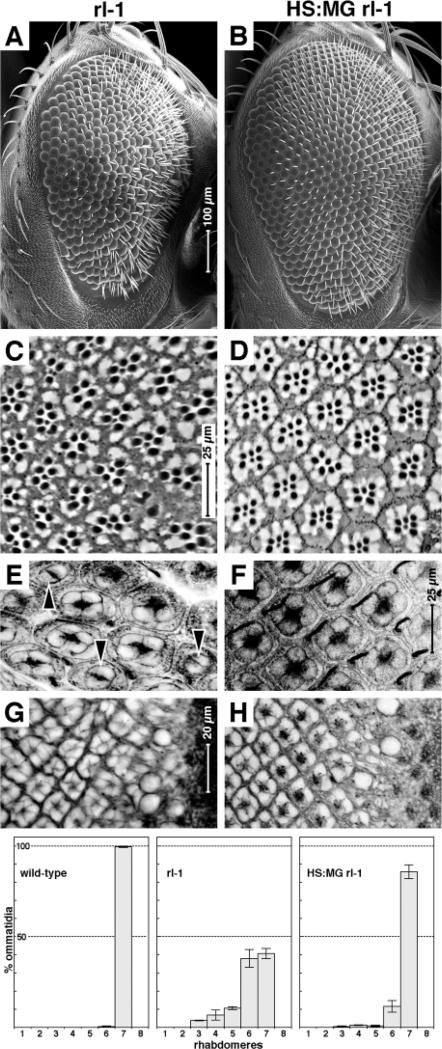
Do the MG and NMG fusion proteins retain MAPK function in the developing fly? We observed a strong suppression of the rl1 eye defects in flies of the genotype: rl1 HS:MG (Fig. 5). rl1 homozygotes have a rough eye (Fig. 5A) (Biggs et al., 1994), which in section can be seen to contain fewer rhabdomeres (Fig. 5C, mean=6.04, s.d.=1.09, 235 ommatidia counted in three individuals) compared with wild type (mean=6.99, s.d.=0.07, 188 ommatidia counted in three individuals), have reduced numbers of cone cells at the pupal stage (Fig. 5E) and disordered development at the third instar (Fig. 5G). All of these defects are strongly suppressed when HS:MG is also present, particularly at 29°C (Fig. 5B,D,F,H). In this case the number of rhabdomeres is closer to the normal number (mean=6.80, s.d.=0.57, in 327 ommatidia counted in three individuals). We also examined three other versions of the Rolled/MAPK fusion protein by external phenotype [with and without the Gal4/VP16 domains, and with and without the SV40 NLS (HS:M, HS:NM and HS:NMG)] and found that these three suppress rl1 to the same degree as HS:MG described above (data not shown). We placed three independent insertions of the GMR:MG and GMR:NMG transgenes in trans to the dominant gain-of-function rlSem allele and observed an enhancement of the dominant rough eye phenotype of rlSem (data not shown). Thus, we conclude that even when fused to Gal4-Vp16, the Rolled sequence retains at least some MAPK function.
Fig. 5.
MAP kinase-Gal4/Vp16 fusion protein expression suppresses the phenotype of a MAP kinase loss-of-function mutant. (A,B) Adult compound eyes. (C,D) Adult retinal tangential sections. (E,F) Forty-two-hour-old pupal retina stained for F-actin. (G,H) Third larval instar retina stained for F-actin. In all panels, anterior rightwards and dorsal is upwards. Genotypes as indicated. (Below) Histograms showing analysis of numbers of rhabdomeres per ommatidium in wild type, rl1 homozygotes and HS:MG rl1 double homozygotes. All animals were raised at 29°C.
MAPK-GAL4/VP16 drives reporter gene expression in all emerging cell types in the third-instar retina, in the order that they are specified after founder cell specification
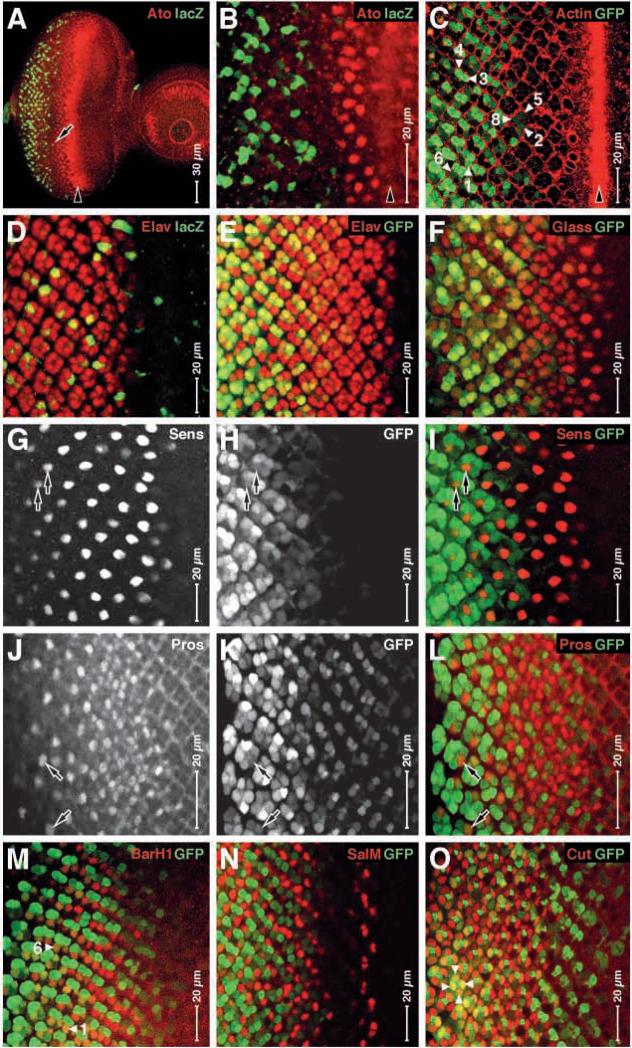
We colocalized reporter gene expression with markers for the developing cell types in the developing eye (Fig. 6). We observe that β-galactosidase reporter expression (driven by HS:MG) first appears after Atonal expression is lost (Fig. 6A,B). We can clearly observe GMR:MG-driven GFP reporter expression in R2/5, R3/4 and R1/6 by morphology and in that order (Fig. 6C). Significantly, this is the same order in which the cells are specified after the R8 founder cell (Tomlinson and Ready, 1987). This result is confirmed for both reporters with a marker for neurons (Elav, Fig. 6D,E) (Robinow and White, 1991), for photoreceptors (Glass, Fig. 7F) (Moses and Rubin, 1991; Ellis et al., 1993), for R8 cells, which express the reporter reliably only late on (Sens, Fig. 6G-I) (Frankfort et al., 2001), for the R7 cell (Pros, Fig. 6J-L) (Xu et al., 2000), for R1 and 6 (BarH1, Fig. 6M) (Higashijima et al., 1992a), and for the cone cells (Cut, Fig. 6O) (Blochlinger et al., 1993). We do not observe contemporaneous expression of the GFP reporter with an early marker for R3 and 4 (SalM, Fig. 6N) (Mollereau et al., 2001). However, we attribute this to a difference in the time of expression as we see reporter expression in R3 and 4 by morphology later on (Fig. 6C).
Fig. 6.
MAP kinase-Gal4/Vp16 fusion protein drives reporter gene expression in all cell types in the developing larval eye. Eye discs are shown; anterior rightwards, dorsal upwards. (A,B,D) HS:MG driver; (C,E-O) GMR:MG driver. Reporter expression shown green (white in H,K) and other antigen in red (or white in G,J) as indicated. Note that reporter expression in A,B follows downregulation of Atonal; in C, it can be detected in the first seven types of photoreceptor cells (arrowheads); in D, it colocalizes with Elav in a subset of cells; in E, it colocalizes with Elav in many more cells, but from a later time; in F, it colocalizes with Glass (similar to Elav); in G-I, it is detected in late R8 cells together with Sens (two examples indicated by arrows); in J-L, it is detected in some R7 cells together with Pro (two examples indicated by arrows); in M it is detected in some R1 and R6 cells together with BarH1 (two examples indicated by arrowheads); in N, it is detected in strongly in R3 and R4 cells, but later than SalM; and in O, it is detected in some cone cells together with Cut (four examples indicated by arrowheads).
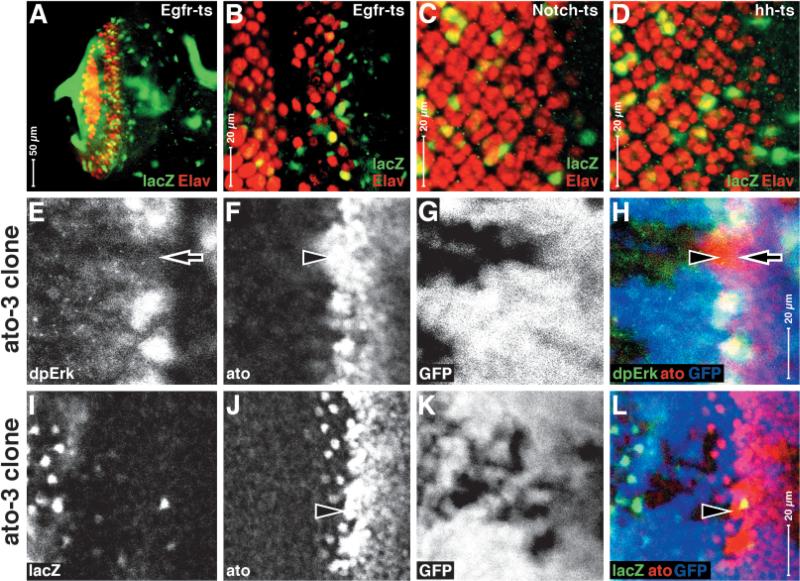
Fig. 7.
MAP kinase-Gal4/Vp16 nuclear translocation activity is not upregulated by loss of Egfr, Notch, hedgehog or atonal function. Eye discs are shown; anterior rightwards, dorsal upwards. HS:MG activity detected by the UAS:lacZ reporter in the genetic background of temperature-sensitive mutations for Egfr (A,B), Notch (C) and hh (D) at restrictive temperature, and in ato3 (loss-of-function, antigen positive) clones (I-L). (E-H) In ato3 clones, MAP kinase phosphorylation is lost (dpErk antigen, as previously reported) (Chen and Chien, 1999). Note that reporter gene expression is not de-repressed in any of these genetic conditions.
Egfr and atonal are Not Genetically Upstream of MAPK-GAL4/VP16 nuclear translocation
Which signal transduction pathway(s) might be directly upstream of MAPK nuclear translocation? A simple hypothesis is that some factor acts to anchor MAPK in the cytoplasm in the intermediate groups, and some pathway is used to release the sequestered MAPK at later stages. We removed Egfr signaling using the Egfrtsla mutant and drove reporter gene expression with HS:MG (Fig. 7A,B). We observe Egfr mutant phenotypes and thus we are confident that we have eliminated Egfr signaling. However, we continue to see reporter gene expression close to the furrow in short-term experiments (not shown) or in longer, 24 shifts (Fig. 7A,B). This suggests that MG nuclear translocation is not wholly dependent on Egfr signaling. This does not show that MAPK nuclear translocation is not dependent on phosphorylation, only that it is not solely dependent on this particular receptor. We were unable to observe release of MG activity to the nucleus in similar experiments using temperature-sensitive alleles of Notch (Fig. 7C) and hedgehog (Fig. 7D).
We also tested the hypothesis that Atonal expression might be upstream of MAPK nuclear translocation. This possibility follows from the observation that where Atonal is expressed, dpErk antigen is not nuclear (Fig. 1). To test this, we derived atonal loss-of-function clones (Fig. 7E-L) and although the clones do show the expected developmental defects (including loss of dpErk antigen, Fig. 9E) (Chen and Chien, 1999), they do not fill with reporter gene expression, as would be expected if Atonal is antagonistic to MAPK translocation.
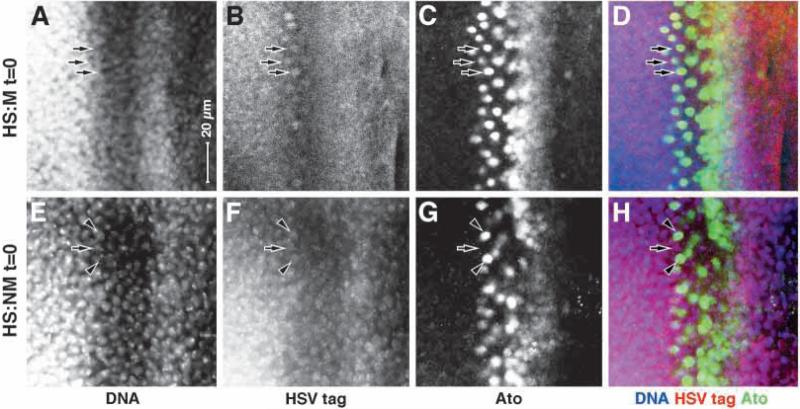
Fig. 9.
Nuclear localization of MAPK detected directly with an epitope tag. Eye discs are shown; anterior rightwards, dorsal upwards. All stained immediately after 1 hour of heat induction. (A-D) Homozygous HS:MG larvae; (E-H) HS:NM larvae. (A,E) Stained to show DNA; (B,F) stained for the HSV epitope tag; (C,G) stained for Atonal; (D,H) Merges. Note that in HS:M, the epitope is expressed at a general and low level and does not appear to be nuclear, except in the final two columns of Atonal-positive cells, where is does appear to be nuclear (three examples indicted by arrows in A-D). Also note that in HS:NM the epitope is expressed at a uniform, low level, and does appear to be nuclear in both Atonal-positive (arrowheads) and -negative (arrow) cells.
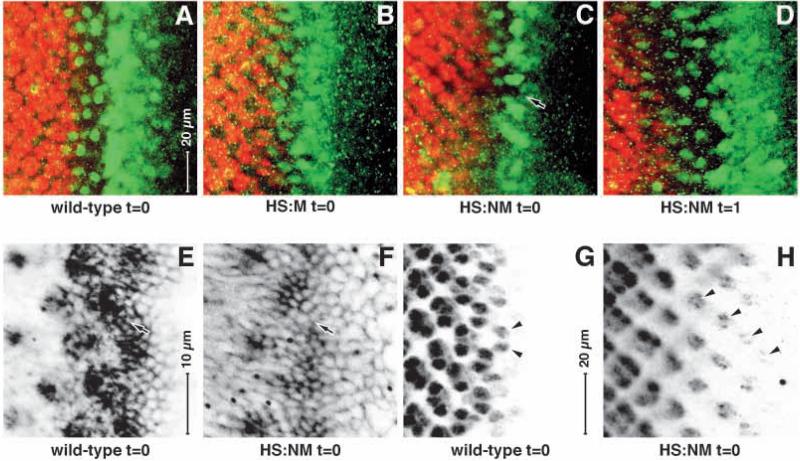
Cytoplasmic hold of activated MAPK is necessary for normal patterning in the furrow
Does this block have any function in normal development in phase 1? To address this, we induced the expression of the HS:M (MAPK alone) and HS:NM (SV40 NLS fused to MAPK) constructs in the morphogenetic furrow. Late larvae of control wild type, as well as three independent transgenic lines each of HS:M and HS:NM were raised at 25°C and eye discs were dissected and stained for Atonal expression as well as Elav (to reveal the developing neurons) and were normal (not shown). We then shifted wild-type, HS:M and HS:NM larvae to 37°C for 1 hour to induce expression and stained immediately after this induction (t=0) and then allowed the larvae to recover at 25°C and dissected more at t=1, 2, 4, 6 and 8 hours.
We see no major effect on Atonal or Elav expression at t=0 for the control wild-type or HS:M (Fig. 8A,B), or any later time point (not shown). However, we do see a clear change in the HS:NM larvae: Atonal expression is reduced ahead of the furrow where it normally ramps up, in the furrow itself, where gaps appear in Atonal expression between the intermediate groups (arrow in Fig. 8C) and after the furrow where Atonal is reduced or undetectable in the single R8 founder cells. In addition, the normal actin driven tight constriction of cells apical surfaces in the furrow (Fig. 8E) is much reduced (Fig. 8F). Furthermore, although Elav normally first appears in pairs of cells (the future R2 and R5 photoreceptors, Fig. 8G) in HS:NM larvae at t=0, Elav is ectopically expressed (at a low level) in earlier columns, in the R8 cells (which have lost their Atonal expression). At t=1 hour in HS:NM larvae the ectopic Elav expression is lost and Atonal expression rebounds, even beyond the levels and extent normally seen (Fig. 8D). At t=2 hours and later in HS:NM, the expression patterns of both Atonal and Elav recover and are indistinguishable from the controls (data not shown). In all cases, the pattern of dpErk antigen is indistinguishable from wild-type (data not shown).
Fig. 8.
Expression of MAPK directed to the nucleus affects development in the morphogenetic furrow. Eye discs are shown; anterior rightwards, dorsal upwards. A-D are at the same magnification; E,F are at the same magnification; G,H are at the same magnification. (AD) Atonal (green), Elav (red). (E,F) F-actin expression. (G,H) Elav expression. Note that at the end of the induction period (t=0) in HS:NM (C), Atonal expression is markedly reduced in the furrow (arrow in C), compared with wild type (A) or HS:M (B). Note that in HS:NM (D), 1 hour later Atonal expression has recovered. In addition, in HS:NM at t=0 the tight apical localization of F-actin markedly reduced in the furrow (arrow in F), compared with wild type (E, arrow). Elav expression appears precociously in the proto-R8 cells (arrowheads in H) compared with its normal first appearance in the proto-R2 and R5 cells (arrowheads in G).
We also stained for directly for the expression of the M and NM fusion proteins immediately after the heat induction (t=0, Fig. 9). In the case of HS:M we observe a general low-level expression of the HSV epitope tag that does not appear to be nuclear (compare Fig. 9A with 9B), except in one interesting place: in the nuclei of late Atonal-positive cells (see arrows in Fig. 9A-D). This is consistent with the UAS reporter experiments described above: in normal development Rolled/MAPK is cytoplasmic in the furrow, despite the high level of phosphorylation in the intermediate groups, and it enters the nucleus only later (see Fig. 3). Furthermore, this time point (the last stages of Atonal expression) is precisely consistent with our observed first expression of the UAS reporters (with our calibration for lag, see Fig. 4). Thus, we can see this developmentally regulated nuclear translocation directly (via the epitope tag) for a Rolled/MAPK protein that lacks the Gal4/VP16 domains. In the case of HS:NM, we observe a general low-level expression of the HSV epitope tag that does appear to be nuclear (compare Fig. 9E with 9F), confirming the function of the SV40 NLS.
Thus, we observe clear consequences in the developing furrow of adding a strong NLS to MAPK: Atonal expression is lost and R8 cells begin to differentiate precociously as neurons (they express Elav early). This is consistent with a crucial function for the cytoplasmic hold of activated MAPK in the morphogenetic furrow of the developing fly eye and others data showing genetic interactions between atonal and Ras pathway signaling (Dokucu et al., 1996; Chanut et al., 2002). This pair of experiments is controlled for other variables (promoter strength and so on) that may have a bearing on expression levels. We cannot claim that cytoplasmic hold can block pathway signaling at any expression level, and thus the observations of ourselves and others that pathway gain-of-function can block Atonal expression may be due to different levels of expression in those conditions (Kumar et al., 1998; Chen and Chien, 1999).
DISCUSSION
We previously reported that phosphorylated MAPK (dpErk) antigen is retained in the cytoplasm for several hours in the intermediate groups of cells in the morphogenetic furrow (Kumar et al., 1998). We suggested that the Egfr pathway signal may be blocked in these cells at the level of nuclear translocation of MAPK after its phosphorylation. To test this, we developed fusion protein reagents to reveal the subcellular location of MAPK independently of its phosphorylation state. By fusing MAPK to an exogenous transcription factor, and then visualizing reporter gene expression during development, we were able to observe the temporal and spatial context of MAPK nuclear translocation during eye development. We have described here that this fusion protein retains MAPK function in vivo, and that it drives reporter gene expression only in a subset of cells in which it is expressed. We have also observed MAPK nuclear translocation more directly using an epitope tag and these results are consistent with those seen with the transcription factor fusion.
Consistent with our suggestion that the pathway signal is blocked in the intermediate groups (phase 1), we see reporter gene expression only later, in the future R8 cells in the last two columns of Atonal expression and later in cells as they are recruited into the assembling ommatidia (phase 2). This is consistent with normal Egfr pathway activity in the downregulation of Atonal at the end of phase 1 and then again at later stages (phase 2), when successive Ras pathway signals recruit each cell type that follows the founding R8 cell (which is specified by other means). Although we cannot detect MAPK nuclear translocation early in the furrow (in the intermediate groups) with either of our reagents (the transcription factor fusion or the epitope tag) we cannot formally exclude the possibility that there is some lower level of nuclear MAPK at these stages that is below the limits of our two detection systems. Similarly, we cannot exclude the possibility that there are cytoplasmic functions for phosphorylated MAPK at these stages. However, our results are consistent with two Egfr pathway functions in the developing R8 cells at this time (as Atonal expression ends): for the maintained expression of differentiation markers (Boss and Elav) and for later cell survival (Kumar et al., 1998).
Furthermore, through the addition of a constitutive NLS, we have driven MAPK into the nuclei of cells in phase 1, thus overcoming MAPK cytoplasmic hold. This results in a rapid downregulation of Atonal and the precocious neural differentiation of the R8 photoreceptors (Fig. 10). Taken together with our observation of the first nuclear translocation of MAPK as Atonal is downregulated in normal development (above), we suggest that the Egfr/Ras pathway may normally contribute to the end of phase 1 by ending Atonal expression.
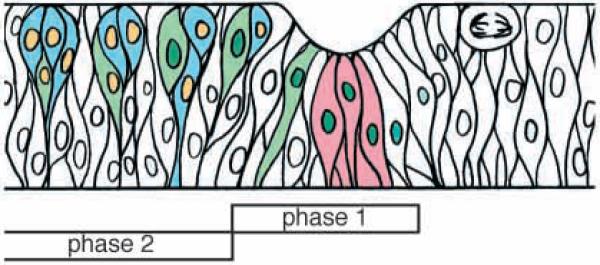
Fig. 10.
MAPK cytoplasmic hold in the furrow. A diagram showing a section through the furrow, apical upwards and anterior rightwards. The furrow is moving from left to right. Phase 1 and phase 2 are indicated. Green nuclei are Atonal positive, yellow nuclei are Elav positive. Green cells are fated to become R8 photoreceptors, blue cells will become other photoreceptors. Red cells are the intermediate group, and have both high levels of Ras pathway activity and also MAPK cytoplasmic hold.
Others have suggested that Egfr pathway loss-of-function normally functions to downregulate Atonal expression at and after the intermediate group stage (Chen and Chien, 1999; Baonza et al., 2001; Yang and Baker, 2001). We did not observe this using our conditional mutation, Egfrtsla (Kumar et al., 1998); however, as this experiment did not include a clone boundary we could not have detected a short delay (such as one column). It may be that the pathway functions at this point through a much lower level signal (below the level of detection of our reagents) or it may be that it functions through cytoplasmic targets of phosphorylated MAPK.
What is the developmental purpose of this block of MAPK signaling in the furrow? Anterior to the furrow, MAPK cytoplasmic hold cannot function, or it would prevent the MAPK signaling required for the G1/S transition and thus halt cell proliferation. Perhaps this is one reason why all cells in phase 1 exit the cell cycle (Ready et al., 1976; Wolff and Ready, 1991; Thomas et al., 1994). However, new data suggest that the Egfr pathway does function in the furrow to maintain G1 arrest (visualized as increased cyclin B expression) (Yang and Baker, 2003). This could be mediated through some low level of nuclear MAPK at this stage or possibly through cytoplasmic targets for MAPK signaling. However, although cyclin B expression is elevated posterior to the furrow in all cells other than R8 in Egfr pathway loss-of-function mutant clones, the leading edge of cyclin B expression does not advance (it is not expressed earlier) (Yang and Baker, 2003). Thus, it may be that the role of Egfr pathway signals in maintaining G1 arrest is later than the end of Atonal expression (i.e. in phase 2, not in phase 1).
We suggest that the founder cells have a special developmental function to fulfill in phase 1: they must act as organizing centers for lateral inhibition to produce the spaced pattern of R8 cells. If the founder cells did not inhibit their neighbors most or all cells in phase 1 might rapidly differentiate as photoreceptors, resulting in disorder. This type of disorder is observed when the Egfr/MAPK pathway is ectopically activated ahead of the furrow, when photoreceptor differentiation becomes independent of Atonal and R8 fate (Baonza et al., 2001). Our model may also explain the loss of ommatidia seen in EgfrElp gain-of-function mutants (Baker and Rubin, 1989; Baker and Rubin, 1992; Zak and Shilo, 1992; Lesokhin et al., 1999). Excess Ras/MAPK pathway signals may reduce Atonal expression and thus the number of R8 founder cells. Our results lead us to predict that G1 cell-cycle arrest may be found in other cases in which a subset of progenitor cells is selected by lateral inhibition through active Notch pathway signaling and repression of Ras/MAPK signaling. In summary, our data are consistent with a model in which Egfr/MAPK signaling functions in ommatidial assembly but not directly in founder cell specification. We propose that MAPK cytoplasmic hold is restricted to the morphogenetic furrow, and does not happen anterior to the furrow (the proliferative phase) or posterior (during ommatidial assembly, or phase 2). It appears to be coincident with the regulated G1 arrest seen in the furrow.
It is interesting to note that our observed pattern of MAPK-Gal4/VP16 is very different from our (and others) observation of the pattern of MAPK phosphorylation (dpErk antigen); indeed, they are almost exclusive. We and many others observe that the predominant expression of dpErk in the developing eye is in the intermediate groups in the furrow, and yet we (and, we believe, the data of others) show little detectable signal function there. Furthermore, we and others have shown in many ways that MAPK signaling is absolutely required for ommatidial assembly posterior to the furrow, yet no one can detect much dpErk at that stage. Perhaps MAPK cytoplasmic hold can explain this paradox as well: where MAPK is anchored in the cytoplasm (in the furrow) it can be phosphorylated by MEK but is protected from abundant phosphatase activity that waits in the nucleus. Thus, the pathway is blocked, there is no negative feedback and the antigen builds up to high (and easily detected) levels for several hours. Later (during ommatidial assembly) it is possible be that there is no cytoplasmic hold, so MAPK passes rapidly to the nucleus after its phosphorylation by MEK, where the signal is passed, negative feedback is triggered and the antigen is cleared by phosphatase. Thus, in vivo the dpErk stain may actually be a stain not for pathway activity per se, but predominantly for MAPK cytoplasmic hold!
Our findings indicate that MAPK nuclear translocation is regulated in vivo by some mechanism in addition to, and regulated separately from, its phosphorylation state. We do not propose that MAPK phosphorylation is not required for MAPK nuclear translocation, only that phosphorylation is not always sufficient. What might the mechanism for MAPK cytoplasmic hold be? The simplest hypothesis is that some anchoring factor sequesters activated MAPK in the cytoplasm until a second developmental signal permits its release. This could provide for a point of signal transduction pathway integration. However, we suggest an alternative model: activated MAPK cannot translocate to the nucleus in the intermediate groups not because it is held fast by some negative anchoring factor, but because it lacks some specific positive factor, such as an import factor.
In summary, the results presented here demonstrate that there is the dual regulation of MAPK signal transduction, both through its phosphorylation by MEK and independently through the control of nuclear translocation. Such dual regulation may be important in many developmental events through which a subset of founder cells must first be specified. As such events involve lateral inhibition and cell contact, this mechanism may not be observable in tissue culture systems.
Acknowledgments
We thank J. Fischer, S. L. Zipursky and D. Gailey for Drosophila stocks; G. Mardon, C. Doe, K. Saigo, A. Jarman and R. Schuh for antibodies; G. Struhl and S. L. Zipursky for plasmids; and C. Desplan for advice. This work was funded by a grant from the National Eye Institute at NIH (EY12537).
REFERENCES
- Assoian RK, Schwartz MA. Coordinate signaling by integrins and receptor tyrosine kinases in the regulation of G1 phase cell-cycle progression. Curr. Opin. Genet. Dev. 2001;11:48–53. doi: 10.1016/s0959-437x(00)00155-6. [DOI] [PubMed] [Google Scholar]
- Baker NE, Rubin GM. Effect on eye development of dominant mutations in Drosophila homologue of the EGF receptor. Nature. 1989;340:150–153. doi: 10.1038/340150a0. [DOI] [PubMed] [Google Scholar]
- Baker NE, Rubin GM. Ellipse mutations in the Drosophila homologue of the EGF receptor affect pattern formation, cell division, and cell death in eye imaginal discs. Dev. Biol. 1992;150:381–396. doi: 10.1016/0012-1606(92)90250-k. [DOI] [PubMed] [Google Scholar]
- Baker NE, Zitron AE. Drosophila eye development: Notch and Delta amplify a neurogenic pattern conferred on the morphogenetic furrow by scabrous. Mech. Dev. 1995;49:173–189. doi: 10.1016/0925-4773(94)00314-d. [DOI] [PubMed] [Google Scholar]
- Baker NE, Yu SY. The R8-photoreceptor equivalence group in Drosophila: fate choice precedes regulated Delta transcription and is independent of Notch gene dose. Mech. Dev. 1998;74:3–14. doi: 10.1016/s0925-4773(98)00054-9. [DOI] [PubMed] [Google Scholar]
- Baker NE, Yu SY. The EGF receptor defines domains of cell cycle progression and survival to regulate cell number in the developing Drosophila eye. Cell. 2001;104:699–708. doi: 10.1016/s0092-8674(01)00266-5. [DOI] [PubMed] [Google Scholar]
- Baker NE, Yu S, Han D. Evolution of proneural atonal expression during distinct regulatory phases in the developing Drosophila eye. Curr. Biol. 1996;6:1290–1301. doi: 10.1016/s0960-9822(02)70715-x. [DOI] [PubMed] [Google Scholar]
- Baonza A, Casci T, Freeman M. A primary role for the epidermal growth factor receptor in ommatidial spacing in the Drosophila eye. Curr. Biol. 2001;11:396–404. doi: 10.1016/s0960-9822(01)00125-7. [DOI] [PubMed] [Google Scholar]
- Baonza A, Murawsky CM, Travers AA, Freeman M. Pointed and Tramtrack69 establish an EGFR-dependent transcriptional switch to regulate mitosis. Nat. Cell Biol. 2002;4:976–980. doi: 10.1038/ncb887. [DOI] [PubMed] [Google Scholar]
- Basler K, Hafen E. Dynamics of Drosophila eye development and temporal requirements of sevenless expression. Development. 1989;107:723–731. doi: 10.1242/dev.107.4.723. [DOI] [PubMed] [Google Scholar]
- Bell LR, Horabin JI, Schedl P, Cline TW. Positive autoregulation of Sex-lethal by alternative splicing maintains the female determined state in Drosophila. Cell. 1991;65:229–239. doi: 10.1016/0092-8674(91)90157-t. [DOI] [PubMed] [Google Scholar]
- Biggs WHD, Zipursky SL. Primary structure, expression, and signal-dependent tyrosine phosphorylation of a Drosophila homolog of extracellular signal-regulated kinase. Proc. Natl. Acad. Sci. USA. 1992;89:6295–6299. doi: 10.1073/pnas.89.14.6295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggs WHR, Zavitz KH, Dickson B, van der Straten A, Brunner D, Hafen E, Zipursky SL. The Drosophila rolled locus encodes a MAP kinase required in the sevenless signal transduction pathway. EMBO J. 1994;13:1628–1635. doi: 10.1002/j.1460-2075.1994.tb06426.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blochlinger K, Jan LY, Jan YN. Postembryonic patterns of expression of cut, a locus regulating sensory organ identity in Drosophila. Development. 1993;117:441–450. doi: 10.1242/dev.117.2.441. [DOI] [PubMed] [Google Scholar]
- Bonner JJ, Parks C, Parker-Thornburg J, Mortin MA, Pelham HRB. The use of promoter fusions in Drosophila genetics: isolation of mutations affecting the heat shock response. Cell. 1984;37:979–991. doi: 10.1016/0092-8674(84)90432-x. [DOI] [PubMed] [Google Scholar]
- Brunner D, Oellers N, Szabad J, Biggs WHR, Zipursky SL, Hafen E. A gain-of-function mutation in Drosophila MAP kinase activates multiple receptor tyrosine kinase signaling pathways. Cell. 1994;76:875–888. doi: 10.1016/0092-8674(94)90362-x. [DOI] [PubMed] [Google Scholar]
- Cagan RL, Ready DF. Notch is required for successive cell decisions in the developing Drosophila retina. Genes Dev. 1989;3:1099–1112. doi: 10.1101/gad.3.8.1099. [DOI] [PubMed] [Google Scholar]
- Chanut F, Woo K, Pereira S, Donohoe TJ, Chang SY, Laverty TR, Jarman AP, Heberlein U. Rough eye is a gain-of-function allele of amos that disrupts regulation of the proneural gene atonal during Drosophila retinal differentiation. Genetics. 2002;160:623–635. doi: 10.1093/genetics/160.2.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C-K, Chien C-T. Negative regulation of atonal in proneural cluster formation of Drosophila R8 photoreceptors. Proc. Natl. Acad. Sci. USA. 1999;96:5055–5060. doi: 10.1073/pnas.96.9.5055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R-H, Sarnecki C, Blenis J. Nuclear localization and regulation of erk and rsk-encoded protein kinases. Mol. Cell. Biol. 1992;12:915–927. doi: 10.1128/mcb.12.3.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng M, Boulton TG, Cobb MH. ERK3 is a constitutively nuclear protein kinase. J. Biol. Chem. 1996;271:8951–8958. doi: 10.1074/jbc.271.15.8951. [DOI] [PubMed] [Google Scholar]
- Cobb MH, Goldsmith EJ. Dimerization in MAP-kinase signaling. Trends Biochem Sci. 2000;25:7–9. doi: 10.1016/s0968-0004(99)01508-x. [DOI] [PubMed] [Google Scholar]
- Dickson BJ. Photoreceptor development: breaking down the barriers. Curr. Biol. 1998;8:R90–R92. doi: 10.1016/s0960-9822(98)70054-5. [DOI] [PubMed] [Google Scholar]
- Dokucu ME, Zipursky LS, Cagan RL. Atonal, Rough and the resolution of proneural clusters in the developing Drosophila retina. Development. 1996;122:4139–4147. doi: 10.1242/dev.122.12.4139. [DOI] [PubMed] [Google Scholar]
- Dominguez M. Dual role for Hedgehog in the regulation of the proneural gene atonal during ommatidia development. Development. 1999;126:2345–2553. doi: 10.1242/dev.126.11.2345. [DOI] [PubMed] [Google Scholar]
- Dominguez M, Wasserman JD, Freeman M. Multiple functions for the EGF receptor in Drosophila eye development. Curr. Biol. 1998;8:1039–1048. doi: 10.1016/s0960-9822(98)70441-5. [DOI] [PubMed] [Google Scholar]
- Ellis MC, O'Neill EM, Rubin GM. Expression of Drosophila glass protein and evidence for negative regulation of its activity in non-neuronal cells by another DNA-binding protein. Development. 1993;119:855–865. doi: 10.1242/dev.119.3.855. [DOI] [PubMed] [Google Scholar]
- Frankfort BJ, Nolo R, Zhang Z, Bellen H, Mardon G. senseless repression of rough is required for R8 photoreceptor differentiation in the developing Drosophila eye. Neuron. 2001;32:403–414. doi: 10.1016/s0896-6273(01)00480-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman M. Reiterative use of the EGF receptor triggers differentiation of all cell types in the Drosophila eye. Cell. 1996;87:651–660. doi: 10.1016/s0092-8674(00)81385-9. [DOI] [PubMed] [Google Scholar]
- Freeman M. Cell determination strategies in the Drosophila eye. Development. 1997;124:261–270. doi: 10.1242/dev.124.2.261. [DOI] [PubMed] [Google Scholar]
- Freeman M. Complexity of EGF receptor signaling revealed in Drosophila. Curr. Opin. Genet. Dev. 1998;8:407–411. doi: 10.1016/s0959-437x(98)80110-x. [DOI] [PubMed] [Google Scholar]
- Gabay L, Seger R, Shilo B-Z. In situ activation pattern of Drosophila EGF receptor pathway during development. Science. 1997;277:1103–1106. doi: 10.1126/science.277.5329.1103. [DOI] [PubMed] [Google Scholar]
- Halfar K, Rommel C, Stocker H, Hafen E. Ras controls growth, survival and differentiation in the Drosophila eye by different thresholds of MAP kinase activity. Development. 2001;128:1687–1696. doi: 10.1242/dev.128.9.1687. [DOI] [PubMed] [Google Scholar]
- Hay BA, Maile R, Rubin GM. P element insertion-dependent gene activation in the Drosophila eye. Proc. Natl. Acad. Sci. USA. 1997;94:5195–5200. doi: 10.1073/pnas.94.10.5195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashijima S.-i., Kojima T, Michiue T, Ishimaru S, Emori Y, Saigo K. Dual Bar homeo box genes of Drosophila required in two photoreceptor cells, R1 and R6, and primary pigment cells for normal eye development. Genes Dev. 1992a;6:50–60. doi: 10.1101/gad.6.1.50. [DOI] [PubMed] [Google Scholar]
- Higashijima S.-i., Michiue T, Emori Y, Saigo K. Subtype determination of Drosophila embryonic external sensory organs by redundant homeo box genes BarH1 and BarH2. Genes Dev. 1992b;6:1005–1018. doi: 10.1101/gad.6.6.1005. [DOI] [PubMed] [Google Scholar]
- Howe AK, Aplin AE, Juliano RL. Anchorage-dependent ERK signaling – mechanisms and consequences. Curr. Opin. Genet. Dev. 2002;12:30–35. doi: 10.1016/s0959-437x(01)00260-x. [DOI] [PubMed] [Google Scholar]
- Jarman AP, Grau Y, Jan LY, Jan YN. atonal is a proneural gene that directs chordotonal organ formation in the Drosophila peripheral nervous system. Cell. 1993;73:1307–1321. doi: 10.1016/0092-8674(93)90358-w. [DOI] [PubMed] [Google Scholar]
- Jarman AP, Grell EH, Ackerman L, Jan LY, Jan YN. atonal is the proneural gene for Drosophila photoreceptors. Nature. 1994;369:398–400. doi: 10.1038/369398a0. [DOI] [PubMed] [Google Scholar]
- Jarman AP, Sun Y, Jan LY, Jan YN. Role of the proneural gene, atonal, in formation of the Drosophila chordotonal organs and photoreceptors. Development. 1995;121:2019–2030. doi: 10.1242/dev.121.7.2019. [DOI] [PubMed] [Google Scholar]
- Jones SM, Kazlauskas A. Growth factor-dependent signaling and cell cycle progression. FEBS Lett. 2001;490:110–116. doi: 10.1016/s0014-5793(01)02113-5. [DOI] [PubMed] [Google Scholar]
- Kalderon D, Richardson WD, Markham AF, Smith AE. Sequence requirements for nuclear location of simian virus 40 large-T antigen. Nature. 1984a;311:33–38. doi: 10.1038/311033a0. [DOI] [PubMed] [Google Scholar]
- Kalderon D, Roberts BL, Richardson WD, Smith AE. A short amino acid sequence able to specify nuclear location. Cell. 1984b;39:499–509. doi: 10.1016/0092-8674(84)90457-4. [DOI] [PubMed] [Google Scholar]
- Karess RE, Rubin GM. Analysis of P transposable element functions in Drosophila. Cell. 1984;38:135–146. doi: 10.1016/0092-8674(84)90534-8. [DOI] [PubMed] [Google Scholar]
- Khokhlatchev AV, Canagarajah B, Wilsbacher J, Robinson M, Atkinson M, Goldsmith E, Cobb MH. Phosphorylation of the MAP Kinase ERK2 promotes its homodimerization and nuclear translocation. Cell. 1998;93:605–615. doi: 10.1016/s0092-8674(00)81189-7. [DOI] [PubMed] [Google Scholar]
- Kuhnlein RP, Frommer G, Friedrich M, Gonzalez-Gaitan M, Weber A, Wagner-Bernholz JF, Gehring WJ, Jackle H, Schuh R. spalt encodes an evolutionarily conserved zinc finger protein of novel structure which provides homeotic gene function in the head and tail region of the Drosophila embryo. EMBO J. 1994;13:168–179. doi: 10.1002/j.1460-2075.1994.tb06246.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar JP, Tio M, Hsiung F, Akopyan S, Gabay L, Seger R, Shilo B-Z, Moses K. Dissecting the roles of the Drosophila EGF receptor in eye development and MAP kinase activation. Development. 1998;125:3875–3885. doi: 10.1242/dev.125.19.3875. [DOI] [PubMed] [Google Scholar]
- Kurada P, White K. Ras promotes cell survival in Drosophila by downregulating hid expression. Cell. 1998;95:319–329. doi: 10.1016/s0092-8674(00)81764-x. [DOI] [PubMed] [Google Scholar]
- Lenormand P, Sardet C, Pages G, L'Allemain G, Brunet A, Pouyssegur J. Growth factors induce nuclear translocation of MAP kinases (p42mapk and p44mapk) but not of their activator MAP kinase kinase (p45mapkk) in fibroblasts. J. Cell Biol. 1993;122:1079–1088. doi: 10.1083/jcb.122.5.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesokhin AM, Yu SY, Katz J, Baker NE. Several levels of EGF Receptor signaling during photoreceptor specification in wild-type, Ellipse, and null mutant Drosophila. Dev. Biol. 1999;205:129–144. doi: 10.1006/dbio.1998.9121. [DOI] [PubMed] [Google Scholar]
- Levis R, Hazelrigg T, Rubin GM. Effects of genomic position on the expression of transduced copies of the white gene of Drosophila. Science. 1985;229:558–561. doi: 10.1126/science.2992080. [DOI] [PubMed] [Google Scholar]
- Ma C, Zhou Y, Beachy PA, Moses K. The segment polarity gene hedgehog is required for progression of the morphogenetic furrow in the developing Drosophila eye. Cell. 1993;75:927–938. doi: 10.1016/0092-8674(93)90536-y. [DOI] [PubMed] [Google Scholar]
- Matz MV, Lukyanov KA, Lukyanov SA. Family of the green fluorescent protein: Journey to the end of the rainbow. BioEssays. 2002;24:953–959. doi: 10.1002/bies.10154. [DOI] [PubMed] [Google Scholar]
- Mollereau B, Dominguez M, Webel R, Colley NJ, Keung B, de Celis JF, Desplan C. Two-step process for photoreceptor formation in Drosophila. Nature. 2001;412:911–913. doi: 10.1038/35091076. [DOI] [PubMed] [Google Scholar]
- Moses K, Ellis MC, Rubin GM. The glass gene encodes a zinc-finger protein required by Drosophila photoreceptor cells. Nature. 1989;340:531–536. doi: 10.1038/340531a0. [DOI] [PubMed] [Google Scholar]
- Moses K, Rubin GM. glass encodes a site-specific DNA-binding protein that is regulated in response to positional signals in the developing Drosophila eye. Genes Dev. 1991;5:583–593. doi: 10.1101/gad.5.4.583. [DOI] [PubMed] [Google Scholar]
- Newsome TP, Asling B, Dickson BJ. Analysis of Drosophila photoreceptor axon guidance in eye-specific mosaics. Development. 2000;127:851–860. doi: 10.1242/dev.127.4.851. [DOI] [PubMed] [Google Scholar]
- Pignoni F, Zipursky SL. Induction of Drosophila eye development by Decapentaplegic. Development. 1997;124:271–278. doi: 10.1242/dev.124.2.271. [DOI] [PubMed] [Google Scholar]
- Ready DF, Hanson TE, Benzer S. Development of the Drosophila retina, a neurocrystalline lattice. Dev. Biol. 1976;53:217–240. doi: 10.1016/0012-1606(76)90225-6. [DOI] [PubMed] [Google Scholar]
- Rebay I. Keeping the receptor tyrosine kinase signaling pathway in check: lessons from Drosophila. Dev. Biol. 2002;251:1–17. doi: 10.1006/dbio.2002.0806. [DOI] [PubMed] [Google Scholar]
- Robinow S, White K. Characterization and spatial distribution of the ELAV protein during Drosophila melanogaster development. J. Neurobiol. 1991;22:443–461. doi: 10.1002/neu.480220503. [DOI] [PubMed] [Google Scholar]
- Robinson MJ, Xu Be BE, Stippec S, Cobb MH. Different domains of the mitogen-activated protein kinases ERK3 and ERK2 direct subcellular localization and upstream specificity in vivo. J. Biol. Chem. 2002;277:5094–5100. doi: 10.1074/jbc.M110935200. [DOI] [PubMed] [Google Scholar]
- Rubin GM, Spradling AC. Genetic transformation of Drosophila with transposable element vectors. Science. 1982;218:348–353. doi: 10.1126/science.6289436. [DOI] [PubMed] [Google Scholar]
- Sadowski I, Ma J, Triezenberg S, Ptashne M. GAL4-VP16 is an unusually potent transcriptional activator. Nature. 1988;335:563–564. doi: 10.1038/335563a0. [DOI] [PubMed] [Google Scholar]
- Seifert HS, Chen EY, So M, Heffron F. Shuttle mutagenesis: a method of transposon mutagenesis for Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA. 1986;83:735–739. doi: 10.1073/pnas.83.3.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shellenbarger DL, Mohler JD. Temperature-sensitive mutations of the Notch locus in Drosophila melanogaster. Genetics. 1975;81:143–162. doi: 10.1093/genetics/81.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer SA, Powell PA, Miller DT, Cagan RL. Regulation of EGF receptor signaling establishes pattern across the developing Drosophila retina. Development. 1998;125:4777–4790. doi: 10.1242/dev.125.23.4777. [DOI] [PubMed] [Google Scholar]
- Struhl G, Adachi A. Nuclear access and action of Notch in vivo. Cell. 1998;93:639–648. doi: 10.1016/s0092-8674(00)81193-9. [DOI] [PubMed] [Google Scholar]
- Thomas BJ, Gunning DA, Cho J, Zipursky SL. Cell cycle progression in the developing Drosophila eye: roughex encodes a novel protein required for the establishment of G1. Cell. 1994;77:1003–1014. doi: 10.1016/0092-8674(94)90440-5. [DOI] [PubMed] [Google Scholar]
- Tio M, Moses K. The Drosophila TGFα homolog Spitz acts in photoreceptor recruitment in the developing retina. Development. 1997;124:343–351. doi: 10.1242/dev.124.2.343. [DOI] [PubMed] [Google Scholar]
- Tomlinson A. Cellular interactions in the developing Drosophila eye. Development. 1988;104:183–193. doi: 10.1242/dev.104.2.183. [DOI] [PubMed] [Google Scholar]
- Tomlinson A, Ready DF. Neuronal differentiation in the Drosophila ommatidium. Dev. Biol. 1987;120:366–376. doi: 10.1016/0012-1606(87)90239-9. [DOI] [PubMed] [Google Scholar]
- Tomlinson A, Struhl G. Delta/Notch and Boss/Sevenless signals act combinatorially to specify the Drosophila R7 photoreceptor. Mol. Cell. 2001;7:487–495. doi: 10.1016/s1097-2765(01)00196-4. [DOI] [PubMed] [Google Scholar]
- Tsuda L, Nagaraj R, Zipursky S, Banerjee U. An EGFR/Ebi/Sno pathway promotes Delta expression by inactivating Su(H)/SMRTER repression during inductive Notch signaling. Cell. 2002;110:625–637. doi: 10.1016/s0092-8674(02)00875-9. [DOI] [PubMed] [Google Scholar]
- Wang Z, Harkins PC, Ulevitch RJ, Han J, Cobb MH, Goldsmith EJ. The structure of mitogen-activated protein kinase p38 at 2.1-A resolution. Proc. Natl. Acad. Sci. USA. 1997;94:2327–2332. doi: 10.1073/pnas.94.6.2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White NM, Jarman AP. Drosophila atonal controls photoreceptor R8-specific properties and modulates both Receptor Tyrosine Kinase and Hedgehog signalling. Development. 2000;127:1681–1689. doi: 10.1242/dev.127.8.1681. [DOI] [PubMed] [Google Scholar]
- Wolff T, Ready DF. The beginning of pattern formation in the Drosophila compound eye: the morphogenetic furrow and the second mitotic wave. Development. 1991;113:841–850. doi: 10.1242/dev.113.3.841. [DOI] [PubMed] [Google Scholar]
- Wolff T, Ready DF. Chapter 22: Pattern formation in the Drosophila retina. In: Bate M, Martinez-Arias A, editors. The Development of Drosophila melanogaster. Vol. 2. Cold Spring Harbor Laboratory Press; Cold Spring Harbor , NY: 1993. pp. 1277–1325. [Google Scholar]
- Xu C, Kauffmann RC, Zhang J, Kladny S, Carthew RW. Overlapping activators and repressors delimit transcriptional response to receptor tyrosine kinase signals in the Drosophila eye. Cell. 2000;103:87–97. doi: 10.1016/s0092-8674(00)00107-0. [DOI] [PubMed] [Google Scholar]
- Xu T, Rubin GM. Analysis of genetic mosaics in developing and adult Drosophila tissues. Development. 1993;117:1223–1237. doi: 10.1242/dev.117.4.1223. [DOI] [PubMed] [Google Scholar]
- Yang L, Baker NE. Role of the EGFR/Ras/Raf pathway in specification of photoreceptor cells in the Drosophila retina. Development. 2001;128:1183–1191. doi: 10.1242/dev.128.7.1183. [DOI] [PubMed] [Google Scholar]
- Yang L, Baker NE. Cell cycle withdrawal, progression, and cell survival regulation by Egfr and its effectors in the differentiating Drosophila eye. Dev Cell. 2003;4:359–369. doi: 10.1016/s1534-5807(03)00059-5. [DOI] [PubMed] [Google Scholar]
- Zak NB, Shilo B-Z. Localization of DER and the pattern of cell divisions in wild-type and Ellipse eye imaginal discs. Dev. Biol. 1992;149:448–456. doi: 10.1016/0012-1606(92)90299-v. [DOI] [PubMed] [Google Scholar]
- Zhang F, Strand A, Robbins D, Cobb MH, Goldsmith EJ. Atomic structure of the MAP kinase ERK2 at 2.3 A resolution. Nature. 1994;367:704–711. doi: 10.1038/367704a0. [DOI] [PubMed] [Google Scholar]